Abstract
The intermembrane space of mitochondria contains the specific mitochondrial intermembrane space assembly (MIA) machinery that operates in the biogenesis pathway of precursor proteins destined to this compartment. The Mia40 component of the MIA pathway functions as a receptor and binds incoming precursors, forming an essential early intermediate in the biogenesis of intermembrane space proteins. The elements that are crucial for the association of the intermembrane space precursors with Mia40 have not been determined. In this study, we found that a region within the Tim9 and Tim10 precursors, consisting of only nine amino acid residues, functions as a signal for the engagement of substrate proteins with the Mia40 receptor. Furthermore, the signal contains sufficient information to facilitate the transfer of proteins across the outer membrane to the intermembrane space. Thus, here we have identified the mitochondrial intermembrane space sorting signal required for delivery of proteins to the mitochondrial intermembrane space.
INTRODUCTION
Mitochondria pose a great challenge for the proper delivery of proteins because of their complex architecture. Mitochondrial precursors must find their way to one of the four mitochondrial subcompartments: the outer membrane, intermembrane space, inner membrane, or matrix. As a direct consequence of this complexity, several machineries for the translocation and sorting of mitochondrial precursors have evolved. Interplay between these machineries and specific signals present in the precursors drive different protein targeting pathways (Schatz and Dobberstein, 1996; Emanuelsson and von Heijne, 2001; Jensen and Johnson, 2001; Endo et al., 2003; Koehler, 2004; Oka and Mihara, 2005; Dolezal et al., 2006; Neupert and Herrmann, 2007; Bolender et al., 2008). Initially, mitochondrial precursors are recognized in a signal-dependent manner by specific receptors and are transferred across the barrier of outer mitochondrial membrane by using the translocase of the outer membrane (TOM) complex. On the trans-side of the outer mitochondrial membrane, sorting machineries decode specific signals in precursors, and this results in the branching of protein import pathways. The most well characterized is the presequence pathway across the inner membrane driven by a cleavable and positively charged signal sequence, called a presequence, and the translocase of the inner membrane (TIM) 23 complex Endo et al., 2003; Oka and Mihara, 2005; Neupert and Herrmann, 2007; Bolender et al., 2008). The presequence is cleaved off by a specific protease liberating the mature protein. However, other mitochondrial signals are not proteolytically removed and remain as part of the native mitochondrial protein. One example is the recently identified β-signal that is recognized by the sorting and assembly machinery (SAM) complex to sort β-barrel proteins to the mitochondrial outer membrane (Kutik et al., 2008). In other mitochondrial membrane proteins, the membrane domains, anchors, and their surrounding regions are used to some extent for selection of a membrane, integration, and in adopting a correct orientation (Rehling et al., 2004; Walther and Rapaport, 2008).
The import and biogenesis of precursors targeted to the intermembrane space (IMS) depends on their cysteine residues, which are used in dithiol-disulfide exchange reactions along the mitochondrial intermembrane space assembly (MIA) pathway. The MIA machinery consists of two essential components, Mia40/Tim40 (Chacinska et al., 2004; Naoé et al., 2004; Terziyska et al., 2005) and the sulfhydryl oxidase Erv1 (Allen et al., 2005; Mesecke et al., 2005; Rissler et al., 2005). Initially, the incoming IMS precursors engage with Mia40 on the trans-side of the outer membrane via a transient intermolecular disulfide bond (Chacinska et al., 2004; Mesecke et al., 2005). Erv1 cooperates with Mia40 in the transfer of disulfide bonds to precursors in two ways. Erv1 associates with Mia40 and precursor protein in a transient ternary complex, thereby directly promoting the transfer of multiple disulfide bonds (Stojanovski et al., 2008a). On completion of disulfide exchange, precursor monomers are released in an oxidized state capable of assembly into mature complexes (Curran et al., 2002; Lu et al., 2004; Webb et al., 2006; Müller et al., 2008). Reduced Mia40 is reoxidized by Erv1, which results in the activation of Mia40 for the next round of precursor binding and import (Mesecke et al., 2005; Grumbt et al., 2007). Additional components contribute in the execution of the MIA pathway, including cytochrome c and cytochrome c peroxidase, which play a role in electron flow from Erv1, and the zinc-binding Hot13 that promotes the reoxidation of Mia40 by Erv1 (Curran et al., 2004; Bihlmaier et al., 2007; Dabir et al., 2007; Mesecke et al., 2008).
Mia40 is able to distinguish its own substrates from other cysteine-rich, but otherwise unrelated, proteins in the formation of disulfide-bonded intermediates (Milenkovic et al., 2007a). This facilitates precursor entry into the IMS. The cysteine residues of Mia40, in particular the first Cys-X-Cys pair, have been shown to be essential for function (Naoé et al., 2004; Grumbt et al., 2007; Terziyska et al., 2009). However, our and others' analyses of the classical IMS precursor proteins Tim9 and Tim10 (Koehler, 2004) revealed a differential role of cysteine residues in precursor proteins (Milenkovic et al., 2007a; Sideris and Tokatlidis, 2007). Although each of the four cysteines within these precursors was required for the oxidative folding before complex assembly, only the most amino-terminal cysteine residue was critical for precursor interaction with Mia40 in the initial biogenesis stage. Up to now, the nature of the precursor recognition site for Mia40 has remained unknown. In this study, we attempted to identify and characterize elements that would serve as a signal for Mia40 in the import of IMS precursors. We constructed various small Tim mutants and chimeric substrates and examined their ability to bind to Mia40 and to enter the IMS. Our extensive analysis has led to the identification of the mitochondrial intermembrane space sorting (MISS) signal, which is crucial for the association of precursors with Mia40 and their correct localization to the mitochondrial IMS.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
The wild-type strain YPH499 (MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801; Sikorski and Hieter, 1989) and its derivatives were used in this study. The temperature-sensitive mutant strains mia40-3 and mia40-4 have been characterized previously (Chacinska et al., 2004; Milenkovic et al., 2007a; Müller et al., 2008; Stojanovski et al., 2008a). The strains expressing Mia40His and a conserved part of Mia40 (Mia40core) were reported previously (Rissler et al., 2005; Milenkovic et al., 2007a; Stojanovski et al., 2008a; Chacinska et al., 2008). Yeast strains were grown at 19°C in YPG medium containing 1% (wt/vol) yeast extract, 2% (wt/vol) bacto-peptone, and 3% (vol/vol) glycerol. Differential centrifugation was used for isolation of mitochondria (Meisinger et al., 2006).
To produce Egd1FLAG and SignalEgd1FLAG, coding regions of Egd1 and chimeric Tim9(29-38)-Egd1 (SNLVERCFTDEgd1) were cloned in plasmid pESC-URA (Stratagene, La Jolla, CA) downstream of GAL10 promoter and upstream of the sequence coding for the FLAG epitope giving rice to DS25 and DS26-A, respectively. Plasmids harboring these genes were transformed into YPH499. The strains were grown on synthetic medium containing 2% galactose. After the subcellular fractionation total, mitochondrial and postmitochondrial fractions were analyzed by Western blot with anti-FLAG (M2 antibody; Sigma-Aldrich, St. Louis, MO), anti-Egd1 antibody and marker proteins.
Generation of Mutant Precursor Proteins
The following plasmids: Tim9ΔC20, Tim9ΔC33, Tim9ΔC49, Tim9ΔN30, Tim9ΔN31, Tim9ΔN32, Tim9ΔN38, Tim10ΔN43, and Tim12ΔN33 were constructed for in vitro transcription/translation procedures. The respective coding regions of Tim9, Tim10, or Tim12 were cloned into a pGEM4Z vector downstream of the SP6 promoter. Tim9ΔC39 was generated using Tim9ΔC33 as a template for polymerase chain reaction (PCR). To generate mutants Tim9ΔN25, Tim9ΔN26, Tim9ΔN27, Tim9ΔN28, and Tim9ΔN29, the templates for in vitro transcription/translation were obtained by PCR from yeast genomic DNA. For the Tim10(35-43)- Tim9ΔN38 and NLVAACAAATim9ΔN38, DNA for in vitro transcription/translation was obtained from PCR reactions using pGEM4Z-Tim9ΔN38 as template. Versions of Rpl24a and Egd1 were obtained by PCR amplification from yeast genomic DNA. The primers used for the amplification of Tim10(35-43)-Tim9ΔN38, NLVAACAAATim9ΔN38, Tim9(30-38)-Rpl24a (SignalRpl24a) and Tim9(30-38)-Egd1 (SignalEgd1) contained the sequence coding for a signal. Single, double, and triple Tim9 and Tim10 amino acid substitutions were generated using site-directed mutagenesis (QuikChange; Stratagene). For the constructs with modification of the amino terminus, methionine was added at position one. In some cases, an additional two to three methionine residues were added to the C terminus of the precursors for better labeling.
Plasmid DNA was used for coupled transcription/translation (TNT SP6 Quick; Promega, Madison, WI). PCR-generated DNA was used for in vitro transcription (mMESSAGE mMACHINE kit; Ambion, Austin, TX) followed by the translation of proteins in rabbit reticulocyte lysate (GE Healthcare, Chalfont St. Giles, Buckinghamshire, United Kingdom). Precursor proteins, synthesized in the presence of [35S]methionine (GE Healthcare), were precipitated using saturated ammonium sulfate solution and denatured in urea buffer (8 M urea, 30 mM 3-(N-morpholino)propanesulfonic acid [MOPS]/KOH, pH 7.2, and 10–20 mM dithiothreitol). The amount of 35S-labeled precursors used in the import experiments was adjusted to the same intensity to allow the comparison.
In Vitro Import Experiments
For in vitro import experiments, isolated mitochondria were incubated with radiolabeled precursor proteins, at 30°C in import buffer (250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 5 mM methionine, 10 mM KH2PO4, and 10 mM MOPS/KOH, pH 7.2). Reactions were stopped on ice by addition of 50 mM iodoacetamide. Where indicated, samples were subjected to 50 μg/ml proteinase K (Prot. K) treatment. Mitochondria were reisolated, lysed in Laemmli buffer, and resolved by SDS-polyacrylamide gel electrophoresis (PAGE) or Tricine-SDS-PAGE (Schägger and von Jagow, 1987). For nonreducing conditions, reducing agent was omitted from Laemmli buffer. For native analysis, mitochondria were solubilized in digitonin buffer [1% (wt/vol) digitonin, 20 mM Tris-HCl, 0.5 mM EDTA, 10% (vol/vol) glycerol, and 50 mM NaCl, pH 7.4], and protein complexes were resolved by Blue Native PAGE (Schägger and von Jagow, 1991).
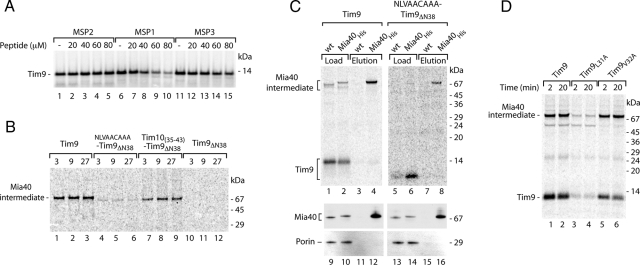
For peptide competition assay, synthetic peptides MSP1 (SNLVERCFTD), MSP2 (SNLVERSFTD) and MSP3 (SNAAERCFTD) were blocked N-terminally by acetylation and C-terminally by amidation. Peptides were dissolved in 50 mM phosphate buffer, pH 4.2. Mitochondria were incubated with the peptides (200 μM or as otherwise indicated) or phosphate buffer (for mock control) in import buffer for 3 min before addition of 35S-labeled precursors or after import of 35S-labeled Tim9 in chase experiments.
Pull-down experiments were performed using wild-type and Mia40His mitochondria essentially as described previously (Milenkovic et al., 2007a). After import of 35S-labeled precursors, mitochondria were isolated and solubilized in digitonin-containing buffer [1% (wt/vol) digitonin, 20 mM Tris-HCl, 0.5 mM EDTA, 10% (vol/vol) glycerol, 50 mM NaCl, and 50 mM iodoacetamide, pH 7.4]. After a clarifying spin, mitochondrial extracts were subjected to nickel-nitrilotriacetic acid (Ni-NTA)-agarose affinity chromatography. After washing [20 mM Tris-HCl, 10% (vol/vol) glycerol, 100 mM NaCl, 10 mM iodoacetamide, and 20 mM imidazole, pH 7.4] proteins were eluted from the beads [20 mM Tris-HCl, 10% (vol/vol) glycerol, 100 mM NaCl, 50 mM iodoacetamide, and 400 mM imidazole, pH 7.4] and analyzed by nonreducing SDS electrophoresis.
Miscellaneous
Homology searches were performed at the Swiss Institute of Bioinformatics by using the BLAST network service. The gels were analyzed by digital autoradiography (Storm Imaging System; GE Healthcare) with the use of ImageQuant software. In some figures, nonrelevant gel parts were excised digitally. Western blot analysis was performed using enhanced chemiluminescence.
RESULTS
Identification of the Region within Tim9 That Functions as a Signal for Mia40
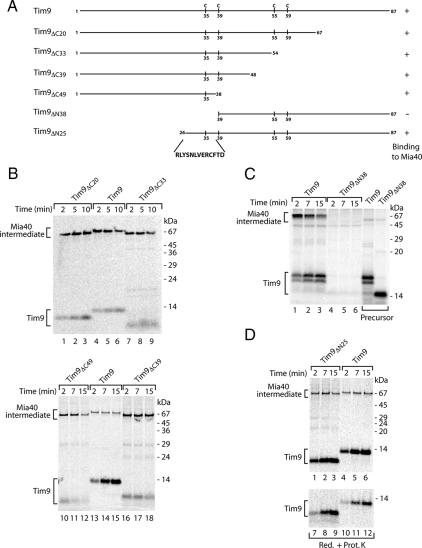
The most amino-terminal cysteine residues of Tim9 and Tim10 (Cys35 and Cys40, respectively) are required for formation of a transient disulfide-bonded intermediate with Mia40 (Milenkovic et al., 2007a). To screen for targeting elements, we generated amino- and carboxy-terminal truncations of Tim9 (Figure 1A) and monitored import of the radiolabeled precursors into isolated mitochondria. The reactions were analyzed under nonreducing conditions to detect disulfide-bonded conjugates. The Tim9 precursors with the carboxy-terminal truncations of 20, 33, and 39 residues were able to bind to Mia40 (Figure 1B). Even Tim9 lacking 49 residues on its carboxy terminus, and containing exclusively Cys35, was efficiently recognized by Mia40. Next, we generated amino-terminal deletions of Tim9. The truncation of 38 residues, which included Cys35, resulted in the complete loss of binding to Mia40 (Figure 1C). In contrast, the absence of 25 amino-terminal residues did not affect the formation of the Mia40–Tim9 conjugate, and this precursor was also efficiently imported into the protease-protected location of the IMS (Figure 1D). We conclude that the region between amino acids 26 and 38 of Tim9 contains the information necessary for recognition and binding to Mia40.
Figure 1.
The formation of the Tim9–Mia40 intermediate depends on the Tim9 region between residues 26 and 38. (A) Schematic representation of Tim9 truncation constructs. Positions of cysteine residues are indicated. (B) Isolated wild-type mitochondria were incubated with 35S-labeled Tim9 and truncation mutant precursors for the indicated times. Import analysis was performed using nonreducing SDS electrophoresis. (C) 35S-labeled Tim9 and Tim9ΔN38 precursors were analyzed directly or imported into mitochondria. Samples were separated by nonreducing SDS electrophoresis. (D) The Tim9 and Tim9ΔN25 precursors were imported into mitochondria. Samples were analyzed either by nonreducing electrophoresis or treated with Prot. K before reducing SDS electrophoresis.
To specify the region required for binding to Mia40, we performed a residue per residue truncation of Tim9 between amino acids Arg26 and Val32 (Figure 2A). The truncated versions of Tim9 were generated as radiolabeled precursors and incubated with isolated mitochondria to monitor association with Mia40. The samples were subsequently analyzed by nonreducing SDS-PAGE for the formation of Mia40-bound species or were subjected to protease treatment after import to determine translocation into the IMS. Tim9 with amino-terminal deletions of 26, 27, 28, and 29 residues formed a conjugate with Mia40 (Figure 2, B and C, top). Consequently, the precursors that were efficiently recognized and bound by Mia40 were also transported into the IMS (Figure 2, B and C, bottom). However, in the case of Tim9ΔN30 formation of the Mia40 conjugate was severely impaired (Figure 2C, lanes 4–6), and the import of this precursor was inhibited (Figure 2C, lanes 13–15). Further deletion of residues 31 and 32 resulted in the abolishment of binding to Mia40 (Figure 2D). The association of precursors with Mia40 represents an intermediate stage before assembly into the mature hexameric Tim9–Tim10 complex and the fraction of Tim9–Tim10 associated with the TIM22 complex (Chacinska et al., 2004; Koehler, 2004). Assembly of Tim9ΔN30, Tim9ΔN31, and Tim9ΔN32 was assayed using native electrophoresis. Tim9ΔN30, Tim9ΔN31, and Tim9ΔN32 neither interacted with Mia40 nor assembled into mature complexes. Tim9ΔN29 formed the Mia40 intermediate (Figure 2E) in agreement with the results obtained using nonreducing analysis (Figure 2C). In summary, the region of Tim9 between residues N30 and D38 serves as a sorting signal for Mia40 and is referred to as the MISS signal.
Figure 2.
Identification of the Tim9 signal for binding to Mia40. (A) Schematic representation of Tim9 and amino terminal truncation mutants. The identified signal is indicated by the gray box. (B–D) 35S-labeled Tim9 and its amino-terminal truncation mutants were imported into mitochondria. Samples were treated with Prot. K as indicated and were further analyzed by nonreducing or reducing SDS electrophoresis. (E) The Tim9, Tim9ΔN29, Tim9ΔN30, Tim9ΔN31, and Tim9ΔN32 precursors were imported into mitochondria. Mitochondrial extracts were lysed in digitonin and were separated by native electrophoresis.
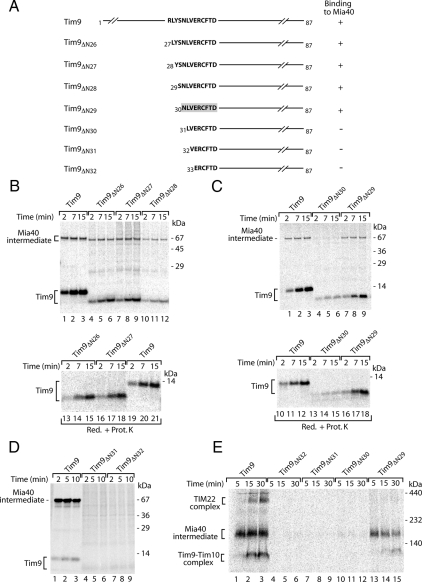
The Signal Peptide Inhibits Binding of Precursors by Mia40
To independently confirm the crucial function of the identified MISS signal for the biogenesis of IMS proteins, we used a peptide inhibition approach. We asked whether a peptide, which mimics the Tim9 signal, was able to bind to Mia40 and thereby block its interaction with precursors targeted to the IMS. Two peptides, MSP1 (SNLVERCFTD), corresponding to the Tim9 MISS signal; and MSP2 (SNLVERSFTD), a variant lacking the critical Cys residue, were used. We imported Tim9 in the presence of the two peptides and analyzed its biogenesis by using native electrophoresis. The signal peptide MSP1 inhibited the formation of the Tim9–Mia40 intermediate and subsequent assembly into the mature complexes (Figure 3A). In contrast, the peptide MSP2, lacking the Cys residue, did not affect the assembly pathway of Tim9 (Figure 3A). To rule out the possibility that the signal peptide affects the release of precursors from Mia40 or their assembly into mature complexes, we undertook the following approaches. First, we directly examined the formation of the Tim9–Mia40 intermediate under nonreducing conditions. The MISS peptide MSP1 was able to efficiently compete with Tim9 for binding to Mia40 (Figure 3B). Second, we performed such analysis in mitochondria isolated from the temperature-sensitive mia40-4 mutant, in which the release of substrate proteins is delayed resulting in their entrapment at the Mia40-bound stage and defective complex assembly (Chacinska et al., 2004; Milenkovic et al., 2007a; Stojanovski et al., 2008a). In a similar manner as in wild-type mitochondria, the signal peptide MSP1 prevented the formation of the Tim9–Mia40 conjugate in mia40-4 mitochondria as determined by both nonreducing SDS-PAGE (Figure 3C, compare lanes 4–6 and 10–12) and native electrophoresis (Figure 3D, compare lanes 4–6 and 7–9). Third, we accumulated the Tim9–Mia40 intermediate and performed a chase experiment in the presence of MISS peptide to directly assess the effect of the peptide on complex assembly. No significant difference in the formation of mature Tim9 complexes was detected (Figure 3E). Altogether, these data demonstrate that the MISS peptide exerted an inhibitory effect at the stage of Tim9 binding to Mia40. We also analyzed the effect of both peptides on the translocation of Tim9 into mitochondria by protease-protection assays. Import of Tim9 was efficiently inhibited by the signal peptide MSP1, and not by the MSP2 peptide lacking the Cys residue (Figure 3F). Thus, the MISS signal peptide is able to saturate Mia40 binding sites preventing association and ultimately retention of Tim9 in the IMS.
Figure 3.
The signal peptide inhibits recognition of Tim9 by Mia40. (A) Before import of 35S-labeled Tim9, wild-type mitochondria were incubated with phosphate buffer (Mock) or synthetic peptides MSP1 (SNLVERCFTD) and MSP2 (SNLVERSFTD). Native analysis of import reactions was performed. (B) 35S-labeled Tim9 was imported into mitochondria in the presence or absence of MSP1 peptide and Mia40-bound intermediate was analyzed by nonreducing SDS electrophoresis. (C) Wild-type and mia40-4 mitochondria were incubated with Tim9 precursor in the presence or absence of MSP1 peptide. Formation of the Mia40 conjugate was monitored by nonreducing SDS electrophoresis. (D) Wild-type and mia40-4 mitochondria were analyzed as described in A. (E) 35S-labeled Tim9 was imported into wild-type and mia40-4 mitochondria for 3 min at 25°C. Mitochondria were reisolated and incubated in the presence or absence of MSP1 peptide and analyzed by native electrophoresis (Chase). (F) Import reactions performed as described for A were subsequently treated with proteinase K and subjected to reducing SDS electrophoresis.
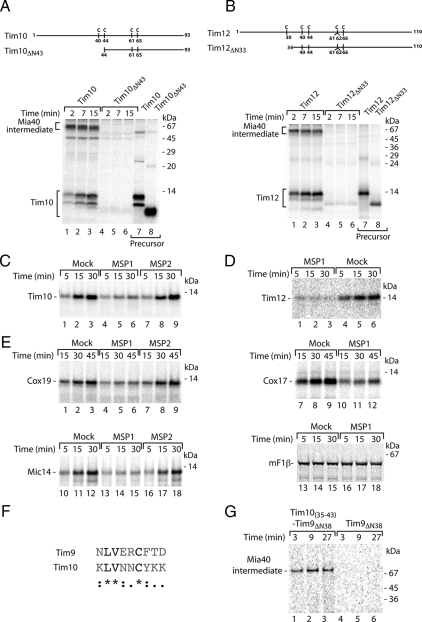
Next, we examined other essential small Tim proteins (Koehler, 2004; Milenkovic et al., 2007a; Gebert et al., 2008) for their translocation to the IMS. Interestingly, they somewhat vary in their mode of binding to Mia40. Tim10, similarly to Tim9, relies on the first cysteine residue (Cys40) of the conserved Cys-X3-Cys motif (Milenkovic et al., 2007a; Sideris and Tokatlidis, 2007), and indeed the removal of 43 amino-terminal residues including Cys40 abolished binding of Tim10 to Mia40 (Figure 4A). The association of Tim12 with Mia40 was dependent on the region around its first cysteine residue (Cys30) (Lionaki et al., 2008; Figure 4B). However, unlike in other small Tim proteins, this residue is not a part of the Cys-X3-Cys motif. The import of tested precursors was efficiently inhibited by the signal peptide derived from Tim9 (Figure 4, C and D). We performed an analysis with other MIA substrates that do not belong to the small Tim family and contain Cys-X9-Cys motifs (Chacinska et al., 2004; Gabriel et al., 2007). The MISS peptide was able to efficiently compete with Cox17, Cox19, and Mic14 substrates for binding to Mia40, which resulted in the inhibition of import for these substrates (Figure 4E). In contrast, the import of F1-ATPase subunit β (F1β), which depends on a presequence and follows the TIM23 pathway, was not affected by the MISS peptide (Figure 4E). This suggests that Mia40 uses the common recognition site for different precursors, which can be blocked by MISS.
Figure 4.
Characterization of signals recognized by Mia40. (A and B) Schematic representation of wild-type and amino-terminal truncation mutants of Tim10 (A) and Tim12 (B). Positions of cysteine residues are indicated. 35S-labeled precursors were analyzed directly or imported into mitochondria for the indicated times. Nonreducing SDS electrophoresis was performed. 35S-labeled Tim10 (C) and 35S-labeled Tim12 (D) were imported into wild-type mitochondria in the presence of MSP1 and MSP2 peptides or buffer (Mock). After the treatment with proteinase K, import reactions were analyzed by reducing SDS electrophoresis. (E) 35S-labeled Cox19, Cox17, Mic14, and F1-ATPase subunit β were analyzed as described in C. Mature F1-ATPase subunit β, mF1β. (F) Sequence alignment of Tim9 and Tim10 reveals the signal conservation. (G) Truncation constructs of Tim9ΔN38 and Tim10(35-43)–Tim9ΔN38 were imported into isolated wild-type mitochondria for the indicated time periods and treated with proteinase K. Mia40-bound intermediates were resolved using nonreducing SDS electrophoresis.
To provide further evidence for the existence of a recognition signal for Mia40, we aligned the amino acid sequences of Tim9 and Tim10 and searched for common motifs. The analysis revealed a region of Tim10 between K35 and K43 with high similarity to the Tim9 recognitions signal (Figure 4F), which was consistent with the dependence on Cys40 for binding of Tim10 to Mia40 (Milenkovic et al., 2007a; Sideris and Tokatlidis, 2007). We generated a construct where the potential signal region (KLVNNCYKK) of Tim10 was fused to the amino terminus of Tim9ΔN38, a mutant lacking its endogenous signal that is not recognized by Mia40 (Figure 1C). The resulting Tim10(35-43)–Tim9ΔN38 protein chimera was able to form the Mia40 conjugate (Figure 4G), with an efficiency comparable with wild-type Tim9 (Figure 6B, compare lanes 1–3 and 7–9). Thus, signals present within a primary structure of IMS precursors are interchangeable and use a universal mechanism for binding to Mia40.
Figure 6.
The Leu residue plays an important role in the signal for Mia40. (A) Mitochondria were incubated with 35S-labeled Tim9 for 20 min in the presence of MSP1 (SNLVERCFTD), MSP2 (SNLVERSFTD), or MSP3 (SNAAERCFTD) peptide. After proteinase K treatment, import reactions were analyzed by reducing SDS electrophoresis. (B) Import reactions of Tim9 versions were treated with proteinase K and analyzed by nonreducing SDS electrophoresis. (C) Tim9 and NLVAACAAATim9ΔN38 were imported into wild-type and Mia40His mitochondria for 20 min at 30°C, and mitochondrial extracts were subjected to Ni-NTA purification. Load (5%) and elution (100%) fractions were separated by nonreducing SDS electrophoresis and analyzed by autoradiography or immunoblotting with antibodies specific to Mia40 and porin. (D) Wild-type Tim9, Tim9L31A, and Tim9V32A were imported into mitochondria and analyzed by nonreducing SDS electrophoresis.
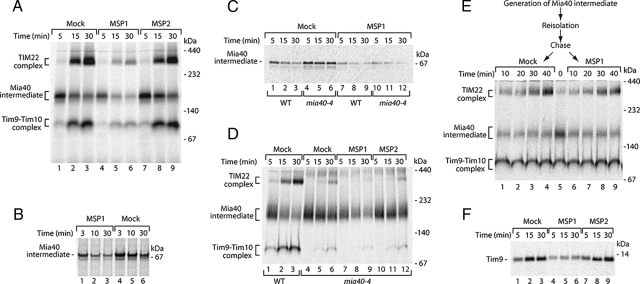
Crucial Role of Leu in the Signal Recognized by Mia40
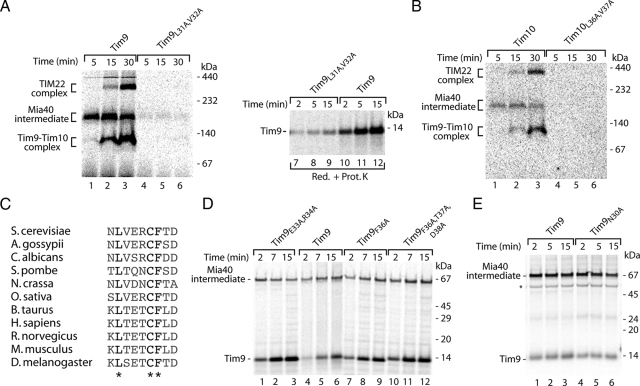
We undertook a systematic mutational analysis to address the role of individual residues in the MISS signal. The alignment of yeast Tim9 and Tim10 (Figure 4D) revealed the presence of two conserved hydrophobic residues within the signal (Leu31 and Val32 for Tim9; Leu36 and Val37 for Tim10) upstream of the critical cysteine. Indeed, mutation of both residues to alanine abolished the association with Mia40 as well as further assembly into mature complexes for both Tim9 and Tim10 (Figure 5, A and B). In agreement with this finding, the translocation of Tim9 into the IMS was also impaired (Figure 5A, lanes 7–9). A homology search among Tim9 proteins from different species revealed that Phe36, in addition to Leu31 and Cys35, is a universally conserved residue within the Tim9 signal (Figure 5C). However, the replacement of Phe36 did not affect binding of Tim9 to Mia40 (Figure 5D, lanes 7–12). The systematic replacement of further residues within the Tim9 signal did not reveal any additional crucial elements required for recognition by Mia40 (Figure 5, D and E).
Figure 5.
Contribution of single amino acid residues to the signal. (A) After import of 35S-labeled Tim9 and Tim9L31A,V32A precursors, mitochondria were subjected to native analysis or were treated with Prot. K and analyzed by reducing SDS electrophoresis. (B) Assembly of Tim10 and mutant Tim10L36A,V37A in wild-type mitochondria was analyzed by native electrophoresis. (C) Sequence similarity of the Tim9 signal among eukaryotic species. (D and E) Various Tim9 precursors were imported into wild-type mitochondria for the indicated times. Samples were treated by proteinase K (D) or applied directly (E) to nonreducing SDS electrophoresis. Asterisk, nonspecific band.
To verify the importance of Leu and Val residues within the signal, we undertook two independent approaches. First, we performed the peptide competition assays of Tim9 by using the MSP3 peptide (SNAAERCFTD), in which leucine and valine corresponding to Leu31 and Val32 of the MISS signal of Tim9 were replaced by alanines. Remarkably, the MSP3 peptide did not efficiently inhibit the translocation of Tim9 into the IMS, similarly to the peptide lacking the critical cysteine (MSP2) and in contrast to the MISS signal peptide (MSP1) (Figure 6A). Thus, exchange of Leu and Val residues inhibited the function of the synthetic signal. In an alternative approach, we introduced an artificial amino acid sequence consisting of NLVAACAAA at the amino terminus of Tim9ΔN38 (Leu31 and Val32 were retained in the native context of Asn30 and Cys35, whereas other amino acids were exchanged for alanines). On import the chimera was capable of forming a disulfide-bonded conjugate, albeit with a reduced efficiency compared with wild-type Tim9 (Figure 6B, lanes 1–3 vs. 4–6). To verify whether this species reflected the Mia40-bound form, we imported NLVAACAAATim9ΔN38 into isolated mitochondria harboring a tagged version of Mia40 (10-histidine tag at the carboxy terminus of Mia40). After import, mitochondrial extracts were subjected to Ni-NTA affinity purifications. Mia40His was efficiently purified as determined with a Mia40-specific antibody in contrast to the control protein, porin (Figure 6C, lanes 9–16). A conjugate of NLVAACAAATim9ΔN38 was found in the eluate fraction of Mia40His. The enrichment of this substrate in the purification procedure was in the similar range as for wild-type Tim9 (Figure 6C, lanes 5–8 vs. 1–4), demonstrating that the NLVAACAAATim9ΔN38 conjugate represented a Mia40-bound form. To further dissect the contribution of Leu31 and Val32 to the MISS signal, we generated Tim9 mutants in which either Leu31 or Val32 were exchanged for alanine. After import into mitochondria only Tim9L31A (but not Tim9V32A) was defective in binding to Mia40 (Figure 6D), demonstrating a specific role of this hydrophobic and conserved (Figure 5C) amino acid in the recognition by Mia40. In summary, based on the mutational analysis of substrates, import inhibition by synthetic peptides, and the behavior of chimeric substrate, we conclude that the Leu31 residue upstream of the critical Cys35 is an essential component of the MISS signal recognized by Mia40.
The MISS Signal Directs Nonmitochondrial Proteins into Mitochondria
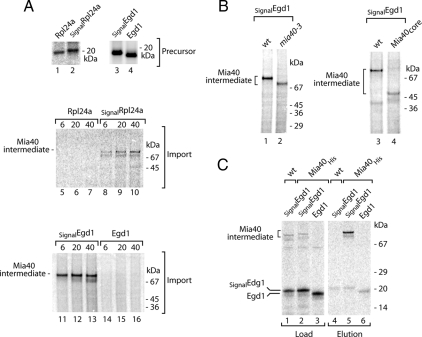
We asked whether the identified MISS signal was sufficient to direct nonmitochondrial proteins into the IMS via an interaction with Mia40. We have chosen two proteins that are localized in the cytosol: Rpl24a, a bona fide component of the large ribosomal subunit (Dresios et al., 2000); and Egd1, a subunit of the nascent polypeptide-associated complex NAC (George et al., 1998; Fünfschilling and Rospert, 1999). When incubated with isolated mitochondria, radiolabeled Rpl24a and Egd1 were not translocated into mitochondria and did not interact with Mia40 (Figure 7A). We fused the coding sequence for the MISS signal derived from Tim9 (NLVERCFTD) upstream of the Rpl24a and Egd1 coding sequences resulting in Tim9(30-38)-Rpl24a and Tim9(30-38)-Egd1, referred to as SignalRpl24a and SignalEgd1, respectively. Import experiments followed by treatment with proteinase K revealed that both chimeras formed protease-protected species (Figure 7A). To demonstrate that these species represented Mia40-bound conjugates, we used mitochondria harboring different versions of Mia40 in combination with SignalEgd1. First, we used the temperature-sensitive mia40-3 mutant. The Mia40-3 mutant protein displays a slightly faster mobility during electrophoresis and reduced efficiency of substrate binding (Chacinska et al., 2004; Müller et al., 2008; Stojanovski et al., 2008a). The SignalEgd1 conjugate formed in the mia40-3 mitochondria displayed both decreased intensity and faster mobility when compared with wild type (Figure 7B, left). Second, we imported SignalEgd1 into mitochondria isolated from the strain expressing Mia40core, a functionally conserved part of Mia40, which is much smaller than native Mia40 (23 vs. 55–60 kDa in the migration) (Chacinska et al., 2008). Indeed, a large change in the molecular weight of the SignalEgd1 conjugate was observed (Figure 7B, lane 4). Third, after the import of SignalEgd1, we performed Ni-NTA affinity purification of Mia40His. The SignalEgd1 conjugate was efficiently enriched in the Mia40His eluate, in contrast to the eluate obtained from mitochondria harboring nontagged Mia40 or from the control experiment with Egd1 lacking the signal (Figure 7C). We conclude that the cytosolic proteins Rpl24a and Egd1, engage with Mia40 and are transferred to the IMS due to the addition of only the nine amino acids MISS signal at their amino termini.
Figure 7.
Chimeras of the signal for Mia40 fused to cytosolic proteins are imported into mitochondria. (A) Cytosolic proteins Rpl24a and Egd1 as well as fusion proteins SignalRpl24a and SignalEgd1 were synthesized as 35S-labeled precursors. After import into isolated wild-type mitochondria and proteinase K treatment, nonreducing SDS electrophoresis analysis was performed. (B) Egd1 and SignalEgd1 were imported into wild-type and mia40-3 or Mia40core mitochondria. Samples were subjected to nonreducing SDS electrophoresis. (C) Egd1 and SignalEgd1 were imported into wild-type and Mia40His mitochondria, solubilized, and subjected to Ni-NTA chromatography. Nonreducing SDS electrophoresis was used for the analysis of load (5%) and elution (100%) fractions.
To directly demonstrate that MISS can facilitate import of proteins into the IMS in vivo, a fusion protein SignalEgd1 with a C-terminal FLAG tag was produced in yeast in large amounts using an inducible GAL10 promoter (Figure 8A). On fractionation, a significant amount of SignalEgd1-FLAG was found in mitochondria, in contrast to Egd1-FLAG (Figure 8B). Thus, MISS is capable of directing a nonnative cytosolic protein to the mitochondrial IMS in the cell.
Figure 8.
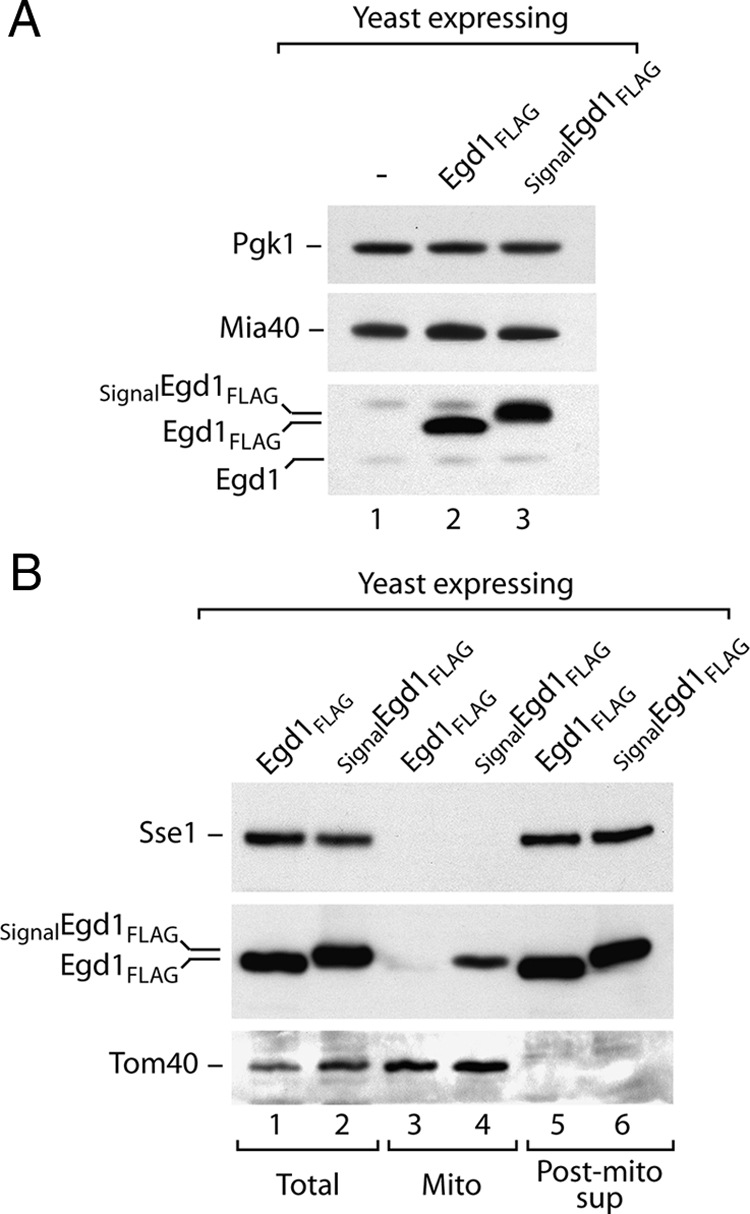
SignalEgd1 is directed to mitochondria in vivo. (A) Total proteins from control strain transformed with empty plasmid, and strains overproducing Egd1-FLAG and SignalEgd1-FLAG were analyzed by immunodecoration with anti-Egd1 antibody and antibodies specific to Mia40 and 3-phosphoglycerate kinase Pgk1. (B) Equal volumes of the fractions after subcellular fractionation were analyzed by immunodecoration with anti-FLAG and control antibodies against cytosolic chaperone Sse1 and mitochondrial Tom40.
DISCUSSION
During recent years, several new components and pathways involved in the recognition, translocation, and sorting of mitochondrial precursor proteins have been identified. However, the signals within mitochondrial precursors, which are recognized by the import components, are not fully understood. The most recently discovered is the MIA pathway, which uses the transfer of disulfide bonds for the import and maturation of proteins targeted to the mitochondrial IMS (reviewed in Tokatlidis, 2005; Herrmann and Köhl, 2007; Hell, 2008; Koehler and Tienson, 2008; Stojanovski et al., 2008b). A crucial role in dictating specificity for protein import into the IMS was assigned to Mia40, one of the two essential components of the MIA machinery (Milenkovic et al., 2007a; Sideris and Tokatlidis, 2007). However, the molecular basis of precursor recognition by Mia40 has remained elusive. In this study, we addressed the selective association of Mia40 with precursor proteins. Our results revealed the existence of MISS signal that is essential for binding of precursors to Mia40.
The systematic import analysis of the MIA substrate Tim9 and its mutated variants allowed us to specify the MISS signal to only nine amino acids. Using peptide competition assays, we found that the synthetic MISS signal requires the presence of a cysteine residue to efficiently interact and saturate Mia40 and thereby block import of other substrates. Thus, MISS–Mia40 intermediate is covalently linked via a disulfide bridge, strengthening previous findings on the important role of the intermolecular disulfides between Mia40 and a precursor (Chacinska et al., 2004; Mesecke et al., 2005; Grumbt et al., 2007; Milenkovic et al., 2007a; Sideris and Tokatlidis, 2007). The mutational analysis uncovered an additional important determinant of the signal, a leucine residue upstream of the cysteine residue. This hydrophobic residue in the MISS signal was essential for efficient disulfide formation between Tim9 and Mia40. Even in a simplified version, when Leu31 remained surrounded by only two native amino acid residues, MISS was still functional in directing an artificial substrate to Mia40 supporting a prevailing role for this residue. However, the efficiency of binding between such a substrate and Mia40 was compromised. In contrast, single mutations of other residues to alanine did not affect recognition by Mia40. Altogether, in addition to the essential Leu and Cys residues, other amino acid residues may also contribute to recognition by Mia40 in a cooperative manner.
We propose the existence of a general mechanism for mitochondrial intermembrane space protein import that is based on the specific recognition and binding of the MISS signal present within IMS proteins by Mia40. We demonstrate that the import of six native MIA substrates is efficiently inhibited by the synthetic MISS peptide. This finding is consistent with a common binding site at Mia40 used by various precursor proteins with MISS signals. Furthermore, the MISS signal functions remarkably well in directing nonnative protein substrates to Mia40. The chimeras of cytosolic proteins fused to the MISS signal were translocated into the IMS of mitochondria. Thus, the MISS signal contains the complete information to drive foreign proteins across the mitochondrial outer membrane through to their engagement with Mia40 in the IMS.
MISS resembles a classical mitochondrial targeting signal, a cleavable and positively charged presequence, with respect to its role in the early stages of protein translocation (Schatz and Dobberstein, 1996; Endo et al., 2003; Oka and Mihara, 2005; Neupert and Herrmann, 2007; Bolender et al., 2008). A presequence is sensed by several receptor and translocation components forming an interaction chain of increasing binding affinities on the way through the TOM complex (Milenkovic et al., 2007b). We hypothesize that the MISS signal, similarly to the presequence, may play multiple functions in the stages of mitochondrial targeting and outer membrane translocation, in addition to precursor sorting on the trans-side of the outer membrane conferred by interaction with Mia40.
In summary, we identified a novel signal involved in the sorting of mitochondrial IMS proteins. Precursor proteins, which use the MIA pathway for their import and biogenesis, possess the MISS signal that is required for the recognition and binding by the Mia40 receptor in the IMS. A transient intermolecular disulfide bond and an additional hydrophobic interaction mediate the signal-driven association of precursors with Mia40. Importantly, the MISS signal is both necessary and sufficient for the transport of proteins into the IMS.
ACKNOWLEDGMENTS
We thank Drs. Nikolaus Pfanner, Bernard Guiard, Nils Wiedemann, Stephan Kutik, and Tomasz Wegierski for discussion. This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 746 and Excellence Initiative of the German Federal & State Governments (EXC 294). D. S. was recipient of an Alexander von Humboldt research fellowship.
Abbreviations used:
- IMS
intermembrane space
- MIA
mitochondrial intermembrane space import and assembly
- MISS
mitochondrial intermembrane space sorting
- PAGE
polyacrylamide gel electrophoresis
- TIM
translocase of inner membrane
- TOM
translocase of outer membrane.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-11-1108) on March 18, 2009.
REFERENCES
- Allen S., Balabanidou V., Sideris D. P., Lisowsky T., Tokatlidis K. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J. Mol. Biol. 2005;353:937–944. doi: 10.1016/j.jmb.2005.08.049. [DOI] [PubMed] [Google Scholar]
- Bihlmaier K., Mesecke N., Terziyska N., Bien M., Hell K., Herrmann J. M. The disulfide relay system of mitochondria is connected to the respiratory chain. J. Cell Biol. 2007;179:389–395. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;9:42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., Sanjuán Szklarz L. K., Schulze-Specking A., Truscott K. N., Guiard B., Meisinger C., Pfanner N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A., Guiard B., Müller J. M., Schulze-Specking A., Gabriel K., Kutik S., Pfanner N. Mitochondrial biogenesis, switching the sorting pathway of the intermembrane space receptor Mia40. J. Biol. Chem. 2008;283:29723–29729. doi: 10.1074/jbc.M805356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran S. P., Leuenberger D., Oppliger W., Koehler C. M. The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J. 2002;21:942–953. doi: 10.1093/emboj/21.5.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran S. P., Leuenberger D., Leverich E. P., Hwang D. K., Beverly K. N., Koehler C. M. The role of Hot13p and redox chemistry in the mitochondrial TIM22 import pathway. J. Biol. Chem. 2004;279:43744–43751. doi: 10.1074/jbc.M404878200. [DOI] [PubMed] [Google Scholar]
- Dabir D. V., Leverich E. P., Kim S. K., Tsai F. D., Hirasawa M., Knaff D. B., Koehler C. M. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 2007;26:4801–4811. doi: 10.1038/sj.emboj.7601909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal P., Likic V., Tachezy J., Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Dresios J., Derkatch I. L., Liebman S. W., Synetos D. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry. 2000;39:7236–7244. doi: 10.1021/bi9925266. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., von Heijne G. Prediction of organellar targeting signals. Biochim. Biophys. Acta. 2001;1541:114–119. doi: 10.1016/s0167-4889(01)00145-8. [DOI] [PubMed] [Google Scholar]
- Endo T., Yamamoto H., Esaki M. Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J. Cell Sci. 2003;116:3259–3267. doi: 10.1242/jcs.00667. [DOI] [PubMed] [Google Scholar]
- Fünfschilling U., Rospert S. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell. 1999;10:3289–3299. doi: 10.1091/mbc.10.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel K., Milenkovic D., Chacinska A., Müller J., Guiard B., Pfanner N., Meisinger C. Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway. J. Mol. Biol. 2007;365:612–620. doi: 10.1016/j.jmb.2006.10.038. [DOI] [PubMed] [Google Scholar]
- Gebert N., Chacinska A., Wagner K., Guiard B., Koehler C. M., Rehling P., Pfanner N., Wiedemann N. Assembly of the three small Tim proteins precedes docking to the mitochondrial carrier translocase. EMBO Rep. 2008;9:548–554. doi: 10.1038/embor.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R., Beddoe T., Landl K., Lithgow T. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc. Natl. Acad. Sci. USA. 1998;95:2296–2301. doi: 10.1073/pnas.95.5.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumbt B., Stroobant V., Terziyska N., Israel L., Hell K. Functional characterization of Mia40p, the central component of the disulfide relay system of the mitochondrial intermembrane space. J. Biol. Chem. 2007;282:37461–37470. doi: 10.1074/jbc.M707439200. [DOI] [PubMed] [Google Scholar]
- Hell K. The Erv1-Mia40 disulfide relay system in the intermembrane space of mitochondria. Biochim. Biophys. Acta. 2008;1783:601–609. doi: 10.1016/j.bbamcr.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Herrmann J. M., Köhl R. Catch me if you can! Oxidative protein trapping in the intermembrane space of mitochondria. J. Cell Biol. 2007;176:559–563. doi: 10.1083/jcb.200611060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. E., Johnson A. E. Opening the door to mitochondrial protein import. Nat. Struct. Biol. 2001;8:1008–1010. doi: 10.1038/nsb1201-1008. [DOI] [PubMed] [Google Scholar]
- Koehler C. M. The small Tim proteins and the twin Cx3C motif. Trends Biochem. Sci. 2004;29:1–4. doi: 10.1016/j.tibs.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Koehler C. M., Tienson H. L. Redox regulation of protein folding in the mitochondrial intermembrane space. Biochim. Biophys. Acta. 2008;1793:139–145. doi: 10.1016/j.bbamcr.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutik S., et al. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- Lionaki E., de Marcos Lousa C., Baud C., Vougioukalaki M., Panayotou G., Tokatlidis K. The essential function of Tim12 in vivo is ensured by the assembly interactions of its C-terminal domain. J. Biol. Chem. 2008;283:15747–15753. doi: 10.1074/jbc.M800350200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Allen S., Wardleworth L., Savory P., Tokatlidis K. Functional TIM10 chaperone assembly is redox-regulated in vivo. J. Biol. Chem. 2004;279:18952–18958. doi: 10.1074/jbc.M313045200. [DOI] [PubMed] [Google Scholar]
- Meisinger C., Pfanner N., Truscott K. N. Isolation of yeast mitochondria. Methods Mol. Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J. M. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Mesecke N., Bihlmaier K., Grumbt B., Longen S., Terziyska N., Hell K., Herrmann J. M. The zinc-binding protein Hot13 promotes oxidation of the mitochondrial import receptor Mia40. EMBO Rep. 2008;9:1107–1113. doi: 10.1038/embor.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic D., Gabriel K., Guiard B., Schulze-Specking A., Pfanner N., Chacinska A. Biogenesis of the essential Tim9-Tim10 chaperone complex of mitochondria: site-specific recognition of cysteine residues by the intermembrane space receptor Mia40. J. Biol. Chem. 2007a;282:22472–22480. doi: 10.1074/jbc.M703294200. [DOI] [PubMed] [Google Scholar]
- Milenkovic D., Müller J., Stojanovski D., Pfanner N., Chacinska A. Diverse mechanisms and machineries for import of mitochondrial proteins. Biol. Chem. 2007b;388:891–897. doi: 10.1515/BC.2007.097. [DOI] [PubMed] [Google Scholar]
- Müller J. M., Milenkovic D., Guiard B., Pfanner N., Chacinska A. Precursor oxidation by Mia40 and Erv1 promotes vectorial transport of proteins into the mitochondrial intermembrane space. Mol. Biol. Cell. 2008;19:226–236. doi: 10.1091/mbc.E07-08-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoé M., Ohwa Y., Ishikawa D., Ohshima C., Nishikawa S., Yamamoto H., Endo T. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J. Biol. Chem. 2004;279:47815–47821. doi: 10.1074/jbc.M410272200. [DOI] [PubMed] [Google Scholar]
- Neupert W., Herrmann J. M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Oka T., Mihara K. A railroad switch in mitochondrial protein import. Mol. Cell. 2005;18:145–146. doi: 10.1016/j.molcel.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Rehling P., Brandner K., Pfanner N. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- Rissler M., Wiedemann N., Pfannschmidt S., Gabriel K., Guiard B., Pfanner N., Chacinska A. The essential mitochondrial protein Erv1 cooperates with Mia40 in biogenesis of intermembrane space proteins. J. Mol. Biol. 2005;353:485–492. doi: 10.1016/j.jmb.2005.08.051. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Schatz G., Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Sideris D. P., Tokatlidis K. Oxidative folding of small Tims is mediated by site-specific docking onto Mia40 in the mitochondrial intermembrane space. Mol. Microbiol. 2007;65:1360–1373. doi: 10.1111/j.1365-2958.2007.05880.x. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A. System of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D., Milenkovic D., Müller J. M., Gabriel K., Schulze-Specking A., Baker M. J., Ryan M. T., Guiard B., Pfanner N., Chacinska A. Mitochondrial protein import: precursor oxidation in a ternary complex with disulfide carrier and sulfhydryl oxidase. J. Cell Biol. 2008a;183:195–202. doi: 10.1083/jcb.200804095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D., Müller J. M., Milenkovic D., Guiard B., Pfanner N., Chacinska A. The MIA system for protein import into the mitochondrial intermembrane space. Biochim. Biophys. Acta. 2008b;1783:610–617. doi: 10.1016/j.bbamcr.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Terziyska N., Lutz T., Kozany C., Mokranjac D., Mesecke N., Neupert W., Herrmann J. M., Hell K. Mia40, a novel factor for protein import into the intermembrane space of mitochondria is able to bind metal ions. FEBS Lett. 2005;579:179–184. doi: 10.1016/j.febslet.2004.11.072. [DOI] [PubMed] [Google Scholar]
- Terziyska N., Grumbt B., Kozany C., Hell K. Structural and functional roles of the conserved cysteine residues of the redox-regulated import receptor Mia40 in the intermembrane space of mitochondria. J. Biol. Chem. 2009;284:1353–1363. doi: 10.1074/jbc.M805035200. [DOI] [PubMed] [Google Scholar]
- Tokatlidis K. A disulfide relay system in mitochondria. Cell. 2005;121:965–967. doi: 10.1016/j.cell.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Walther D. M., Rapaport D. Biogenesis of mitochondrial outer membrane proteins. Biochim. Biophys. Acta. 2008;1793:42–51. doi: 10.1016/j.bbamcr.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Webb C. T., Gorman M. A., Lazarou M., Ryan M. T., Gulbis J. M. Crystal structure of the mitochondrial chaperone TIM9-10 reveals a six-bladed α-propeller. Mol. Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]