Abstract
Clostridium perfringens type A, is both a ubiquitous environmental bacterium and a major cause of human gastrointestinal disease, which usually involves strains producing C. perfringens enterotoxin (CPE). The gene (cpe) encoding this toxin can be carried on the chromosome or a large plasmid. Interestingly, strains carrying cpe on the chromosome and strains carrying cpe on a plasmid often exhibit different biological characteristics, such as resistance properties against heat. In this study, we investigated the genetic properties of C. perfringens by PCR-surveying 21 housekeeping genes and genes on representative plasmids and then confirmed those results by Southern blot assay (SB) of five genes. Furthermore, sequencing analysis of eight housekeeping genes and multilocus sequence typing (MLST) analysis were also performed. Fifty-eight C. perfringens strains were examined, including isolates from: food poisoning cases, human gastrointestinal disease cases, foods in Japan or the USA, or feces of healthy humans. In the PCR survey, eight of eleven housekeeping genes amplified positive reactions in all strains tested. However, by PCR survey and SB assay, one representative virulence gene, pfoA, was not detected in any strains carrying cpe on the chromosome. Genes involved in conjugative transfer of the cpe plasmid were also absent from almost all chromosomal cpe strains. MLST showed that, regardless of their geographic origin, date of isolation, or isolation source, chromosomal cpe isolates, i) assemble into one definitive cluster ii) lack pfoA and iii) lack a plasmid related to the cpe plasmid. Similarly, independent of their origin, strains carrying a cpe plasmid also appear to be related, but are more variable than chromosomal cpe strains, possibly because of the instability of cpe-borne plasmid(s) and/or the conjugative transfer of cpe-plasmid(s) into unrelated C. perfringens strains.
Introduction
Clostridium perfringens, an anaerobic Gram-positive bacterium, is ubiquitous in the intestinal flora of human and animals, and is also commonly isolated from environmental materials such as soil and water [1], [2]. Moreover, C. perfringens is an extremely important pathogen of human and domestic animals. In a commonly used classification scheme, C. perfringens is divided into five toxinotypes (A to E) based on the production of four toxins (alpha, beta, epsilon, and iota); however, this bacterium also produces ten other toxins such as C. perfringens enterotoxin (CPE), beta2 toxin, and theta toxin (also known at perfringolysin O or PFO) [2].
Of these many toxins, CPE is extremely important for human gastrointestinal diseases such as food poisoning and antibiotic-associated diarrhea [2]. Interestingly, the gene encoding this toxin (cpe) is highly conserved, although the cpe gene can be present on the chromosome or on a large plasmid(s) in type A strains [3]. Plasmid cpe-carrying strains can cause C. perfringens food poisoning outbreaks, but chromosomal cpe isolates are responsible for the majority of foodborne illnesses [3]–[5]. In contrast, nearly all CPE-associated human AAD is caused by plasmid cpe isolates. In addition, there are several reports of biological differences, such as heat resistance and other traits [6]–[8], between chromosomal cpe isolates and plasmid cpe isolates that may suggest these two types of strains possess different genetic backgrounds.
To investigate the genetic background of a bacterium, total genome sequence analysis has been performed in many pathogens. To date, the complete genome sequence of three pathogenic type A strains of C. perfringens (ATCC13124, SM101, and strain 13) has been published [9], [10] and genome sequencing of several animal toxigenic strains (type B, C, and E) is in progress. Thus far, all sequenced C. perfringens isolates share a similar chromosomal genetic organization, although this genome sequencing has tested only a limited number of strains. Multilocus sequence typing (MLST) represents an alternative approach capable of investigating the genetic characterization of many strains of a given species. A MLST approach has been applied to many bacteria, but not yet to compare the genetic relatedness of large numbers of chromosomal cpe isolates vs. plasmid cpe isolates from varied sources [11]–[13].
This study sought to characterize the genetic background of enterotoxigenic C. perfringens strains, firstly using a conventional PCR survey for representative virulence genes on the chromosome or for genes borne on toxin plasmid(s), as well as for several housekeeping genes. After confirming selected PCR results by Southern blot, a MLST analysis based on those PCR/Southern blot results was then applied to characterize the genetic backgrounds of chromosomal cpe isolates vs. plasmid cpe isolates.
Results
PCR survey for representative C. perfringens chromosomal or plasmid-borne genes
To assess genetic diversity amongst enterotoxigenic C. perfringens isolates, a PCR survey was first performed to evaluate the carriage of selected genes including chromosomal toxin genes (plc and pfoA), several chromosomal housekeeping genes, plasmid maintenance genes, and genes related to plasmid transfer (Table 1).
Table 1. The results of PCR survey of the representative genes.
| strain | genotype | genes | ||||||||||||||||||||||
| cpe | cpb2 | gyrB | sigK | sodA | groEL | pgk | nadA | plc | colA | lonB | eno | virS | pfoA | tcpH | tcpF | rep | cna | soj | parB | topA | bcn | |||
| Strain 13 | A | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | + | + | + | + | − | |
| ATCC13124 | A | type strain | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − |
| ATCC3624 | A | Gas gangrene | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − |
| KZ210 | A | BP6K derivative | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + |
| NCTC8239 | A | food poisoning | + | − | + | + | + | + | + | + | + | + | + | − | + | − | − | − | − | − | + | + | + | − |
| NCTC8798 | A | food poisoning | + | − | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − | + |
| OSAKA1 | A | food poisoning | + | − | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | − |
| OSAKA2 | A | food poisoning | + | − | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | − |
| OSAKA3 | A | food poisoning | + | − | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | − |
| OSAKA4 | A | food poisoning | + | − | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | − |
| W4232 | A | food poisoning | + | − | + | + | + | + | + | + | + | + | − | − | + | − | − | − | − | − | + | + | + | − |
| W5837 | A | food poisoning | + | − | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + |
| W6205 | A | food poisoning | + | − | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | + | + | + | − |
| P-1/09/03 | A | food | + | − | + | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + | + | + | − |
| T-1/08/03 | A | food | + | − | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | + | + | + | + | − |
| F4013 | A | sporadic diarrhea | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| F4969 | A | sporadic diarrhea | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − |
| F5603 | A | sporadic diarrhea | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + |
| No.2 | A | food poisoning | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| No.24 | A | food poisoning | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| No.110 | A | food poisoning | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| T1 | A | food poisoning | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | − |
| T16 | A | food poisoning | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | − |
| T102 | A | food poisoning | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | − |
| BL-D1 | A | sepsis | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| DR-T1 | A | diarrhea | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | + | + | + | − |
| BI-D2 | A | cholecystitis | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | + | + | + | − |
| MR1-1 | A | healthy | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | + | + | + | − |
| MR1-2 | A | healthy | − | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | − | − | − | − |
| MR2-2 | A | healthy | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − |
| MR2-3 | A | healthy | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − |
| MR2-4 | A | healthy | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | − |
| MR2-5 | A | healthy | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | + | + | + | + |
| MR2-9 | A | healthy | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| MR2-12 | A | healthy | − | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | − | − | − | − |
| MR2-14 | A | healthy | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − |
| MR2-19 | A | healthy | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − |
| MR2-20 | A | healthy | − | + | + | + | + | + | + | + | + | + | − | + | − | + | + | + | + | + | − | − | − | − |
| JCM1290 | A | ATCC13124 | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − |
| JCM3819 | A | ATCC3629 | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − |
| JCM3816 | A | ATCC3624 | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − |
| NCTC8533 | B | animal disease (lamb) | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| NCTC8081 | C | necrotizing enteritis | + | − | + | + | + | + | + | + | + | + | + | − | − | − | − | + | + | + | + | + | + | − |
| NCTC3182 | C | animal disease (sheep) | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − |
| NCTC8346 | D | animal disease (sheep) | − | − | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | − | − | − | + |
| NCTC8084 | E | animal disease (calf) | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + |
| M-01 | A | food | − | − | + | + | + | + | + | + | + | + | + | − | + | − | − | − | + | + | − | − | − | + |
| M-02 | A | food | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − |
| M-03 | A | food | − | − | + | + | + | + | + | + | + | + | − | − | − | + | − | − | − | − | − | − | − | + |
| M-04 | A | food | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + |
| M-06 | A | food | − | − | + | + | + | + | + | + | + | + | + | − | + | + | − | + | + | + | − | − | − | − |
| M-07 | A | food | − | − | + | + | + | + | + | + | + | + | + | − | + | − | − | − | + | + | − | − | − | + |
| M-08 | A | food | − | − | + | + | + | + | + | + | + | + | + | − | + | − | − | − | + | + | − | − | − | + |
| TM111-C1 | A | food | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| TM111-C6 | A | food | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| TM138C1A | A | food | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| TM178C5 | A | food | + | − | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | − |
The carriage of twelve known (from genome sequencing) C. perfringens chromosomal genes was first evaluated in this PCR survey. For eight representative housekeeping genes, PCR amplified a product of the expected size from all investigated C. perfringens strains. However, PCR for the lonB ORF gave negative reactions in a few investigated strains, indicating either that these strains lack lonB or they have minor nucleotide diversity on the primer site(s). Similarly, PCR reactions for three chromosomal genes (eno, virS, and pfoA) failed to amplify PCR products from chromosomal cpe strains that originated in Europe, Japan, or the USA, as well as from some plasmid cpe-positive strains and cpe-negative strains. These results indicate that eno, virS or pfoA genes are either missing from these strains or there is nucleotide diversity at the primer binding site(s).
PCR assays were then performed to detect the carriage of representative genes present on four completely sequenced C. perfringens plasmids, including two cpe-encoding conjugative plasmids (pCPF4969 and pCPF5603) [14], a beta2 toxin gene-encoding plasmid (pCP13) [9], and a UV inducible bacteriocin gene-encoding plasmid (pIP404) [15] (Table 1). PCR amplified products of the expected size for three cpe- plasmid genes (tcpF, tcpH, and rep) from all surveyed plasmid-cpe positive strains, including sporadic diarrhea strains, food poisoning strains, and food strains, as well as from several isolates originating from feces of healthy humans, and several food isolates. However, PCR assays for these genes did not produce a positive reaction from any surveyed chromosomal cpe strain, except for food strain P-1/09/03. In contrast, a PCR survey for three genes present on pCP13 (soj, parB, and topA) amplified a positive reaction for ten of eleven chromosomal cpe strains and for eleven of twelve plasmid cpe strains. In PCR assays for genes present on both cpe plasmids and pCP13, a cpb2 product was not amplified from any chromosomal cpe strains, but a cna product was obtained for two chromosomal cpe food strains. A PCR assay for the bacteriocin gene (bcn) present on pIP404, was positive for 16 of 58 investigated C. perfringens strains (including both cpe-positive and cpe-negative strains).
Southern blot assay for carriage of chromosomal and plasmid-borne genes
To definitively establish the presence or absence of eno, virS, pfoA tcpH, cna, and soj ORFs Southern blot assays were performed (Table 2) using ten chromosomal cpe strains, twelve plasmid cpe strains, two cpe-negative strains, and also a Clostridium tertium food isolate (as a negative control).
Table 2. The results of Southern blot assay with seven genes.
| strain | cpe | eno | virS | pfoA | tcpH | cna | soj | ||
| NCTC8239 | A | food poisoning | c | + | + | − | − | − | + |
| NCTC8798 | A | food poisoning | c | + | + | − | − | − | − |
| OSA1 | A | food poisoning | c | + | + | − | − | − | + |
| OSAKA2 | A | food poisoning | c | + | + | − | − | − | + |
| OSAKA4 | A | food poisoning | c | + | + | − | − | − | + |
| W4232 | A | food poisoning | c | + | + | − | − | − | + |
| W5837 | A | food poisoning | c | + | + | − | − | − | + |
| W6205 | A | food poisoning | c | + | + | − | − | − | + |
| P-1/09/03 | A | food isolate | c | + | + | − | − | − | + |
| T-1/08/03 | A | food isolate | c | + | + | − | − | − | + |
| F4969 | A | sporadic diarrhea | p | + | + | + | + | + | − |
| F5603 | A | sporadic diarrhea | p | + | + | + | + | + | + |
| No.2 | A | food poisoning | p | + | + | + | + | + | + |
| No.24 | A | food poisoning | p | + | + | + | + | + | + |
| No.110 | A | food poisoning | p | + | + | + | + | + | + |
| T1 | A | food poisoning | p | + | + | − | + | + | + |
| T16 | A | food poisoning | p | + | + | − | + | + | + |
| T102 | A | food poisoning | p | + | + | − | + | + | + |
| TM111-C1 | A | food isolate | p | + | + | + | + | + | + |
| TM138 | A | food isolate | p | + | + | + | + | + | + |
| TM178 | A | food isolate | p | + | + | − | + | + | + |
| MR2-4 | A | human feces isolate | p | + | + | − | NT | NT | NT |
| MR2-12 | A | human feces isolate | − | + | + | + | + | + | − |
| M-08 | A | food isolate | − | + | + | − | − | + | − |
| Clostridium tertium | food isolate | − | + | − | − | − | − | − |
NT: not tested
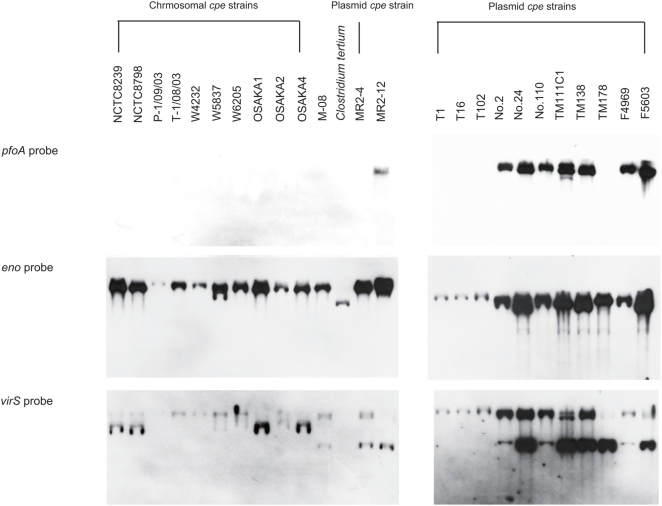
For two tested housekeeping genes (eno and virS), Southern blot assays showed a positive reaction with all tested C. perfringens strains (Fig. 1). Interestingly, the positive results obtained with both PCR and Southern blot assays suggest the eno gene of C. tertium was homologous with the C. perfringens eno gene.
Figure 1. Southern blot assay of chromosomal- or plasmid-cpe strains.
DNA digested with PstI from cpe-positive and cpe-negative (M-08 and MR2-12) type A strains was subjected to 1% agarose electrophoresis prior to Southern blotting and hybridization with a DIG-labeled, pfoA-, virS-, or eno-specific probe.
However, Southern blot assay for pfoA showed a negative result for all ten chromosomal cpe strains and for four of eleven plasmid cpe strains (Fig. 1), all of which also showed negative results for pfoA in our PCR assay.
PCR assay for the pfoA region in pfoA-negative, cpe-positive strains
To investigate whether any portion of the pfoA region is still present in cpe-positive, pfoA-negative strains, we first performed a bioinformatics comparison between the recently completed sequences of C. perfringens strains SM101 (pfoA-negative) and ATCC13124 (type strain, pfoA-positive) [10]. This analysis indicated that the upstream pfoR gene and a gene downstream of pfoA that encodes a conserved hypothetical protein are present on the chromosome of both strains (Fig. 2A).
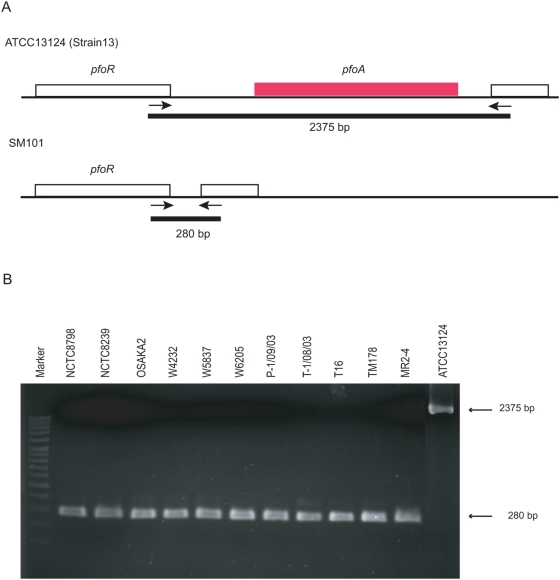
Figure 2. PCR assay for the pfoA region in pfoA-negative, cpe-positive strains.
(A) Schematic representation of the pfoA region in C. perfringens. Genetic organization of the pfoA region is shown for pfoA-positive, cpe-negative ATCC13124 and pfoA-negative, cpe-positive SM101 strains (accession number: CP000312, and NC008261). The arrows depict the position of primers for the pfoA genotyping assay. The black bars show the predictive PCR products. (B) PCR results of cpe-positive, pfoA-negative strains investigated by the pfoA PCR genotyping assay. An ∼2,375 bp PCR product was obtained by pfoA-positive, cpe-negative ATCC13124 strain. An ∼280 bp PCR products was obtained from pfoA-negative, cpe-positive strains.
To further investigate the pfoA region amongst pfoA-negative, cpe-positive strains, primers to pfoR or the downstream gene were constructed for use in a pfoA-region genotype PCR assay. In this assay, when pfoA is simply deleted from the chromosome of a strain, the PCR product should be 280 bp. With this pfoA-genotype PCR assay, a 280 bp product was detected for eleven of twelve pfoA-negative, cpe-positive strains (Fig. 2B). These results indicate that portions of the pfoA region are conserved even among pfoA-negative type A enterotoxigenic C. perfringens strains, including both chromosomal cpe strains and plasmid cpe strains. Interestingly, type C human necrotizing enteritis cpe-positive strain, NCTC8081, also did not produce any specific pfoA region PCR products (data not shown).
Multi-locus sequence typing (MLST) analysis of C. perfringens
The results from our PCR survey and Southern blot assay suggested that chromosomal cpe strains might share a common genetic background. To further compare the genetic backgrounds of chromosomal cpe strains versus plasmid cpe strains collected from various geographical origins (Japan, Europe and the US) or sources (food, food poisoning, and nonfdoodborne diarrhea patients), MLST analysis was performed with eight representative housekeeping genes.
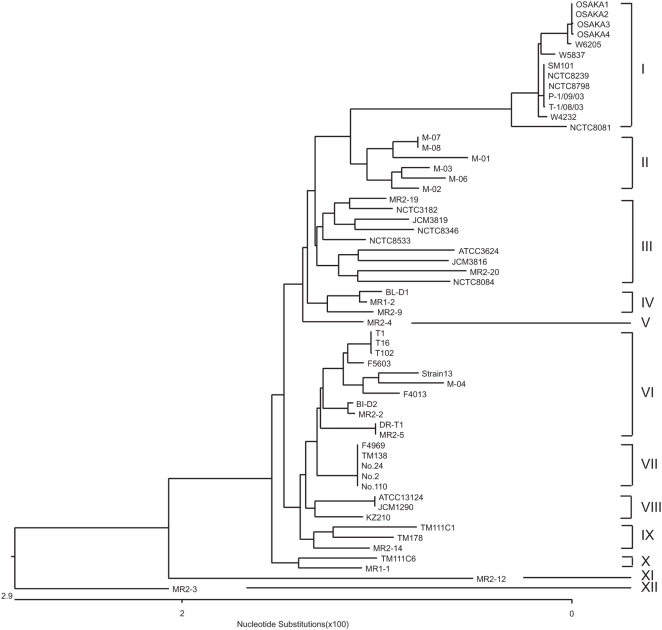
Phylogenetic analysis by the Clustal W program was then performed on our MLST results for fifty-eight strains, including eleven chromosomal cpe strains and thirteen plasmid cpe strains. This analysis identified twelve main groups, designated Cluster I to XII, (Fig. 3). Several strains showed completely conserved sequences for all eight genes surveyed in this MLST. Of these strains, OSAKA1 to 4 were each isolated from the same outbreak and had been reported to show the same PFGE pattern [16]. T1, T16, and T102 strains were also isolated from a single food poisoning outbreak [17], as were No.2, No.24, and No.110 strains [18]. These MLST results suggest that strains isolated in three different food poisoning outbreaks share the same genetic backgrounds. JCM1290 is a derivative of ATCC13124 (a cpe-negative type strain), while SM101 is a derivative of European food poisoning isolate NCTC8798. M-07 and M-08 strains were isolated from the same meat sample so these pairs of strains might also share the same genetic backgrounds. However, thirty-seven strains gave unique individual MLST patterns in this study. Collectively, these results confirm that MLST is useful to investigate the genetic relationship between C. perfringens strains.
Figure 3. Phylogenetic relationships among 58 cpe-positive or cpe-negative C. perfringens strains.
The phylogenetic tree was constructed by Clustal W analysis based on the concatenated nucleotide sequence of eight housekeeping genes. The phylogenetic clusters are given on the right.
Nine chromosomal cpe strains were assigned to Cluster I (Fig. 3). These strains included two European food poisoning strains (NCTC8239 and NCTC8798, both isolated in the 1950s) and several Japanese food poisoning strains, including: OSAKA1, OSAKA2,OSAKA3,and OSAKA4 (each isolated in the Osaka area in 1997), W4232 (isolated from the Kanto area in 1995), and W5837 and W6205 (isolated from the Kanto area in 2000). Cluster I also included two American chromosomal cpe isolates (P-1/09/03,T-1/08/03) obtained from retail foods not associated with a food poisoning outbreak. These genetic findings showing relatedness between chromosomal cpe nonoutbreak food strains and chromosomal cpe food poisoning strains were consistent with previous findings indicating that food strains carrying cpe on the chromosome form similarly heat resistant spores as do food poisoning strains [19]. The conserved nucleotide diversity of six genes was also evident at the translational level, where there were one (pgk, sod, gyrB), two (nadA), or four (plc, sigK) amino acid substitutions. Overall, our MLST results strongly suggest that, regardless of their geographical origins, date of isolation, or origination from nonoutbreak foods or food poisoning outbreaks, type A chromosomal cpe strains in Japan, Europe, and USA share a common genetic background and belong to a cluster distinct from most other C. perfringens isolates. The one exception was that Cluster I also includes the type C cpe-positive NCTC8081 strain that was isolated from human necrotizing enteritis in Europe.
In contrast to the chromosomal cpe isolates localizing to Cluster I, three isolates from a 2001 food poisoning outbreak in Toyama area (T1,T16,and T102), each carrying a plasmid with an IS1151 sequence downstream of the cpe gene [4], were assigned to Cluster VI (Fig. 3). This Cluster VI also included other plasmid cpe strains, with a downstream IS1151 sequence, which had been obtained from human sporadic diarrhea cases occurring in Europe (F5603, and F4013) [20]. These findings could suggest that plasmid cpe isolates with downstream IS1151 sequence share a common genetic relationship. However, this Cluster VI also included two cpe-negative strains.
Three food poisoning isolates from a 1980 outbreak occurring in the Toyama area of Japan (No2,No24,and No110), each carrying a plasmid with an IS1470-like sequence downstream of the cpe gene, belonged to Cluster VII, a neighboring cluster to Cluster VI (Fig. 3). This Cluster VII also included two other isolates with an IS1470-like cpe plasmid (F4969 and TM138). These two other cpe plasmid strains have the exact same sequence for the eight investigated housekeeping genes as found in the three Toyama food poisoning strains belonging to this cluster. This finding strongly suggested that strains in Cluster VII also share a similar genetic background.
Another three plasmid cpe strains (including two nonoutbreak food isolates [TM111C1, and TM178] and one plasmid cpe human feces isolate, MR2-4) belonged to different clusters. This genetic variability may have resulted from the conjugative nature of cpe- plasmids, with this plasmid transferring into unrelated C. perfringens strains.
Out of eleven nonoutbreak food isolates from Wakayama city in Japan that were investigated in this study (Table 1), eight isolates did not carry cpe. Six of those eight isolates (M-01,M-02,M-03,M-06,M-07,and M-08) belonged to Cluster II, one isolate (M-04) aligned with Cluster VI, and the final isolate (TM111C6)belonged to Cluster X. While six of these food isolates were assigned to Cluster II, no plasmid cpe strains from food belonged to Cluster II or VI. This finding further suggested that plasmid cpe-positive strains often share a similar genetic background.
Veterinary toxigenic type B to E strains (type B: NCTC8533, type C: NCTC3182,type D: NCTC8346,and type E: NCTC8084) isolated from European animals suffering from C. perfringens diseases were assigned to Cluster III (Fig. 3). Cluster III also included MR2-19, obtained from the feces of a healthy human, but not any meat isolates. This result might suggest that C. perfringens strains acting as animal pathogens have a relatively specific genetic background, but further study of additional type B-E strains is warranted.
While many strains from each source belonged to the same cluster, eleven fecal strains isolated in 2000 from healthy people in Japan belonged to eight different clusters (Fig. 3). Two isolates (MR2-19,MR2-20) belonged to Cluster III, two isolates (MR1-2,MR 2-9) aligned with Cluster IV, MR2-4 with a cpe plasmid carrying a downstream IS1470-like sequence was assigned to Cluster V, two isolates (MR2-2,MR 2-5) belonged to Cluster VI, MR2-14 aligned with Cluster IX, MR1-1 was assigned to Cluster X, MR2-12 belonged to Cluster XI, MR2-3 to Cluster XII. These findings suggested that healthy humans carry C. perfringens strains with various genetic backgrounds.
Discussion
Clostridium perfringens strains producing enterotoxin (CPE) are the causative agent for several human GI diseases, including food poisoning, antibiotic-associated diarrhea, and sporadic diarrhea [2]. The gene encoding CPE (cpe) is found in a small population of type A C. perfringens (approximately 1 to 5%) [2], where it can reside on the chromosome or large transferable plasmids [3], [20]. Strains carrying cpe on the chromosome usually possess higher resistance properties against heat, cold, and nitrates than strains carrying cpe on a plasmid [6]–[8]. In addition, the chromosomal cpe strains typically grow faster at optimal temperature, and have a broader growth temperature range, compared to plasmid cpe strains or other C. perfringens isolates [8]. These complex differences in biological properties, which are likely relevant for foodborne disease, may reflect broad genetic variations between chromosomal cpe isolates and other C. perfringens isolates.
Therefore, the current study investigated the genetic background of type A enterotoxigenic C. perfringens, by surveying fifty-eight cpe-positive and cpe-negative strains from various sources. Previous genetic studies have identified four groups of type A enterotoxigenic C. perfringens: i) food and food poisoning isolates that carry cpe on the chromosome [2], [19], ii) isolates that carry a plasmid-borne cpe gene with a downstream IS1470-like sequence [2], [20], iii) isolates that carry a plasmid-borne cpe gene with a downstream IS1151 sequence [2], [20] and iv) isolates that carry a cpe gene but produced no PCR product with a cpe-genotyping PCR assay [21].
The results of our PCR survey and Southern blot assay for toxin genes (plc, colA, pfoA), remarkably found the θ toxin gene (pfoA) is missing from all ten surveyed chromosomal cpe strains and from five of twelve plasmid cpe strains. A study conducted back in the 1960's [22] had identified some PFO-negative, heat-resistant enterotoxigenic strains of C. perfringens, but localization of the cpe gene to the chromosome or plasmids was not possible at that time. Our finding that most chromosomal cpe isolates are pfoA-negative, significantly extends the genome sequencing observation that chromosomal cpe isolate SM101 is pfoA-negative [10].
However, our PCR analyses also indicated that the region that normally flanks pfoA is still present in cpe-positive, pfoA-negative strains, except for the type C NCTC8081 strain (Fig 1A, 1B).These findings suggest that most pfoA-negative chromosomal cpe, and some plasmid cpe isolates appear to have undergone a specific deletion of their pfoA gene.
Concerning housekeeping genes, our initial PCR survey for eno, virS, and lonB ORFs did not amplify products from many chromosomal cpe strains, but did for most other strains, including plasmid cpe strains. The presence of eno and virS genes in chromosomal cpe strains was confirmed by Southern blot assay (Table 2, Fig. 1). PCR surveys for genes present on the representative plasmids pCPF4969, pCP13, or pIP404, suggested that almost all chromosomal cpe strains carry a pCP13 and/or pIP404 related plasmid(s), but not a plasmid related to cpe plasmids such as pCPF4969.
In epidemiological studies of C. perfringens food poisoning, the relationships among C. perfringens isolates have primarily been investigated by pulsed-field gel electrophoresis (PFGE) [16], [23]. Although useful, PFGE lacks the precision of MLST, in which gene fragments are amplified and sequenced from several loci spread across the whole genome [11]–[13]. Three previous studies of C. perfringens strains using MLST analysis have been reported [24]–[26]. Two of those studies focused on C. perfringens animal disease isolates (type B to E) [25], [26]. The other MLST report examined a broad range of C. perfringens, isolates, including a limited number of cpe-positive food poisoning strains and suggested that food poisoning isolates form a distinct cluster of C. perfringens isolates.
However, that previous study examined only a few food poisoning isolates, mostly American and all carrying a chromosomal cpe gene, as well as only three plasmid cpe isolates, all nonfoodborne GI disease isolates from Europe. Since that earlier study, it has become clear that some food poisoning isolates carry their cpe gene on large transferable plasmids rather than on the chromosome and that chromosomal cpe isolates can be recovered from some retail meats. Therefore, MLST analysis using housekeeping genes present on the chromosome and cpe plasmid was employed in the current study to explore i) the relatedness among a larger collection of cpe-positive isolates, including many from Japan and ii) the similarity of these isolates to cpe-negative isolates. The cpe location, ability to produce CPE and spore heat resistance of all cpe-positive strains included in this study had been previously determined [6], [19], [27]. Our MLST scheme with eight housekeeping genes involves 5,274 bp (0.17% of the genome) of analyzed sequence versus the 3,918 bp of sequence included in the previous MLST applied to some cpe-positive C. perfringens [24]. Moreover, our MLST includes several genes likely contributing to survival (sod, and groEL) and propagation (plc, colA, pgk, nadA) in foods, and genes related to spore formation (sigK), which can also contribute to survival in foods.
From our MLST analysis, the most remarkable finding was that, regardless of their source their geographic origin, or date of isolation, all surveyed chromosomal cpe strains share a common genetic background and belong to the distinct Cluster I. In particular, these surveyed chromosomal cpe strains all possess three common features, 1) absence of the pfoA gene, but retention of neighboring sequences in the pfoA locus, 2) lack of plasmid-borne major-toxin genes (including cpe) [28]–[30], although they sometimes carried a plasmid encoding the putative toxin CPB2 [9], and 3) the presence in many housekeeping genes of conserved nucleotide differences, often resulting in amino acid substitutions, compared to the homolog genes present in other plasmid cpe-positive and/or cpe-negative strains.
Interestingly, cluster I also included type C strain NCTC8081, which carries cpe and was isolated from a patient suffering from necrotizing enteritis (Pigbel) in Europe. Human necrotizing enteritis is a rare disease and not fully understood with respect to its pathogenesis, although the plasmid-encoded β-toxin clearly plays a major role in the enteric virulence of type C isolates [31]. For research, relatively few cpe-positive type C strains are available from strain collections in Japan, USA, and Europe. Further investigation, if possible, using more cpe-positive type C strains should evaluate the genetic relationship between type A chromosomal cpe strains and type C cpe-positive strains.
Cluster VI included five strains carrying a plasmid with IS1151 sequences located downstream of the cpe gene. These isolates included three food poisoning strains from an outbreak in Japan in 2000 [4], and two strains isolated from the patients with sporadic diarrhea in Europe in 1900s [20]. Of these five strains, four strains showed a very close relationship upon MLST analysis, suggesting these IS1151 cpe-genotype strains share a similar genetic background that facilitates their ability to cause human GI disease.
Of eight surveyed strains carrying a plasmid with IS1470-like sequences downstream of the cpe gene, five strains belonged to Cluster VII. However, the other three strains belonged to Cluster V or Cluster IX. These results suggested that while IS1470-like cpe-genotype strains often share a similar genetic background, they are more variable than IS1151-genotype plasmid strains.
Collectively, our results suggest for plasmid cpe isolates that, 1) food poisoning outbreaks involving plasmid cpe strains often involve clonal expansion rather than plasmid transfer, 2) IS1151 cpe genotype strains are closely related, but can also share a genetic relationship with some cpe negative isolates which may have lost the cpe plasmid or which represent potential future hosts for the IS1151 cpe plasmid, 3) IS1470-like cpe genotype strains also share genetic relationships but are more variable than the IS1151 cpe genotype strains. In addition, the current results conclusively demonstrate that chromosomal cpe isolates, whether originating from food poisoning or nonoutbreak-associated foods, do not share a close genetic linkage with plasmid cpe food poisoning strains (or other plasmid cpe strains).
Strains isolated from feces of healthy humans were found to distribute into many clusters, i.e. these strains have varied genetic backgrounds. This variability of fecal strains might be attributable, in part, to dietary differences or personal factors such as age or economic status. The investigated type B to E livestock origin strains all formed one cluster, which was distinguished from cpe-positive human strains (even from a type C human strain), and also from cpe-positive and cpe-negative food strains. Further MLST analyses of non-type A strains is warranted to confirm these conclusions.
Finally, the common and distinct genetic background of chromosomal cpe isolates provides one explanation for previous phenotypic studies [6], [7] that revealed substantial differences between the vegetative cells and spores of chromosomal cpe isolates versus other C. perfringens isolates. A shared genetic background by most or all chromosomal cpe isolates is also consistent with our previous studies identifying a variant small acid soluble protein, that is made by most chromosomal cpe isolates [32]. Regarding the evolution of chromosomal cpe isolates, it has been proposed [33] that these bacteria arose from integration of a composite transposon named Tn5565 onto the C. perfringens chromosome. If so, our MLST findings could suggest this chromosomal transposon integration occurred only a limited number of times (perhaps only once) in a C. perfringens type A isolate(s) possessing a genetic background favorable for growth and survival in the food environment. Acquiring the ability to produce a potent enterotoxin, upon Tn5565 integration, thus created a formidable food poisoning agent. Since C. perfringens isolates multiply to very high levels in the GI tract during food poisoning, t is possible that acquiring the ability to produce CPE is advantageous by facilitating the dissemination of these bacteria so the infectious cycle can be repeated in other hosts.
Materials and Methods
Bacterial strains
Fifty-eight strains of C. perfringens were included in this study, including type A cpe-positive strains, type A cpe-negative strains and type B to E strains. A breakdown of C. perfringens strains with their various origins is shown in Table 3. Briefly, the investigated strains included the type strain, several reference strains and type A chromosomal cpe-positive strains from food poisoning outbreaks occurring in Japan [16] or Europe [20], and isolated from foods in USA [19]. In addition, the surveyed type A plasmid cpe-positive strains were isolated from food poisoning outbreaks in Japan [4], [17], [18], from foods in Japan [27], from patients with sporadic diarrhea in Europe [20], or from feces of healthy humans in Japan [34]. Type B to E reference strains (one human necrotizing enteritis strain and four animal strains) were provided by NCTC.
Table 3. Clostridium perfringens strains used in this study.
| type | location of cpe | source | Date and region | Reference | |
| Strain 13 | A | cpe negative | Gas gangrene | 9) | |
| ATCC13124 | A | cpe negative | type strain | 10) | |
| JCM1290 | A | cpe negative | ATCC13124 derivative | this study | |
| ATCC3624 | A | cpe negative | Gas gangrene | ||
| JCM3816 | A | cpe negative | ATCC3624 derivative | this study | |
| JCM3819 | A | cpe negative | ATCC3629 derivative | this study | |
| KZ210 | A | cpe negative | BP6K derived | 1940s, USA | this study |
| SM101 | A | Chromosome | NCTC8798 derivative | 10) | |
| NCTC8239 | A | Chromosome | food poisoning | 1950s, Europe | 21) |
| NCTC8798 | A | Chromosome | food poisoning | 1950s, Europe | 21) |
| OSAKA1 | A | Chromosome | food poisoning | 1997, Japan | 21) |
| OSAKA2 | A | Chromosome | food poisoning | 1998, Japan | 21) |
| OSAKA3 | A | Chromosome | food poisoning | 1999, Japan | 21) |
| OSAKA4 | A | Chromosome | food poisoning | 2000, Japan | 21) |
| W4232 | A | Chromosome | food poisoning | 1995, Japan | 21) |
| W5837 | A | Chromosome | food poisoning | 2000, Japan | 21) |
| W6205 | A | Chromosome | food poisoning | 2000, Japan | 21) |
| F5603 | A | Plasmid | sporadic diarrhea | 1990s, Europe | 3) |
| F4013 | A | Plasmid | sporadic diarrhea | 1990s, Europe | 3) |
| F4969 | A | Plasmid | sporadic diarrhea | 1990s, Europe | 3) |
| No.2 | A | Plasmid | food poisoning | 1980s, Japan | 18) |
| No.24 | A | Plasmid | food poisoning | 1980s, Japan | 18) |
| No.110 | A | Plasmid | food poisoning | 1980s, Japan | 18) |
| T1 | A | Plasmid | food poisoning | 2001, Japan | 17) |
| T16 | A | Plasmid | food poisoning | 2001, Japan | 17) |
| T102 | A | Plasmid | food poisoning | 2001, Japan | 17) |
| BL-D1 | A | cpe negative | sepsis | 2001, Japan | this study |
| DR-T1 | A | cpe negative | diarrhea | 2001, Japan | this study |
| BI-D2 | A | cpe negative | cholecystitis | 2001, Japan | this study |
| MR1-1 | A | cpe negative | healthy | 2000, Japan | this study |
| MR1-2 | A | cpe negative | healthy | 2000, Japan | this study |
| MR2-2 | A | cpe negative | healthy | 2000, Japan | this study |
| MR2-3 | A | cpe negative | healthy | 2000, Japan | this study |
| MR2-4 | A | Plasmid | healthy | 2000, Japan | 21) |
| MR2-5 | A | cpe negative | healthy | 2000, Japan | this study |
| MR2-9 | A | cpe negative | healthy | 2000, Japan | this study |
| MR2-12 | A | cpe negative | healthy | 2000, Japan | this study |
| MR2-14 | A | cpe negative | healthy | 2000, Japan | this study |
| MR2-19 | A | cpe negative | healthy | 2000, Japan | this study |
| MR2-20 | A | cpe negative | healthy | 2000, Japan | this study |
| NCTC8533 | B | cpe negative | animal disease (lamb) | 1950s, Europe | |
| NCTC8081 | C | Plasmid | necrotizing enterocolitis | 1940s, Europe | |
| NCTC3182 | C | cpe negative | animal disease (sheep) | 1930s, Europe | |
| NCTC8346 | D | cpe negative | animal disease (sheep) | 1950s, Europe | |
| NCTC8084 | E | cpe negative | animal disease (calf) | 1940s, Europe | |
| M-01 | A | cpe negative | food isolate | 2006, Japan | 28) |
| M-02 | A | cpe negative | food isolate | 2006, Japan | 28) |
| M-03 | A | cpe negative | food isolate | 2006, Japan | 28) |
| M-04 | A | cpe negative | food isolate | 2006, Japan | 28) |
| M-06 | A | cpe negative | food isolate | 2006, Japan | 28) |
| M-07 | A | cpe negative | food isolate | 2006, Japan | 28) |
| M-08 | A | cpe negative | food isolate | 2006, Japan | 28) |
| TM111C1 | A | Plasmid | food isolate | 2006, Japan | 28) |
| TM111C6 | A | cpe negative | food isolate | 2006, Japan | 28) |
| TM138 | A | Plasmid | food isolate | 2006, Japan | 28) |
| TM178 | A | Plasmid | food isolate | 2006, Japan | 28) |
| P-1/09/03 | A | Chromosome | food isolate | 2003, USA | 19) |
| T-1/08/03 | A | Chromosome | food isolate | 2003, USA | 19) |
Bacterial culture and DNA preparation
An aliquot of a cooked meat medium [Difco] stock culture of each C. perfringens strain was inoculated into 5 ml of fluid thioglycolate medium(FTG [Becton Dickinson]) and then incubated overnight at 37°C. An aliquot of that overnight FTG culture was inoculated into 10 ml of TGY broth (3% Trypticase soy [Difco],2% D-glucose [WAKO],1% yeast extract [Difco],0.1% L-cystein) and then incubated overnight at 37°C. DNA for PCR and multilocus sequence typing analysis was prepared from 200 µl of overnight TGY culture with the InstaGene matrix kit [Bio-Rad] according to the manufacture's instructions. To reduce the chance of cross-contamination, DNA templates was prepared with the InstaGene matrix kit, because it requires only two tubes with three steps for preparing DNA templates. DNA materials for Southern blot assays were prepared according to methods described previously [20].
PCR survey for housekeeping genes on the chromosome or on plasmids carrying cpe and/or cpb2
For this PCR survey, eleven housekeeping genes were selected, including: phospholipase C (alpha toxin) gene (plc, a ubiquitous gene of C. perfringens), DNA gyrase B gene (gyrB), one of the sporulation sigma factors (sigK, involved in regulating C. perfringens enterotoxin production [35]), three stress response genes (superoxide dismutase gene (sodA), heat shock protein gene (groEL, lonB), genes encoding enzymes involved in energy production from glucose (phosphofructokinase gene; pgk, enolase gene; eno), a nucleotide metabolism gene (nadA, quinolinate synthetase), a collagenase gene (colA, a possible virulence gene), a regulator gene (virS, a two component regulator gene) and also theta toxin gene (pfoA).
For plasmid-encoded genes, this PCR survey tested for: two toxin genes (the CPE gene (cpe) and the beta2 toxin gene (cpb2)), a putative collagen adhesion protein gene (cna) [9], plasmid transfer genes for cpe-carrying and antibiotics-resistant gene-carrying plasmid (tcpF, tcpH) [36], a replication gene (rep) on transferable plasmids [36], putative plasmid maintenance genes (soj, parB, top) present on pCP13, which is found in the first completely sequenced strain, strain 13 [9], and UV-induced bacteriocin gene (bcn) on pIP404 [15], from which most of C. perfringens shuttle vectors were derived.
PCR primer pairs were principally designed based on genes annotated in the C. perfringens strain 13 genome sequence [9], or genes on cpe-borne plasmids [14], or pIP404 [15] (Table 4). PCR reactions for all genes were performed under the same reaction conditions. Each PCR mixture contained 4 µl of template DNA preparations, 0.5 µl of Taq DNA polymerase [Promega], 2 µl of 2 µM NTP, 4 µl of 25 mM MgCl2, 5 µl of PCR buffer, 2 µl of each primer pair (1 µM final concentration). The reaction mixtures, with a total volume of 50 µl, were placed in a thermal cycler [MiniCycler, MJ] and subjected to the following amplification conditions: 1 cycle at 94°C for 2 min; 35 cycles at 94°C for 30 s, 55°C for 60 s, 68°C for 60 s, and a single extension of 68°C for 8 min. PCR products were then electrophoresed on a 1.5% agarose gel, which were stained with ethidium bromide.
Table 4. Primers used in this study.
| Gene | Primers | Sequence (5′-3′) | Amplicon size (bp) | Analysed size (bp) | Reference |
| House keeping genes | |||||
| gyrB | gyrB-F | ATTGTTGATAACAGTATTGATGAAGC | 905 | 735 | This study |
| gyrB-R | ATTTCCTAATTTAGTTTTAGTTTGCC | This study | |||
| sigK | sigK-F | CAATACTTATTAGAATTAGTTGGTAG | 643 | 589 | This study |
| sigK-R | CTAGATACATATGATCTTGATATACC | This study | |||
| sodA | sod-F | CAAAAAAAGTCCATTAATGTATCCAG | 663 | 554 | This study |
| sod-R | TTATCTATTGTTATAATATTCTTCAC | This study | |||
| groEL | groEL-F | TACAAGATTTATTACCATTACTTGAG | 901 | 685 | This study |
| groEL-R | CATTTCTTTTTCTGGAATATCTGC | This study | |||
| pgk | pgk-F | GACTTTAACGTTCCATTAAAAGATGG | 830 | 681 | This study |
| pgk-R | CTAATCCCATGAATCCTTCAGCGATG | This study | |||
| nadA | nadA-F | ATTAGCACATTATTATCAAATTCCTG | 821 | 689 | This study |
| nadA-R | TTATATGCCTTTAATCTTAAATCCTC | This study | |||
| colA | colA-F | ATTAGAAAGTTTATGTACAATAGGTG | 816 | 670 | This study |
| colA-R2 | AAGACATTCTATTATTTCTATCGTAAGC | This study | |||
| plc | plc-F | AGGAACTCATGCTATGATTGTAACTC | 725 | 671 | This study |
| plc-R | GGATCATTACCCTCTGATACATCGTG | This study | |||
| lonB | lonB-F | ATATATATGAGCAAGTCCTTTGCGAG | This study | ||
| lonB-R | TTTTCTAATCTCTTCAACAGTTAGCC | This study | |||
| eno | eno-F | GCAGTACCTTCAGGAGCTTCAACAGG | This study | ||
| eno-R | CTTCAGCCATACCATCTTCAATTGAG | This study | |||
| virS | virS-F | CATTGTAATAATAATTTTTTCTGTC | This study | ||
| virS-R | TTTCCTTCAATACAGGCTATGTG | This study | |||
| pfoA | pfoA-F | CAAGTATTGCAATGGCTTTATGTCTG | This study | ||
| pfoA-R | CTTTATAAGAGCTTTGAAAGCAGCTTG | This study | |||
| Genes on the representative plasmids | |||||
| cpe | |||||
| can | CAN-F | GTAGGGGAATTGATAGAACAAGACTTC | 14) | ||
| CAN-R | CTTTTATTTGAGTATCAACCATTTCAGC | 14) | |||
| cpb2 | cpb2MPRC | CAATAACCCTCACCAAATACTC | 14) | ||
| cpb2MPFC | AGATTTTAAATATGATCCTAACC | 14) | |||
| tcpF | ORF15-HF | GACTATAGGAACTAGTGCTATAGTTGC | 14) | ||
| ORF15-HR | CGCTGGATTTACTACATAGTCCTCTG | 14) | |||
| tcpH | ORF16-HF | GTTAATCCAGGATATGAATATTGGTGC | 14) | ||
| ORF16-HR | GTCTCTATTATAATTAGAGTTAGCAGG | 14) | |||
| rep | repCPEF | CTTAAATCAAATCGAATATAAAGAGTC | This study | ||
| repCPER | AATTTCTTTCTGTAAAGTTTGGTAGAG | This study | |||
| soj | soj-F | GGAGTTGCTAAAACAACGTCTACTGC | This study | ||
| soj-R | CTTCAAATGTACTTTCTACTACC | This study | |||
| parB | parB-F | GAAATAGTGGATATTGAATCTCTTGCAG | This study | ||
| parB-R | CCTTGTTCTATAACTGCTTTTAACTCTGG | This study | |||
| topA | topA-F | CATATATATTCTTGCCACAACGAGG | This study | ||
| topA-R | GATAGTAAGATAGAAAGTCATAGTGCC | This study | |||
| bcn | bcnF2 | GTTTCCGCCAAATGCAGTAGTAAGAG | This study | ||
| bcnR2 | GTTCATCACCAACTACCTCTGCATTG | This study | |||
| pfoA genotyping assay | |||||
| pfoAR-F | AAAATACATACAGTAGATGAGATACGTGG | This study | |||
| pfoAR-R | AAATCTGCTCTTAAAATCAATGCCTCAGC | This study | |||
Southern blot assays for the presence of eno, pfoA, tcpH, cna, and soj genes in C. perfringens strains
To further investigate PCR-negative results, ten chromosomal cpe strains, six plasmid cpe food poisoning strains, three plasmid cpe food strains, two human sporadic diarrhea strains, were investigated by Southern blot analyses (Table 2). Plasmid cpe human feces isolate (MR2-4) [15], cpe-negative human feces isolate (MR2-12) and food isolate in Japan (M-08) were also examined. As a negative control, Clostridium tertium isolate, identified based on 16 rRNA gene sequence, from Japanese retail food was used.
DNA was prepared from strains F4969 or F5603 and that DNA was used with a PCR DIG-labeling kit [Roche] [34] to prepare DIG-labeled probes for eno, virS, pfoA, cna, tcpH, and soj. For Southern blot assays, DNA sample of each strains, prepared with the methods previously described [34], were digested with PstI overnight at 37°C and then electrophoresed on a 1% agarose gel with 8 mA constant current, for 16 to 18 hours and then transferred to nylon membranes [Roche] with a vacuum blotter [Bio-Rad] with manufactures' instructions. The membranes were hybridized with one of the gene probes as described previously [34]. Fluorescence signals were detected with X-ray film [Fuji-Film].
PCR analysis of the pfoA region in pfoA-negative, cpe-positive C. perfringens strains
Since PCR survey and Southern blot assay results indicated that all ten tested chromosomal cpe strains and five plasmid cpe strains do not carry pfoA, the genetic organization of upstream and downstream region of pfoA in pfoA-negative strains was investigated. First, a bioinformatic investigate was performed on the complete genome sequenced pfoA-positive and pfoA-negative strains, ATCC13124 and SM101 [10], respectively. From the GenBank database, both chromosomal cpe-positive SM101 and cpe-negative ATCC13124 carry upstream pfoR and downstream hypothetical protein gene on the pfoA region. To investigate whether this simple pfoA-specific deletion might be common among pfoA-negative, cpe-positive strains, primer pairs were constructed inside of the upstream pfoR ORF and a downstream hypothetical protein ORF, based on sequence information for ATCC13124 and SM101. In this pfoA-genotype assay, the estimated size of a PCR product should be 2,375 bp for a pfoA-positive strain (such as ATCC13124) but only 280 bp for a pfoA-negative strain (such as SM101). In this pfoA-genotype assay, PCR was performed with twelve pfoA-negative, cpe-positive, type A strains under the same conditions as cpe-genotying assay [20], but with a different Taq polymerase, PrimeSTAR GXL DNA polymerase [Takara], that is suitable for long-range PCR.
Multilocus sequencing typing (MLST) analysis
To thoroughly investigate the genetic background of enterotoxigenic C. perfringens strains, MLST was performed using eight housekeeping genes, related to survival in foods, bacterial proliferation in foods, or spore formation (CPE is formed during sporulation). The genes used in our MLST analysis contained genes for toxin genes (plc, colA), stress response (sodA, groEL), sigma factor for sporulation (sigK), putative metabolism genes (pgk, nadA) and genes in DNA replication (gyrB). PCR products were purified with a QIA quick PCR purification kit [QIAGEN], and then sequenced with ABI PRISM®BigDye™terminator Cycle Sequencing Ready Reaction Kits (Version 1.1 and 3.1) according to the manufacture's instructions. All sequence data were concatenated to produce an in-frame 5,274 bp, according to genome arrangement of strain 13, plc, colA, nadA, sodA, pgk, sigK, groEL, and gyrB, (approximately 0.17% of strain 13 whole genome). Sequence information of these eight genes from the three completely sequenced C. perfringens strains, i.e., Strain13,ATCC13124,SM101 (derivative from NCTC8798), were also included in this survey. Concatenated sequence data were applied to phylogenetic analysis with Clustal W format by using Lasergene software Ver. 6 [DNASTAR].
Nucleotide sequence accession numbers
The sequences determined in this study have been deposited in the GenBank under accession AB477535-AB477966.
Whole genome sequence information for strain 13, SM101, and ATCC13124 is available according to the following accession numbers, NC003366, CP000312, NC008261, respectively.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported in part by grant 2005-53201-15387 from the Ensuring Food Safety Program of the United States Department of Agriculture and by R37AI19844 from the National Institute of Allergy and Infectious Diseases. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Matches JR, Liston J, Curran D. Clostridium perfringens in the environment. Appl Microbiol. 1974;28:655–660. doi: 10.1128/am.28.4.655-660.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClane BA. Clostridial enterotoxins. In: Durre P, editor. Handbook on Clostridia. Boca Raton: CRC Press; 2005. pp. 385–406. [Google Scholar]
- 3.Collie RE, McClane BA. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J Clin Microbiol. 1998;36:30–36. doi: 10.1128/jcm.36.1.30-36.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka D, Kimata K, Shimizu M, Isobe J, Watahiki M, et al. Genotyping of Clostridium perfringens isolates collected from food poisoning outbreaks and healthy individuals in Japan based on the cpe locus. Jpn J Infect Dis. 2007;60:68–610. [PubMed] [Google Scholar]
- 5.Lahti P, Heikinheimo A, Johansson T, Korkeala H. Clostridium perfringens type A isolates carrying the plasmid-borne enterotoxin gene (genotypes IS1151-cpe or IS1470-like-cpe) are a common cause of food poisonings. J Clin Microbiol. 2007;46:371–373. doi: 10.1128/JCM.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarker MR, Shivers RP, Sparks SG, Juneja VK, McClane BA. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfrinens isolates carrying plasmid versus chromosomal enterotoxin genes. Appl Environ Microbiol. 2000;66:3234–3240. doi: 10.1128/aem.66.8.3234-3240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Li J, McClane BA. Comparative effects of osmotic, sodium nitrate-induced, and pH-induced stress on growth and survival of Clostridium perfringens type A isolates carrying chromosomal or plasmid-borne enterotoxin genes. Appl Environ Microbiol. 2006;72:7620–7625. doi: 10.1128/AEM.01911-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, McClane BA. Further comparison of temperature effects on growth and survival of Clostridium perfringens type A isolates carrying a chromosomal or plasmid-borne enterotoxin gene. Appl Environ Microbiol. 2006;72:4561–4568. doi: 10.1128/AEM.00177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, et al. Complete genome sequence of Clostridium perfringens,an anaerobic flesh-eater. Proc Natl Acad Sci U S A. 2002;99:996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myers GS, Rasko DA, Cheung JK, Ravel J, Seshadri R, et al. Skewed genomic variability in strains of the toxinogenic bacterial pathogen, Clostridium perfringens. Genome Res. 2006;16:1031–1040. doi: 10.1101/gr.5238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enright MC, Sporatt BG. Multilocus sequence typing. Trends Microbiol. 1999;7:482–487. doi: 10.1016/s0966-842x(99)01609-1. [DOI] [PubMed] [Google Scholar]
- 12.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, et al. Multilocus sequence typing: a portable approach to the identification of clones within population of pathogenetic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urwin R, Maiden MC. Multi-locus sequence typing: a tool for global epidemiology. Trend Microbiol. 2003;11:479–487. doi: 10.1016/j.tim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Miyamoto K, Fisher DJ, Li J, Sayeed S, Akimoto S, McClane BA. Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J Bacteriol. 2006;188:1585–1598. doi: 10.1128/JB.188.4.1585-1598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garnier T, Cole ST. Complete nucleotide sequence and genetic organization of the bacteriocinogenic plasmid, pIP404, from Clostridium perfringens. Plasmid. 1988;19:134–150. doi: 10.1016/0147-619x(88)90052-2. [DOI] [PubMed] [Google Scholar]
- 16.Kawai T, Tsukamoto T, Asao T, Nishioka M, Yoshioka M, et al. An epidemiological analysis of a food-borne outbreak caused by Clostridium perfringens using Pulse-field Gel Electrophoresis. OSAKA Furitu Koshu Eisei Kenkyusho Nenpo. 1998;36:191–194 in Japanese. [Google Scholar]
- 17.Tanaka D, Isobe J, Hosorogi S, Kimata K, Shimizu M, et al. An outbreak of food-borne gastroenteritis caused Clostridium perfringens carrying the cpe gene on a plasmid. Jpn J Infect Dis. 2003;56:137–139. [PubMed] [Google Scholar]
- 18.Yamagishi T, Sakamoto K, Sakurai S, Konishi K, Daimon Y, et al. A nosocomial outbreak of poisoning caused by enterotoxigenic Clostridium perfringens. Microbiol Immunol. 1983;27:291–296. doi: 10.1111/j.1348-0421.1983.tb03591.x. [DOI] [PubMed] [Google Scholar]
- 19.Wen Q, McClane BA. Detection of enterotoxigenic Clostridium perfringens type A isolates in American retail foods. Appl Environ Microbiol. 2004;70:2685–2691. doi: 10.1128/AEM.70.5.2685-2691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamoto K, Wen Q, McClane BA. Multiplex PCR genotyping assey that distinguishes between isolates of Clostridium perfringens type A carrying a chromosonal enterotoxin gene (cpe) locus,a plasmid cpe locus with an IS1470-Like sequence,or a plasmid cpe locus with an IS1151 sequence. J Clin Microbiol. 2004;42:1552–1558. doi: 10.1128/JCM.42.4.1552-1558.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Sayeed S, McClane BA. Prevalence of enterotoxigenic Clostridium perfringens isolates in Pittsburgh (Pennsylvania) area soil and home kitchens. 2007;73:7218–7224. doi: 10.1128/AEM.01075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutton RGA, Hobbes BC. Food poisoning caused by heat-sensitive Clostridium welchii. A report of five recent outbreaks. J Hyg. 1965;66:135–146. doi: 10.1017/s0022172400041000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson A, Aspan A, Bagge E, Baverud V, Engstrom B, et al. Genetic diversity of Clostridium perfringens type A isolates from animals, food poisoning outbreaks and sludge. BMC Microbiol. 2006;6:47. doi: 10.1186/1471-2180-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rooney AP, Swezey JL, Friedman R, Hecht DW, Maddox CW. Analysis of core housekeeping and virulence genes reveals cryptic lineages of Clostridium perfringens that are associated with distinct disease presentations. Genetics. 2006;172:2081–2092. doi: 10.1534/genetics.105.054601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jost BH, Trinh HT, Songer JG. Clonal relationship among Clostridium perfringens of porcine origin as determined by multilocus sequence typing. Vet Microbiol. 2006;116:158–165. doi: 10.1016/j.vetmic.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 26.Chalmers G, Bruce HL, Hunter DB, Parreira VR, Kulkarni RR, et al. Multilocus sequence typing analysis of Clostridium perfringens isolates from necrotic enteritis outbreaks in broiler chickens. J Clin Microbiol. 2008;46:3957–3964. doi: 10.1128/JCM.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miki Y, Miyamoto K, Kaneko-Hirano I, Fujiuchi K, Akimoto S. Prevalence and characterization of enterotoxin gene carrying Clostridium perfringens from retail meat products in Japan. Appl Environ Microbiol. 2008;74:5366–5372. doi: 10.1128/AEM.00783-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brynestad S, Sarker MR, McClane BA, Granum PE, Rood JI. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect Immun. 2000;69:3483–3487. doi: 10.1128/IAI.69.5.3483-3487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meredith L, Poon R, Adams V, Sayeed S, Saputo J, et al. Epsilon-toxin plasmids of Clostridium perfringens type D are conjugative. J Bacteriol. 2007;189:7531–7538. doi: 10.1128/JB.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parson JA, Bannam TL, Devenish RJ, Rood JI. TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 in Clostridium perfringens. J Bacteriol. 2007;189:7782–7790. doi: 10.1128/JB.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayeed S, Uzal FA, Fisher DJ, Saputo J, Vidal JE, et al. Beta toxin is essential for the intestinal virulence of Clostridium perfringens type C disease isolate CN3685 in a rabbit ileal loop model. Molec Microbiol. 2008;67:15–30. doi: 10.1111/j.1365-2958.2007.06007.x. [DOI] [PubMed] [Google Scholar]
- 32.Li J, McClane BA. A novel small acid solble protein variant is important for spore resistance of most Clostridium perfringens food poisoning isolates. PLos Pathogens. 2008;4:e1000056. doi: 10.1371/journal.ppat.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brynestad S, Granum PE. Evidence that Tn5565, which includes the enterotoxin gene in Clostridium perfringens, can have a circular form which may be a transposition intermediate. FEMS Microbiol Lett. 1999;170:281–286. doi: 10.1111/j.1574-6968.1999.tb13385.x. [DOI] [PubMed] [Google Scholar]
- 34.Miyamoto K, Chakrabarti G, Morino Y, McClane BA. Organization of the plasmid cpe locus in Clostridium perfringens type A isolates. Infect Immun. 2002;70:4261–4272. doi: 10.1128/IAI.70.8.4261-4272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harry KH, Zhou R, Kroos L, Melville SB. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific sigma factors SigE and SigK in Clostridium perfringens. J Bacteriol. 2009;191:2728–2742. doi: 10.1128/JB.01839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bannam TL, Teng WL, Bulach D, Lyras D, Rood JI. Functional identification of conjugative and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J Bacteriol. 2006;188:4942–4951. doi: 10.1128/JB.00298-06. [DOI] [PMC free article] [PubMed] [Google Scholar]