Abstract
Responses to prolonged low-temperature treatment of imbibed seeds (vernalization) were examined in barley (Hordeum vulgare). These occurred in two phases: the perception of prolonged cold, which occurred gradually at low temperatures, and the acceleration of reproductive development, which occurred after vernalization. Expression of the VERNALIZATION1 gene (HvVRN1) increased gradually in germinating seedlings during vernalization, both at the shoot apex and in the developing leaves. This occurred in darkness, independently of VERNALIZATION2 (HvVRN2), consistent with the hypothesis that expression of HvVRN1 is induced by prolonged cold independently of daylength flowering-response pathways. After vernalization, expression of HvVRN1 was maintained in the shoot apex and leaves. This was associated with accelerated inflorescence initiation and with down-regulation of HvVRN2 in the leaves. The largest determinant of HvVRN1 expression levels in vernalized plants was the length of seed vernalization treatment. Daylength did not influence HvVRN1 expression levels in shoot apices and typically did not affect expression in leaves. In the leaves of plants that had experienced a saturating seed vernalization treatment, expression of HvVRN1 was higher in long days, however. HvFT1 was expressed in the leaves of these plants in long days, which might account for the elevated HvVRN1 expression. Long-day up-regulation of HvVRN1 was not required for inflorescence initiation, but might accelerate subsequent stages of inflorescence development. Similar responses to seed vernalization were also observed in wheat (Triticum aestivum). These data support the hypothesis that VRN1 is induced by cold during winter to promote spring flowering in vernalization-responsive cereals.
Keywords: Barley, floral development, FT, MADS box gene, photoperiod, vernalization, VRN1, VRT2, VRN2, wheat
Introduction
Prolonged cold treatment of imbibed seeds, or vernalization, promotes flowering of many temperate cereals, including varieties of wheat, barley, oat, and rye. When grown without cold pre-treatment, the same varieties grow vegetatively for extended periods and often fail to flower altogether. Thus, these cereal varieties have a ‘Kaltbedurfnis’, or cold requirement, that must be met for rapid transition to reproductive growth (Gassner 1918). Although originally defined as prolonged cold treatment of seeds, vernalization also accelerates flowering when applied to plants during the vegetative growth phase (Gott, 1957; Flood and Halloran, 1984).
Flowering of temperate cereals is also accelerated by long days (see Purvis, 1934). Studies in Arabidopsis have shown that the long-day flowering response is controlled by FLOWERING LOCUS T (FT) (Kardailsky et al., 1999; Kobayashi et al., 1999). The FT protein is produced in leaves in long days and transported to the shoot apex to promote reproductive development (Corbesier et al., 2007). FT-like 1 (TaFT1 in wheat or HvFT1 in barley) is an orthologue of FT likely to fulfil a similar role in temperate cereals (Turner et al., 2005).
The requirement for vernalization suppresses the long-day flowering response (Hemming et al., 2008). This ensures that flowering is delayed before winter, reducing the risk of frost damage (Mahfoozi et al., 2001; Limin and Fowler, 2006). Many varieties flower rapidly without vernalization, however, and this natural variation in vernalization requirement has been useful to adapt varieties to warmer growing conditions. Genes controlling natural variation in vernalization requirement have been identified in temperate cereals: VERNALIZATION1 (VRN1), VRN2, and VRN3 (reviewed in Trevaskis et al., 2007a; Distalfeld et al., 2009).
VRN1 is a FRUITFULL-like MADS box transcription factor that is essential for flowering in temperate cereals (Danyluk et al., 2003; Murai et al., 2003; Trevaskis et al., 2003; Yan et al., 2003; Preston and Kellog, 2006; Shitsukawa et al., 2007). In varieties that require vernalization, VRN1 is activated by prolonged exposure to low temperatures (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003). In other varieties, VRN1 is expressed without vernalization, reducing or removing the requirement for vernalization (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003). Mutations in the promoter or deletions within the first intron of the VRN1 gene are associated with increased VRN1 expression in these varieties (Yan et al., 2003; Fu et al., 2005).
VRN2, or HvVRN2 in barley, is a floral repressor (Takahashi and Yasuda, 1971; Dubcovsky et al., 1998; Yan et al., 2004). VRN2 is active in long days (Karsai et al., 2005; Dubcovsky et al., 2006; Trevaskis et al., 2006), where it represses FT1 to delay the long-day flowering response until plants are vernalized (Hemming et al., 2008). VRN1 represses expression of VRN2 (Loukoianov et al., 2005; Trevaskis et al., 2006; Hemming et al., 2008), so when plants are vernalized VRN1 is likely to repress VRN2 to allow long-day induction of FT1 (Trevaskis et al., 2007a; Hemming et al., 2008). Loss-of-function mutations at the VRN2 locus allow expression of FT1 without prior vernalization, causing rapid flowering in long days (Yan et al., 2004; Karsai et al., 2005; Hemming et al., 2008). This requires an active PHOTOPERIOD1 gene (Hemming et al., 2008), which promotes long-day induction of HvFT1 (Turner et al., 2005). Similarly, dominant alleles of VRN3 elevate FT1 expression levels to accelerate flowering and bypass the vernalization requirement (Yan et al., 2006; Faure et al., 2007). VRN3 has been mapped to the FT1 sequence, suggesting that FT1 is the VRN3 gene (Yan et al., 2006). Dominant VRN3 alleles in wheat are associated with an insertion in the promoter of the TaFT1 gene (Yan et al., 2006) and, although there is an association between polymorphisms in the HvFT1 gene and dominant alleles of VRN3 in barley (Yan et al., 2006), the molecular basis for the increased expression of HvFT1 associated with dominant VRN3 alleles in barley is not clear (Hemming et al., 2008; Stracke et al., 2009).
In barley plants that lack HvVRN2, expression of HvVRN1 is induced in long days without prior cold treatment (Yan et al., 2003; Trevaskis et al., 2006; Hemming et al., 2008). HvVRN1 is also expressed without cold treatment in barleys plants with dominant alleles of VRN3 (Yan et al., 2006). In these genotypes, HvFT1 is expressed without prior cold treatment and the FT1 protein might activate VRN1 through interactions with FD-like proteins (Li and Dubcovsky, 2008). It is unclear whether this is a feature of vernalization-induced flowering, where low-temperature induction of VRN1 precedes long days (Hemming et al., 2008). Furthermore, the potential for low-temperature and long-day activation of VRN1 complicates analysis of the vernalization response in cereals. For example, although expression of VRN1 increases in leaves when wheat plants are vernalized in long days (Yan et al., 2003; Preston and Kellog 2008), it is unclear whether this is a response to low temperatures or to the combined effects of low temperatures and long days. Analysis of gene expression in plants vernalized in short days has been useful to determine the discrete effects of low temperature on expression of the vernalization genes in cereals (von Zitzewitz et al., 2005; Dubcovsky et al., 2006; Trevaskis et al., 2006; Fu et al., 2007; Hemming et al., 2008), but such studies have not yet examined responses to cold in specific organs.
The seed vernalization response first investigated by Gassner (1918) provides an ideal experimental system to resolve the effects of low temperature and daylength cues on expression of flowering-time genes during vernalization-induced flowering. Seeds can be exposed to long cold treatments in darkness then sown in different daylengths. In this study the molecular responses to seed vernalization are examined by assaying the expression of flowering-time genes in different organs during and after seed vernalization.
Materials and methods
Seeds of the winter barley (Hordeum vulgare) variety cv. Sonja, or the winter wheat cv. Norstar, were imbibed, planted in foil-covered pots, and exposed to cold treatments of different durations (e.g. 0, 1, 2, 3, 4, 5, 7, and 9 weeks) at 2±1 °C. After 9 weeks in these conditions, plants reached an average coleoptile length of 4 cm. At each time point, embryos or germinating seedlings were harvested for RNA extraction and apex dissection. The developing roots and leaves were also isolated after 4 weeks cold treatment. Following cold treatment, seedlings were transferred to glasshouses (temperature at 18±2 °C) and grown in either short (8 h light/16 h dark) or long (16 h light/8 h dark) days. Supplementary light was provided when natural levels dropped below 200 μE. Plants were harvested at the third leaf stage (Z=13,21, Zadoks et al., 1974), ∼2 weeks after the end of the cold treatment, and leaves and shoot apices were harvested for RNA extraction. Final leaf number (FLN), and days to heading were recorded for each treatment. Similarly, seeds of a barley which lacks HvVRN2 (Line 347, ΔHvVRN2/HvVRN1/PPD-H1; Hemming et al. 2008) were imbibed and exposed to cold for 9 weeks, then transferred to glasshouse temperatures in short days. Plants were harvested for apex dissection and RNA extraction after 9 weeks of cold treatment, and from glasshouses at the third leaf stage, ∼2 weeks after the end of the cold treatment.
Apex dissection and flowering time measurements
Apices were isolated under a binocular dissecting microscope and then digitally photographed on a Leica M8 digital camera. Apex samples included the apex, the base of the apex, and any leaf primordia <0.2 mm in length. Leaves were numbered sequentially and plants were grown until the flag leaf emerged to determine FLN. Heading date was measured as the day when the head first emerged from the sheath on the main shoot. FLN and heading date were measured for 9–18 plants for each data point. Average FLN and days to heading are presented, and error bars show the standard error (SE).
Gene expression analysis
Total RNA was extracted using the method of Chang et al. (1993). An oligo(T) primer (T18[G/C/A]) was used to prime first-strand cDNA synthesis from 5 μg of total RNA using the SuperScript III reverse transcriptase enzyme (Invitrogen) according to the manufacturer's instructions. A single reverse transcription reaction was performed for each RNA sample. Quantitative reverse transcription-PCR (qRT-PCR) was performed on a Rotor-Gene 3000 real-time cycler (Corbett Research). The primers used for HvVRN1, HvVRN2 (HvZCCTb), HvFT1, Barley MADS1 (BM1), BM10, Hordeum vulgare VEGETATIVE to REPRODUCTIVE TRANSITION 2 (HvVRT2), and ACTIN have been described previously (Trevaskis et al., 2006, 2007b). For analysis of VRN1 and VRN2 expression in wheat (cv. Norstar), the primer sets described above, which are predicted to amplify all three genomes from wheat, were used. The following primers were used for other MADS box genes: BM3 5′-GCCGTCACCAGCACAAGCAA-3′ and 5′-CCCCATTCACCCTGTAGCAAAGA-3′; BM7 5′-GCTTGACCAGATAGAGAACCAAATAG-3′ and 5′-GCTGGTGGTGGTGGTGTCTTGC3′; BM8 5′-CGCACAGCAGCCGACACCTA-3′ and 5′-TGCCTTTGGGGGAGAAGACG-3′; and BM9 5′-TCGTCCTTGAAGCACATTAGAAC-3′ and 5′-GGGTTCACCAGCAGCATCAAGGG-3′. Primer pairs amplify cDNA-specific DNA products. qRT-PCR was performed using Platinum Taq DNA polymerase (Invitrogen). Cycling conditions were 4 min at 94 °C, 50 cycles of 10 s at 95 °C, 15 s at 60 °C, and 20 s at 72 °C. This was followed by a melting curve program (72–95 °C with a 5 s hold at each temperature). Fluorescence data were acquired at the 72 °C step and during the melting curve program. Expression levels of genes of interest were calculated relative to ACTIN using the comparative quantification analysis method (Rotogene-5; Corbett Research), which takes into account the amplification efficiency of each primer set. Quantification for each primer set and cDNA template combination was performed in quadruplicate, and included a no-template control, to ensure results were not influenced by primer–dimer formation or DNA contamination. Data presented are the average and SE from four RT-PCR quantifications (gene of interest versus ACTIN) for each RNA sample; each sample was extracted from 30–50 apices, 3–5 leaves or 3–5 seedlings. Similar results were obtained in a second set of samples from the same experiments. Where Student's t-tests show that expression levels for a gene of interest differed significantly between two treatments (Figs 2F and 6A, B), differences of similar significance levels were found in both sample sets.
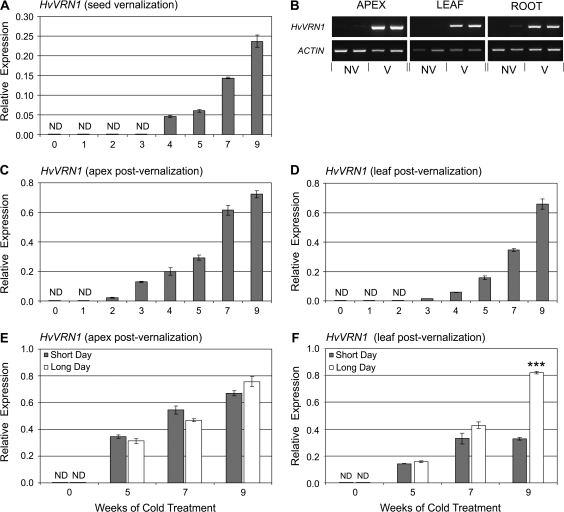
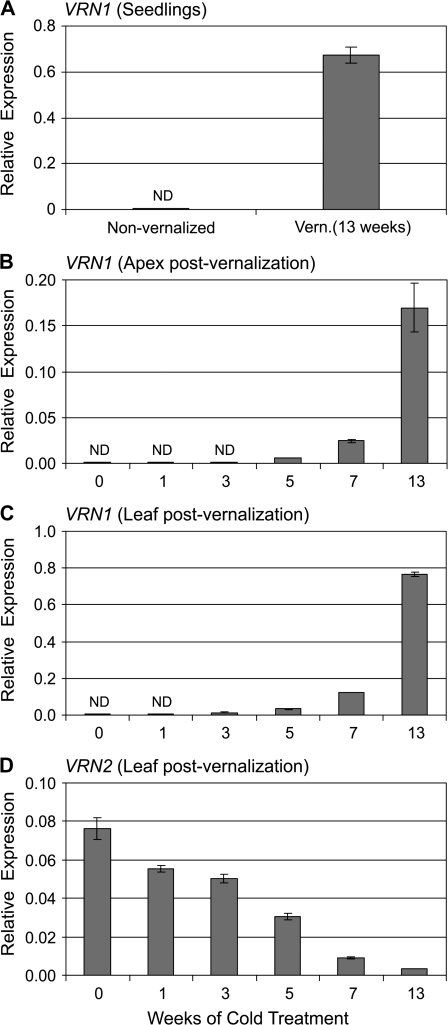
Fig. 2.
The effect of seed vernalization treatment on expression of HvVRN1. (A) Expression of HvVRN1 in seedlings at different time points (weeks) during seed vernalization. (B) RT-PCR analysis of HvVRN1 expression (35 cycles) in different organs during seed vernalization. Expression of ACTIN (25 cycles) is shown as a positive control, and two biological repeats are shown for each data point. NV refers to samples from non-vernalized plants and V refers to samples from vernalized plants. (C) Expression of HvVRN1 in the shoot apex at the third leaf stage, in non-vernalized plants (0) compared with plants that received seed vernalization treatments of different durations (1–9 weeks). (D) Expression of HvVRN1 in leaves (fully expanded second leaf) at the third leaf stage in the same experiment. (E) HvVRN1 expression in shoot apex in short versus long days at the third leaf stage, in non-vernalized plants (0) or after seed vernalization treatments of different durations (5, 7, or 9 weeks). (F) Comparison of HvVRN1 expression in leaves (fully expanded second leaf) in the same experiment. Expression of HvVRN1 was assayed by quantitative RT-PCR and is shown relative to ACTIN. ND denotes no expression detected. Asterisks indicate P-values of Student's t-test: ***P <0.001.
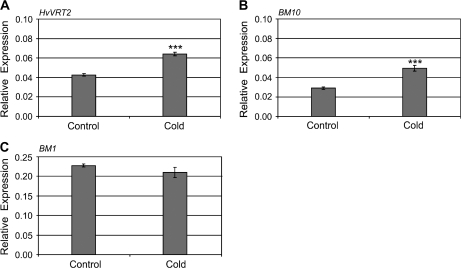
Fig. 6.
The influence of seed vernalization on the transcript levels of other MADS box genes in barley. Expression of MADS box genes was assayed in seedlings at the 7 week vernalization time point (cold), and compared with non-vernalized plants at a similar stage of development (control). Expression of HvVRT2 (A), BM10 (B), and BM1 (C) was assayed by quantitative RT-PCR and is shown relative to ACTIN. Asterisks indicate P-values of Student's t-test: ***P <0.001.
Results
Prolonged cold treatment of imbibed seeds promotes flowering by accelerating inflorescence initiation
Flowering time, as indicated by FLN and days to heading, was assayed in plants grown from seeds germinated with or without cold treatment. In short days, control plants (no cold treatment) did not flower within 150 d (FLN >25), but plants grown from seeds that had experienced >4 weeks of cold during germination flowered within 130 d (FLN=14). Longer cold treatments caused no further reduction in FLN in short days and little further reduction in the days to heading (Fig. 1A, B).
Fig. 1.
The effect of seed vernalization treatment on flowering time of a winter barley. (A) Final leaf number (FLN), in short or long days, of non-vernalized (0) plants of a winter barley (cv. Sonja) versus plants subjected to seed vernalization treatments of different durations (1–9 weeks). NF denotes no flowering observed. (B) Days to flowering (heading) for the same treatments. (C) Images of representative shoot apices (50× magnification) from: imbibed seeds (0 seed), cold-treated seeds (9W seed), and plants at the third leaf stage in either short day or long day conditions (+SD or +LD), ∼2 weeks after the end of seed vernalization treatments of different durations (0, 5, 7, or 9 weeks). For example, 5W+SD refers to plants that were vernalized for 5 weeks in darkness then grown in short days.
In long days, control plants (no cold treatment) flowered within 120 d (FLN=20). Plants grown from cold-treated seeds flowered earlier than control plants. Short-term cold treatments accelerated flowering. One week cold treatment reduced FLN to 18, for example, and longer cold treatments further reduced flowering time and FLN. The 9 week cold treatment had the greatest impact on flowering time, reducing the time taken to flower to 47 d (FLN=10) (Fig. 1A, B). Beyond 9 weeks, longer cold treatments had no additional impact on flowering time or FLN (FLN=10 for plants subjected to 11 weeks cold treatment). Thus, 9 weeks cold treatment of seeds completely fulfils the vernalization requirement of this barley (vernalization saturation point).
Cold treatment of imbibed seeds accelerated flowering by promoting early stages of shoot apex development (Fig. 1C), consistent with the reduced FLN of plants grown from cold-treated seeds. The shoot apex remained vegetative at the end of even the longest vernalization treatments (Fig. 1C), so accelerated floral development occurred after vernalization during growth at normal glasshouse temperatures. Shoot apex development was accelerated to a greater extent when vernalized plants were grown in long days (Fig. 1A, C). Stem elongation and post-initiation (double ridge stage, when floral primordia develop at the shoot apex) inflorescence development also occurred rapidly when vernalized plants were grown in long days (Fig. 1C). In short days, the shoot apex elongated more than in control plants and the apex developed to the double ridge stage, but further inflorescence development and stem elongation occurred slowly. Consequently flowering (head emergence) was delayed in short days in comparison with plants grown in long days, after equivalent vernalization treatment.
Cold treatment induces expression of HvVRN1 in germinating seeds
Expression of HvVRN1 was assayed during germination at low temperatures (seed vernalization). HvVRN1 expression was first detected after 4 weeks cold treatment and was induced to higher levels with longer cold treatments (Fig. 2A). Expression of HvVRN1 increased in the developing leaves, roots, and the shoot apex (Fig. 2B). Elevated HvVRN1 transcript levels were maintained during subsequent development at normal glasshouse temperatures. At the third leaf stage, ∼2 weeks after the end of cold treatment, HvVRN1 was expressed in both shoot apices and leaves (Fig. 2C, D). In both tissue types, HvVRN1 transcript levels correlated with the length of cold treatment applied during germination (Fig. 2C, D).
Daylength did not influence expression of HvVRN1 in the shoot apex of vernalized plants (Fig. 2E). Similarly, daylength did not influence expression of HvVRN1 in the leaves of plants grown from seeds vernalized for 5 or 7 weeks. In plants grown from seeds subjected to 9 weeks of seed vernalization, expression of HvVRN1 was ∼2-fold higher in the leaves in long days than in short days (Fig. 2F).
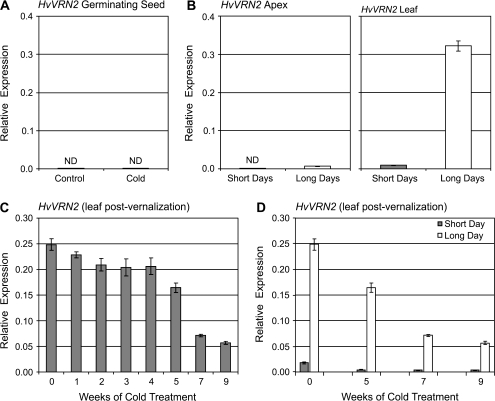
HvVRN2 is down-regulated in the leaves of vernalized plants but is not required for the low-temperature flowering response
Expression of the floral repressor HvVRN2 was not detected in seeds germinating in the dark, regardless of temperature (Fig. 3A). HvVRN2 was expressed in the leaves of plants in long days (Fig. 3B) and, although HvVRN2 expression did not change during vernalization, expression of HvVRN2 was lower in the leaves of plants grown from cold-treated seeds. The extent to which HvVRN2 was down-regulated correlated with the length of cold treatment applied (Fig. 3C). There was an inverse relationship between HvVRN1 and HvVRN2 expression levels in leaves in plants grown in long days (compare Figs 2C and 3C), consistent with the suggestion that HvVRN1 down-regulates HvVRN2 in vernalized plants (Trevaskis et al., 2006, 2007a). This relationship was not observed in short days, where HvVRN2 was expressed at uniformly low levels in leaves irrespective of cold treatment or HvVRN1 expression levels (Fig. 3D).
Fig. 3.
The effect of seed vernalization treatment on HvVRN2 expression. (A) HvVRN2 expression in non-vernalized seedlings (control) versus seedlings germinated at low temperature for 7 weeks (cold). (B) Expression of HvVRN2 in the shoot apex or second leaf for non-vernalized plants at the third leaf stage, in short versus long days. (C) Relative expression of HvVRN2 in leaves (fully expanded second leaf) at the third leaf stage in long days, in non-vernalized plants (0), or after seed vernalization for different durations (1–9 weeks). (D) Relative expression levels of HvVRN2 in leaves (fully expanded second leaf) from plants at the third leaf stage in short or long days. The comparison shows non-vernalized plants (0) versus plants grown from seeds vernalized for 5, 7, or 9 weeks. Expression of HvVRN2 was assayed by quantitative RT-PCR and is shown relative to ACTIN. ND denotes no expression detected.
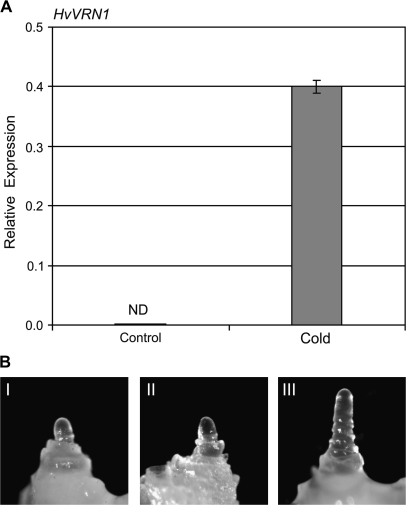
The influence of seed vernalization on HvVRN1 transcript levels and shoot apex development was assayed in a barley which lacks HvVRN2. In this barley, expression of HvVRN1 was low during germination but was induced by prolonged cold treatment (Fig. 4A). Cold treatment also accelerated inflorescence initiation during subsequent development at normal glasshouse temperatures, and at the third leaf stage the shoot apex of cold-treated plants had progressed to the double ridge stage whereas the apices of control plants remained vegetative (Fig. 4B).
Fig. 4.
The response to seed vernalization in a barley that lacks HvVRN2. (A) Expression of HvVRN1 in seedlings of barley lacking HvVRN2 (HvVRN1, ΔHvVRN2/PPD-H1), exposed to low-temperature treatment for 9 weeks during germination (cold) versus seedlings at the same stage of development that were germinated at normal growth temperatures (control). Expression of HvVRN1 was assayed by quantitative RT-PCR and is shown relative to ACTIN. (B) The morphology of the shoot apex after 9 weeks of seed vernalization treatment (I), at the third leaf stage in either non-vernalized plants (II) or plants grown from seeds that were vernalized for 9 weeks (III) (×50 magnification). As this barley lacks HvVRN2, plants were grown in short days to prevent activation of flowering by the long-day response.
Seed vernalization allows long-day induction of HvFT1 in leaves
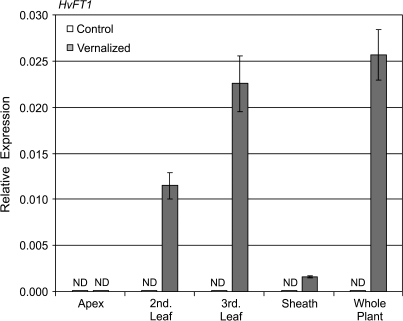
Expression of HvFT1 was assayed during and after seed vernalization treatments of different durations and in non-vernalized control plants at the same stage of development. HvFT1 expression was not detected in seedlings germinating in darkness, regardless of temperature. Following germination, at the third leaf stage, HvFT1 was not expressed in plants grown from seeds germinated without cold treatment but was expressed in the leaves of plants grown in long days from seeds that were vernalized for 9 weeks (Fig. 5). No expression of HvFT1 was detected in plants grown in short days, as has been reported previously for this barley variety (Hemming et al., 2008).
Fig. 5.
The effect of seed vernalization treatment on expression of HvFT1. (A) Plants subjected to different vernalization treatments were grown in long days until the third leaf stage when different organs were isolated for gene expression analysis. Expression of HvFT1 was assayed in the apex, second leaf, third leaf, or the sheath and compared with expression in whole plants. The comparison shows HvFT1 expression in control (non-vernalized) versus vernalized plants (9 weeks seed vernalization). Expression of HvFT1 was assayed by quantitative RT-PCR and is shown relative to ACTIN. ND denotes no expression detected.
The influence of low-temperature treatment on expression of other MADS box genes in barley
It has been suggested that the SHORT VEGETATIVE PHASE-like gene HvVRT2 is repressed by cold and that this allows expression of HvVRN1 to increase during vernalization (Kane et al., 2005). Expression of HvVRT2 increased slightly in vernalized seeds, as did the related gene BM10 (Fig. 6A, B). Transcript levels of a third SVP-like MADS box gene, BM1, did not change during seed vernalization (Fig. 6C).
In addition to HvVRN1, there are two other FRUITFULL-like MADS box genes in barley: BM3 and BM8. Expression of these genes was assayed before and after seed vernalization to determine whether these genes play a similar role to HvVRN1 in the low-temperature flowering response. No expression of either BM3 or BM8 was detected in germinating seeds, and neither gene was activated by low-temperature treatment (Supplementary Fig. S1 available at JXB online). Similarly, no expression of the SEPALLATA-like MADS box genes BM7 and BM9 (Schmitz et al., 2000) was detected in germinating seeds irrespective of temperature (Supplementary Fig. S1).
The seed vernalization response is similar in wheat and barley.
To determine whether low temperature and long day cues regulate expression of flowering-time genes in a similar manner in other temperate cereals, expression of VRN1 and VRN2 was examined in the winter wheat cultivar Norstar during and after seed vernalization treatments. For these experiments, primers predicted to amplify all three copies (A, B, and D genomes) of the target genes were used, to assay the sum expression levels of VRN1 or VRN2. Seed vernalization promoted flowering in this wheat (Supplementary Fig. S2 at JXB online). Prolonged cold treatment induced expression of VRN1 in imbibed wheat seeds, and expression of VRN1 was maintained in the shoot apex and leaves of wheat plants after vernalization treatment (Fig. 7A–C). VRN2 was expressed in leaves in long days, but in vernalized plants expression of VRN2 in the leaves was lower than in control plants that had not experienced cold (Fig. 7D). The extent to which VRN2 was down-regulated in the leaves of vernalized plants correlated with the length of cold treatment during germination (Fig. 7D). Thus, the molecular responses to seed vernalization appear to be similar in barley and wheat.
Fig. 7.
The seed vernalization response of wheat. (A) Expression of VRN1 in seeds/seedlings of the winter wheat Norstar before and after seed vernalization. (B) Expression of VRN1 in the shoot apex at the third leaf stage, after seed vernalization treatments of different durations. (C) Expression of VRN1 in leaves (second leaf) at the third leaf stage, in long days, after seed vernalization treatments of different durations. (D) Expression of VRN2 in leaves (second leaf) at the third leaf stage, in long days, after seed vernalization treatments of different durations. VRN1 and VRN2 transcript levels were assayed by quantitative RT-PCR and are shown relative to ACTIN. ND denotes no expression detected.
Discussion
The molecular responses to seed vernalization parallel those observed in plants vernalized in short days; HvVRN1 is activated by low temperatures to a degree that correlates with the length of cold treatment, while HvVRN2 and HvFT1 are not expressed (Fig. 2; von Zitzewitz et al., 2005; Trevaskis et al., 2006; Hemming et al., 2008). This suggests that the pathways controlling perception of cold in germinating seeds are similar to those in growing plants, and support the hypothesis that activation of VRN1 mediates the low-temperature flowering response in cereals (see Trevaskis et al., 2007a).
Compared with previous studies using plants vernalized in short days (von Zitzewitz et al., 2005; Trevaskis et al., 2006), analysis of the vernalization response in seeds has further clarified how HvVRN1 is regulated. Induction of HvVRN1 in seeds occurred in darkness, demonstrating that a low-temperature response pathway can activate expression of HvVRN1 independently of light or daylength. This pathway is unlikely to involve HvVRN2, which is not expressed in seeds during vernalization (Fig. 3) and is not required for low-temperature induction of HvVRN1 (Hemming et al., 2008; Fig. 4). Similarly, induction of HvVRN1 is unlikely to involve HvFT1, which is not expressed in seeds during vernalization. Low-temperature induction of HvVRN1 is not limited to any particular organ type in barley seeds/seedlings (Fig. 2B).
Low-temperature induction of HvVRN1 at the shoot apex precedes changes in shoot apex morphology (Fig. 1C), and so cannot be a consequence of floral development. Similar observations have been made by Yan et al. (2003) in einkorn wheat plants (Triticum monococcum) vernalized in long days. Preston and Kellog (2008) found that in oat (Avena sativa) induction of VRN1 at the shoot apex occurs after vernalization, at the same time as floral development. These conflicting findings might reflect differences between oats versus barley and wheat, or might be due to the different sensitivities of the gene expression analysis techniques used; in situ hybridization (Preston and Kellog, 2008) versus qRT-PCR (Yan et al., 2003; this study).
Unlike HvVRN1, two related AP1/FRUITFULL-like genes, BM3 and BM8, were not activated by seed vernalization. So while the oat (A. sativa) orthologue of BM8, AsFUL2, is induced in leaves when plants are vernalized in long days (Preston and Kellog, 2008), this might not be a general feature of the vernalization response in cereals. Expression of HvVRT2, another MADS box gene which has been suggested to play a role in the low-temperature response (Kane et al., 2005), increased slightly during seed vernalization (Fig. 6A). This is similar to previous observations in vernalized plants (Trevaskis et al., 2007b), and it seems unlikely that HvVRT2 mediates low-temperature induction of HvVRN1 in the manner suggested.
Following vernalization, HvVRN1 expression levels remained high in the shoot apex and leaves (Fig. 2). This was associated with more rapid inflorescence initiation (Fig. 1A, C), consistent with genetic data that show that expression of HvVRN1 promotes inflorescence initiation (Hemming et al., 2008). This might be a specific consequence of elevated HvVRN1 levels in the shoot apex. Expression of HvVRN1 is also associated with reduced expression of HvVRN2 and with induction of HvFT1 in long days (Hemming et al., 2008). Here it has been shown that down-regulation of HvVRN2 occurs in leaves, and is a quantitative response; longer cold treatments are associated with greater down-regulation of HvVRN2. These findings have also been extended to wheat (Fig. 7). Overall, these data are consistent with the proposed role of VRN1 in repressing VRN2 to activate the long-day flowering response in leaves (Trevaskis et al., 2007a).
The main determinant of HvVRN1 expression levels in the leaves and shoot apices was the length of seed vernalization treatment (Fig. 2), similar to the response observed in whole plants (von Zitzewitz et al., 2005; Trevaskis et al., 2006; Hemming et al., 2008). Daylength only influenced expression of HvVRN1 in the leaves of plants grown from seeds vernalized for 9 weeks, where expression of HvVRN1 was ∼2-fold higher in long days (Fig. 2). This might be caused by elevated expression of HvFT1 (Fig. 5), through interactions with FD-like proteins (Teper-Bamnolker and Samach, 2005; Li and Dubcovksy, 2008). Long-day induction of HvVRN1 in leaves was not critical for inflorescence initiation, which occurred irrespective of daylength in vernalized plants, but might accelerate later stages of inflorescence development.
In general, the daylength flowering responses of vernalized barley plants are similar to those in rye (Purvis 1934); prolonged exposure to cold accelerates inflorescence initiation, but long days are required for rapid inflorescence development and for stem elongation. The present findings can explain the molecular events underlying these physiological responses; HvVRN1 is induced by cold to accelerate inflorescence initiation, then long days induce HvFT1 and further activate HvVRN1 to accelerate subsequent stages of inflorescence development and stem elongation.
In summary, the seed vernalization treatment first described by Gassner (1918) provides a simple experimental system to analyse the vernalization response of cereals. Plants are vernalized in the dark, so artificial daylength conditions are not required during vernalization, and vernalized plants are easily matched to non-vernalized control plants at the same stage of development. Analysis of the expression patterns of flowering-time genes during and after seed vernalization supports the hypothesis that activation of HvVRN1 mediates the low-temperature flowering response in temperate cereals.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Expression of APETALA1-like and SEPALLATA-like MADS box genes in seeds at normal or vernalizing temperatures.
Fig. S2. Flowering time of the winter wheat cv. Norstar after seed vernalization treatments, in long days.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the support of the Commonwealth Scientific and Industrial Research Organisation, the Ministry of Science, Research and Technology of Iran, the Agricultural Research, Extension, and Education Organization (AREEO) of Iran, and the Grains Research and Development Corporation. We would also like to thank Dr Jose Barrero Sanchez, Dr Marc Ellis, Dr Jayne Griffiths, Dr Alfonso Cuesta-Marcos, Dr Luis Cistue and Professor Pat Hayes for the insightful comments and helpful suggestions contributed during the preparation of this manuscript.
References
- Chang S, Puryear J, Cairney K. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. FT protein movement contributes to long-distance signalling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiology. 2003;132:1849–1860. doi: 10.1104/pp.103.023523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J. Regulation of flowering in temperate cereals. Current Opinion in Plant Biology. 2009 doi: 10.1016/j.pbi.2008.12.010. in press doi:10.1016/j.pbi.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G. Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theoretical and Applied Genetics. 1998;97:968–975. [Google Scholar]
- Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L. Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Molecular Biology. 2006;60:469–480. doi: 10.1007/s11103-005-4814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S, Higgins J, Turner A, Laurie DA. The FLOWERING LOCUST-like gene family in barley (Hordeum vulgare) Genetics. 2007;176:599–609. doi: 10.1534/genetics.106.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood RG, Halloran GM. The nature and duration of gene action for vernalization response in wheat. Annals of Botany. 1984;53:363–368. [Google Scholar]
- Fu D, Dunbar M, Dubcovsky J. Wheat VIN3-like PHD finger genes are up-regulated by vernalization. Molecular Genetics and Genomics. 2007;277:301–313. doi: 10.1007/s00438-006-0189-6. [DOI] [PubMed] [Google Scholar]
- Fu D, Szucs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Molecular Genetics and Genomics. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- Gassner G. Beitraege zur physiologischen Charakteristik sommer- und winterannueller Gewaechse in besondere der Getreidepflanzen. Zeitschrift für Botanik. 1918;10:27–476. [Google Scholar]
- Gott MB. Vernalization of green plants of a winter wheat. Nature. 1957;180:714–715. [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B. Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiology. 2008;147:355–366. doi: 10.1104/pp.108.116418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane NA, Danyluk J, Tardif G, Ouellet F, Laliberte JF, Limin AE, Fowler DB, Sarhan F. TaVRT-2, a member of the StMADS-11 clade of flowering repressors, is regulated by vernalization and photoperiod in wheat. Plant Physiology. 2005;138:2354–2363. doi: 10.1104/pp.105.061762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Karsai I, Szucs P, Meszaros K, Filichkina T, Hayes PM, Skinner JS, Lang L, Bedo Z. The Vrn-H2 locus is a major determinant of flowering time in a facultative×winter growth habit barley (Hordeum vulgare L.) mapping population. Theoretical and Applied Genetics. 2005;110:1458–1466. doi: 10.1007/s00122-005-1979-7. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Li C, Dubcovsky J. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. The Plant Journal. 2008;55:543–554. doi: 10.1111/j.1365-313X.2008.03526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limin AE, Fowler DB. Low-temperature tolerance and genetic potential in wheat (Triticum aestivum L.): responses to photoperiod, vernalization and plant development. Planta. 2006;224:360–366. doi: 10.1007/s00425-006-0219-y. [DOI] [PubMed] [Google Scholar]
- Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J. Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiology. 2005;138:2364–2373. doi: 10.1104/pp.105.064287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfoozi S, Limin AE, Fowler DB. Developmental regulation of low temperature tolerance in winter wheat. Annals of Botany. 2001;87:751–757. [Google Scholar]
- Murai K, Miyamae M, Kato H, Takumi S, Ogihara Y. WAP1, a wheat APETALA1 homolog, plays a central role in the phase transition from vegetative to reproductive growth. Plant Cell Physiology. 2003;44:1255–1265. doi: 10.1093/pcp/pcg171. [DOI] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA. Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae) Genetics. 2006;174:421–437. doi: 10.1534/genetics.106.057125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA. Discrete developmental roles for temperate cereal grass VERNALIZATION1/FRUITFULL-Like genes in flowering competency and the transition to flowering. Plant Physiology. 2008;146:265–276. doi: 10.1104/pp.107.109561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis ON. An analysis of the influence of temperature during germination on the subsequent development of certain winter cereals and its relation to the effect of length of day. Annals of Botany. 1934;48:919–955. [Google Scholar]
- Schmitz J, Franzen R, Ngyuen TH, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W. Cloning, mapping and expression analysis of barley MADS-box genes. Plant Molecular Biology. 2000;42:899–913. doi: 10.1023/a:1006425619953. [DOI] [PubMed] [Google Scholar]
- Shitzukawa N, Ikari C, Shimada S, et al. The einkorn wheat (Triticum monococcum) mutant, maintained vegetative phase, is caused by a deletion in the VRN1 gene. Genes and Genetic Systems. 2007;82:167–170. doi: 10.1266/ggs.82.167. [DOI] [PubMed] [Google Scholar]
- Stracke S, Haseneyer G, Veyrieras JB, Geiger HH, Sauer S, Graner A, Piepho HP. Association mapping reveals gene action and interactions in the determination of flowering time in barley. Theoretical and Applied Genetics. 2009;118:259–273. doi: 10.1007/s00122-008-0896-y. [DOI] [PubMed] [Google Scholar]
- Takahashi R, Yasuda S. Genetics of earliness and growth habit in barley. In: Nilan RA, editor. Barley Genetics II. Proceedings of the Second International Barley Genetics Symposium. Pullman, WA: Washington State University Press; 1971. pp. 388–408. [Google Scholar]
- Teper-Bamnolker P, Samach A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. The Plant Cell. 2005;17:2661–2675. doi: 10.1105/tpc.105.035766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. MADS box genes control vernalization-induced flowering in cereals. Proceedings of the National Academy of Sciences, USA. 2003;100:13099–13104. doi: 10.1073/pnas.1635053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Dennis ES, Peacock WJ. The molecular basis of vernalization-induced flowering in cereals. Trends in Plant Science. 2007a;12:353–357. doi: 10.1016/j.tplants.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES. HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiology. 2006;140:1397–1405. doi: 10.1104/pp.105.073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Tadege M, Hemming MN, Peacock WJ, Dennis ES. Short vegetative phase-like MADS-box genes inhibit floral meristem identity in barley. Plant Physiology. 2007b;143:225–235. doi: 10.1104/pp.106.090860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- von Zitzewitz J, Szücs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen THH, Hayes PM, Skinner JS. Molecular and structural characterization of barley vernalization genes. Plant Molecular Biology. 2005;59:449–467. doi: 10.1007/s11103-005-0351-2. [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proceedings of the National Academy of Sciences, USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, San Miguel P, Bennetzen JL, Echenique V, Dubcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proceedings of the National Academy of Sciences, USA. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF. A decimal code for the growth stages of cereals. Weed Research. 1974;14:415–421. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.