Summary
Current therapies do not eradicate HIV from infected patients. Indeed, HIV hides in a latent form insensitive to these therapies. Thus, one priority is to purge these latent reservoirs. But what mechanisms are responsible for latency and what are the reservoirs of latently infected cells? The present knowledge in terms of HIV latency is still incomplete and current therapeutic strategies fail to eradicate completely latently infected cells. What could the future bring?
Keywords: HIV latency, transcriptional interference, HDAC, Tat, NF-κB, HMBA, SAHA, Valproic Acid, PTB, Prostratin, IL-2, IL-7
INTRODUCTION
The major obstacle in curing AIDS is a reservoir of cells, in which HIV resides in a latent form. In these cells, replication-competent virus persists even in patients treated for long periods of time with highly active antiretroviral therapy (HAART). The main source of latent virus is thought to be a small pool (~106 per individual) of latently infected resting memory CD4+ T-lymphocytes [1]. This is a critical point as these resting CD4+ T-lymphocytes, contrary to their mature infected counterparts, have a long life-span. Studies demonstrated that the decay rate of the pool of latently infected cells is extremely slow with a half-life of 44 months meaning that ~70 years of HAART treatment would be required for the eradication of the latent reservoir [2]. Although resting CD4+ T-lymphocytes are the major source of latent virus there are other cell types that carry hidden virus, e.g. microglia in the central nervous system (CNS) [3], dendritic cells [4], resting monocytes and macrophages [5, 6]. Studies have also demonstrated that new cells were being infected in patients undergoing HAART [7–9]. These studies suggest that low-levels of on-going HIV replication and/or occasional activation of latently infected cells may contribute to the persistence of virus in resting CD4+ T-lymphocytes.
Regarding the integration state of viral DNA, two forms of latency are observed: pre-integration and post-integration latency. Although non-integrated DNA is the predominant form of HIV DNA in resting CD4+ T-lymphocytes, pre-integration latency does not contribute greatly to the viral reservoir due to its labile nature. The half-life of non-integrated viral DNA is short due to the susceptibility of reverse transcripts to intracellular degradation [10]. This latency results from the block at the level of HIV DNA integration. In resting cells, levels of ATP are probably too low for the energy dependent nuclear import of viral cDNA together with the large pre-integration complex [11]. However, upon activation of resting cells by antigens or other stimulatory signals, viral cDNA integrates into the host genome and produces new virions [12].
Post-integration latency arises when infected CD4+ T-lymphocytes with integrated HIV DNA revert to a resting memory state. These cells do not produce detectable levels of virions [13]. Thus, they are not recognized by the immune system and are one reason for unsuccessful HAART. Post-integration latency is established and maintained by several different mechanisms. The most important ones at the level of transcription depend on integration sites and the chromatin environment, lack of key host transcription factors (TFs), transcriptional interference (TI) and lack of viral activator Tat. Of minor importance are mechanisms that inhibit post-transcriptional processes: RNA interference, lack of viral protein Rev and lack of host polypyrimidine tract binding protein (PTB).
MECHANISMS OF LATENCY
Latency as a result of inefficient transcriptional initiation and/or elongation
Repressive chromatin environment
The levels of viral expression are influenced by the chromatin environment of cellular DNA at the site of integration of the provirus. Reports on sites of integration of viral DNA are controversial. Jordan et al. demonstrated that latent proviruses integrate preferentially into centromeric heterochromatin [14]. However, other studies indicate that the HIV genome is inserted into intronic regions of actively transcribed genes [15]. The difference in integration sites probably reflects the method and/or cells used. While in the first study, a T lymphocyte cell line was infected in vitro with a full-length viral genome expressing green fluorescent protein and latently infected clones were selected, in the second study the population of resting CD4+ T-cells that do not produce virus was isolated from individuals on HAART. Thus, the later experiment ex vivo presumably reproduces a more relevant picture on viral DNA integration sites. Three studies where T-cells were infected in vitro without selection also demonstrated integration into actively transcribed genes [16–18]. In the study of Lewinski et al. [19] Jurkat T cells were infected with an HIV-based vector and cells were separated into populations with different expression levels from the HIV promoter. They found that in cells with low-level expression, proviral genome integrates into gene deserts, centromeric heterochromatin and very highly expressed cellular genes [19].
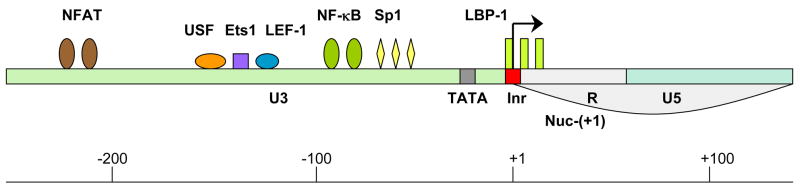
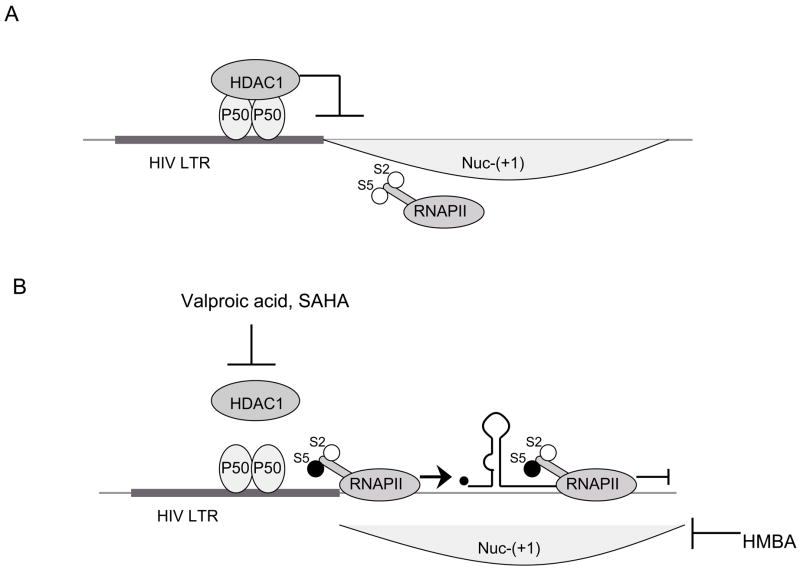
Despite the preferential integration into actively transcribed genes, there are several factors that could form repressive chromatin structure on the HIV long terminal repeat (LTR) which encompasses the viral HIV promoter. In the transcriptionally silent state, two nucleosomes are found on the HIV-LTR. Nuc-(0) spans the region from positions −415 to −255 and Nuc-(+1) from positions +1 to +155 with respect to the transcription start site of HIV genome. The region covered by and between these two nucleosomes contains recognition elements for the sequence-specific host TFs [20], the important ones for transcription of the HIV genome being nuclear factor kappa B (NF-κB), nuclear factor of activated T cell (NFAT), upstream stimulating factor (USF), Ets1 (a protein that binds ETS - a winged helix-turn-helix domain), lymphoid enhancer binding factor-1 (LEF-1), stimulatory protein 1 (Sp1) and leader binding protein-1 (LBP-1) (Fig. 1). The availability of certain cellular proteins directs the binding of chromatin remodeling complexes and consequently the formation of the preinitiation complex. One of the TFs, which plays a crucial role in the repression/de-repression of local chromatin structure surrounding the HIV LTR, is NF-κB. The active form of NF-κB is composed of heterodimers p50 and RelA. When the levels of free p50:RelA in the cell are too low, the same binding sites are occupied by p50:p50 homodimer which binds the histone deacetylase 1 (HDAC1). HDAC1 promotes deacetylation of surrounding histones, compaction of chromatin and consequently suppression of gene expression [21] (Fig. 2A). Williams et al. demonstrated that the recruitment of p50:RelA heterodimer to the HIV LTR relieves this repressive chromatin environment by removing HDAC1 [22]. In this situation, histone acetyltransferases (HATs) bind to the LTR and cause histone acetylation thus inducing the relaxation of chromatin structure and activation of gene transcription [23].
Fig. 1. Binding sites for the critical TFs within the HIV LTR.
LTR is composed of three regions: 3′ untranslated region (U3) (green), transcription regulatory region (R) (grey) and 5′ untranslated region (U5) (blue). The following host TFs which bind to the HIV LTR are designated: NFAT (brown), USF (orange), Ets1 (purple), LEF-1 (blue), NF-kB (green), Sp1 (yellow) and LBP-1 (yellow-green). TATA box and initiator element are marked in grey and red, respectively. Nucleosome 1 location is from +1 to +155 (Nuc-(+1)). Arrow marks the transcription start site.
Fig. 2. Repressive chromatin structure suppresses transcription from the HIV LTR.
A) p50 homodimer bound to the LTR recruits HDAC1 which promotes deacetylation of histones. Thus, nucleosomes form compact chromatin structure and prevent transcription from the LTR. B) Inhibitors of HDAC1 (Valproic Acid and SAHA) which inhibit deacetylation of histones and HMBA relieve this repressive effect. Relaxation of chromatin structure allows the recruitment of preinitiation complex and phosphorylation of serine 5 of peptide repeats on CTD of RNAPII (S5). In this form, RNAPII successfully initiates transcription. However, transcriptionally active chromatin does not suffice for RNAPII to proceed to transcriptional elongation. For this step, RNAPII has to be phosphorylated on serine 2 of CTD peptide repeats (S2).
Lack of key transcription factors
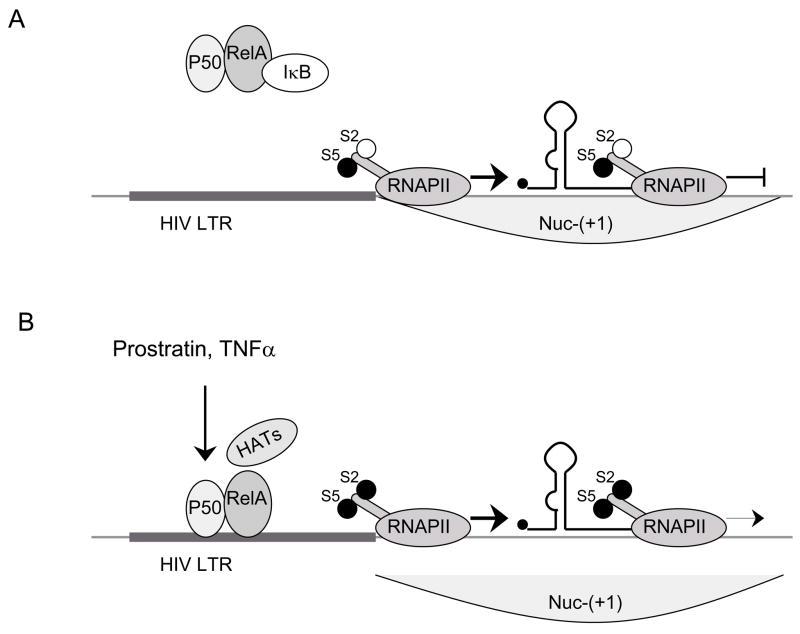
Relaxed chromatin is necessary but not sufficient for the initiation of transcription. Other important factors are levels of some key cellular transcriptional regulators that vary depending on cellular activation. Two of the key TFs, upon which successful transcription depends on are NF-κB [24] and NFAT [25]. Levels of these proteins vary depending on cellular activation. In the cytoplasm of resting memory CD4+ T-lymphocytes, inactive NF-κB (p50:RelA heterodimer) is bound to the inhibitor IκB (Fig. 3A). Interestingly, host protein Murr1 decreases NF-κB activity by increasing the concentrations of IκB. Indeed, genetic knockdown of Murr1 increases the replication of HIV in resting CD4+ T lymphocytes [26].
Fig. 3. The role of host TFs in transcription from the HIV LTR.
A) LTR is accessible for the recruitment of preinitiation complex, S5 is phosphorylated and RNAPII initiates transcription. Lack of positive TFs (non-active NF-κB is bound in the cytoplasm by IκB) results in insufficient transcriptional elongation. S2 is not phosphorylated and HATs are not recruited to the promoter (deacetylated histones in Nuc-(+1)). B) Prostratin and TNF-α increase levels of active NF-κB which results in the recruitment of HATs to the LTR. Possibly, P-TEFb is recruited to the promoter and it phosphorylates S2. Acetylation of histones by HATs relieve the transcriptional elongation block and leads to a suboptimal activation of transcriptional elongation.
When concentrations of active NF-κB are too low, transcription can be initiated successfully but there is still a block at the level of transcriptional elongation. The transcriptosome forms efficiently on the promoter, but RNA polymerase II (RNAPII) stops transcribing the gene soon after it clears the promoter (Fig. 3A). Stimulation by IL-2 or TNFα releases NF-κB, which is then transported to the nucleus where it binds to several promoters including the HIV LTR [24]. In this situation, NF-κB recruits HATs for the relaxation of chromatin structure [27] (Fig. 3B). In addition, NF-κB activation could result in the recruitment of the positive transcription elongation factor b (P-TEFb) to the HIV LTR which has been demonstrated to occur for the IL-8 promoter [28] [29]. This recruitment of P-TEFb to the HIV LTR may lead to a suboptimal activation of transcriptional elongation. NFAT, which synergizes with NF-κB to activate transcription from HIV LTR, is of lesser importance for HIV transcription [25].
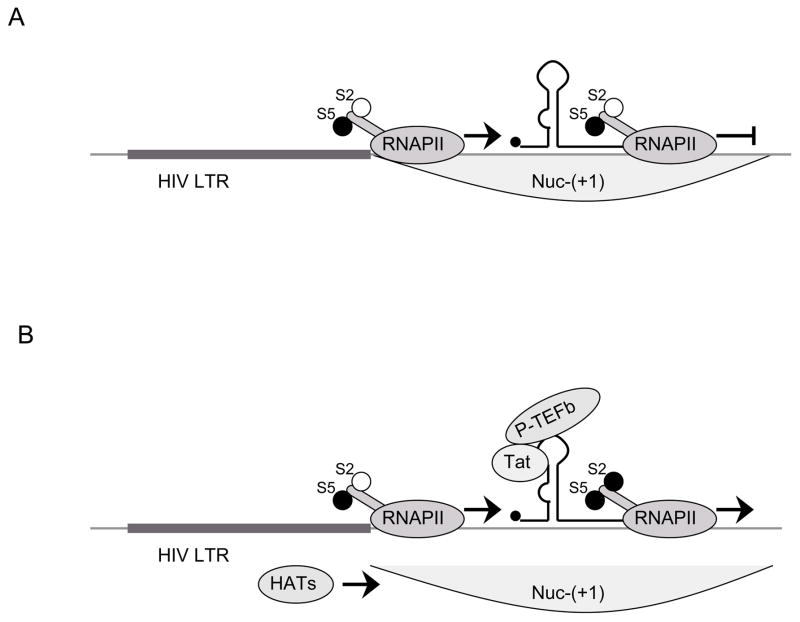
Copying is followed by the synthesis of viral protein Tat, which recruits P-TEFb to the close proximity of C-terminal domain (CTD) of RNAPII via the transactivation response element (TAR) that forms a stable RNA stem loop at the 5′ end of all viral transcripts. [30] (Fig. 4B). As a consequence, optimal RNAPII elongation of viral transcription ensues. Thus, levels of cellular NF-κB alone are not sufficient for productive transcriptional elongation. Rather, a threshold concentration of Tat determines the elongation of RNAPII through the HIV genome. Too low levels of Tat in the cell or mutations in the Tat gene are possible additional reasons for maintaining latency in infected cells (Fig. 4A).
Fig. 4. The role of viral protein Tat in transcription from the HIV LTR.
A) LTR is accessible for the recruitment of preinitiation complex, S5 is phosphorylated and RNAPII initiates transcription. Binding of NF-κB to the HIV LTR leads to the proceeding of RNAPII through HIV genome to some extent, however, lack of viral protein Tat prevents productive transcriptional elongation. B) Tat binds to TAR and recruits P-TEFb which in turn phosphorylates S2. Also, Tat recruits some of the HATs (e.g. PCAF) which acetylate histones following collapse of Nuc-(+1). This results in successful transcriptional elongation.
Transcriptional interference
TI is a mechanism that arises as a result of a relative close proximity of two promoters. Ongoing transcription from an upstream promoter interferes or suppresses initiation of transcription from a downstream promoter if RNAPII ‘reads through’ into downstream gene. The first report on TI was described for prophage lambda promoters which inhibited the expression of the downstream gal operon [31]. Authors named the phenomenon ‘promoter occlusion’ and observed that its effect was complete (the downstream promoter was fully inhibited) when the distance between the promoters was small (less than 10 kb).
TI can be overcome by preventing RNAPII from continuing to transcribe into the downstream gene or by activating to a greater extent the downstream promoter.
By inserting the strong termination signal between two HIV promoters integrated into the genome of HeLa cells Greger et al. prevented the occlusion of the downstream promoter. They also demonstrated that the binding of transcriptional activator Sp1 to the occluded promoter was reduced and this effect was overcome when the promoter was protected by a terminator [32]. TI was described in several biological systems ex vivo. However, because of the integration of HIV genome into introns of actively transcribed genes, TI is probably a common cause for latency in vivo (Fig. 5A).
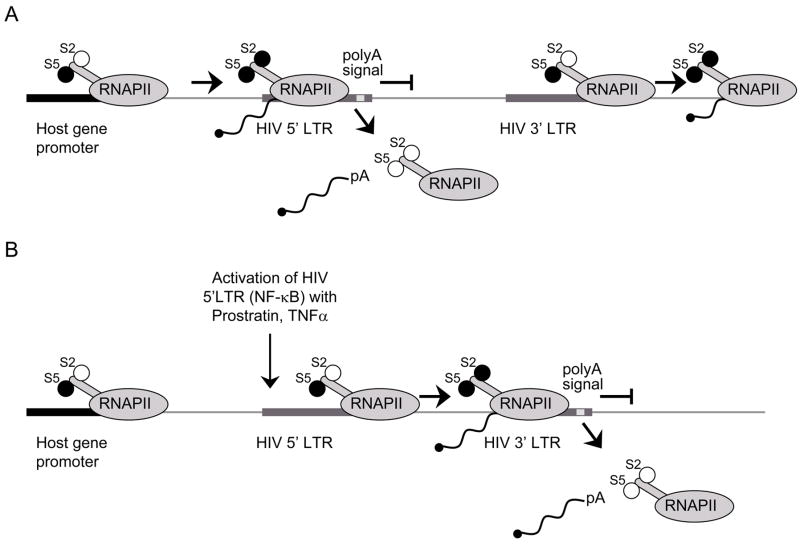
Fig. 5. TI is a possible mechanism that directs latency.
A) Active transcription from a host gene promoter interferes with transcription from downstream HIV 5′LTR. Polymerase ‘reads through’ the 5′LTR and dissociates from DNA at the polyadenylation signal in the 5′LTR resulting in a truncated form of mRNA. 3′ LTR is not occluded and is available for the recruitment of the preinitiation complex and proceeding of the transcription into the host gene. Transcription from the 3′LTR again results in a truncated form of mRNA. B) Activation of the 5′LTR (NF-κB, Tat) overcomes TI. Transcription is successfully initiated from 5′LTR. RNAPII proceeds through the HIV genome and terminates at the polyadenylation signal in 3′LTR. In this case, 3′LTR is occluded and there is no transcription from this LTR.
Post-transcriptional blocks
Inefficient export of genomic RNA
By binding to a target sequence (Rev response element, RRE) located within the viral RNA region coding for the envelope protein, the viral protein Rev exports unspliced HIV RNA from the nucleus of infected cells. In CD4+ positive HeLa cells, which express constitutively a Rev-deficient virus, a critical threshold of Rev is required for highly productive HIV replication [33]. Thus, low levels of Rev in infected cells might potentiate viral latency (Fig. 6).
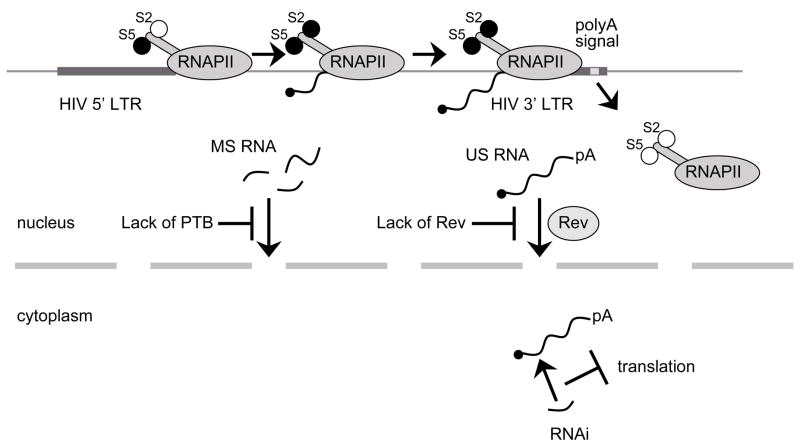
Fig. 6. Latency resulting from post-transcriptional block.
Viral genome is efficiently transcribed into full-length polyadenylated RNA. Aberrant localization of US and MS RNA: (i) without viral protein Rev US RNA are not protected from splicing and consequently, the full-length HIV RNA is not exported from the nucleus into the cytoplasm, (ii) too low levels of host protein PTB result in nuclear localization of MS RNA. Degradation of viral RNA: in the nucleus or in the cytoplasm viral RNA can be degraded by viral or host miRNA.
Inefficient export of multiply spliced RNA species
Lately, a similar block to inefficient genomic RNA export has been described [34]. In this scenario, an aberrant localization of multiply spliced (MS) HIV RNAs causes a post-transcriptional block in viral gene expression. These RNA species encode positive regulators Rev and Tat, which are crucial for the expression of the viral genome. It has been demonstrated that in resting CD4+ T-lymphocytes, MS RNA species are localized to the nucleus and are not exported to the cytoplasm. Interestingly, the overexpression of host PTB, a protein involved in post-transcriptional regulation of gene expression (reviewed in [35]), resulted in cytoplasmic localization of MS RNA species in latently infected cells from patients on HAART, thus reverting this post-transcriptional block (Fig. 6) [34].
RNA interference
Recently, microRNAs (miRNAs) that encode sequences embedded in the HIV genome and virus-derived miRNAs have been identified. Interestingly, host cellular factor TAR RNA-binding protein (TRBP) which is a component of dsRNA-specific endoribonuclease (Dicer) -mediated dsRNA processing was found to be a cofactor in Tat:TAR interactions. Possibly, miRNAs bind to the HIV RNA which is in turn degraded and thus not translated into viral proteins. One can not exclude the possibility that HIV exploits the cellular RNAi machinery to maintain a latent infection (Fig. 6, reviewed in [36]).
OTHER RESERVOIRS OF LATENTLY INFECTED CELLS
Although the most important source of latent reservoirs are the resting memory CD4+ T-lymphocytes present in lymph nodes and in peripheral blood, there are some other cells that contribute to the reservoir. In lymphoid tissues, follicular dendritic cells play important roles in the transmission of HIV to CD4+ lymphocytes. By producing a subsequent systematic spread of the virus, these cells represent a major reservoir for HIV in lymphoid tissues [37]. Dendritic cells retain viral particles on their surface, but they are not permissive for HIV infection. The virus is trapped on their surfaces and can be transmitted to T-cells in trans [4].
Macrophages contribute significantly to the viral reservoir in patients on HAART in the later phase when CD4+ T-lymphocytes are depleted [5]. Of importance, virions persist within monocyte-derived macrophages for weeks [6]. It has to be noticed that potential reservoirs for HIV are located in various locations in the body and not just in lymph nodes and peripheral blood. Latent virus was detected in CD4+ T-lymphocytes and macrophages in the semen [38], in the male urogenital tract [39], in the liver (Kupffer cells) [40] and in the CNS [41]. CNS is an important reservoir for the latent virus, which most probably accesses the CNS from the blood stream by ingress of infected monocytes and macrophages. In the CNS, the principal cellular targets for HIV are macrophages and microglial cells [3].
ERADICATION OF HIV LATENTLY INFECTED CELLS
How to purge the reservoir of latently infected cells?
Strategies have been designed to reactivate HIV from latency or to directly target this reservoir of resting memory CD4+ T-lymphocytes (See Table 1 for a summary of the different drugs that could reactivate HIV from latency). In the first approach, once reactivated, HIV expresses its genes, replicates and thus becomes susceptible to immune elimination and HAART. Eventually, host cells bearing the integrated HIV genome, which have a relatively short half-life in vivo, die. The second strategy aims at purging the reservoir by coupling immunotoxins to molecules recognizing specific markers.
Table 1.
| Compound | Effects on cells and roles in HIV latency | IC50 | Ref |
|---|---|---|---|
| IL-2 |
|
106 units per day for 5 days | [49, 50] |
| IL-7 |
|
10 to 20 ng/ml (R&D) | [53] |
| Prostratin |
|
0.1 μM | [54–57] |
| HMBA |
|
1 mM | [46] |
| EMBA |
|
100 μM | [67] |
| SAHA |
|
1 μM | [68] |
| Valproic Acid |
|
500 mg x2 daily in patients | [43] |
| Tat |
|
1 μg/ml of recombinant Tat protein exogenously | [59–61] |
Overcoming chromatin compaction
As chromatin remodeling has been shown to play a key role in HIV silencing in cell line models of latency, inhibitors of chromatin compaction were tested (Fig. 2B) [42, 43]. Valproic acid, an HDAC inhibitor previously shown to stimulate the release of virus from latently infected CD4+ T-lymphocytes in vitro [44] has recently been tested in 4 patients who also received enfuvirtide as a complement for their therapy. Analysis of the latent reservoir showed a decrease of 68 to 84 % in the number of latently infected memory CD4+ T-lymphocytes in 3 out of 4 patients [43]. Valproic acid may also lower HIV-induced cytotoxicity in the central nervous system [45]. More recently HMBA (Hexamethylene Bisacetamide), a compound of the same family as SAHA (Suberoylamide Hydroxamic Acid), another HDAC inhibitor, has been implicated as a chromatin remodeling factor. Indeed, although it does not induce any histone acetylation, it can displace nucleosome 1, thus activating HIV replication in chronically infected cell lines [46]. HMBA is not an HDAC inhibitor and acts through an as yet unknown mechanism. Our results suggest a role for Akt and P-TEFb in this activation as HMBA releases it from its regulatory factor HEXIM1 (Contreras et al., submitted). HMBA is of particular interest as it also inhibits HIV replication ex-vivo in PBMCs. However, clinical trials have shown that HMBA may be too toxic and labile to use in patients [47, 48]. In addition, structurally related compounds such as polymethylene bisacetamide and diethyl bis-(pentamethylene-N,N-dimethylcarboxamide) malonate demonstrate effects similar to the one of HMBA on cell metabolism and might be further considered for new approaches aiming at purging the reservoir of latently infected cells.
Activation of transcription
The following approaches help to overcome blocks involving lack of transcription factors. They also prevent TI by increasing transcription at the HIV LTR (Fig. 3B, 5B). Initial trials involved the use of broad activators of T-lymphocytes, that activate resting CD4+ T-lymphocytes, like anti-CD3 antibodies and IL-2 in individuals receiving HAART to reactivate the latent pool of HIV infected cells, and also to stimulate the exhausted immune system [49]. Although the anti-CD3 approach revealed itself unusable because of its toxicity, the IL-2 approach resulted in a marked decrease of the latent pool of HIV but failed to eradicate the infection [49, 50]. Indeed, this treatment may induce susceptibility to HIV infection in a wide number of cells and possibly fails to activate all latently infected cells.
Milder activators of T-lymphocytes were studied in a second approach. IL-7 is an important cytokine for T-lymphocyte homeostasis. It promotes memory CD4+ T-lymphocytes maintenance, turnover, and, importantly, it also activates HIV replication. It was also tested as a complement to HAART [51, 52]. IL-7 reactivated HIV replication with minimal effects on T-lymphocyte phenotype [51]. Importantly, the analysis of viral genomes after reactivation demonstrated that IL-7 induces the replication of different viral isolates compared to stimulation with IL-2/Phytohemagglutinin (PHA). These results suggested that a combination using IL-2 and IL-7 might be complimentary [53]. Prostratin, a phorbol ester that activates protein kinases C (PKCs), is another promising compound. It activates cells without induction of proliferation [54, 55]. Studies in PBMCs from infected individuals as well as on lymphoid tissue ex-vivo have demonstrated that prostratin reactivates viral replication in latently infected cells with minimal side effects as well as inhibits de novo infection [55, 56]. Prostratin acts on activated cells by inducing the transcription factor NF-κB, which is excluded from the nucleus in quiescent T-lymphocytes [57]. NF-κB (p65) has three main effects: it recruits HATs, removes the p50 subunit involved in chromatin remodeling at the HIV LTR and could recruit P-TEFb to increase transcriptional elongation.
Most of the latent reservoir is established and maintained at the level of transcription. Inefficient or no elongation of transcription represents one of the most important mechanisms whereby integrated proviruses are not expressed in vivo [58]. As Tat recruits P-TEFb to the HIV LTR via TAR, it has been proposed as a possible treatment alternative to reactivate HIV from latency (Fig. 4B) [59]. In this study, transgenic mice that secrete a Tat.GFP fusion protein from the β-cells of the pancreas were created. The secreted Tat.GFP chimera spread to all tissues and targeted latently infected cells ex-vivo. As expected, Tat was able to enter cells [60] and to activate viral transcription. In addition, the extracellular Tat has also been shown to interact with several membrane receptors and thus stimulate signaling pathways including the NF-κB pathway [61]. These effects may also participate in the further activation of latent pools of HIV infected cells. Importantly, Tat had no deleterious effects in this system, as well as when used in vaccine trials. Thus Tat, either added exogenously or secreted by transduced cells, could be tested further for its effects on the reactivation of HIV from latency.
Overcoming the block at the level of mRNA export
Over-expressing PTB in PBMCs from treated patients has been shown to reactivate HIV replication [34]. Unraveling the mechanisms of regulation of PTB expression in resting memory CD4+ T-lymphocytes might give new insights into the design of new approaches to eradicate the latent virus. Importantly, molecules involved in the differentiation of CD4+ T-lymphocytes and able to induce expression of PTB could be used as therapeutic agents (Fig. 6). To bypass the need for multiply-spliced mRNAs to be exported from the nucleus, another possibility would be to introduce Rev and Tat proteins produced by these mRNAs into latently infected cells.
Directly depleting the reservoir
HIV latently infected resting CD4+ T-lymphocytes are transcriptionally silent. Thus integrated proviruses are under perfect covert, no specific antigen can be used as a marker for this population. Attempts to deplete directly this reservoir have focused on the use of antibodies against CD45RO, a specific marker of resting memory CD4+ T-lymphocytes. Coupled to ricin, an immunotoxin, these antibodies kill specifically memory CD4+ T-lymphocytes, that bear most of the latent HIV genomes [62]. These studies conducted ex-vivo have shown a significant reduction in the number of latently infected cells obtained from HIV-infected individuals with viremia below the limit of detection. However, such an approach would severely compromise the memory T-lymphocyte population of the patient.
FUTURE PERSPECTIVES
One of the key issues in HIV latency research is to create relevant models. Obviously, primary cells from treated patients are not easy to obtain or handle. Some cell lines are used as models for latency. However, these cells enter the cell cycle and are not quiescent, contrary to latently infected primary cells which are blocked in the Go state of the cell cycle. One of the main goals for future studies in HIV latency is to establish a reliable model.
To study factors involved in the reactivation of HIV replication, one could analyze in detail the pattern of expression of genes differentially expressed in latently infected cells. Such efforts have already been realized using microarray approaches and have identified two potential candidates involved in viral latency: IRF8 and NCoA3 [63, 64], suggesting new strategies might be investigated targeting these factors. Importantly, memory CD4+ T-lymphocytes, the major reservoir of latent HIV, and mechanisms involved in their stimulation should be further studied. Such work would provide new approaches to reactivate the latent reservoir.
What can be realized in terms of therapy? Probably the most promising results have been obtained using the HDAC inhibitor Valproic Acid which could lower by 70% the number of latently infected cells. But, all latently infected cells must be purged. The design and testing of new HDAC inhibitors may improve this therapy. However, as different mechanisms account for latency, it is highly probable that a combination of compounds will be required. This combination, including HDAC inhibitor and activators of viral transcription such as Tat or prostratin, may be able to reactivate a wider population of latent proviruses.
As an alternative to reactivating the virus from latency, all HIV reservoirs may be targeted by molecules recognizing the viral genome, the only marker of HIV-latently infected cells. PNA (polyamide nucleic acid), or zinc finger based proteins could be used in therapy [64]. These molecules coupled to DNA endonucleases or bleomycin, for example, could induce the degradation of the targeted viral genomes.
However, the assessment that memory CD4+ T-lymphocytes bear most of the latent virus is not certain. Other reservoirs of HIV infected cells, notably in the brain and semen [3, 38, 41], may also represent another threat. In this respect, further work should be performed in monkey models of AIDS to analyze the presence of integrated SIV genomes in different organs after HAART. Furthermore, the penetrance and efficiency of current therapies to these reservoirs is not very well known and should be investigated.
CONCLUSION
Since the advent of HAART, viral latency has been considered the main barrier to the eradication of HIV. Latency is established mainly in memory CD4+ T-lymphocytes. Several mechanisms may account for viral silencing in this reservoir including chromatin remodeling, lack of transcription factors and TI. Further work is needed to understand fully the mechanisms involved in HIV latency in vivo.
Combination of therapies aiming at reactivating HIV from different states of latency may help to purge the virus from its reservoirs. However, eradicating HIV may prove to be quite a challenge. Other strategies may aim at restoring immuno-competence and avoid immune exhaustion. Recent publications pointed out the crucial role of PD-1, a negative regulator involved in immune exhaustion during chronic HIV infection [65, 66]. Strategies inhibiting this pathway may prove useful to maintain the integrity of the immune system during viral infection and lead to its clearance after reactivation.
Executive summary
Epidemiology
HIV is a scourge that hits 40 million people worldwide.
HAART is a powerful therapy that can lower viral load to undetectable levels.
Despite its potency, HIV is not cleared from infected individuals, HIV remains in hidden reservoirs, mainly in a latent form insensitive to HAART.
Reservoirs of HIV
HIV is present in a latent form mainly in memory resting CD4+ T-lymphocytes.
Chromatin remodeling is an important mechanism accounting for this latent state by preventing accessibility of the HIV genome
HIV is also silenced at the level of transcription by two other mechanisms: transcriptional interference and lack of transcription factors (e.g. NF-κB)
Other blocks at a post-transcriptional level might support HIV latency
Other reservoirs host HIV latent forms insensitive to therapy: Macrophages, Follicular dendritic cells, dendritic cells, Kupfer cells, urogenital tract, semen CNS.
Purging latent HIV
Different approaches are considered to reactivate HIV from this latent state thus rendering it sensitive to therapies.
Valproic Acid is an HDAC inhibitor that gave encouraging results in patients
HMBA reactivates HIV replication from latency in an HDAC independent manner.
Interleukins (2/7) have been tested for their ability to activate lymphocytes and restore immune system functions.
Prostratin, a promising agent, activates HIV from latency through the PKC/NF-κB pathway.
The memory CD4+ T-lymphocytes could also be purged using immunotoxins specific for this subpopulation.
Future Directions
The HIV Tat protein itself could be considered as a therapeutic agent for its role in HIV transcription as well as for its ability to penetrate any cell in the organism.
Other chromatin remodeling agents should be tested.
Endonucleases coupled to specific molecules may target and destroy HIV genomes.
Combination of therapies might be the only way to go as it could reactivate viruses from different kind of latent states.
More work has to be performed to clearly identify the HIV latent reservoirs.
References
- 1**.Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1(12):1284–90. doi: 10.1038/nm1295-1284. The first study in which stable form of latent HIV was found in purified resting CD4+ T-lymphocytes. [DOI] [PubMed] [Google Scholar]
- 2*.Finzi D, Blankson J, Siliciano JD, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5(5):512–7. doi: 10.1038/8394. Demonstrates the importance of latent infection of resting CD4+ T cells for viral persistence and predicts the long time required for eradication of the latent reservoir. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Wesselingh SL, Griffin DE, McArthur JC, Johnson RT, Glass JD. Localization of HIV-1 in human brain using polymerase chain reaction/in situ hybridization and immunocytochemistry. Ann Neurol. 1996;39(6):705–11. doi: 10.1002/ana.410390606. [DOI] [PubMed] [Google Scholar]
- 4.Geijtenbeek TB, Torensma R, van Vliet SJ, et al. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100(5):575–85. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 5.Igarashi T, Imamichi H, Brown CR, Hirsch VM, Martin MA. The emergence and characterization of macrophage-tropic SIV/HIV chimeric viruses (SHIVs) present in CD4+ T cell-depleted rhesus monkeys. J Leukoc Biol. 2003;74(5):772–80. doi: 10.1189/jlb.0503196. [DOI] [PubMed] [Google Scholar]
- 6.Sharova N, Swingler C, Sharkey M, Stevenson M. Macrophages archive HIV-1 virions for dissemination in trans. Embo J. 2005;24(13):2481–9. doi: 10.1038/sj.emboj.7600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci U S A. 1998;95(15):8869–73. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hockett RD, Kilby JM, Derdeyn CA, et al. Constant mean viral copy number per infected cell in tissues regardless of high, low, or undetectable plasma HIV RNA. J Exp Med. 1999;189(10):1545–54. doi: 10.1084/jem.189.10.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Chung C, Hu BS, et al. Genetic characterization of rebounding HIV-1 after cessation of highly active antiretroviral therapy. J Clin Invest. 2000;106(7):839–45. doi: 10.1172/JCI10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swiggard WJ, O’Doherty U, McGain D, Jeyakumar D, Malim MH. Long HIV type 1 reverse transcripts can accumulate stably within resting CD4+ T cells while short ones are degraded. AIDS Res Hum Retroviruses. 2004;20(3):285–95. doi: 10.1089/088922204322996527. [DOI] [PubMed] [Google Scholar]
- 11.Bukrinsky MI, Sharova N, Dempsey MP, et al. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc Natl Acad Sci U S A. 1992;89(14):6580–4. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12*.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61(2):213–22. doi: 10.1016/0092-8674(90)90802-l. Demonstrates for the first time that pre-integration latency is a labile form of latency. [DOI] [PubMed] [Google Scholar]
- 13.Chun TW, Justement JS, Lempicki RA, et al. Gene expression and viral prodution in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals. Proc Natl Acad Sci U S A. 2003;100(4):1908–13. doi: 10.1073/pnas.0437640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. Embo J. 2003;22(8):1868–77. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Han Y, Lassen K, Monie D, et al. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol. 2004;78(12):6122–33. doi: 10.1128/JVI.78.12.6122-6133.2004. The first ex-vivo study that demonstrates the preferential integration of the HIV genome into actively transcribed host genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(4):521–9. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 17.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300(5626):1749–51. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 18.Lewinski MK, Yamashita M, Emerman M, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2(6):e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewinski MK, Bisgrove D, Shinn P, et al. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J Virol. 2005;79(11):6610–9. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steger DJ, Workman JL. Stable co-occupancy of transcription factors and histones at the HIV-1 enhancer. Embo J. 1997;16(9):2463–72. doi: 10.1093/emboj/16.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9(3):625–36. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 22.Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. Embo J. 2006;25(1):139–49. doi: 10.1038/sj.emboj.7600900. Describes a specific mechanism of latency where p50 dimers recruit HDAC1 to the HIV-1 LTR. The proximity of HDAC1 probably results in a repressed chromatin structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4(4):276–84. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 24**.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326(6114):711–3. doi: 10.1038/326711a0. The pioneer studies demonstrating that NF-κB regulates HIV transcription. [DOI] [PubMed] [Google Scholar]
- 25.Kinoshita S, Su L, Amano M, Timmerman LA, Kaneshima H, Nolan GP. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity. 1997;6(3):235–44. doi: 10.1016/s1074-7613(00)80326-x. [DOI] [PubMed] [Google Scholar]
- 26.Ganesh L, Burstein E, Guha-Niyogi A, et al. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature. 2003;426(6968):853–7. doi: 10.1038/nature02171. [DOI] [PubMed] [Google Scholar]
- 27.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1(5):661–71. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 28*.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8(2):327–37. doi: 10.1016/s1097-2765(01)00314-8. Identifies binding of NF-kB with P-TEFb and its recruitment to the IL-8 promoter. The study is important as a potential mechanism for the recruitment of P-TEFb to the HIV LTR. [DOI] [PubMed] [Google Scholar]
- 29.Luecke HF, Yamamoto KR. The glucocorticoid receptor blocks P-TEFb recruitment by NFkappaB to effect promoter-specific transcriptional repression. Genes Dev. 2005;19(9):1116–27. doi: 10.1101/gad.1297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92(4):451–62. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 31**.Adhya S, Gottesman M. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell. 1982;29(3):939–44. doi: 10.1016/0092-8674(82)90456-1. The first report on transcriptional interference described for prophage lambda promoters which inhibited the expression of the downstream gal operon. [DOI] [PubMed] [Google Scholar]
- 32*.Greger IH, Demarchi F, Giacca M, Proudfoot NJ. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res. 1998;26(5):1294–301. doi: 10.1093/nar/26.5.1294. Demonstrates that there is a lack of transcriptional regulator Sp1 on the HIV promoter which is occluded by an upstream promoter and investigates the possibilities of overcoming transcriptional interference. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Pomerantz RJ, Seshamma T, Trono D. Efficient replication of human immunodeficiency virus type 1 requires a threshold level of Rev: potential implications for latency. J Virol. (3) 1992;66:1809–13. doi: 10.1128/jvi.66.3.1809-1813.1992. The first study in which Rev protein is described as a crucial protein for the HIV mRNA expression and thus, as a factor contributing to post-transcriptional latency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006;2(7):e68. doi: 10.1371/journal.ppat.0020068. Demonstrates for the first time that not only unspliced but also multiply spliced HIV RNA species are retained in the nucleus and most probably contribute to the post-transcriptional latency. The study also identifies a PTB protein which rescues the retention of multiply spliced mRNA in resting CD4+ T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spellman R, Rideau A, Matlin A, et al. Regulation of alternative splicing by PTB and associated factors. Biochem Soc Trans. 2005;33(Pt 3):457–60. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- 36.Weinberg MS, Morris KV. Are viral-encoded microRNAs mediating latent HIV-1 infection? DNA Cell Biol. 2006;25(4):223–31. doi: 10.1089/dna.2006.25.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spiegel H, Herbst H, Niedobitek G, Foss HD, Stein H. Follicular dendritic cells are a major reservoir for human immunodeficiency virus type 1 in lymphoid tissues facilitating infection of CD4+ T-helper cells. Am J Pathol. 1992;140(1):15–22. [PMC free article] [PubMed] [Google Scholar]
- 38.Quayle AJ, Xu C, Mayer KH, Anderson DJ. T lymphocytes and macrophages, but not motile spermatozoa, are a significant source of human immunodeficiency virus in semen. J Infect Dis. 1997;176(4):960–8. doi: 10.1086/516541. [DOI] [PubMed] [Google Scholar]
- 39.Zhu T, Wang N, Carr A, et al. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70(5):3098–107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lefkowitch JH. The liver in AIDS. Semin Liver Dis. 1997;17(4):335–44. doi: 10.1055/s-2007-1007210. [DOI] [PubMed] [Google Scholar]
- 41.Glass JD, Johnson RT. Human immunodeficiency virus and the brain. Annu Rev Neurosci. 1996;19:1–26. doi: 10.1146/annurev.ne.19.030196.000245. [DOI] [PubMed] [Google Scholar]
- 42.Ylisastigui L, Coull JJ, Rucker VC, et al. Polyamides reveal a role for repression in latency within resting T cells of HIV-infected donors. J Infect Dis. 2004;190(8):1429–37. doi: 10.1086/423822. [DOI] [PubMed] [Google Scholar]
- 43**.Lehrman G, Hogue IB, Palmer S, et al. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet. 2005;366(9485):549–55. doi: 10.1016/S0140-6736(05)67098-5. First proof of the effect of an HDAC inhibitor in-vivo on the pool of latently infected T-CD4+ lymphocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. Aids. 2004;18(8):1101–8. doi: 10.1097/00002030-200405210-00003. [DOI] [PubMed] [Google Scholar]
- 45.Schifitto G, Peterson DR, Zhong J, et al. Valproic acid adjunctive therapy for HIV-associated cognitive impairment: a first report. Neurology. 2006;66(6):919–21. doi: 10.1212/01.wnl.0000204294.28189.03. [DOI] [PubMed] [Google Scholar]
- 46.Klichko V, Archin N, Kaur R, Lehrman G, Margolis D. Hexamethylbisacetamide remodels the human immunodeficiency virus type 1 (HIV-1) promoter and induces Tat-independent HIV-1 expression but blunts cell activation. J Virol. 2006;80(9):4570–9. doi: 10.1128/JVI.80.9.4570-4579.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andreeff M, Stone R, Michaeli J, et al. Hexamethylene bisacetamide in myelodysplastic syndrome and acute myelogenous leukemia: a phase II clinical trial with a differentiation-inducing agent. Blood. 1992;80(10):2604–9. [PubMed] [Google Scholar]
- 48.Ward FT, Kelley JA, Roth JS, et al. Phase I bioavailability and pharmacokinetic study of hexamethylene bisacetamide (NSC 95580) administered via nasogastric tube. Cancer Res. 1991;51(7):1803–10. [PubMed] [Google Scholar]
- 49*.Chun TW, Engel D, Mizell SB, et al. Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat Med. 1999;5(6):651–5. doi: 10.1038/9498. Determines the effect of IL-2 on the pool of latently infected cells in HAART treated patients. [DOI] [PubMed] [Google Scholar]
- 50.Stellbrink HJ, van Lunzen J, Westby M, et al. Effects of interleukin-2 plus highly active antiretroviral therapy on HIV-1 replication and proviral DNA (COSMIC trial) Aids. 2002;16(11):1479–87. doi: 10.1097/00002030-200207260-00004. [DOI] [PubMed] [Google Scholar]
- 51.Scripture-Adams DD, Brooks DG, Korin YD, Zack JA. Interleukin-7 induces expression of latent human immunodeficiency virus type 1 with minimal effects on T-cell phenotype. J Virol. 2002;76(24):13077–82. doi: 10.1128/JVI.76.24.13077-13082.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Wang FX, Xu Y, Sullivan J, et al. IL-7 is a potent and proviral strain-specific inducer of latent HIV-1 cellular reservoirs of infected individuals on virally suppressive HAART. J Clin Invest. 2005;115(1):128–37. doi: 10.1172/JCI22574. Demonstrates that the effect of IL-7 is distinct and potentially complimentary to the effect of IL-2 on latently infected cells fromm HAART treated patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nunnari G, Pomerantz RJ. IL-7 as a potential therapy for HIV-1-infected individuals. Expert Opin Biol Ther. 2005;5(11):1421–6. doi: 10.1517/14712598.5.11.1421. [DOI] [PubMed] [Google Scholar]
- 54.Korin YD, Brooks DG, Brown S, Korotzer A, Zack JA. Effects of prostratin on T-cell activation and human immunodeficiency virus latency. J Virol. 2002;76(16):8118–23. doi: 10.1128/JVI.76.16.8118-8123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55**.Kulkosky J, Culnan DM, Roman J, et al. Prostratin: activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood. 2001;98(10):3006–15. doi: 10.1182/blood.v98.10.3006. Describes a potential role for prostratin in HIV reactivation from latency. [DOI] [PubMed] [Google Scholar]
- 56.Biancotto A, Grivel JC, Gondois-Rey F, et al. Dual role of prostratin in inhibition of infection and reactivation of human immunodeficiency virus from latency in primary blood lymphocytes and lymphoid tissue. J Virol. 2004;78(19):10507–15. doi: 10.1128/JVI.78.19.10507-10515.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57*.Williams SA, Chen LF, Kwon H, et al. Prostratin antagonizes HIV latency by activating NF-kappaB. J Biol Chem. 2004;279(40):42008–17. doi: 10.1074/jbc.M402124200. Describes the signaling pathway used by prostratin to activate NF-kappaB and thus HIV transcription. [DOI] [PubMed] [Google Scholar]
- 58.Adams M, Sharmeen L, Kimpton J, et al. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc Natl Acad Sci U S A. 1994;91(9):3862–6. doi: 10.1073/pnas.91.9.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59*.Lin X, Irwin D, Kanazawa S, et al. Transcriptional profiles of latent human immunodeficiency virus in infected individuals: effects of Tat on the host and reservoir. J Virol. 2003;77(15):8227–36. doi: 10.1128/JVI.77.15.8227-8236.2003. Describes a role for Tat in reactivation of HIV from latency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vendeville A, Rayne F, Bonhoure A, Bettache N, Montcourrier P, Beaumelle B. HIV-1 Tat enters T cells using coated pits before translocating from acidified endosomes and eliciting biological responses. Mol Biol Cell. 2004;15(5):2347–60. doi: 10.1091/mbc.E03-12-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rubartelli A, Poggi A, Sitia R, Zocchi MR. HIV-I Tat: a polypeptide for all seasons. Immunol Today. 1998;19(12):543–5. doi: 10.1016/s0167-5699(98)01351-6. [DOI] [PubMed] [Google Scholar]
- 62**.Saavedra-Lozano J, McCoig C, Xu J, et al. An anti-CD45RO immunotoxin kills latently infected human immunodeficiency virus (HIV) CD4 T cells in the blood of HIV-positive persons. J Infect Dis. 2002;185(3):306–14. doi: 10.1086/338565. Novel approach aimed at depleting the cells that host most of the latent reservoir of latently infected cells. [DOI] [PubMed] [Google Scholar]
- 63.Munier S, Delcroix-Genete D, Carthagena L, Gumez A, Hazan U. Characterization of two candidate genes, NCoA3 and IRF8, potentially involved in the control of HIV-1 latency. Retrovirology. 2005;2:73. doi: 10.1186/1742-4690-2-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sergeyev DS, Godovikova TS, Zarytova VF. Catalytic site-specific cleavage of a DNA-target by an oligonucleotide carrying bleomycin A5. Nucleic Acids Res. 1995;23(21):4400–6. doi: 10.1093/nar/23.21.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65**.Trautmann L, Janbazian L, Chomont N, et al. Upregulation of PD-1 expression on HIV-specific CD8 + T cells leads to reversible immune dysfunction. Nat Med. 2006 doi: 10.1038/nm1482. First to identify a role for PD-1 in immune dysfunction during HIV infection. [DOI] [PubMed] [Google Scholar]
- 66.Day CL, Kaufmann DE, Kiepiela P, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443(7109):350–4. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 67.Richon VM, Webb Y, Merger R, et al. Second generation hybrid polar compounds are potent inducers of transformed cell differentiation. Proc Natl Acad Sci U S A. 1996;93(12):5705–8. doi: 10.1073/pnas.93.12.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marks PA, Richon VM, Kelly WK, Chiao JH, Miller T. Histone deacetylase inhibitors: development as cancer therapy. Novartis Found Symp. 2004;259:269–81. discussion 281–8. [PubMed] [Google Scholar]