Abstract
We have characterized 28 zebrafish lines with heterozygous mutations in ribosomal protein (rp) genes, and found that 17 of these are prone to develop zebrafish malignant peripheral nerve sheath tumors (zMPNST). Heterozygotes from the vast majority of tumor-prone rp lines were found to be growth-impaired, though not all growth-impaired rp lines were tumor-prone. Significantly, however, the rp lines with the greatest incidence of zMPNSTs all displayed a growth impairment. Furthermore, heterozygous cells from one tumor-prone rp line were out-competed by wild-type cells in chimeric embryos. The growth impairment resulting from heterozygosity for many rp genes suggests that a global defect in protein translation exists in these lines, raising the possibility that a translation defect that precedes tumor development is predictive of tumorigenesis.
Keywords: ribosomal proteins, growth, translation, cancer, zebrafish
Introduction
Control of cell growth and cell proliferation are intimately linked. While it has long been known that protein translation is upregulated in many tumors, this has been assumed to be a consequence of increased cell proliferation. However, a direct causal role for aberrant translation in cancer is beginning to be elucidated (Clemens, 2004; Bilanges and Stokoe, 2007). There is evidence to suggest that many translation factors may be proto-oncogenes, most notably eIF2, eIF4E, and eIF4G. Overactivity or overexpression of these proteins has been demonstrated to cause malignant transformation (Donzé et al., 1995; Fukuchi-Shimogori et al., 1997; Lazaris-Karatzas et al., 1990). Increased eIF4E activity, for example, selectively enhances the translation of transcripts with complex 5’ untranslated regions, many of which encode proteins that promote cell survival and growth (DeBenedetti and Graff, 2004). Upregulation of the mTOR pathway contributes to tumorigenesis, in part through activation of both ribosome biogenesis and cap-dependent translation (Guertin and Sabatini, 2007). Oncogenic signaling by Ras and Akt has also been shown to lead directly to a preferential recruitment of certain mRNAs to the ribosome (Rajasekhar et al., 2003). Furthermore, changes in mRNAs associated with polysomes were found to be more significant than changes at the transcriptional level in a cell model of metastatic progression of colorectal cancer (Provenzani et al., 2006), suggesting that alterations in translation may play a role not only in tumor initiation, but also progression.
There is reason to believe that disruption of the core components of the translation machinery itself, specifically mutation of ribosomal protein (rp) genes, can lead to cancer. In humans, heterozygous mutation of RPS19 is found in 25% of cases of Diamond-Blackfan anemia (DBA) (Draptchinskaia et al., 1999). DBA patients suffer not only from anemia due to a block in differentiation of erythroid precursors, but are also prone to the development of leukemias (Willig et al., 2000). A small number of patients also develop solid tumors; for example, a statistically significant association of DBA with osteogenic sarcoma has been demonstrated (Lipton et al., 2001). More recently, two other ribosomal protein genes, RPS24 and RPS17, have also been found to be linked to DBA (Gazda et al., 2006; Cmelja et al., 2007). Moreover, another human disease marked by a defect in erythroid differentiation and predisposition to leukemia, 5q− syndrome, has been attributed to haploinsufficiency of RPS14 (Ebert et al., 2008). Thus, mounting evidence suggests that a deficiency of ribosomal function, rather than the function of any one particular rp, may play a key role in these diseases. However, to our knowledge, there are no data specifically addressing the cancer incidence of DBA patients who do or do not carry rp mutations, and thus it is unclear if rp mutations play a direct role in carcinogenesis in these hematopoietic disorders. A more direct connection between rp mutations and tumorigenesis was our surprising finding that numerous ribosomal protein (rp) genes are haploinsufficient tumor suppressors in zebrafish (Amsterdam et al., 2004). Although fish from several rp mutant lines did not develop cancer, the large number of lines that were tumor-prone suggested that the mechanism of tumorigenesis might involve a common function of the rps, the most obvious being their role in translation.
Ribosomal protein mutations have been studied in other metazoan species, most extensively in Drosophila where nearly all of the 65 dominant Minute mutations have been mapped to ribosomal protein genes (Lambertsson, 1998; Marygold et al., 2007). Minute flies have short thin bristles and may also have pleiotropic defects of varying severity, such as reduced viability, developmental delay, and smaller body size. Aberrant regulation of growth of particular organs and structures is also observed in some heterozygotes. For example, adult flies heterozygous for RpL5 and RpL38 have wings that are larger than normal, due to an increase in the size of cells comprising these structures (Marygold et al., 2005). Additionally, homozygous mutation of RpS6 and RpS21, though lethal, causes an overgrowth of the hematopoietic organs in the larvae (Török et al., 1999; Watson et al., 1992), suggesting that at least some rps may play a role in suppressing a tumor-like phenotype in flies. Similar to many of the Drosophila Minutes, mice heterozygous for point mutations in either RpL24 or RpS19 display growth impairment, among other developmental abnormalities (Oliver et al., 2004; McGowan et al., 2008); however, an increased predisposition to cancer has not yet been demonstrated in either of these mouse mutants.
Here we present evidence that many rp mutations in zebrafish lead to a growth impairment, i.e., that we have identified the first examples of a Minute-like phenotype in fish. While these fish have no other detectable developmental abnormalities, they are prone to tumor development later in life. Furthermore, we show that the growth defect is manifested at the level of rp heterozygous cells. Since growth defects are often associated with defects in translation in other species, the correlation between growth impairment and cancer suggests that an impairment in translation may play a role in driving tumorigenesis in rp heterozygous fish.
Results
Heterozygous Mutation of 17 of 28 rp Genes Predisposes Zebrafish to Malignant Peripheral Nerve Sheath Tumors
We previously reported that heterozygotes in 11 of 16 lines bearing mutagenic retroviral insertions in 16 different rp genes are predisposed to zebrafish malignant peripheral nerve sheath tumors (zMPNST) at an average penetrance of 45%, although zMPNST incidence per line varied from 7% to 100% (Amsterdam et al., 2004). The wild-type rp gene is not lost in these tumors, indicating that it is haploinsufficiency for these genes which leads to tumorigenesis (Amsterdam et al., 2004). To further explore this effect, we went on to analyze all 28 of the rp mutant lines in our collection and found that, in total, 17 of them are tumor-prone (Table 1). As found previously, the predominant tumor type in these rp mutant lines is zMPNST. Our original control data set included only 152 fish. In order to further demonstrate the significance of the elevated incidence of zMPNSTs in the rp heterozygotes, we examined approximately 10,000 additional fish from 437 transgenic lines (heterozygous carriers of non-rp mutations). The tumor incidence in the control population was found to be approximately 5% (Table 1C), the predominant tumor types being seminomas and bile duct adenomas. As the vast majority of the seminomas were benign neoplasias, the incidence of malignant cancer in the control population was less than 3%. Significantly, amongst these 10,000 control fish, only 5 had zMPNSTs, reinforcing the notion that zMPNSTs are extremely rare within the control population (approximately 1 in 2000 fish).
Table 1. Incidence of zMPNSTs in zebrafish rp-heterozygous lines.
rp-heterozygous fish were collected as tumors became apparent, or as healthy fish at the maximum age specified (Age Range). Number Lost indicates those that either died before the appearance of external symptoms or were lost from their tanks. Control fish from the colony were selected without regard to gross appearance. Incidence rates are based on the number of fish examined histologically (i.e., excluding lost fish). (A) Lines prone to developing zMPNSTs; (B) Lines with no zMPNSTs observed; (C) Various control lines (heterozygous carriers of non-rp mutations) from the colony. NA = not applicable. ND = not determined.
| Gene | Line | Age Range (in months) | Initial Number of Fish | Number Lost | Number of Fish Examined | Number of Grossly-Apparent Tumors | Number of Tumor-Bearing Fish | Number of zMPNSTs | % of Fish Examined with zMPNSTs | |
|---|---|---|---|---|---|---|---|---|---|---|
| A | rpL35 | hi258 | up to 21.5 | 13 | 7 | 6 | 5 | 6 | 6 | 100% |
| rpL14 | hi823 | up to 24.5 | 26 | 4 | 22 | 20 | 21 | 16 | 73% | |
| rpS15a | hi2649 | up to 18 | 9 | 2 | 7 | 4 | 6 | 5 | 71% | |
| rpL36 | hi1807 | up to 23 | 62 | 11 | 51 | 34 | 40 | 33 | 65% | |
| rpS3a | hi1290 | up to 23.5 | 41 | 10 | 31 | 24 | 25 | 20 | 65% | |
| rpL36a | hi10 | up to 22 | 14 | 6 | 8 | 4 | 5 | 5 | 63% | |
| rpS8 | hi1974 | up to 22 | 19 | 6 | 13 | 6 | 9 | 8 | 62% | |
| rpS11 | hi2799 | up to 23.5 | 23 | 8 | 15 | 6 | 9 | 7 | 47% | |
| rpL7 | hi1061 | up to 22.5 | 28 | 6 | 22 | 11 | 11 | 10 | 45% | |
| rpS5 | hi577B | up to 23 | 27 | 4 | 23 | 7 | 12 | 10 | 43% | |
| rpS7 | hi1034B | up to 22 | 19 | 4 | 15 | 3 | 6 | 5 | 33% | |
| rpS28 | hi3883 | up to 23 | 25 | 7 | 18 | 6 | 6 | 6 | 33% | |
| rpL13 | hi1016 | up to 23 | 15 | 4 | 11 | 2 | 5 | 3 | 27% | |
| rpS29 | hi2903 | up to 22 | 18 | 3 | 15 | 4 | 4 | 4 | 27% | |
| rpS18 | hi1026 | up to 24 | 59 | 13 | 46 | 9 | 12 | 8 | 17% | |
| rpL23a | hi2582 | up to 22.5 | 71 | 8 | 63 | 5 | 7 | 6 | 10% | |
| rpL19 | hi1987 | up to 23 | 55 | 16 | 39 | 2 | 4 | 3 | 8% | |
| B | rpL12 | hi3076 | up to 23.5 | 24 | 2 | 22 | 2 | 3 | 0 | 0% |
| rpS12 | hi1227 | all at 22 | 14 | 1 | 13 | 0 | 1 | 0 | 0% | |
| rpL9 | hi1422 | all at 22.5 | 23 | 8 | 15 | 0 | 1 | 0 | 0% | |
| rpL3 | hi2437 | all at 23 | 19 | 2 | 17 | 0 | 1 | 0 | 0% | |
| rpLP1 | hi1444 | up to 22.5 | 18 | 0 | 18 | 1 | 1 | 0 | 0% | |
| rpL6 | hi3655B | up to 24.5 | 42 | 1 | 41 | 0 | 2 | 0 | 0% | |
| rpSa | hi1479 | up to 24.5 | 29 | 7 | 22 | 0 | 1 | 0 | 0% | |
| rpL28 | hi3893 | up to 24.5 | 34 | 8 | 26 | 0 | 1 | 0 | 0% | |
| rpL11 | hi3820B | up to 23.5 | 29 | 2 | 27 | 1 | 1 | 0 | 0% | |
| rpL24 | hi1284 | all at 26 | 18 | 0 | 18 | 0 | 0 | 0 | 0% | |
| rpS15 | hi2430 | all at 23 | 16 | 0 | 16 | 0 | 0 | 0 | 0% | |
| C | NA | Controls | 20 to 26 | ND | ND | 9988 | 167 | 473 | 5 | 0.05% |
Since the incidence of zMPNSTs among the control population is so rare, even lines such as hi1987, in which three of 39 fish in one cohort developed zMPNSTs, can be considered significantly tumor-prone (Table 1A). Thus, we have now classified the rp lines in our collection into only two categories: (1) “tumor-prone” lines, which develop zMPNSTs (these include lines heterozygous for rpS3a, rpS5, rpS7, rpS8, rpS11, rpS15a, rpS18, rpS28, rpS29, rpL7, rpL13, rpL14, rpL19, rpL23a, rpL35, rpL36, and rpL36a) (Table 1A); and (2) “non-tumor-prone” lines, which have a tumor incidence and spectrum that is indistinguishable from the controls (these include lines heterozygous for rpSa, rpS12, rpS15, rpL3, rpL6, rpL9, rpL11, rpL12, rpL24, rpL28, and rpLP1) (Table 1B). Based on this distinction, the hi3076 (rpL12) line is considered “non-tumor-prone” because, even though tumors were observed, only one of the three tumors was malignant (a lymphoma) and the other two were more common benign seminomas (Table 1B and data not shown). As reported previously, there are some rp lines that are extremely tumor-prone, in which the incidence of zMPNSTs exceeds 60%. Our distinction between “high-tumor” and “medium-tumor” lines, however, is now considered somewhat arbitrary since the incidence of zMPNSTs among the 17 tumor-prone rp lines ranges continuously from 8 to 100% (Table 1A). We have observed small fluctuations in tumor incidence and onset between different cohorts of each line, but the tendency for any given line to be extremetly tumor-prone or less tumor-prone has repeated over multiple generations (data not shown).
Degree of rp Message Knockdown in rp Homozygous Mutant Embryos Does Not Correlate with Incidence of zMPNSTs in rp Heterozygous Adults
Upon finding that some rp mutant lines are tumor-prone and some are not, we wanted to determine the molecular basis for this difference in tumor susceptibility. One possible explanation is that the retroviral insertion in the tumor-prone lines causes a more severe reduction of the rp mRNA level, leading to a greater loss of function of the rp in these lines than in the non-tumor-prone lines. None of the insertions we have in rp genes disrupt exons; most are in the first intron (positions of all of the mutagenic insertions can be found online at http://web.mit.edu/hopkins/). Our experience with this provirus as a mutagen is that the level of steady state mRNA for the affected gene can be either reduced or completely abrogated (Amsterdam et al., 2004b; Golling et al., 2002). In order to conveniently assess the impact of the insertions on gene expression, we chose to examine rp mRNA levels in homozygous mutant embryos, where the mutation of both alleles would make this easier to measure. We previously determined that the rp message level is reduced to varying extents in homozygous rp mutant embryos from three tumor-prone lines (Amsterdam et al., 2004). Analysis of additional lines, including non-tumor-prone lines, revealed that the degree of rp message knockdown generally correlated with the severity of the homozygous mutant phenotype (Table S1). However, there was no correlation between the amount of rp mRNA knockdown in homozygous embryos and the incidence of zMPNSTs in the adult heterozygotes (Table S1). For example, in the hi2430 line, the rpS15 message is reduced by 2000-fold in homozygous mutants compared to their wild-type siblings, though the heterozygous adults are not tumor-prone. On the other hand, homozygotes from the hi1807 line showed a reduction of rpL36 message of only 10-fold, yet heterozygotes from this line were highly tumor-prone (Table S1). Thus, it appears that merely the degree of reduction in expression of any rp does not account for the cancer phenotype. At some level, this is not particularly surprising since the wild-type copy of the rp gene is presumably still expressed at a normal level in rp heterozygotes. Thus, the wide range of reduced expression of the mutant alleles, as measured in homozygotes, likely reduces rp message level over a much a smaller range in heterozygotes, perhaps between 50 and 60% of the wild-type level.
Heterozygous Embryos from a Tumor-Prone rp Line Are Developmentally Delayed
Heterozygosity for many rp mutations in flies, and for two rp mutations in mice, leads to a growth impairment phenotype. Thus, we wanted to determine whether there was any growth impairment in rp heterozygous zebrafish. As an initial test of this hypothesis, we sorted a clutch of embryos from an outcross of a hi258 (rpL35) heterozygote and a T-AB wild-type fish at 4 days post-fertilization (dpf) for the inflation of the swim bladder (SB), which is an excellent reporter for developmental delay since it appears suddenly on day 4 and is highly visible. We subsequently sorted each group on the basis of embryo size by lining up the embryos and ranking them by length, selecting the top and bottom quartiles. Beginning with 122 embryos, we selected the 32 largest of those with an inflated swim bladder (large+SB) and the 32 smallest of those lacking one (small-SB). If rp heterozygosity impairs growth, we would expect enrichment of wild-types in the large+SB group and of heterozygotes in the small-SB group. Alternatively, if there is no effect on growth, wild-types and heterozygotes should be equally represented in each group. Notably, genotyping the embryos supports the first hypothesis (Figure 1): 32/32 of the large+SB embryos were wild-type while 24/32 of the small-SB embryos were heterozygotes. While 8/32 of the smaller swim bladder-lacking embryos were wild-type, it should be noted that in a clutch of purely wild-type embryos some embryos have not yet inflated their swim bladder on day 4, reflecting some variation in developmental rate. It is thus especially significant that whereas wild-type embryos may be small and lack a swim bladder due to normal variation, no hi258 heterozygotes were found that were large and had a swim bladder. The strong enrichment for the predicted genotype in both groups was highly significant (Chi-square test, p<0.0001 for large+SB, p<0.005 for small-SB). Thus, we conclude that heterozygosity for rpL35 causes a developmental delay in early embryos. Notably, this is a time before embryos need to be fed, so this growth impairment cannot be due to differential accessibility to food.
Figure 1. Heterozygous mutant embryos from the tumor-prone hi258 (rpL35) line are growth-impaired.
122 embryos from an outcross of a hi258 heterozygote were sorted into two groups by the presence or absence of a swim bladder at 4 days of age then further sorted by embryo length. The 32 largest, swim bladder-positive embryos were predicted to be wild-types (“W”), and the 32 smallest, swim bladder-negative embryos were predicted to be hi258 heterozygotes (“H”). Individual embryos were genotyped by PCR revealing the presence or absence of the hi258 transgenic (258 Tg) band. Primers for Wnt5 were used concurrently to control for the efficiency of the PCR. Genotyping (“Actual”) revealed that the correct prediction was made in 32 of 32 wild-type calls (Chi-square test, p<0.0001) and 24 of 32 heterozygote calls (Chi-square test, p<0.005). * and ** represent non-specific background bands and primer-dimers, respectively.
Tumor-Prone rp Lines Are Growth-Impaired
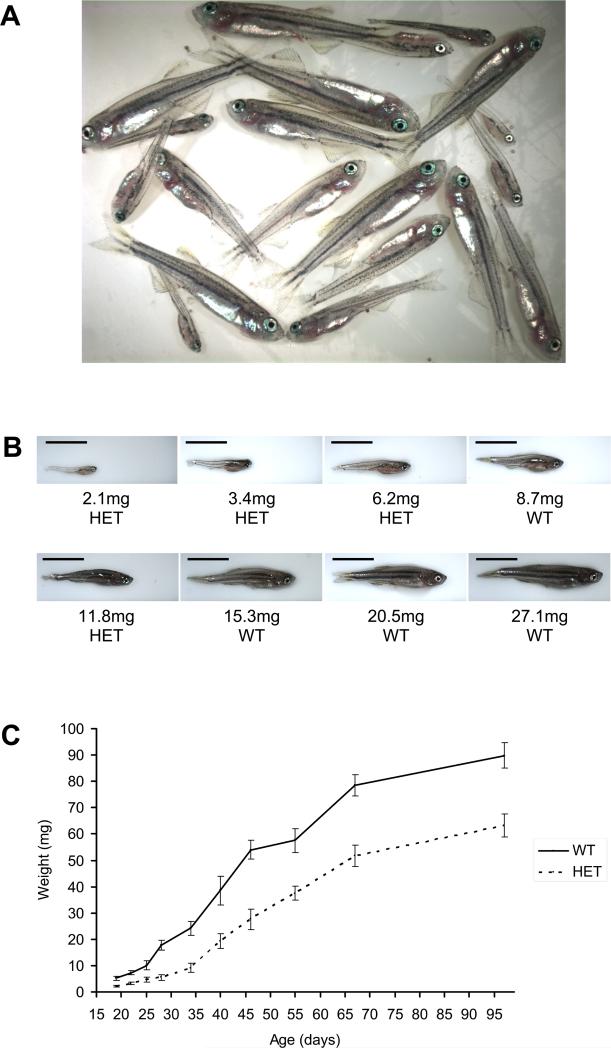
Given the finding that rpL35 heterozygotes show an embryonic developmental delay, we sought to determine whether rp heterozygous fish were growth-impaired later in life. We found that 28-day-old offspring generated from an outcross mating of a hi258 heterozygote to a T-AB wild-type fish varied greatly in size (Figure 2A), with the largest fish weighing over 10 times more than the smallest fish (Figure 2B). Upon genotyping the fish individually, the rp heterozygotes were found to be generally smaller than their wild-type siblings (Figure 2B). Significantly, the largest fish were all wild-type and the smallest fish were all hi258 heterozygous; only in the middle of the range of weights was there some overlap in the genotypes (Figure 2B and Table S2).
Figure 2. Heterozygous fish from the tumor-prone hi258 (rpL35) line are growth-impaired.
(A) The offspring of an outcross of a hi258 (rpL35) heterozygote, expected to be 50% hi258 heterozygous and 50% wild-type, are widely variable in size at 28 days of age. The fish were housed in a single tank.
(B) hi258 heterozygotes (HET) are generally smaller than their wild-type siblings (WT). Representative fish from the group depicted in (A) are shown with their weights and genotype. Scale bar in each image is 5 mm. The weights of all fish in (A) are listed in Table S2.
(C) hi258 heterozygotes were outcrossed to T-AB wild-type fish, and their progeny were housed in the same tank. At selected time points, fish were weighed and genotyped. hi258 heterozygotes are significantly smaller than their wild-type siblings at every age examined. Error bars are ±2 SEM.
We then conducted a carefully controlled experiment with the hi258 line, in which the offspring of a hi258 outcross, i.e., both heterozygous and wild-type siblings, were housed together in the same tank with ample food. At selected time points, all the fish in each tank were weighed and genotyped. It became clear that, on average, the hi258 heterozygous fish were significantly smaller than their wild-type siblings at every time point (Figure 2C). The hi258 heterozygous fish are growth-impaired early on and do not catch up in size to their wild-type siblings. The growth rate of the heterozygotes, however, does appear to be comparable to that of the wild-type siblings by 67 days of age, as the increase in size to the 97-day time point is similar for both groups (Figure 2C).
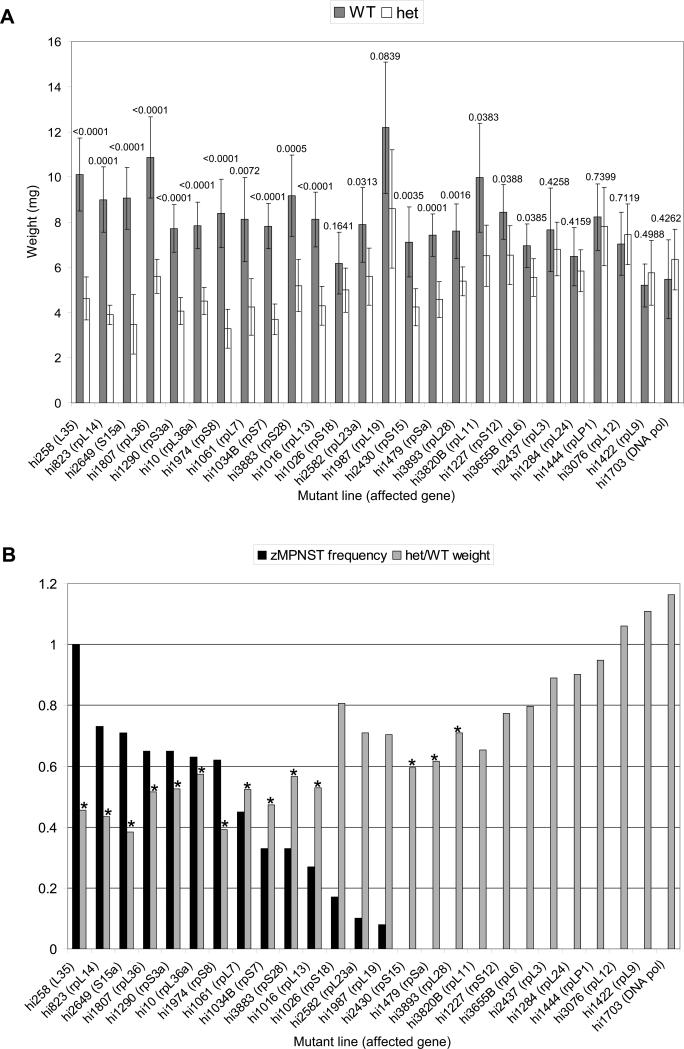
Next, we wanted to determine whether all of our rp mutant lines displayed a growth impairment and whether there is any relationship with the tumor phenotype. Since the growth defect was evident by day 25 in the hi258 line, we performed the same experiment at this time point for most of the rp lines, both tumor-prone and non-tumor-prone. This analysis revealed that heterozygotes in a large subset of the rp lines showed a significant growth impairment (Figure 3A). Strikingly, when the rp lines are ordered by the incidence of zMPNSTs, it is evident that the 11 most tumor-prone lines included in this analysis (>25% incidence of zMPNSTs) are also the most growth-impaired (Figure 3B). Among the tumor-prone lines, only the three with the lowest incidence of zMPNSTs, hi1026 (rpS18), hi2582 (rpL23a), and hi1987 (rpL19), did not display a statistically significant (unpaired t-test, p<.01) growth impairment. For the most part, the non-tumor-prone lines did not show a statistically significant growth impairment, similar to the control non-rp line, hi1703, a mutant for a DNA polymerase subunit. There were, however, several exceptions: hi2430 (rpS15), hi1479 (rpSa), and hi3893 (rpL28) were growth-impaired but not tumor-prone (Figure 3).
Figure 3. Heterozygous fish from high-tumor rp mutant lines are growth-impaired.
(A) Outcrosses of each rp line were performed, and their progeny were weighed and genotyped at day 25. Grey bars represent the average weight (mg) of heterozygotes (het) whereas white bars represent the average weight (mg) of the wild-type (WT) siblings for each rp line; error bars are ±2 SEM. The p value (unpaired t-test) between heterozygotes and wild-type siblings is indicated above each set of bars. Lines are ordered along the x-axis by decreasing incidence of zMPNSTs.
(B) The growth defect determined in (A), represented as the ratio of average heterozygote weight to average wild-type weight (grey bars), compared to the frequency of zMPNSTs (black bars), as determined in Table 1. The most highly tumor-prone lines appear to be the most severely growth-impaired. * indicates a statistically significant difference (p<0.01 by unpaired t-test). Lines are ordered along the x-axis the same as in (A) for ease of comparison.
rp Mutants Are Growth-Impaired at the Cellular Level
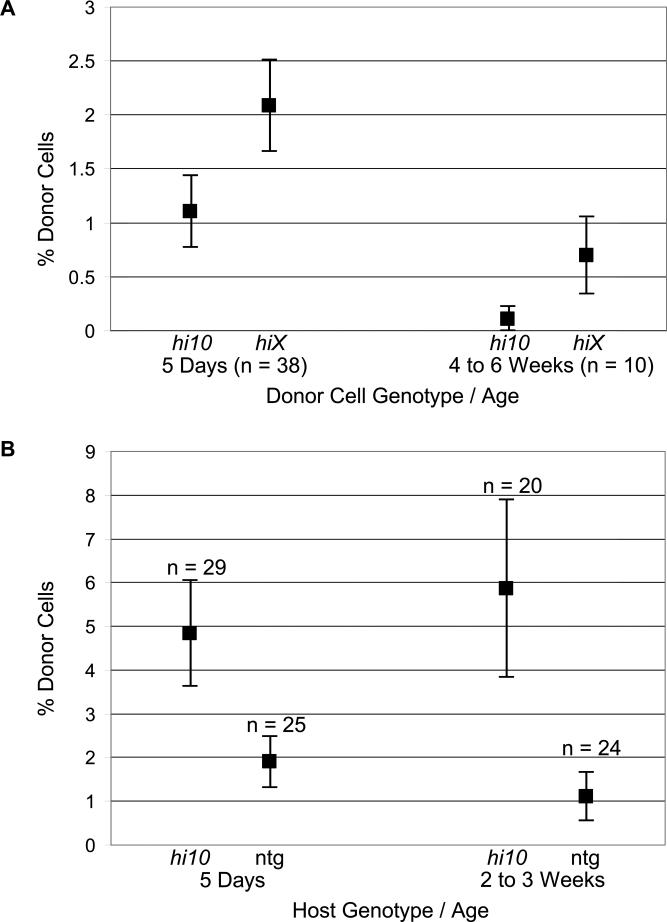
In order to determine whether rp heterozygosity leads to a growth impairment at the cellular level, we generated chimeric fish and analyzed the ability of rp heterozygous or wild-type cells to compete in hosts of each genotype. Real-time quantitative PCR was used to measure the relative contribution of cells of each genotype. In order to track the wild-type cells, a line bearing a non-mutagenic insertion, termed hiX, was identified. Heterozygotes from a high-tumor line (and therefore a line that displays a growth defect at the organismal level), hi10 (rpL36a), were used for the chimera analysis.
In the first experiment, cells were harvested from embryos produced by a cross of a hi10 heterozygote and a hiX heterozygote. The expected ratio of genotypes amongst the cells was 25% hi10 only, 25% hi10 and hiX, 25% hiX only, and 25% non-transgenic. This mixture of cells was injected into wild-type host embryos, which were allowed to grow until the fish were homogenized for quantitative PCR analysis. Five days after injection, the contribution of cells bearing the hi10 insertion was significantly less (2-fold) than that of cells bearing the nonmutagenic hiX insertion (Figure 4A). At 4 to 6 weeks after injection, the contribution of hi10 cells was reduced even more relative to the contribution of hiX cells (Figure 4A). Moreover, in 7 of the 10 fish analyzed at this latter time point, the hi10 allele was completely undetectable by PCR, suggesting that the growth disadvantage is amplified over time.
Figure 4. Heterozygous mutation of an rp gene impairs growth at the cellular level.
(A) Chimeric embryos were generated by injecting into wild-type hosts a mixture of cells isolated from the progeny of an intercross between hi10 (rpL36a) and hiX heterozgyotes. hi10 is a high-tumor rp line, and hiX is a line that bears a non-mutagenic insertion. The relative contribution of cells carrying each insertion was determined by real-time PCR at 5 days and 4 to 6 weeks post injection. In both cases, the cells bearing the hiX insertion comprise a greater portion of the embryo than cells bearing the hi10 insertion.
(B) Cells from the progeny of a hiX outcross were injected into hi10 (rpL36a) heterozygous or non-transgenic (ntg) host embryos. Contribution of cells carrying the hiX insertion was determined by real-time PCR at 5 days and 2 to 3 weeks post injection. The phenotypically wild-type hiX cells appear to be able to contribute better to hi10 heterozygous hosts than to wild-type hosts.
To determine whether wild-type cells can compete better in an rp heterozygous background than in a wild-type background, the converse experiment was performed. Cells were harvested from embryos produced by mating a hiX heterozygote and a non-transgenic fish and injected into either hi10 heterozygotes or their wild-type siblings. Thus, all of the cells were expected to be phenotypically wild-type, and 50% of them carried the hiX insertion that was being tracked. At five days after injection, the contribution of wild-type (hiX) cells was significantly higher in hi10 heterozygous hosts than in wild-type hosts (Figure 4B). This held true when the fish were analyzed at 2 to 3 weeks post injection (Figure 4B). The data presented thus strongly suggest that the rp mutations cause a defect in growth, both at the cellular and organismal levels.
Heterozygous Embryos from a Tumor-Prone rp Line Are Not Overtly Impaired in Translation
While it is formally possible that some extraribosomal function of many of the rps is disrupted in the tumor-prone lines, a more likely explanation is that the mechanism of tumorigenesis involves the role of these proteins in their known common function, translation. In order to test whether certain rp lines have a defect in translation, polysome fractionation analysis was performed to determine the proportion of ribosomal subunits that are free or bound in monosomes or polysomes. Embryos from an outcross of a hi258 heterozygote were sorted into wild-type-enriched and hi258 heterozygote-enriched samples as in Figure 1. When the mixtures were analyzed by polysome fractionation, the shapes of the profiles of the wild-type-and heterozygote-enriched samples were very similar, including the number of higher-order polysomes observed (Figure S1A and B). In contrast, homozygous rp mutant embryos showed a drastic reduction in the number of higher-order polysomes, as well as preferential loss of the ribosomal subunit containing the mutant rp (Figure S1C and D). While this result suggests that there is no drastic translation defect in the hi258 heterozygous embryos, it does not rule out the possibility that an effect may be too subtle to be detected by this method, especially since the sample populations were likely not completely pure (i.e., the heterozygote-enriched sample likely contained some wild-type embryos, Figure 1). Future studies will require a more sensitive assay to determine the extent of any alteration in the rate of translation in the rp heterozygous fish.
Discussion
We have now characterized the tumor phenotype of all 28 zebrafish lines in our collection with heterozygous mutations in ribosomal protein genes and found that 17 of these are tumor-prone, displaying an incidence of zMPNSTs that is far higher than that of wild-type fish. As noted previously (Amsterdam et al., 2004), these tumor-prone lines include mutants for proteins in both the large and small ribosomal subunits. We did not find a correlation between the severity of the rp mRNA reduction in the homozygous rp mutants and the tumor incidence of the adult heterozygotes. As noted, however, a large difference in the degree of rp message reduction between homozygotes from two rp lines may correspond to a much smaller difference between the respective heterozygotes. Since it has been shown that mRNAs encoding certain rps are normally produced in excess (Bortoluzzi et al., 2001; Thorrez et al., 2008), a reduction of the mRNA encoding such rps to just over half the wild-type level in heterozygous fish may have no adverse consequences.
Intriguingly, we found that the most highly tumor-prone lines have a significant impairment of growth. In one high-tumor line, hi258 (rpL35), the growth defect was apparent as early as the embryonic stages. Furthermore, this growth impairment is detected at the cellular level, as rp heterozygous cells from another high-tumor line, hi10 (rpL36a), are out-competed by wild-type cells in chimeric embryos. This defect in growth of the hi10 (rpL36a) heterozygous cells appeared to worsen over time, suggesting that the growth impairment observed in day 4 heterozygous embryos that persists into adulthood reflects an intrinsic defect at the cellular level. The observed defect in growth, coupled with the fact that a ribosomal protein gene is mutated, strongly suggests that the underlying defect is an impairment in protein translation.
A number of rp heterozygous Minute flies and RpL24 heterozygous mice are smaller at the organismal level but do not have smaller cells (Oldham et al., 2000; Oliver et al., 2004). Similarly, an analysis of cell size in wild-type and rp heterozygous zebrafish showed a greater variation within fish of the same genotype than between genotypes (data not shown). This suggests that rp heterozygotes are smaller because they have fewer cells, not smaller cells, which is consistent with our finding that rp heterozygous cells are out-competed by wild-type cells. One possible connection between the observed growth defect and tumorigenesis is that a mutation that would not normally confer a growth advantage might do so amongst a population of growth-impaired cells (Bilousova et al., 2005). Thus, the growth defect might simply make these cells more susceptible to oncogenic mutations.
On the other hand, tumorigenesis might be a direct result of a translation defect. While we have thus far been unable to directly demonstrate experimentally that protein translation is reduced in the rp heterozygotes [nor is overall protein synthesis reduced in the tumors (MacInnes et al., 2008)], there is precedent to suggest that even subtle defects in global translation can have dramatic consequences. For example, tumor cells treated with the rapamycin analog RAD001 are impaired only slightly in global translation, but this is enough to reduce the translation of p21 such that its induction is inhibited after DNA damage (Beuvink et al., 2005). In addition, it is possible that a translation defect could be manifested in the rp heterozygotes by a selective translation of mRNAs; i.e., under conditions of impaired translational capacity, certain messages may be translated at the expense of others that would normally also be translated (Rajasekhar et al., 2003). Interestingly, zMPNSTs from rp heterozygous fish contain p53 mRNA but do not express detectable levels of p53 protein, even under conditions (DNA damage, proteasome inhibition) which should prevent its degradation (MacInnes et al., 2008). It is possible that certain cell types, such as the cell of origin (presumably nerve sheath cells) that give rise to zMPNSTs, may be more susceptible to a translation defect than other cell types. Alternatively, it may be that translation in the cell of origin is less profoundly affected than in other cells. In this model, most of the cells in the organism could have a global decrease in protein synthesis, resulting in the organismal growth defect, while nerve sheath cells might only lose expression of select messages (such as p53) without compromising their growth properties.
Either of these models might also explain why the correlation between growth impairment and tumor susceptibility is not absolute. As noted, three lines displayed a growth impairment, but were not tumor-prone [hi2430 (rpS15), hi1479 (rpSa), and hi3893 (rpL28)]. In addition, there were three other lines that were prone to develop zMPNSTs, but were not significantly growth-impaired [hi1026 (rpS18), hi2582 (rpL23a), and hi1987 (rpL19)] (Figure 3). These data suggest the possibility that while a general growth impairment predisposes to tumor development, tumorigenesis also depends on some unknown modifying factors that may be cell-type specific. As different rps are expressed at different levels in different cell types (Bortoluzzi et al., 2001; Thorrez et al., 2008), it is possible that an alteration of a given rp gene dosage that affects protein synthesis in most cells sufficiently to cause an organismal growth phenotype might not result in a tumor-promoting level of translation impairment in the nerve sheath cells.
Another possible explanation for the finding that three rp heterozygous lines had a moderate incidence of zMPNSTs but no observable growth impairment is that there may be stochastic variation in the severity of the translation defect among individual fish. It is important to realize that the average weight of a population of rp heterozygotes was used to determine whether the line had an overall growth impairment. As shown for the hi258 line, there certainly exists a large variation in size among the heterozygotes (Figure 2B). It is currently not known, and will be important to determine, if the fish that are smallest at a young age are the ones that will eventually develop tumors. It will also be interesting to determine whether the size of the fish correlates with the time to onset of cancer. Thus, there may exist an even finer correlation between growth and tumorigenicity than we have discovered thus far.
Regardless of the precise mechanism of tumor promotion, our data show that heterozygosity of numerous rp genes causes growth impairment at the organismal level reminiscent of the Minute phenotype of rp mutant flies and mice. This growth defect is predictive of a predisposition to develop zMPNSTs and clearly precedes tumorigenesis. In addition, these mutant lines may be useful to use as recipients in cell transplant experiments where it is desirable to have a greater expansion of the donor cell population, similar to the use of Minute strains in Drosophila.
Experimental Procedures
Fixation and Histology
Adult fish were euthanized in ice water or 250mg/L Tricaine and fixed in 10% neutral buffered formalin or Bouin's fixative. Embedding in paraffin and sectioning were performed as previously described (Amsterdam et al., 2004; Moore et al., 2002).
RNA Analysis
Degree of rp message reduction in the rp homozygous mutants was determined by PCR on serial dilutions of cDNA prepared from rp homozygous mutant embryos and their wild-type siblings, as previously described (Amsterdam et al., 2004). Relative levels of myc gene expression were determined using real time PCR with Sybr Green Master Mix (ABI) on cDNA prepared from either pools of 5 day 5 embryos or individual day 25 or day 35 fish. Standard curves were established for each primer set with cDNA dilutions and all samples were run in triplicate and normalized with primers for gapdh. All primer sequences are available upon request.
Analysis of Growth of rp Heterozygous Mutants
rp heterozygous mutant fish were outcrossed to T-AB wild-type fish. Randomly selected, un-genotyped fish were housed in tanks at an average density of 20 fish per tank. Two tanks, or approximately 40 fish, were selected for analysis at each time point. Fish were euthanized in ice water, blotted dry, and weighed on a Mettler AE50 balance. Genotyping PCRs were performed on DNA isolated from tail clippings or the entire fish, with insert specific primers for each line to detect heterozygotes. Primers for Wnt5 were used concurrently to control for the efficiency of the PCR (Lin et al., 1994).
Generation and Real-Time PCR Analysis of Chimeras
Chimeras were generated basically as previously described (Lin et al., 1992). Briefly, a pool of 100−200 embryos was dissociated at the 1000-cell stage with a Pasteur pipette in 1mL Holtfreter's solution. Cells were spun down and washed in 10mL Holtfreter's solution, followed by 10mL 1:1 (v:v) PBS:Holtfreter's solution. The final cell pellet was resuspended in 50−100μL 2:1 (v:v) PBS:Holtfreter's solution. Approximately 50 cells (range: 30−100 cells) were injected into 1000-stage embryo hosts.
Real-time PCR analysis was performed as previously described (Amsterdam et al., 1999). In order to determine the percentage contribution of the hi10 and hiX alleles, a standard curve was generated by mixing known quantities of pure DNA samples from hi10 or hiX embryos into non-transgenic DNA, and analyzing the samples by real-time PCR.
Polysome Fractionation
Fifty to 200 embryos at 3 dpf were washed in PBS and Dounce-homogenized in ice-cold lysis buffer containing 110mM potassium acetate, 2mM magnesium acetate, 10mM HEPES pH 7.6, 100mM potassium chloride, 10mM magnesium chloride, 0.1% NP-40, 2mM dithiothreitol, and 40U/mL RNase inhibitor. The dithiothreitol and RNase inhibitor were added immediately prior to use. The lysate was spun down at 1000×g for 10 min. at 4°C to pellet nuclei and cellular debris. The cytoplasmic extract was layered onto sucrose gradients prepared by layering 17% sucrose solution over 50% sucrose solution in a 13.2mL Beckmann ultracentrifuge tube (Cat. No. 344059) and laying the Parafilm-sealed tube on its side overnight. The sucrose solutions also contained 110mM potassium acetate, 2mM magnesium acetate, and 10mM HEPES pH 7.6. The extract was fractionated by centrifuging at 40,000×g for 2 hrs. at 4°C in an Sw41Ti rotor. Fractions were collected by injecting 60% sucrose into the bottom of the tube on an ISCO Model 640 Density Gradient Fractionator, and absorbance at 280nm was measured continuously using a Pharmacia LKB Uvicord SII detector. Profiles were generated with a Pharmacia LKB 19−8003−01 chart recorder.
Supplementary Material
Figure S1: Polysome profiles do not reveal an overt translation defect in hi258 (rpL35) heterozygous embryos.
(A) Polysome profile generated from an embryo mixture enriched for wild-type embryos. Peaks representing the small ribosomal subunit (40S), the large ribosomal subunit (60S), the monosomes (80S), and the polysomes are detected.
(B) Polysome profile of an embryo mixture enriched for hi258 (rpL35) heterozygotes appears very similar to that of the wild-type-enriched embryo mixture.
(C) Homozygous mutants for hi258 (rpL35) display a general reduction in all peaks. The decrease of the 60S peak is relatively most severe.
(D) Homozygous mutants for hi1974 (rpS8) display a loss of the 40S peak, a relative increase in the 60S peak, and a reduction in the 80S and polysome peaks.
Table S1: Degree of rp message reduction in rp homozygous mutant embryos correlates with severity of homozygous mutant phenotype but does not correlate with incidence of zMPNSTs in adult rp heterozygotes.
Incidence of zMPNSTs in adult rp heterozygotes was determined in Table 1. RNA was isolated from homozygous rp mutant embryos and their wild-type siblings. PCR was performed using serial dilutions of cDNA to detect the rp message and estimate the level of knockdown in the homozygous mutants compared to their wild-type siblings. The severity of the homozygous mutant phenotype was rated on a scale as follows:
+++ = severe defects, including very obvious brain necrosis on 1 dpf
++ = moderate defects, including some brain necrosis on 1 dpf
+ = mild defects: no visible necrosis on 1 dpf; mild necrosis and body curvature on 3 dpf
Note that while the fold rp message reduction in rp homozygotes generally correlates with the severity of the homozygous mutant phenotype, it does not correlate with the incidence of zMPNSTs in the adult rp heterozygotes.
Table S2: Heterozygous fish from the tumor-prone hi258 (rpL35) line are growth-impaired.
The fish depicted in Figure 2A represent one tank of offspring from an outcross of a hi258 (rpL35) heterozygous fish. The weights of all fish and their genotypes are listed below. The smallest fish are hi258 heterozygous (HET), the largest fish are wild-type (WT), and those in the middle of the weight range are either hi258 heterozygous or wild-type.
Acknowledgements
We thank Kirsten Sadler for use of her previously published data, included in Table 1. We thank Kate Anderson for maintenance of the mutant lines of fish, and Tim Angelini and Sam Farrington for maintenance of the zebrafish colony. We thank the Histology Facility of the MIT Koch Institute for Integrative Cancer Research, especially Alicia Caron and Mike Brown, for sample processing and sectioning. K.L. was supported by a fellowship from the Anna Fuller Fund. This work was supported by an NIH-NCI grant (CA106416) to N.H. and J.A.L. J.A.L. is a Ludwig Scholar at MIT.
Grant sponsor: NIH-NCI; Grant number: CA106416
References
- Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:690–698. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004b;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuvink I, Boulay A, Fumagalli S, Zilbermann F, Ruetz S, O'Reilly T, Natt F, Hall J, Lane HA, Thomas G. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell. 2005;120:747–759. doi: 10.1016/j.cell.2004.12.040. [DOI] [PubMed] [Google Scholar]
- Bilanges B, Stokoe D. Mechanisms of translational deregulation in human tumors and therapeutic intervention strategies. Oncogene. 2007;26:5973–5990. doi: 10.1038/sj.onc.1210431. [DOI] [PubMed] [Google Scholar]
- Bilousova G, Marusyk A, Porter CC, Cardiff RD, DeGregori J. Impaired DNA replication within progenitor cell pools promotes leukemogenesis. PLoS Biol. 2005;3:2135–2147. doi: 10.1371/journal.pbio.0030401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortoluzzi S, d'Alessi F, Romualdi C, Danieli GA. Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics. 2001;17:1152–1157. doi: 10.1093/bioinformatics/17.12.1152. [DOI] [PubMed] [Google Scholar]
- Clemens MJ. Targets and mechanisms for the regulation of translation in malignant transformation. Oncogene. 2004;23:3180–3188. doi: 10.1038/sj.onc.1207544. [DOI] [PubMed] [Google Scholar]
- Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;28:1178–1182. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- DeBenedetti AD, Graff JR. eIF4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- Donzé O, Jagus R, Koromilas AE, Hershey JWB, Sonenberg N. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 1995;14:3828–3834. doi: 10.1002/j.1460-2075.1995.tb00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig T-N, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D, Mohandas N, Carlsson B, Dahl N. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, Golub TR. Identification of RPS14 as a 5q− syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Shimogori T, Ishii I, Kashiwagi K, Mashiba H, Ekimoto H, Igarashi K. Malignant transformation by overproduction of translation initiation factor eIF4G. Cancer Res. 1997;57:5041–5044. [PubMed] [Google Scholar]
- Gazda HT, Grabowska A, Merida-Long LB, Latawiec E, Schneider HE, Lipton JM, Vlachos A, Atsidaftos E, Ball SE, Orfali KA, Niewiadomska E, Da Costa L, Tchernia G, Niemeyer C, Meerpohl JJ, Stahl J, Schratt G, Glader B, Backer K, Wong C, Nathan DG, Beggs AH, Sieff CA. Ribosomal protein S24 is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79:1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, Lin S-Y, Nissen RM, Hopkins N. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Lambertsson A. The Minute genes in Drosophila and their molecular functions. Adv Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5' cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- Lin S, Long W, Chen J, Hopkins N. Production of germ-line chimeras in zebrafish by cell transplants from genetically pigmented to albino embryos. Proc Natl Acad Sci USA. 1992;89:4519–4523. doi: 10.1073/pnas.89.10.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Gaiano N, Culp P, Burns JC, Friedmann T, Yee JK, Hopkins N. Integration and germ-line transmission of a pseudotyped retroviral vector in zebrafish. Science. 1994;265:666–669. doi: 10.1126/science.8036514. [DOI] [PubMed] [Google Scholar]
- Lipton JM, Federman N, Khabbaze Y, Schwartz CL, Hilliard LM, Clark JI, Diamond-Blackfan Anemia Registry Osteogenic sarcoma associated with Diamond-Blackfan anemia: A report from the Diamond-Blackfan Anemia Registry. J Pediatr Hematol Oncol. 2001;23:39–44. doi: 10.1097/00043426-200101000-00009. [DOI] [PubMed] [Google Scholar]
- MacInnes AW, Amsterdam A, Whittaker CA, Hopkins N, Lees JA. Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proc Natl Acad Sci USA. 2008;105:10408–10413. doi: 10.1073/pnas.0805036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, Coelho CMA, Leevers SJ. Genetic analysis of RpL38 and RpL5, two Minute genes located in the centric heterochromatin of chromosome 2 of Drosophila melanogaster. Genetics. 2005;169:683–695. doi: 10.1534/genetics.104.034124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, Roote J, Reuter G, Lambertsson A, Ashburner M, Millburn GH, Harrison PM, Yu Z, Kenmochi N, Kaufman TC, Leevers SJ, Cook KR. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007;8:R216.1–R216.26. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, Attardi LD, Barsh GS. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–70. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JL, Aros M, Steudel KG, Cheng KC. Fixation and decalcification of adult zebrafish for histological, immunocytochemical, and genotypic analysis. BioTechniques. 2002;32:300–304. doi: 10.2144/02322st03. [DOI] [PubMed] [Google Scholar]
- Oldham S, Böhni R, Stocker H, Brogiolo W, Hafen E. Genetic control of size in Drosophila. Phil Trans R Soc Lond B. 2000;355:945–952. doi: 10.1098/rstb.2000.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver ER, Saunders TL, Tarlé SA, Glaser T. Ribosomal protein L24 defect in Belly spot and tail (Bst), a mouse Minute. Development. 2004;131:3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzani A, Fronza R, Loreni F, Pascale A, Amadio M, Quattrone A. Global alterations in mRNA polysomal recruitment in a cell model of colorectal cancer progression to metastasis. Carcinogenesis. 2006;27:1323–1333. doi: 10.1093/carcin/bgi377. [DOI] [PubMed] [Google Scholar]
- Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol Cell. 2003;12:889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- Thorrez L, Van Deun K, Tranchevent L-C, Van Lommel L, Engelen K, Marchal K, Moreau Y, Van Mechelen I, Schuit F. Using ribosomal protein genes as a reference: A tale of caution. PLoS ONE. 2008;3:e1854. doi: 10.1371/journal.pone.0001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Török I, Herrmann-Horle D, Kiss I, Tick G, Speer G, Schmitt R, Mechler BM. Down-regulation of RpS21, a putative translation initiation factor interacting with P40, produces viable Minute imagos and larval lethality with overgrown hematopoietic organs and imaginal discs. Mol Cell Biol. 1999;19:2308–2321. doi: 10.1128/mcb.19.3.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson KL, Konrad KD, Woods DF, Bryant PJ. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc Natl Acad Sci USA. 1992;89:11302–11306. doi: 10.1073/pnas.89.23.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willig TN, Gazda H, Sieff CA. Diamond-Blackfan anemia. Curr Opin Hematol. 2000;7:85–94. doi: 10.1097/00062752-200003000-00003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Polysome profiles do not reveal an overt translation defect in hi258 (rpL35) heterozygous embryos.
(A) Polysome profile generated from an embryo mixture enriched for wild-type embryos. Peaks representing the small ribosomal subunit (40S), the large ribosomal subunit (60S), the monosomes (80S), and the polysomes are detected.
(B) Polysome profile of an embryo mixture enriched for hi258 (rpL35) heterozygotes appears very similar to that of the wild-type-enriched embryo mixture.
(C) Homozygous mutants for hi258 (rpL35) display a general reduction in all peaks. The decrease of the 60S peak is relatively most severe.
(D) Homozygous mutants for hi1974 (rpS8) display a loss of the 40S peak, a relative increase in the 60S peak, and a reduction in the 80S and polysome peaks.
Table S1: Degree of rp message reduction in rp homozygous mutant embryos correlates with severity of homozygous mutant phenotype but does not correlate with incidence of zMPNSTs in adult rp heterozygotes.
Incidence of zMPNSTs in adult rp heterozygotes was determined in Table 1. RNA was isolated from homozygous rp mutant embryos and their wild-type siblings. PCR was performed using serial dilutions of cDNA to detect the rp message and estimate the level of knockdown in the homozygous mutants compared to their wild-type siblings. The severity of the homozygous mutant phenotype was rated on a scale as follows:
+++ = severe defects, including very obvious brain necrosis on 1 dpf
++ = moderate defects, including some brain necrosis on 1 dpf
+ = mild defects: no visible necrosis on 1 dpf; mild necrosis and body curvature on 3 dpf
Note that while the fold rp message reduction in rp homozygotes generally correlates with the severity of the homozygous mutant phenotype, it does not correlate with the incidence of zMPNSTs in the adult rp heterozygotes.
Table S2: Heterozygous fish from the tumor-prone hi258 (rpL35) line are growth-impaired.
The fish depicted in Figure 2A represent one tank of offspring from an outcross of a hi258 (rpL35) heterozygous fish. The weights of all fish and their genotypes are listed below. The smallest fish are hi258 heterozygous (HET), the largest fish are wild-type (WT), and those in the middle of the weight range are either hi258 heterozygous or wild-type.