Abstract
Increased expression of genes, silenced by methylation of their promoters, could have relevance for increasing effects of not only interferons (IFNs) but also APO2L/TRAIL, cytotoxics and immunotherapeutics for melanoma and other malignancies. A resistant melanoma cell line, A375, lacked APO2L/TRAIL or apoptosis induction by either IFN-α2 or IFN-β. However, apoptosis was induced by IFNs in A375 cells by 5-aza, 2′deoxycytidine, evaluated based upon the postulate that promoter methylation might be silencing pro-apopoptotic IFN-stimulated genes (ISGs). RASSF1A, commonly methylated at high frequency in many tumors including melanoma, which we discovered to be also an IFN-regulated gene, was increased by 5-Aza-dC. RASSF1A was important in enhancing apoptotic effects of not only IFNs and APO2L/TRAIL but also cisplatin. Unraveling epigenetic regulatory mechanisms, as yet only partially identified, will result in new biological insights and improved strategies for therapeutic use of IFNs or ISGs such as APO2L/TRAIL.
Keywords: APO2L/TRAIL, azacytidine, apoptosis
Introduction
Despite use over the past 15 years, interferons (IFNs) have yet to reach their full therapeutic potential in oncology. To do so, 1) augmentation of function and regulation of induced genes and proteins must be attained, 2) interventions to overcome cellular resistance developed, and 3) integration with other therapeutic modalities improved. From the initial introduction of IFNs into clinical trials, melanoma has been a prototype for applications in oncology [1–10]. Approximately 75,000 individuals in the United States will be newly diagnosed with melanoma this year; 60% of those who die will be individuals under 60 years of age who would otherwise have had a life expectancy of 25 years or more. Unfortunately, since the advances marked by identification of IFNs and IL-2 as human proteins therapeutically active in melanoma, the past 15 years has been notable more for negative Phase III trials than for clinically significant reduction in mortality from systemic metastatic disease [11–13]. While sorafenib and antibodies to the T cell ligand, CTLA-4, hold new promise, improved therapies for the high risk primary and metastatic patient are much needed.
One approach to potentially enhance effects of IFNs is to reverse epigenetic silencing of gene expression, since methylation of gene promoters occurs commonly in malignancies. Thus, in the spirit of the Milstein awards of encouraging research to improve clinical outcomes, this article will describe rationale and results of one specific ongoing investigational approach. It however grows out of research on gene regulation and melanoma that has involved ideas and collaborations with many individuals including friends and other Milstein award winners, Paula Pitha-Rowe, Jordan Gutterman, Susan Krown, John Kirkwood, Bryan Williams, Robert Silverman, Ganes Sen, and George Stark.
Interferons and stimulated genes in melanoma
Although no systemic modality has substantial activity, IFN-α2 has resulted in response rates in metastatic melanoma equivalent to the best of any single agent. It results in metastatic disease regression in 15% of patients in summarized data from clinical studies [2,3,5,9]. Approximately 3% of patients will have a complete response of significant duration. Prolongation of disease-free survival and a strong trend toward prolonged overall survival have emerged from use of IFN-α2 as an adjuvant to surgery for high-risk patients with primary melanoma [6–8,13]. Though randomized studies, meta-analyses and cost benefit analyses all support usage in these patients with Stage IIc and III disease, clinical adoption has been slowed by perceptions of a marginally beneficial therapeutic index. Thus ways to enhance gene expression, particularly with improved tolerance, could be of substantial clinical benefit.
Protein products, encoded by interferon-stimulated genes (ISGs), underlie the changes in cellular function that result in clinical effects. ISG expression translates into alterations of the state of differentiation, rate of proliferation, and functional activity of endothelial cells, immune effector cells, and tumor cells. Changes in ISGs vary both quantitatively and qualitatively depending upon tissue and IFN type. Building upon the identification of transcriptional induction of genes by IFNs [10,14,15], our prior studies have focused on the functional significance of over a dozen different ISGs ([6–24]. In clinical studies, this work has identified differences in IFN types and helped guide considerations of schedule, route, and dose.
After receptor binding by IFNs-α or IFN-β, the tyrosine kinases, tyk2 and JAK-1, are phosphorylated; these subsequently phosphorylate STAT (signal transducers and activators of transcription) 1 and/or STAT 2 leading to activated dimers [14,15]. Receptor signaling is suppressed by the phosphatases SHP-1 and SHP-2 and by the induced SOCS and PIAS proteins [1,14,25]. The phosphorylated ISGF-3 complex is translocated to the nucleus and forms (with the addition of a third subunit, p48 or IRF-9), a DNA binding complex specific for the IFN-stimulated response element.
Suppression of the IFN response system of the host in and/or by malignant cells has emerged as a potentially significant contributor to development of clinical disease. Mutation in an ISG, RNase L, increased prostate cancer risk [26,27]. Gene expression profiling and cytogenetic analyses have identified homozygous and heterozygous deletion or decreased expression of ISGs in melanoma, colon, breast, hematologic and other malignancies [28–60]..
Another important mechanism of ISG suppression may be epigenetic silencing with potential influence on tumor development [45,51–60]. Decreases in ISGs has been identified in transformed compared to diploid cells of varying histogenesis [45,54,58,61]. Inhibition of normal ISG expression could be the basis for effectiveness of IFNs and/or inducers in suppressing tumor emergence in carcinogen-induced murine tumors [61, 62]. Activation of the IFN system, as has been used chronic hepatitis to decrease risk of hepatocellular carcinoma [63–67], suggests IFNs or inducers could eventually play a role in chemoprevention.
Postulating that suppressed cellular responses to IFNs in tumors could in part result from aberrant methylation of the promoters of ISGs, we have examined inhibitors of epigenetic alteration in gene expression. 5-aza,2′deoxycytidine (5-Aza-dC) is a nucleoside analogue, which after incorporation into DNA, inhibits DNA methyltransferase 1 (DNMT1) by covalent binding. DNMT1 is thus trapped and not available at the DNA replication fork to copy methylation patterns from mother to daughter strand. Similar but more specific inhibition of DNMT1 can result from antisense oligonucleotides to DNMT1. Both have been utilized in our initial studies to identify changes in expression of pro-apoptotic genes, RASSF1A and XAF1[57, 58], two which are decreased in expression in patients with melanoma.
Pro-apoptotic genes and cellular effects of IFNs
The pathway that led to a focus on epigenetic silencing of gene expression emerged from studies of genes and pathways that regulate IFN-induced apoptosis. We and others had consistently identified greater antiproliferative effects for IFN-β when compared to IFNs-α [68–78]. Despite their binding to the same heterodimeric receptor and a signal transduction pathway with common elements, this occurs presumably as a result of differing three-dimensional ligand conformations [79,80]. We undertook studies which were among the first to identify IFNs through TNF-related apoptosis-inducing ligand or Apo2L (APO2L/TRAIL) induction as activating the extrinsic apoptotic cascade in melanoma and other cell lines [72,73,81].
By both TUNEL and Annexin V assays, apoptosis was identified with up to 50% apoptotic cells. Although IFN-α2 and IFN-β have equivalent antiviral activity per mg of protein, IFN-β was >5x more potent for apoptosis induction [72]. Antiviral specific activity remains the WHO-recommended standard for determining potency of an IFN. Thus to confirm labeled biological activity of the pharmaceutical grade IFNs, the two were confirmed as equivalent in the same cell line [72]. This work has suggested that antiviral specific activity may not be the best standard for IFNs for clinical antitumor applications [82].
To identify ISGs that might be involved, RNA samples from WM9 cells and WM35 melanoma cell lines were treated with IFN-α2 or IFN-β and assessed on a 10k Affymetrix oligonucleotide array. For 95% of genes assessed, IFN-β was more potent than IFN-α2 in inducing ISG expression; 910 genes were identified as induced by IFN-β at 500 units and 260 ISGs as significantly induced by IFN-β at both 50 and 500 units. Of these 260 ISGs, 209 were new ISGs based upon the array analysis [83]. Increased expression of 28 genes was further confirmed by Northern blot or semi-quantitative or quantitative RT-PCR analysis.
Nearly half of the 260 genes were functionally categorized as encoding growth regulatory proteins. Of the 104 with described growth regulatory function, 71 were induced more than 3x by 500 units and 2x by 50 units of IFN-β; 48 were new or not previously considered as ISGs. Included in this latter category were APO2L/TRAIL, XIAP-Associated Factor 1 (XAF1), galectin 9, a cyclin E binding protein, amphiphysin 1, MyD88, and ubiquitin pathway genes. Direct comparison of the gene modulatory potential of IFN-α2 and IFN-β was possible for 73 of the new and previously identified ISGs. For these, comparisons in both WM9 and WM35 melanoma cell lines, IFN-β potency was >IFN-α2 for induction of gene expression in 137/146 instances (95%).
Most known and novel ISGs were induced by both IFNs to a similar extent in the apoptosis-sensitive WM9 and apoptosis-resistant WM35 cells. However, some of the potentially apoptosis-related genes were specific to one or the other cell line. For example, APO2L/TRAIL was higher in sensitive WM9 as compared to resistant WM35 melanoma cells. Kinetic studies of induction by IFN-α and IFN-β with specific RNA probes confirmed APO2L/APO2L/TRAIL, XAF-1, K12, MYD88, SP100, AIPC, UBEL6, TSG6, Cyclin E associated, USP18, RIG-G, BST-2, and ISG27; these studies identified genes that peaked in expression early (8h) and late (36h) [83]. APO2L/TRAIL however was sustained at high levels throughout in the sensitive WM9 melanomas (83). The anti-apoptotic XIAP was present at higher levels in resistant cells; XAF1 had been defined as an XIAP interacting protein and thus potentially a tumor suppressor [84,85]. As assessed by caspase inhibitors, IFN-β-induced apoptosis was dependent on activation of the caspase cascade with cleavage of caspases 3, 8, and 9 and of the caspase 3 substrate, poly (ADP-ribose) polymerase. These changes together with the release of cytochrome c from mitochondria to cytoplasm were also identified in response to Apo2L/APO2L/TRAIL (but not for FAS or FASL). Other sensitive melanoma cell lines had a similar induction by IFN-β of APO2L/TRAIL. Antibody to APO2L/TRAIL inhibited IFN-β-induced apoptosis in the WM9 cells sensitive to IFN-α2 and IFN-β (Fig 1). In resistant A375 cells, IFN-β did not induce APO2L/TRAIL expression. Thus, induction of APO2L/TRAIL was critical for IFNs initiated apoptotic cascade.
Fig 1. Neutralization of IFN-β mediated apoptosis by APO2L/TRAIL MAb.
WM9 cells were untreated (left panel), 6% apoptotic cells by Annexin V assay; treated with IFN-β for 72 h (center), 31% apoptotic cells; or IFN-β with a MAb to APO2L/TRAIL (right), 7% apoptotic cells. Controls (MAb alone, isotype control) for each sample gave expected result. Reproduced with permission.
To further probe the mechanism of cellular refractoriness to apoptosis, resistant melanoma cell lines were analyzed for their sensitivity to recombinant APO2L/TRAIL protein [73,86]. As assessed by Annexin V and TUNEL assays, all were also resistant to apoptosis induction by APO2L/TRAIL protein (the Zn based trimer kindly provided by A. Ashkenzi (Genentech). No correlation existed between expression of apoptosis regulators (Apaf1, FLIP, caspase-8, caspase-9, caspase-3, cIAP, Bcl-2 or Bax) and resistance to APO2L/TRAIL induced apoptosis. APO2L/TRAIL activated caspase-8 and caspase-3, but subsequent apoptotic events, such as PARP cleavage and DNA fragmentation, were not observed, suggesting a possible role of inhibitors of apoptosis downstream of caspase-3. Postulating that in addition to APO2L/TRAIL, induction of one or more IFN stimulated genes might sensitize cells to APO2L/TRAIL, melanoma cell lines were treated with IFN-β for 16–24 h prior to APO2L/TRAIL; more than 30% of cells underwent apoptosis. Induction of apoptosis by IFN-β and APO2L/TRAIL correlated with synergistic activation of caspase-9, decrease in mitochondrial potential and cleavage of PARP. Cleavage a p19 fragment from XIAP to an inactive p29 and other fragments, following IFN-β or APO2L/TRAIL occurred in WM9 (Fig 2), FEMX, A375 and WM3211 cells and correlated with apoptosis [86]. Treatment with IFN-β after APO2L/TRAIL did not result in potentiated apoptosis, suggesting induction of a gene(s) by IFN-β that potentiated apoptosis. Thus in vitro IFN-β and APO2L/TRAIL in combination had more potent apoptotic and anti-growth effects compared to either cytokine alone in melanoma cells lines, an effect postulated to result from modulation by IFNs of anti-apoptotic action of IAPs.
Fig 2.
IFN-β and APO2L/TRAIL synergistically induce cleavage of inactivating p29 fragment from intact XIAP following combination treatment of WM9 cells, detected by Western blot; similar results in A375 (after exogenous APO2L/TRAIL) and WM3211 melanoma cells. No cleavage in cells resistant to apoptosis from IFN-β, APO2L/TRAIL or the combination (WM164, WM35 melanoma) (data not shown). Two different APO2L/TRAIL preparations (Preprotech-APO2L/TRAIL-P and Genentech-APO2L/TRAIL-G). Reproduced with permission.
To further probe the role of IAPs in conferring resistance to APO2L/TRAIL-induced apoptosis, melanoma cells were transfected with siRNAs to XIAP or survivin [86]. Since higher expression of inhibitors of apoptosis such as Bcl-2 or FLIP could also play a role, siRNAs to them were also assessed. Compared to both scrambled and mismatch controls, target specific siRNAs (si-Bcl-2, si-XIAP, si-FLIP or si-Surv), followed by APO2L/TRAIL resulted in marked increase in apoptosis. Compared to si-Bcl-2 or si-FLIP, siRNAs against XIAP and survivin were most potent in sensitizing melanoma cells. A substantial increase in apoptosis also occurred in renal carcinoma cells (SKRC-45, Caki-2), following inhibition of either XIAP or survivin by siRNAs. Thus, APO2L/TRAIL resistance could be overcome by interfering with expression of XIAP and survivin and to a lesser extent by inhibitors of the intrinsic (mitochondrial) pathway of apoptosis.
IFNs induced high expression of XAF1 predominantly in cell lines sensitive to the apoptotic effects of IFN-β [87]. In apoptosis-resistant cells, including WM164 and WM35 melanoma, U937 lymphoma and HT1080 fibrosarcoma cells, XAF-1 mRNA was strongly upregulated but XAF1 protein was expressed only weakly or not at all. APO2L/TRAIL is a critical mediator of IFN-β induced apoptosis, but most melanoma cell lines were resistant to exogenous recombinant APO2L/TRAIL protein. A375 melanoma cells, for example, were defective in APO2L/TRAIL induction by IFN-β and were resistant to APO2L/TRAIL-induced apoptosis. Thus, APO2L/TRAIL was necessary for apoptosis induction but only effective in the presence of XAF-1.
Examining the converse interaction, evidence that XAF-1 was a component of IFN-β sensitization for APO2L/TRAIL was provided by constitutively overexpressing XAF1 in A375 cells. A375 cells expressing XAF1 constitutively were more sensitive to APO2L/TRAIL-induced apoptosis as compared to empty vector transfected cells; sensitization correlated with the level of XAF1 expressed. Furthermore, overexpression of only the zinc-finger portion of XAF1 blocked IFN sensitization of A375 melanoma cells to the proapoptotic effects of APO2L/TRAIL [87]. These results suggested that induction of the ISG, XAF1, possibly in part due to its interaction with XIAP, determined directly cellular sensitivity to the proapoptotic actions of APO2L/TRAIL and indirectly IFNs. In melanoma, renal carcinoma and myeloma cells, if both were not induced by IFNs, no apoptosis resulted, an effect confirmed by use of MAb to APO2L/TRAIL and a dominant negative construct of XAF1 [72,87]
Epigenetic silencing of gene expression
Expression profiling of melanoma cell lines when compared to melanocytes identified many ISGs that were constitutively suppressed [45]. Although direct consequences for malignant cell function or survival was not evaluated, gene profiling and expression studies had identified genes in IFN pathways that were epigenetically silenced by hypermethylation of their 5′ regulatory regions [51–56,109,110]. Maintenance of DNA methylation in promoters can result in heritable silencing of genes that control DNA stability, cell proliferation, and apoptosis, and like mutation, are integral to the neoplastic process [90–94]. The relative degree of importance of the maintenance DNA methyltransferase 1 (DNMT1) and de novo DNMTs such as DNMT 3b [95] is still an unresolved issue; however, at least in colon, breast, and lung cancer cell lines, inhibition of DNMT1 by oligonucleotide antisense or siRNA was sufficient for re-expression of silenced genes [96,97].
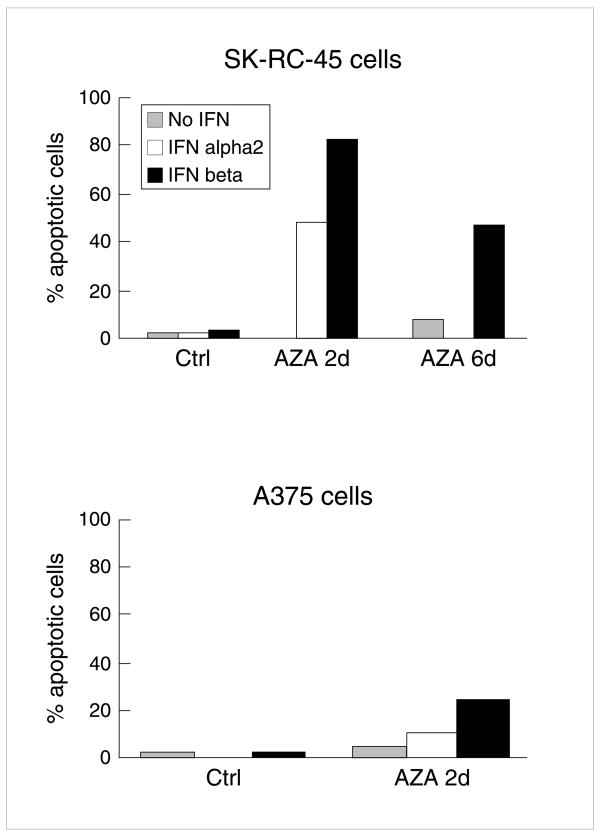
Sensitization to apoptosis in cell lines resistant to IFN-β occurred at 5-Aza-dC concentrations that alone did not result in apoptosis and resulted in an augmented apoptotic response to IFNs (Fig 3) [57]. 5-Aza-dC and also an antisense to DNMT1 (DNMT1 AS) depleted available DNMT1 protein (no similar effect with control oligonucleotide for DNMT1 AS); both DNMT1 AS and 5-Aza-dC resulted in augmented apoptotic response to IFNs in SK-RC-45 renal carcinoma (up to 80% apoptotic cells with IFN-β) and A375 cells, which were otherwise resistant to IFN-induced apoptosis (Fig 3). Confirming these results was an almost 10x reduction with 5-azacytidine in IC50 of two melanoma cell lines resistant to apoptosis induction by IFNs, FEMX and Minors (data not shown). In contrast, 5-Aza-dC caused only slight apoptosis in normal kidney epithelial cells or melanocytes (<10% TUNEL+ cells) [57]. Neither IFN-β, APO2L/TRAIL, 5-AZA-dC, nor the combinations resulted any substantial degree of apoptosis in normal melanocytes; representative data for IFN-β are presented (Fig 4). Thus for induction of apoptosis, treatments were relatively specific for malignant cells [58].
Fig 3. 5-Aza-dC effects on resistance of SK-RC-45 RCC and A375 melanoma cells to IFN-induced apoptosis.
TUNEL assay: (FITC) positive DNA was used to assess apoptosis of cells that had been treated with IFNs over 4 d 16h after plating. Cell lines were resistant to apoptosis induction by up to 1500 U/ml of IFN-α2 or IFN-β. Pretreatment with 200 nM 5-Aza-dC, daily over 2–6 d before IFN overcame resistance to apoptosis induction by 50 – 100 U/ml IFN-α2 or IFN-β, while causing little to moderate apoptosis alone (5–20% TUNEL + cells). Marked reduction in DNMT1 protein was confirmed d after 4d of 5-Aza-dC at 200 nM on western blot in SK-RC-45, and A375 cells. Reproduced with permission (57).
Fig 4. Lack of effect of 5-aza-dC on apoptosis induction for melanocytes or in combination with IFN-β.
Normal human melanocytes were obtained from Cambrex Bioscience, Walkersville MD. Melanocytes were cultured from neonatal foreskins, confirmed for gp75/TRP-1 and L-DOPA activity, maintained in the Clonetics Melanocyte culture system according to manufacturer instructions, and then treated with 0.2 μM 5-aza-dC, 250 units of IFN-β, or the combination for the indicated times and apoptosis quantified by TUNEL assay (contrast to quantitative effects on apoptosis induction in tumor cells in Figs 3, 4–7, 9).
DNMT1 inhibitors reactivated the proapoptotic tumor suppressor gene, RASSF1A, in SKRC45 and in ACHN renal carcinoma cells and A375 melanoma cells by demethylating its promoter [57]. A375 cells treated with IFNs had further augmented RASSF1A protein expression [57]. In IFN sensitive WM9 cells, in which RASSF1A was basally expressed, IFN-β increased RASSF1A protein without 5AZA-dC pretreatment. RASSF1A siRNA reduced IFN-induced apoptosis in WM9 cells and in DNMT1 depleted ACHN RCC cells [57] (Fig 5). Conversely lentiviral overexpression of RASSF1A in the absence of reversal by inhibitors of DNMT1 resulted in IFN-β or APO2L/TRAIL-induced apoptosis [57]. These results defined RASSF1A, commonly silenced by DNA methylation in melanoma and renal carcinoma, as an IFN regulated gene that participates in IFN-induced apoptosis [57]. Primary melanomas and nephrectomy specimens have a high frequency of silencing of RASSF1A by promoter hypermethylation [98]. RASSF1A interacts with the proapoptotic kinase MST1 and scaffolding protein CNK1 to induce apoptosis and has sensitized cells to apoptosis induction and mitosis regulation [98–105].
Fig 5. siRNA inhibition of apoptosis resulting from RASSF1A reactivation.
ACHN RCC were transfected first with an antisense to DNMT1 or mismatch (8d), then treated with siRNA to RASSF1A for 4 hrs, then IFN-β for 16hrs later. Apoptosis by TUNEL assay after 4 additional days Reproduced with permission (57)
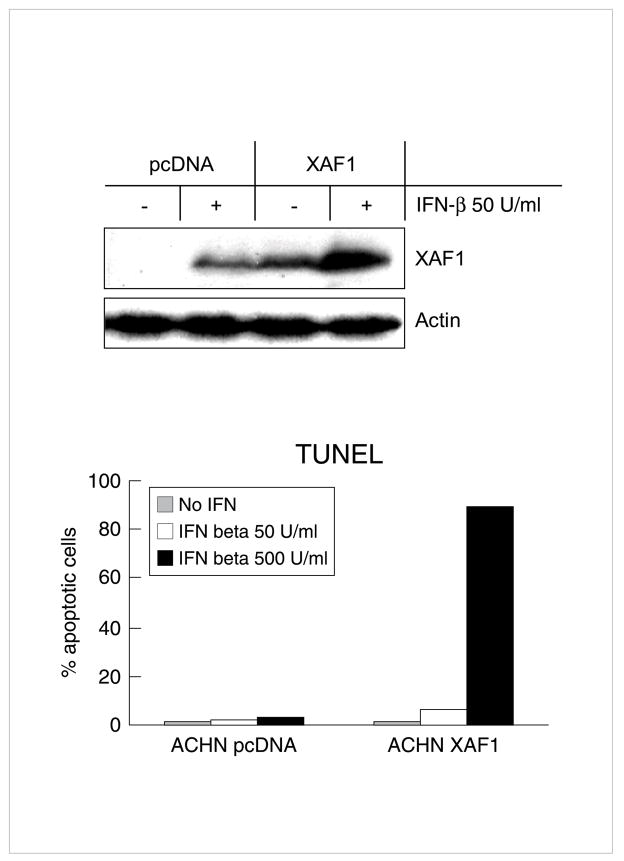
In addition to reduced RASSF1A, XAF1 was also decrease in expression in malignant cells [106]. Treatment of A375 melanoma identified increase in expression by 5-Aza-dC [58]. Methylation specific PCR confirmed demethylation of the 5′regulatory region of XAF1 and DNMT1 depletion and augmented XAF1 protein expression by IFN in A375 melanoma and ACHN cells [58]. XAF1 siRNA reduced IFN-induced apoptosis in 5-Aza-dC treated ACHN cells and in IFN sensitive melanoma cells (A375.S2). Conversely, lentiviral overexpression of XAF1 overcame resistance to apoptosis by IFN-β (Fig 6). In 6/11 melanoma cell lines, greater than 10-fold increase in XAF-1 expression resulted from DNMT1 inhibition (Table 1). Thus expression of XAF1, first identified as an ISG in our array studies, was also epigenetically silenced in melanoma and in renal carcinoma cells. Its re-expression may well contribute to augmentation of apoptosis by interactions with XIAP or by other mechanisms.
Fig 6.
Reactivation of XAF1 in A375 melanoma cells pretreated with 5-Aza-dC at 200 nM daily over 4d levels achieved by DNA demethylation. Similar effect in ACHN cells. Resistance of apoptosis induction by IFN-β overcome by overexpression of XAF1. Overexpression of XAF1plasmid vector in ACHN cells overcame resistance to high doses of IFN-β (500 U/ml, hatched), while lower doses (50 U/ml, solid) had less effect suggesting that reactivation Reproduced with permission. (58)
Table 1.
For qRT-PCR all cell lines were treated with 0.2 μM of 5-AZA-dC for 96 h. Total RNA was extracted using TRIzol Reagent according to the manufacturer’s protocol. Two μg of RNA was used for reverse transcription in 20μl of reaction mixture using a Superscript III kit. For quantitative PCR, a reaction mixture of 25 μl of TaqMan Universal PCR Master Mix, 250 nM of Taqman primer, and 60 ng of cDNA were prepared. Primers were purchased from Applied Biosystems for APO2L/TRAIL R1, APO2L/TRAIL R2, and XAF1. GAPDH for gene normalization was measured by TaqMan Human GAPDH control reagents. MUM-2C, MUM-2B, C918, and OCM-1A are ocular melanoma cell lines, while all others are cutaneous melanoma cell lines.
| Melanoma Cell | Fold Increase in Gene Expression from 5-Aza-dC | ||
|---|---|---|---|
| XAF-1 | APO2L/TRAIL R1 | APO2L/TRAIL R2 | |
| MUM 2C | 150 | 6 | 230 |
| MUM 2B | 60 | <5 | <5 |
| C918 | 20 | <5 | <5 |
| OCM-1 | 120 | <5 | <5 |
| A375 | 70 | <5 | <5 |
| SKMEL 1 | <5 | <5 | <5 |
| SKMEL 3 | <5 | 70 | <5 |
| SKMEL 28 | <5 | 30 | <5 |
| WM164 | <5 | 6 | <5 |
| WM3211 | <5 | <5 | <5 |
| MeWo | 30 | 7 | <5 |
Despite induction of APO2L/TRAIL by IFN-β in SK-MEL28 and SK-MEL3 human melanoma cells, neither underwent apoptosis in response to IFN-β or IFN-α2b (1500 U/ml). However pretreatment with 5-Aza-dC (0.1 μM) over 4d sensitized for IFN-induced apoptosis (>70% TUNEL + cells after 100 U/ml of IFN-β (data not shown). As in prior studies, Westerns confirmed decrease in DNMT1. APO2L/TRAIL R1 RNA was increased by 0.1 μM of 5-Aza-dC in SK MEL 28 and SK MEL 3 when assayed by qRT-PCR (Table 1) with a concomitant increase in protein expression on the cell surface of APO2L/TRAIL R1(unpublished, S. Bae, V. Cheriyath, 2006). Thus resistance to apoptosis from IFNs or APO2L/TRAIL could also be overcome by 5-Aza-dC in these cells through an increase in APO2L/TRAIL R1 expression
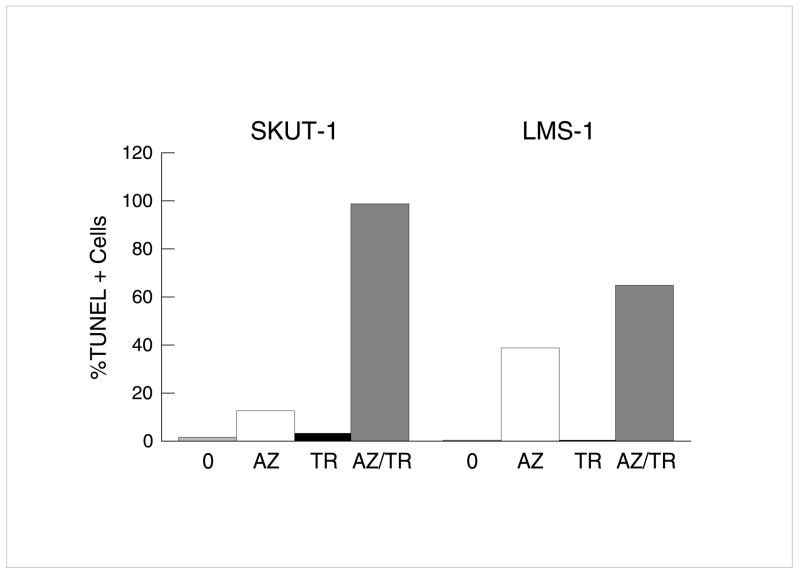
To demonstrate the potential applicability of findings to other histologies, we have extended studies of methylation inhibition to hepatocellular carcinoma (HCC) with IFN-β and leiomyosarcomas (LMS) with APO2L/TRAIL in cells completely resistant to the cytokines For example, in two LMS cell lines fewer than 5% apoptotic cells were identified after APO2L/TRAIL alone but almost 100% apoptotic cells resulted from pretreatment with 5-Aza-dC (Fig 7). In the LMS cells, the role of RASSF1A was suggested by identification of >1000x augmentation of RASSF1A by qRT-PCR after 5-Aza-dC. Similar results occurred with IFN-β in HCC cell lines (Y Ren, E Borden, unpublished, 2006).
Fig 7. Induction of apoptosis in APO2L/TRAIL-resistant leiomyosarcoma by 5-Aza-dC.
SKUT1 and LMS-1 human leiomyosarcoma cell lines, both APO2L/TRAIL resistant, were treated with 5-Aza-dC. Apoptosis was assessed 3d after APO2L/TRAIL (TR) (100ng/ml). Cells were pretreated for 4d with 5-Aza-dC (0.2μM). After wash, APO2L/TRAIL was added. Apoptosis assessed by TUNEL assay 4d later.
Overall summary and perspectives
The focus of our research has been and is to define cellular effects of IFNs and how to thus enable IFNs and their induced genes, such as APO2L/TRAIL, to reach their full clinical antitumor potential. Expression of APO2L/TRAIL proved necessary but not sufficient for apoptosis induction. To identify new therapeutic strategies for IFNs in melanoma, we have sought ways to overcome resistance. The putative tumor suppressor genes, XAF1 and RASSF1A, which are ISGs augmenting apoptosis, have been identified. The low clinical levels in melanomas of XAF1 and RASSF1A (and also possibly APO2L/TRAIL-R1) may reflect epigenetic silencing; our studies of reversal of hypermethylation by 5-Aza-dC have identified markedly augmented gene expression and functional effects. These results have led to design of a Phase I combination trial to establish gene modulation of the combination of 5-Aza-dC with IFNA. In addition to the in vitro studies, we have also established effects of 5-Aza-dC in combination with IFN-β in human tumor xenografts in nude mice [58]. Further rationale for such a trial was increase in global DNA demethylation in peripheral blood mononuclear cells (PBMCs) of patients treated with 5-Aza-dC [107].
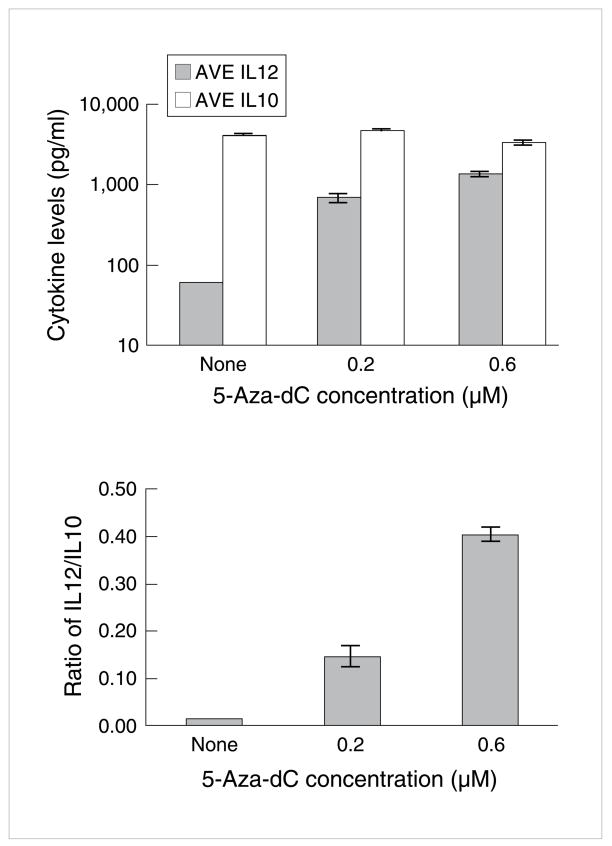
These latter findings on influence of 5-Aza-dC on gene expression in PBMCs are in accord with our findings on influence of 5-Aza-dC on dendritic cell function. Recent papers suggest methylation silencing of genes could have a critical role in host tumor interactions: changes in antigen presentation, recognition, T cell function, and immunosurveillance [45,107,108,109–114]. We have defined an additional effect of methylation silencing of gene expression in dendritic cells (DC). Human myeloid DCs were matured from peripheral blood with CSF-GM and IL-4, treated with 5-Aza-dC (0.1 uM) for 4d, and then with poly I:poly C, as a representative ligand for TLR3. 5-Aza-dC resulted in a 20x augmentation in IL-12 production when compared to cultures not treated with the methylation inhibitor (Fig 8). Furthermore no increase resulted in IL-10 suggesting a potential influence on enhancement of Th1 cell function and suppression of Th2 cell activity (M Whitmore, E Borden, unpublished).
Fig 8. Augmentation of IL-12 production by human dendritic cells by 5-aza-dC.
Human myeloid DCs were matured from peripheral blood with CSF-GM and IL-4, treated with 5-Aza-dC (0.1 uM) for 4d, and then poly I:poly C, as a representative ligand for TLR3.
Increase in expression of other methylation-silenced genes, such as other apoptosis related genes (PTEN, p16), ISGs (IRF7, ISG15, or HLA-DR) or melanoma-associated antigens (MAGEs or membrane proteoglycans) could all also potentiate effects of IFNs, APO2L/TRAIL, immunoaugmenting agents, or cytotoxics such as cisplatin for melanoma and other tumors (Fig 7, Fig 9). Furthermore, clinical findings on reversal of methylation could further stimulate studies of epigenetic silencing in other signaling pathways, such as wnt, hedgehog, steroid hormones, or those influencing angiogenesis, which may be in part each regulated by promoter methylation [115–118].
Fig 9. Augmentation of apoptosis by 5-Aza-Dc (AZ) in cisplatin (CP)-resistant melanoma cells (A375 or SKMEL 28).
Cells were first treated with 5-Aza-dC (0.2uM) for 96 hrs, then cisplatin (10uM) added and apoptosis assessed 96 hrs later by TUNEL assay. In both cell lines, RASSF1A was increased >500x by 5-Aza-dC.
Acknowledgments
This research from my laboratory at Cleveland Clinic Foundation would not have been possible without the dedicated work and innovative ideas of a number of talented individuals, who have or will establish their own independent laboratories, including Mamta Chawla-Sarkar, Douglas Leaman, Frederic Reu, Venu Cheriyath, Soo-In Bae, Yi Ren and Mark Whitmore. Barbara Jacobs has been a critical individual in assuring quality and technical excellence in all of these investigations. Kathy Nagle has provided outstanding secretarial assistance. Supported in part by NIH CA89091 and National Center for Research Resources General Clinical Research Grant M01 RR-018390.
Biography
 Ernest C. Borden, M.D. Ernest C. Borden obtained his medical degree from Duke University and joined Cleveland Clinic in 1998 to direct the Center for Cancer Drug Discovery and Development. In 2005 the 5 laboratories in the Case Western Reserve Cleveland Clinic College of Medicine were integrated to form the Center for Hematology and Oncology Molecular Therapeutics (CHOMT) with Dr. Borden named as Director. He is also a staff member in the Department of Solid Tumor Oncology and a Professor in Cancer Biology in the Lerner Research Institute. In the 1980s, he was amongst the first to initiate clinical trials of interferons, the first human protein effective in stimulating immune mechanisms to fight cancer. In addition to developing improved approaches to clinical assessment of interferons and its inducers, Dr. Borden’s laboratory has focused on function and action of genes stimulated by interferons and anti-tumor effects of other biological therapeutics such as monoclonal antibiodies. In addition to targeted biological therapies, he has an international reputation for research and treatment of melanomas and sarcomas. Dr. Borden has also been listed in Best Doctors of America for the past 10 years and honored by the Milstein Award from the International Society of Interferon and Cytokine Research (ISICR) in 2004 and as an American Cancer Society Professor of Clinical Oncology. Dr. Borden has also served as consultant to several biotechnology companies, including CIBA GEIGY (now Novartis AG in Basel, Switzerland), Ares-Serono (Geneva, Switzerland), IDEC Pharmaceuticals (LaJolla, CA), Ribozyme Pharmaceuticals Inc. (Boulder, CG) and Igeneon Inc. (Vienna, Austria). He holds 4 patents.
Ernest C. Borden, M.D. Ernest C. Borden obtained his medical degree from Duke University and joined Cleveland Clinic in 1998 to direct the Center for Cancer Drug Discovery and Development. In 2005 the 5 laboratories in the Case Western Reserve Cleveland Clinic College of Medicine were integrated to form the Center for Hematology and Oncology Molecular Therapeutics (CHOMT) with Dr. Borden named as Director. He is also a staff member in the Department of Solid Tumor Oncology and a Professor in Cancer Biology in the Lerner Research Institute. In the 1980s, he was amongst the first to initiate clinical trials of interferons, the first human protein effective in stimulating immune mechanisms to fight cancer. In addition to developing improved approaches to clinical assessment of interferons and its inducers, Dr. Borden’s laboratory has focused on function and action of genes stimulated by interferons and anti-tumor effects of other biological therapeutics such as monoclonal antibiodies. In addition to targeted biological therapies, he has an international reputation for research and treatment of melanomas and sarcomas. Dr. Borden has also been listed in Best Doctors of America for the past 10 years and honored by the Milstein Award from the International Society of Interferon and Cytokine Research (ISICR) in 2004 and as an American Cancer Society Professor of Clinical Oncology. Dr. Borden has also served as consultant to several biotechnology companies, including CIBA GEIGY (now Novartis AG in Basel, Switzerland), Ares-Serono (Geneva, Switzerland), IDEC Pharmaceuticals (LaJolla, CA), Ribozyme Pharmaceuticals Inc. (Boulder, CG) and Igeneon Inc. (Vienna, Austria). He holds 4 patents.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Borden EC, Waalen J, Schreiber R, Williams B. Response and resistance to interferons and interacting cytokines. J Natl Cancer Inst. 1995;87:257–264. doi: 10.1093/jnci/87.4.257. [DOI] [PubMed] [Google Scholar]
- 2.Kirkwood J. Cancer immunotherapy: The interferon-alpha experience. Semin Oncol. 2002;29(3 Suppl 7):18–26. doi: 10.1053/sonc.2002.33078. [DOI] [PubMed] [Google Scholar]
- 3.Borden EC. In: Interferons in Cancer Medicine. 7. Holland JF, Kufe D, Pollock R, Weicheselbaum R, Bast R, Gansler T, Frei T, editors. BC Decker Hamilton; Ontario: 2006. pp. 733–743. [Google Scholar]
- 4.Buzaid AC. Management of metastatic cutaneous melanoma. Oncology (Williston Park) 2004;18:1443–1450. [PubMed] [Google Scholar]
- 5.Kirkwood JM, Harris JE, Vera R, Sandler S, Fischer DS, Khandekar J, et al. A randomized study of low and high doses of leukocyte alpha-interferon in metastatic renal cell carcinoma: The American Cancer Society collaborative trial. Cancer Res. 1985;45:863–871. [PubMed] [Google Scholar]
- 6.Kirkwood JM, Manola J, Ibrahim J, Sondak V, Ernstoff MS, Rao U Eastern Cooperative Oncology Group. A pooled analysis of eastern cooperative oncology group and intergroup trials of adjuvant high-dose interferon for melanoma. Clin Cancer Res. 2004;10:1670–1677. doi: 10.1158/1078-0432.ccr-1103-3. [DOI] [PubMed] [Google Scholar]
- 7.Cole BF, Gelber RD, Kirkwood JM, Goldhirsch A, Barylak E, Borden EC. Quality-of-life-adjusted survival analysis of interferon alfa-2b adjuvant treatment of high-rish resected cutaneous melanoma: An Eastern Cooperative Oncology Group study. J Clin Oncol. 1996;14:2666–2673. doi: 10.1200/JCO.1996.14.10.2666. [DOI] [PubMed] [Google Scholar]
- 8.Hillner BE, Kirkwood JM, Atkins MB, Johnson ER, Smith TJ. Economic analysis of adjuvant interferon alfa-2b in high-rish melanoma based on projections from Eastern Cooperative Oncology Group 1684. J Clin Oncol. 1997;15:2351–2358. doi: 10.1200/JCO.1997.15.6.2351. J Clin Oncol. 1997 Jun; 15(6):2351–8. [DOI] [PubMed] [Google Scholar]
- 9.Borden EC, Parkinson D. A perspective on the clinical effectiveness and tolerance of interferon-alpha. Semin Oncol. 1998;25(Suppl 1):3–8. [PubMed] [Google Scholar]
- 10.Kalvakolanu DV, Borden EC. An overview of the interferon system: Signal transduction and mechanisms of action. Cancer Invest. 1996;14:25–53. doi: 10.3109/07357909609018435. [DOI] [PubMed] [Google Scholar]
- 11.Balch C, Houghton A, Sober A, Soong SJ. Cutaneous Melanoma. 4. Quality Medical Publishing; St. Louis, MO: 2003. [Google Scholar]
- 12.Masci P, Borden EC. Malignant Melanoma: treatments emerging, but early detectin is still key. Cleve Clin J Med. 2002;69:529–545. doi: 10.3949/ccjm.69.7.529. [DOI] [PubMed] [Google Scholar]
- 13.Tsao H, Atkins MB, Sober AJ. Management of cutaneous melanoma. N Engl J Med. 2004;351:998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 14.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 15.Pfeffer LM, Dinarello CA, Herberman RB, Williams BR, Borden EC, Bordens R, et al. Biological properties of recombinant alpha-interferons: 40th anniversary of the discovery of interferons. Cancer Res. 1998;58:2489–2499. [PubMed] [Google Scholar]
- 16.Merritt JA, Ball LA, Sielaff KM, Meltzer DM, Borden EC. Modulation of 2′, 5′-oligoadenylate synththetase in patients treated with alpha-interferon: effects of dose, schedule, and route of administration. J Interferon Res. 1986;6:189–198. doi: 10.1089/jir.1986.6.189. [DOI] [PubMed] [Google Scholar]
- 17.Spear GT, Paulnock DM, Jordan RL, Meltzer DM, Merritt JA, Borden EC. Enhancement of monocyte class I and II histocompatibility antigen expression in man by in vivo beta-interferon. Clin Exp Immunol. 1987;69:107–115. [PMC free article] [PubMed] [Google Scholar]
- 18.Borden EC. Augmented tumor-associated antigen expression by interferons. J Natl Cancer Inst. 1988;80:148–149. doi: 10.1093/jnci/80.3.148. [DOI] [PubMed] [Google Scholar]
- 19.Carlin JM, Borden EC, Sondel PM, Byrne GI. Interferon-induced indoleamine 2,3-dioxygenase activity in human mononuclear phagocytes. J Leukoc Biol. 1989;45:29–34. doi: 10.1002/jlb.45.1.29. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein D, Sielaff KM, Storer BE, Brown RR, Datta SP, Witt PL, et al. Human biologic response modification by interferon in the absence of measurable serum concentrations: A comparative trial of subcutaneous and intravenous interferon-beta serine. J Natl Cancer Inst. 1989;81:1061–1068. doi: 10.1093/jnci/81.14.1061. [DOI] [PubMed] [Google Scholar]
- 21.Schiller JH, Horisberger MA, Bittner G, Carlin JM, Storer B, Byrne GI, et al. Effects of combinations of interferon-beta ser and interferon-gamma on interferon-inducible proteins and on the cell cycle. J Biol Response Mod. 1990;9:368–377. [PubMed] [Google Scholar]
- 22.Greiner JW, Guadagni F, Goldstein D, Borden EC, Ritts RE, Jr, Witt P, et al. Evidence for the elevation of serum carcinoembryonic antigen and tumor-associated glycoprotein-72 levels in patients administered interferons. Cancer Res. 1991;51:4155–4163. [PubMed] [Google Scholar]
- 23.D’Cunha J, Ramanujam S, Wagner RJ, Witt PL, Knight E, Jr, Borden EC. In vitro and in vivo secretion of human ISG15, an IFN-induced immunomodulatory cytokine. J Immunol. 1996;157:4100–4108. [PubMed] [Google Scholar]
- 24.D’Cunha J, Knight E, Jr, Haas AL, Truitt RL, Borden EC. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc Natl Acad Sci U S A. 1996;93:211–215. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi T, Pathak MK, Lindner DJ, Ketterer ME, Farver C, Borden EC. Anticancer activity of sodium stibogluconate in synergy with IFNs. J Immunol. 2002;169:5978–5985. doi: 10.4049/jimmunol.169.10.5978. [DOI] [PubMed] [Google Scholar]
- 26.Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, Xu J, et al. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet. 2002;30:181–184. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- 27.Casey G, Neville PJ, Plummer SJ, Xiang Y, Krumroy LM, Klein EA, et al. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat Genet. 2002;32:581–583. doi: 10.1038/ng1021. [DOI] [PubMed] [Google Scholar]
- 28.Diaz MO, Rubin CM, Harden A, Ziemin S, Larson RA, Le Beau MM, et al. Deletions of interferon genes in acute lymphoblastic leukemia. N Engl J Med. 1990;322:77–82. doi: 10.1056/NEJM199001113220202. [DOI] [PubMed] [Google Scholar]
- 29.James CD, He J, Carlbom E, Nordenskjold M, Cavenee WK, Collins VP. Chromosome 9 deletion mapping reveals interferon alpha and interferon beta-1 gene deletions in human glial tumors. Cancer Res. 1991;51:1684–1688. [PubMed] [Google Scholar]
- 30.Fountain JW, Karayiorgou M, Ernstoff MS, Kirkwood JM, Vlock DR, Titus-Ernstoff L, et al. Homozygous deletions within human chromosome band 9p21 in melanoma. Proc Natl Acad Sci U S A. 1992;89:10557–10561. doi: 10.1073/pnas.89.21.10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olopade OI, Buchhagen DL, Malik K, Sherman J, Nobori T, Bader S, et al. Homozygous loss of the interferon genes defines the critical region on 9p that is deleted in lung cancers. Cancer Res. 1993;53(10 Suppl):2410–2415. [PubMed] [Google Scholar]
- 32.Cairns P, Tokino K, Eby Y, Sidransky D. Homozygous deletions of 9p21 in primary human bladder tumors detected by comparative multiplex polymerase chain reaction. Cancer Res. 1994;54:1422–1424. [PubMed] [Google Scholar]
- 33.Green WB, Slovak ML, Chen IM, Pallavicini M, Hecht JL, Willman CL. Lack of IRF-1 expression in acute promyelocytic leukemia and in a subset of acute myeloid leukemias with del(5)(q31) Leukemia. 1999;13:1960–1971. doi: 10.1038/sj.leu.2401596. [DOI] [PubMed] [Google Scholar]
- 34.Johnsen A, France J, Sy MS, Harding CV. Down-regulation of the transporter for antigen presentation, proteasome subunits, and class I major histocompatibility complex in tumor cell lines. Cancer Res. 1998;58:3660–3677. [PubMed] [Google Scholar]
- 35.DeYoung KL, Ray ME, Su YA, Anzick SL, Johnstone RW, Trapani JA, et al. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15:453–457. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 36.Holland EA, Beaton SC, Edwards BG, Kefford RF, Mann GJ. Loss of heterozygosity and homozygous deletions on 9p21–22 in melanoma. Oncogene. 1994;9:1361–1365. [PubMed] [Google Scholar]
- 37.Delp K, Momburg F, Hilmes C, Huber C, Seliger B. Functional deficiencies of components of the MHC class I antigen pathway in human tumors of epithelial origin. Bone Marrow Transplant. 2000;25 Suppl 2:S88–S95. doi: 10.1038/sj.bmt.1702363. [DOI] [PubMed] [Google Scholar]
- 38.Landolfo S, Guarini A, Riera L, Gariglio M, Gribaudo G, Cignetti A, et al. Chronic myeloid leukemia cells resistant to interferon-alpha lack STAT1 expression. Hematol J. 2000;1:7–14. doi: 10.1038/sj.thj.6200004. [DOI] [PubMed] [Google Scholar]
- 39.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka N, Taniguchi T. The interferon regulatory factors and oncogenesis. Semin Cancer Biol. 2000;10:73–81. doi: 10.1006/scbi.2000.0310. [DOI] [PubMed] [Google Scholar]
- 41.Shou J, Soriano R, Hayward SW, Cunha GR, Williams PM, Gao WQ. Expression profiling of a human cell line model of prostatic cancer reveals a direct involvement of interferon signaling in prostate tumor progression. Proc Natl Acad Sci U S A. 2002;99:2830–2835. doi: 10.1073/pnas.052705299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seth A, Kitching R, Landberg G, Xu J, Zubovits J, Burger AM. Gene expression profiling of ductal carcinomas in situ and invasive breast tumors. Anticancer Res. 2003;23:2043–2051. [PubMed] [Google Scholar]
- 43.Cabrera CM, Jimenez P, Cabrera T, Esparza C, Ruiz-Cabello F, Garrido F. Total loss of MHC class I in colorectal tumors can be explained by two molecular pathways: beta2-microglobulin inactivation in MSI-positive tumors and LMP7/TAP2 downregulation in MSI-negative tumors. Tissue Antigens. 2003;61:211–219. doi: 10.1034/j.1399-0039.2003.00020.x. [DOI] [PubMed] [Google Scholar]
- 44.Seliger B, Atkins D, Bock M, Ritz U, Ferrone S, Huber C, et al. Characterization of human lymphocyte antigen class I antigen-processing machinery defects in renal cell carcinoma lesions with special emphasis on transporter-associated with antigen-processing down-regulation. Clin Cancer Res. 2003;9:1721–1727. [PubMed] [Google Scholar]
- 45.Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 46.Pitha-Rowe I, Petty WJ, Feng Q, Koza-Taylor PH, Dimattia DA, Pinder L, et al. Microarray analyses uncover UBE1L as a candidate target gene for lung cancer chemoprevention. Cancer Res. 2004;64:8109–8115. doi: 10.1158/0008-5472.CAN-03-3938. [DOI] [PubMed] [Google Scholar]
- 47.Olson JJ, James CD, Lawson D, Hunter S, Tang G, Billingsley J. Correlation of the response of recurrent malignant gliomas treated with interferon alpha with tumor interferon alpha gene content. Int J Oncol. 2004;25:419–427. [PubMed] [Google Scholar]
- 48.Akyerli CB, Beksac M, Holko M, Frevel M, Dalva K, Ozbek U, et al. Expression of IFITM1 in chronic myeloid leukemia patients. Leuk Res. 2005;29:283–286. doi: 10.1016/j.leukres.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Zimmer R, Thomas P. Expression profiling and interferon-beta regulation of liver metastases in colorectal cancer cells. Clin Exp Metastasis. 2002;19:541–550. doi: 10.1023/a:1020325327461. [DOI] [PubMed] [Google Scholar]
- 50.Stadler WM, Sherman J, Bohlander SK, Roulston D, Dreyling M, Rukstalis D, et al. Homozygous deletions within chromosomal bands 9p21–22 in bladder cancer. Cancer Res. 1994;54:2060–2063. [PubMed] [Google Scholar]
- 51.Katzenellenbogen RA, Baylin SB, Herman JG. Hypermethylation of the DAP-kinase CpG island is a common alteration in B-cell malignancies. Blood. 1999;93:4347–4353. [PubMed] [Google Scholar]
- 52.Lu R, Au WC, Yeow WS, Hageman N, Pitha PM. Regulation of the promoter activity of interferon regulatory factor-7 gene. Activation by interferon snd silencing by hypermethylation. J Biol Chem. 2000;275:31805–31812. doi: 10.1074/jbc.M005288200. [DOI] [PubMed] [Google Scholar]
- 53.Morris AC, Spangler WE, Boss JM. Methylation of class II trans-activator promoter IV: a novel mechanism of MHC class II gene control. J Immunol. 2000;164:4143–4149. doi: 10.4049/jimmunol.164.8.4143. [DOI] [PubMed] [Google Scholar]
- 54.Kulaeva OI, Draghici S, Tang L, Kraniak JM, Land SJ, Tainsky MA. Epigenetic silencing of multiple interferon pathway genes after cellular immortalization. Oncogene. 2003;22:4118–4127. doi: 10.1038/sj.onc.1206594. [DOI] [PubMed] [Google Scholar]
- 55.Lee JS, O’Neill L. Methylation of the HLA-DR alpha gene is positively correlated with expression. Immunogenetics. 1987;26:92–98. doi: 10.1007/BF00345460. [DOI] [PubMed] [Google Scholar]
- 56.Karpf AR, Peterson PW, Rawlins JT, Dalley BK, Yang Q, Albertsen H, et al. Inhibition of DNA methyltransferase stimulates the expression of signal transducer and activator of transcription 1, 2, and 3 genes in colon tumor cells. Proc Natl Acad Sci. 1999;96:14007–14012. doi: 10.1073/pnas.96.24.14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reu FJ, Leaman DW, Maitra RR, Bae SI, Cherkassky L, Fox MW, Rempinski DR, Beaulieu N, MacLeod AR, Borden EC. Expression of RASSF1A, an epigenetically silenced tumor suppressor, overcomes resistance to apoptosis induction by interferons. Cancer Res. 2006;66:2785–2793. doi: 10.1158/0008-5472.CAN-05-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reu FJ, Bae SI, Cherkassky L, Leaman DW, Fox MW, Beaulieu N, MacLeod AR, Borden EC. Overcoming resistance to interferon-induced apoptosis of renal cancer and melanoma cells by DNA demethylation. J Clin Oncol. 2006;24:3771–3779. doi: 10.1200/JCO.2005.03.4074. [DOI] [PubMed] [Google Scholar]
- 59.Liang G, Gonzales FA, Jones PA, Orntoft TF, Thykjaer T. Analysis of gene induction in human fibroblasts and bladder cancer cells exposed to the methylation inhibitor 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:961–966. [PubMed] [Google Scholar]
- 60.Reid Tony R, Merigan Thomas C, Basham Teresa Y. Resistance to Interferon-a in a Mouse B-Cell Lymphona Involves DNA Methylation. J Interferon Res. 1992;12:131–137. doi: 10.1089/jir.1992.12.131. [DOI] [PubMed] [Google Scholar]
- 61.Nakaji M, Yano Y, Ninomiya T, Seo Y, Hamano K, Yoon S, et al. IFN-alpha prevents the growth of pre-neoplastic lesions and inhibits the development of hepatocellular carcinoma in the rat. Carcinogenesis. 2004;25:389–397. doi: 10.1093/carcin/bgh028. [DOI] [PubMed] [Google Scholar]
- 62.Borden EC, Sidky YA, Erturk E, Wierenga W, Bryan GT. Protection from carcinogen-induced murine bladder carcinoma by interferons and an oral interferon-inducing pyrimidinone, bropirimine. Cancer Res. 1990;50:1071–1074. [PubMed] [Google Scholar]
- 63.Oon CJ, Chen WN. Lymphoblastoid alpha-interferon in the prevention of hepatocellular carcinoma (HCC) in high-risk HbsAg-positive resected cirrhotic HCC cases: a 14-year follow-up. Cancer Invest. 2003 Jun;21(3):394–399. doi: 10.1081/cnv-120018231. [DOI] [PubMed] [Google Scholar]
- 64.Yoshida H, Tateishi R, Arakawa Y, Sata M, Fujiyama S, Nishiguchi S, et al. Benefit of interferon therapy in hepatocellular carcinoma prevention for individual patients with chronic hepatitis C. Gut. 2004;53:425–430. doi: 10.1136/gut.2003.030353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kubo S, Nishiguchi S, Hirohashi K, Tanaka H, Shuto T, Kinoshita H. Randomized clinical trial of long-term outcome after resection of hepatitis C virus-related hepatocellular carcinoma by postoperative interferon therapy. Br J Surg. 2002;89:418–422. doi: 10.1046/j.0007-1323.2001.02054.x. [DOI] [PubMed] [Google Scholar]
- 66.Takimoto M, Ohkoshi S, Ichida T, Takeda Y, Nomoto M, Asakura H, et al. Interferon inhibits progression of liver fibrosis and reduces the risk of hepatocarcinogenesis in patients with chronic hepatitis C: A retrospective multicenter analysis of 652 patients. Dig Dis Sci. 2002;47:170–176. doi: 10.1023/a:1013244326874. [DOI] [PubMed] [Google Scholar]
- 67.Kulik LM. Can therapy of hepatitis C affect the development of hepatocellular carcinoma? J Natl Compr Canc Netw. 2006;4:751–757. doi: 10.6004/jnccn.2006.0065. [DOI] [PubMed] [Google Scholar]
- 68.Borden EC, Hogan TF, Voelkel JG. Comparative antiproliferative activity in vitro of natural interferons alpha and beta for diploid and transformed human cells. Cancer Res. 1982 Dec;42(12):4948–4953. [PubMed] [Google Scholar]
- 69.Schiller JH, Willson JK, Bittner G, Wolberg WH, Hawkins MJ, Borden EC. Antiproliferative effects of interferons on human melanoma cells in the human tumor colony-forming assay. J Interferon Res. 1986;6:615–625. doi: 10.1089/jir.1986.6.615. [DOI] [PubMed] [Google Scholar]
- 70.Rosenblum MG, Yung WK, Kelleher PJ, Ruzicka F, Steck PA, Borden EC. Growth inhibitory effects of interferon-beta but not interferon-alpha on human glioma cells: Correlation of receptor binding, 2′,5′-oligoadenylate synthetase and protein kinase activity. J Interferon Res. 1990;10:141–151. doi: 10.1089/jir.1990.10.141. [DOI] [PubMed] [Google Scholar]
- 71.Johns TG, Mackay IR, Callister KA, Hertzog PJ, Devenish RJ, Linnane AW. Antiproliferative potencies of interferons on melanoma cell lines and xenografts: Higher efficacy of interferon beta. J Natl Cancer Inst. 1992;84:1185–1190. doi: 10.1093/jnci/84.15.1185. [DOI] [PubMed] [Google Scholar]
- 72.Chawla-Sarkar M, Leaman DW, Borden EC. Preferential induction of apoptosis by interferon (IFN)-beta compared with IFN-alpha2: correlation with APO2L/TRAIL/Apo2L induction in melanoma cell lines. Clin Cancer Res. 2001;7:1821–1831. [PubMed] [Google Scholar]
- 73.Chawla-Sarkar M, Leaman DW, Jacobs BS, Borden EC. IFN-β pretreatment sensitizes human melanoma cells to APO2L/TRAIL/Apo2 ligand-induced apoptosis. J Immunol. 2002;169:847–855. doi: 10.4049/jimmunol.169.2.847. [DOI] [PubMed] [Google Scholar]
- 74.Chen Q, Gong B, Mahmoud-Ahmed AS, Zhou A, Hsi ED, Hussein M, et al. Apo2L/APO2L/TRAIL and Bcl-2-related proteins regulate type I interferon-induced apoptosis in multiple myeloma. Blood. 2001;98:2183–2192. doi: 10.1182/blood.v98.7.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thyrell L, Erickson S, Zhivotovsky B, Pokrovskaja K, Sangfelt O, Castro J, et al. Mechanisms of Interferon-alpha induced apoptosis in malignant cells. Oncogene. 2002;21:1251–1262. doi: 10.1038/sj.onc.1205179. [DOI] [PubMed] [Google Scholar]
- 76.Sangfelt O, Erickson S, Castro J, Heiden T, Einhorn S, Grander D. Induction of apoptosis and inhibition of cell growth are independent responses to interferon-alpha in hematopoietic cell lines. Cell Growth Differ. 1997;8:343–352. [PubMed] [Google Scholar]
- 77.Damdinsuren B, Nagano H, Sakon M, Kondo M, Yamamoto T, Umeshita K, Dono K, Nakamori S, Monden M. Interferon-beta is more potent than interferon-alpha in inhibition of human hepatocellular carcinoma cell growth when used alone and in combination with anticancer drugs. Ann Surg Oncol. 2003;10:1184–1190. doi: 10.1245/aso.2003.03.010. [DOI] [PubMed] [Google Scholar]
- 78.Sanceau J, Hiscott J, Delattre O, Wietzerbin J. IFN-beta induces serine phosphorylation of Stat-1 in Ewing’s sarcoma cells and mediates apoptosis via induction of IRF-1 and activation of caspase-7. Oncogene. 2000;19:3372–3383. doi: 10.1038/sj.onc.1203670. [DOI] [PubMed] [Google Scholar]
- 79.Lewerenz M, Mogensen KE, Uze G. Shared receptor components but distinct complexes for alpha and beta interferons. J Mol Biol. 1998;282:585–599. doi: 10.1006/jmbi.1998.2026. [DOI] [PubMed] [Google Scholar]
- 80.Jaitin DA, Roisman LC, Jaks E, Gavutis M, Piehler J, Van der Heyden J, Uze G, Schreiber G. Inquiring into the differential action of interferons (IFNs): an IFN-alpha2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-beta. Mol Cell Biol. 2006;26:1888–1897. doi: 10.1128/MCB.26.5.1888-1897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chawla-Sarkar M, Lindner DJ, Liu YF, Williams BR, Sen GC, Silverman RH, et al. Apoptosis and interferons: Role of interferon-stimulated genes as mediators of apoptosis. Apoptosis. 2003 Jun;8(3):237–249. doi: 10.1023/a:1023668705040. [DOI] [PubMed] [Google Scholar]
- 82.Borden EC. Interferons and cancer: where from here? J Interferon Cytokine Res. 2005;25:511–527. doi: 10.1089/jir.2005.25.511. [DOI] [PubMed] [Google Scholar]
- 83.Leaman DW, Chawla-Sarkar M, Jacobs B, Vyas K, Sun Y, Ozdemir A, et al. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-beta compared with IFN-alpha2. J Interferon Cytokine Res. 2003;23:745–756. doi: 10.1089/107999003772084860. [DOI] [PubMed] [Google Scholar]
- 84.Liston P, Fong WG, Kelly NL, Toji S, Miyazaki T, Conte D, Tamai K, Craig CG, McBurney MW, Korneluk RG. Identification of XAF1 as an antagonist of XIAP anti-Caspase activity. Nat Cell Biol. 2001;3:128–133. doi: 10.1038/35055027. [DOI] [PubMed] [Google Scholar]
- 85.Fong WG, Liston P, Rajcan-Separovic E, St Jean M, Craig C, Korneluk RG. Expression and genetic analysis of XIAP-associated factor 1 (XAF1) in cancer cell lines. Genomics. 2000;70:113–122. doi: 10.1006/geno.2000.6364. [DOI] [PubMed] [Google Scholar]
- 86.Chawla-Sarkar M, Bae SI, Reu FJ, Jacobs BS, Lindner DJ, Borden EC. Downregulation of Bcl-2, FLIP or IAPs (XIAP and survivin) by siRNAs sensitizes resistant melanoma cells to Apo2L/APO2L/TRAIL-induced apoptosis. Cell Death Differ. 2004;11:915–923. doi: 10.1038/sj.cdd.4401416. [DOI] [PubMed] [Google Scholar]
- 87.Leaman DW, Chawla-Sarkar M, Vyas K, Reheman M, Tamai K, Toji S, et al. Identification of X-linked Inhibitor of Apoptosis-associated Factor-1 as an Interferon-stimulated Gene That Augments APO2L/TRAIL Apo2L-induced Apoptosis. J Biol Chem. 2002;2(277):28504–28511. doi: 10.1074/jbc.M204851200. [DOI] [PubMed] [Google Scholar]
- 88.Byun DS, Cho K, Ryu BK, Lee MG, Kang MJ, Kim HR, et al. Hypermethylation of XIAP-associated factor 1, a putative tumor suppressor gene from the 17p13. 2 locus, in human gastric adenocarcinomas. Cancer Res. 2003;63:7068–7075. [PubMed] [Google Scholar]
- 89.Hopkins-Donaldson S, Ziegler A, Kurtz S, Bigosch C, Kandioler D, Ludwig C, et al. Silencing of death receptor and caspase-8 expression in small cell lung carcinoma cell lines and tumors by DNA methylation. Cell Death Differ. 2003;10:356–364. doi: 10.1038/sj.cdd.4401157. [DOI] [PubMed] [Google Scholar]
- 90.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 91.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 92.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 93.Jones PA. Overview of cancer epigenetics. Semin Hematol. 2005;42:S3–8. doi: 10.1053/j.seminhematol.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 94.Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- 95.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 96.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, et al. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 97.Suzuki M, Sunaga N, Shames DS, Toyooka S, Gazdar AF, Minna JD. RNA interference-mediated knockdown of DNA methyltransferase 1 leads to promoter demethylation and gene re-expression in human lung and breast cancer cells. Cancer Res. 2004;64:3137–3143. doi: 10.1158/0008-5472.can-03-3046. [DOI] [PubMed] [Google Scholar]
- 98.Spugnardi M, Tommasi S, Dammann R, Pfeifer GP, Hoon DS. Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer Res. 2003;63:1639–1643. [PubMed] [Google Scholar]
- 99.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 100.Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J Biol Chem. 2000;275:35669–35672. doi: 10.1074/jbc.C000463200. [DOI] [PubMed] [Google Scholar]
- 101.Mathe E. RASSF1A, the new guardian of mitosis. Nat Genet. 2004;36:117–118. doi: 10.1038/ng0204-117. [DOI] [PubMed] [Google Scholar]
- 102.Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol. 2002;22:4309–4318. doi: 10.1128/MCB.22.12.4309-4318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Song MS, Song SJ, Ayad NG, Chang JS, Lee JH, Hong HK, et al. The tumour suppressor RASSF1A regulates mitosis by inhibiting the APC-Cdc20 complex. Nat Cell Biol. 2004;6:129–37. doi: 10.1038/ncb1091. [DOI] [PubMed] [Google Scholar]
- 104.Agathanggelou A, Cooper WN, Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497–3508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- 105.Hoon DS, Spugnardi M, Kuo C, Huang SK, Morton DL, Taback B. Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene. 2004;23:4014–4022. doi: 10.1038/sj.onc.1207505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ng KC, Campos EI, Martinka M, Li G. XAF1 expression is significantly reduced in human melanoma. J Inveset Dermatol. 2004;123:1127–1134. doi: 10.1111/j.0022-202X.2004.23467.x. [DOI] [PubMed] [Google Scholar]
- 107.Gollob JA, Sciambi CJ, Peterson BL, Richmond T, thoreson M, Moran K, Dressman HK, Jelinek J, Issa JP. Phase I trial of sequential low-dose 5-aza-2′-deoxycytidine plus high-dose intravenous bolus interleukin-2 in patients with melanoma or renal cell carcinoma. Clin Cancer Res. 2006;12:4619–4627. doi: 10.1158/1078-0432.CCR-06-0883. [DOI] [PubMed] [Google Scholar]
- 108.Aparicio A, Eads CA, Leong LA, Laird PW, Newman EM, Synold TW, Baker SD, Zhao M, Weber JS. Phase I trial of continuous infusion 5-aza-2′deoxycytidine. Cancer Chemother Pharmacol. 2003;51:231–239. doi: 10.1007/s00280-002-0563-y. [DOI] [PubMed] [Google Scholar]
- 109.Hoshino K, Kaisho T, Iwabe T, Takeuchi O, Akira S. Differntial involvement of IFN-beta in Toll-like receptor-stimulated dendritic cell activation. Int Immunol. 2002;14:1225–1231. doi: 10.1093/intimm/dxf089. [DOI] [PubMed] [Google Scholar]
- 110.Renneson J, Salio M, Mazouz N, Goldman M, Marchant A, Cerundolo V. Mature dendritic cells differentiated in the presence of interferon-beta and interleukin-3 prime functional antigen-specific CD8 T cells. Clin Exp Immunol. 2005;139:468–475. doi: 10.1111/j.1365-2249.2005.02700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Breckpot K, Corthals J, Bonehill A, Michiels A, Tuyaerts S, Aerts C, Heirman C, Thielemans K. Dendritic cells differentiated in the presence of IFN- {beta} and IL-3 are potent inducers of an antigen-specific CD8 + T cell response. J Leukoc Bio. 2005;78:898–908. doi: 10.1189/jlb.0105052. [DOI] [PubMed] [Google Scholar]
- 112.Severa M, Remoli ME, Giacomini E, Ragimbeau J, Lande R, Uze G, Pellegrini S, Coccia EM. Differential responsiveness to IFN-alpha and IFN-beta of human mature DC through modulation of IFNAR expression. J Leukoc Bio. 2006;79:1286–1294. doi: 10.1189/jlb.1205742. [DOI] [PubMed] [Google Scholar]
- 113.Shirota H, Ishii KJ, Takakuwa H, Klinman DM. Contribution of interferon-beta to the immune activation induced by double-stranded DNA. Immunology. 2006;118:302–310. doi: 10.1111/j.1365-2567.2006.02367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dunn GP, Bruce AT, Sheehan KC, Shankaran V, Uppaluri R, Bui JD, Diamond MS, Koebel CM, Arthur C, White JM, Schreiber RD. A critical function for type I interferons in cancer immunoediting. Natl Immunol. 2005;6:722–729. doi: 10.1038/ni1213. [DOI] [PubMed] [Google Scholar]
- 115.CpG island methylation and expression of the secreted frizzled-related protein gene family in chronic lymphocytic leukemia. Liu TH, Raval A, Chen SS, Matkovic JJ, Byrd JC, Plass C. Cancer Res. 2006;66:653–658. doi: 10.1158/0008-5472.CAN-05-3712. [DOI] [PubMed] [Google Scholar]
- 116.Ai L, Tao Q, Zhong S, Fields CR, Kim WJ, Lee MW, Cui Y, Brown KD, Robertson KD. Inactivation of Wnt inhibitory factor-w (WIF1 expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis. 2006;27:1341–1348. doi: 10.1093/carcin/bgi379. [DOI] [PubMed] [Google Scholar]
- 117.Yan L, Nass SJ, Smith D, Nelson WG, Herman JG, Davidson NE. Specific inhibition of DNMT1 by antisense oligonucleotides induces res-expression of estrogen receptor-alpha (ER) in ER-negative human breat cancer cell lines. Cancer Biol Ther. 2003;2:552–556. doi: 10.4161/cbt.2.5.469. [DOI] [PubMed] [Google Scholar]
- 118.Mori T, Martinex SR, O’Day SJ, Morton DL, Umetani N, Kitago M, Tanemura A, Nguyen SL, Tran AN, Wang HJ, Hoon DS. Estrogen receptor-alpha methylation predicts melanoma progression. Cancer Res. 2006;66:6692–6698. doi: 10.1158/0008-5472.CAN-06-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]