Abstract
Motivation: KEGG PATHWAY is a service of Kyoto Encyclopedia of Genes and Genomes (KEGG), constructing manually curated pathway maps that represent current knowledge on biological networks in graph models. While valuable graph tools have been implemented in R/Bioconductor, to our knowledge there is currently no software package to parse and analyze KEGG pathways with graph theory.
Results: We introduce the software package KEGGgraph in R and Bioconductor, an interface between KEGG pathways and graph models as well as a collection of tools for these graphs. Superior to existing approaches, KEGGgraph captures the pathway topology and allows further analysis or dissection of pathway graphs. We demonstrate the use of the package by the case study of analyzing human pancreatic cancer pathway.
Availability:KEGGgraph is freely available at the Bioconductor web site (http://www.bioconductor.org). KGML files can be downloaded from KEGG FTP site (ftp://ftp.genome.jp/pub/kegg/xml).
Contact: j.zhang@dkfz-heidelberg.de
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Since its first introduction in 1995, KEGG PATHWAY has been widely used as a reference knowledge base for understanding biological pathways and functions of cellular processes. Over the last years, KEGG PATHWAY has been significantly expanded with the addition of new pathways related to signal transduction, cellular process and disease (Kaneshia et al., 2008), enhancing its popularity built upon featuring traditional metabolic pathways.
Pathways are stored and presented as graphs on the KEGG server side, where nodes are mainly molecules (protein, compound, etc.) and edges represent relation types between the nodes, e.g. activation or phosphorylation. The graph nature of pathways raised our interest to investigate them with powerful tools implemented in R and Bioconductor (Gentleman et al., 2004), e.g. graph, RBGL and Rgraphviz (Carey et al., 2003). While it is barely possible to query the graph characteristics by manual parsing, a native and straightforward client-side tool is currently missing. Packages like KEGG.db and keggorth use information from KEGG, however none of them makes use of the graph information, precluding the option to study pathways from the graph theory perspective (see Section 4 for more details).
To address this problem, we developed the open source software package KEGGgraph, an interface between KEGG pathways and graph-theoretical models as well as a collection of tools to analyze, dissect and visualize these graphs.
2 SOFTWARE FEATURES
KEGGgraph offers the following functionalities:
Parsing: the package parses the regularly updated KGML (KEGG XML) files into graph models maintaining pathway attributes. It should be noted that one ‘node’ in KEGG pathway does not necessarily map to merely one gene product, for example, the node ERK in the human TGF-β signaling pathway contains two homologs, MAPK1 and MAPK3. Therefore, among several parsing options, users can decide whether to expand these nodes topologically. Beyond facilitating the interpretation of pathways in a gene-oriented manner, the approach also assigns unique identifiers to nodes, enabling merging graphs from different pathways.
Graph operations: two common operations on graphs are subset and merge (union). A subgraph of selected nodes and the edges in between are returned when subsetting, while merging produces a new graph that contains nodes and edges of individual ones. Both are implemented in KEGGgraph.
Visualization: KEGGgraph provides functions to visualize KEGG graphs with custom style. Nevertheless, users are not restricted by them, alternatively they are free to render the graph with other tools like the ones in Rgraphviz.
Besides the functionalities described above, KEGGgraph also provides tools for remote KGML file retrieval, graph feature study and other related tasks. We refer interested readers to the vignettes released along the package.
3 EXAMPLE
Software usage is demonstrated by exploring the graph char-acteristics of pancreatic cancer pathway (http://www.genome.jp/dbget-bin/show_pathway?hsa05212), as KEGG provides pathways also of human diseases.
The human pancreatic cancer pathway is linked to eight other pathways as indicated in the KEGG pathway map. To investigate the global network, we merge them into one graph, consisting of 714 nodes and 3196 edges (see Supplementary Material for the complete source code).
Our aim is to computationally identify the most important nodes. To this end we turn to relative betweenness centrality, one of the measures reflecting the importance of a node in a graph relative to other nodes (Aittokallio and Schwikowski, 2006). For a graph G≔(V, E) with n vertices, the relative betweenness centrality C′B(v) is defined by:
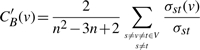 |
(1) |
where σst is the number of shortest paths from s to t, and σst(v) is the number of shortest paths from s to t that pass through a vertex v (Freeman, 1977).
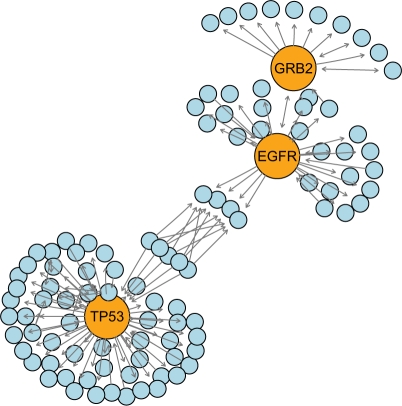
With the function implemented in RBGL package (Brandes, 2001), we identified the most important nodes (Fig. 1) judged by relative betweenness centrality that are TP53 (tumor protein p53), GRB2 (growth factor receptor-bound protein 2) and EGFR (epidermal growth factor receptor). While the oncological roles of TP53 and EGFR are long established in pancreatic carcinoma (Garces et al., 2005), it has only very recently been suggested that the binding of GRB2 to TβR-II is essential for mammary tumor growth and metastasis stimulated by TGF-β (Galliher-Beckley and Schiemann, 2007). No evidence is known to us proving the direct relation between GRB2 and pancreatic cancer. Considering the importance of GRB2 in the network, we suggest to study its role also in this cancer type.
Fig. 1.
Nodes with the highest relative betweenness centrality (in orange) and their interacting partners (blue) in the pancreatic cancer pathway. Relative betweenness centrality estimates the relative importance or role in global network organization.
4 DISCUSSION
Prior to the release of KEGGgraph, several R/Bioconductor packages have been introduced and proved their usefulness in understanding biological pathways with KEGG. However, KEGGgraph is the first package able to parse any KEGG pathways from KGML files into graphs. Existing tools either neglect the graph topology (KEGG.db), or do not parse pathway networks (keggorth), or are specialized for certain pathways (cMAP and pathRender).
Tools have also been implemented on other platforms to use the knowledge of KEGG, e.g. MetaRoute (Blum and Kohlbacher, 2008), Gaggle (Shannon et al., 2006) and Cytoscape (Shannon et al., 2003). To make it unique and complementary to these tools, KEGGgraph allows native statistical and computational analysis of any KEGG pathway based on graph theory in R. Thanks to the variety of Bioconductor packages, KEGGgraph can be built into analysis pipelines targeting versatile biological questions. No active Internet connection is required once the KGML files have been downloaded, reducing the waiting time and network overhead unavoidable in web-service-based approaches. Using tools like KGML-ED (Klukas and Schreiber, 2007), with KEGGgraph it is even possible to explore newly created or edited pathways via KGML files.
Funding: National Genome Research Network (grant number 01GS0864) of the German Federal Ministry of Education and Research (BMBF); International PhD program of the DKFZ (to J.D.Z.).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Aittokallio T, Schwikowski B. Graph-based methods for analysing networks in cell biology. Brief. Bioinform. 2006;7:243–255. doi: 10.1093/bib/bbl022. [DOI] [PubMed] [Google Scholar]
- Blum T, Kohlbacher O. Metaroute: fast search for relevant metabolic routes for interactive network navigation and visualization. Bioinformatics. 2008;24:2108–2109. doi: 10.1093/bioinformatics/btn360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes U. A faster algorithm for betweenness centrality. J. Math. Sociol. 2001;25:163–177. [Google Scholar]
- Carey VJ, et al. Network structures and algorithms in Bioconductor. Bioinformatics. 2005;21:135–136. doi: 10.1093/bioinformatics/bth458. [DOI] [PubMed] [Google Scholar]
- Freeman LC. A set of measures of centrality based on betweenness. Sociometry. 1977;40:35–41. [Google Scholar]
- Galliher-Beckley AJ, Schiemann WP. Grb2 binding to Tyr284 in TβR-II is essential for mammary tumor growth and metastasis stimulated by TGF-β. Carcinogenesis. 2007;29:244–251. doi: 10.1093/carcin/bgm245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcea G, et al. Molecular prognostic markers in pancreatic cancer: a systematic review. Eur. J. Cancer. 2005;41:2213–2236. doi: 10.1016/j.ejca.2005.04.044. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klukas C, Schreiber F. Dynamic exploration and editing of KEGG pathway diagrams. Bioinformatics. 2007;23:344–350. doi: 10.1093/bioinformatics/btl611. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. The Gaggle: an open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006;7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.