Abstract
Cilia and flagella play multiple essential roles in animal development and cell physiology. Defective cilium assembly or motility represents the etiological basis for a growing number of human diseases. Therefore, how cilia and flagella assemble and the processes that drive motility are essential for understanding these diseases. Here we show that Drosophila Bld10, the ortholog of Chlamydomonas reinhardtii Bld10p and human Cep135, is a ubiquitous centriolar protein that also localizes to the spermatid basal body. Mutants that lack Bld10 assemble centrioles and form functional centrosomes, but centrioles and spermatid basal bodies are short in length. bld10 mutant flies are viable but male sterile, producing immotile sperm whose axonemes are deficient in the central pair of microtubules. These results show that Drosophila Bld10 is required for centriole and axoneme assembly to confer cilium motility.
INTRODUCTION
Centrioles lie at the core of centrosomes. They consist of a ninefold symmetrical array of nine triplet microtubules arranged in a cylinder. In most differentiated cell types, centrioles transform into basal bodies, membrane-embedded centrioles that template cilium and flagellum axoneme assembly. The requirement of cilia and flagella for many developmental and physiological processes, together with the growing list of human diseases that result from defects in basal bodies and cilia (Badano et al., 2006; Bisgrove and Yost, 2006; Bettencourt-Dias and Glover, 2007; Fliegauf et al., 2007; Marshall, 2008), drives the need to understand the basic processes involved in centriole, basal body, cilium assembly, and motility.
In Drosophila several approaches have identified evolutionarily conserved centriole proteins required for centriole biogenesis including Sak, Sas4, Sas6, Ana1, Ana2, and Asterless (Bettencourt-Dias et al., 2005; Basto et al., 2006; Goshima et al., 2007; Rodrigues-Martins et al., 2007; Blachon et al., 2008). In flies, mutations in these genes abolish centrosome and cilium/flagellum assembly (except mutations in ana1 and ana2, which have not been described yet). Additional centriole/basal body proteins including Spd-2, Drosophila pericentrin-like protein (D-PLP)/CP309, and uncoordinated (UNC) function in pericentriolar material (PCM) recruitment to centrosomes and/or the assembly of cilia and flagella (Baker et al., 2004; Kawaguchi and Zheng, 2004; Martinez-Campos et al., 2004; Dix and Raff, 2007; Giansanti et al., 2008). Identification of the complete set of centriole components is necessary to define their individual and cooperative roles in the assembly and function of centrioles and cilia.
In Chlamydomonas reinhardtii, mutations in the bld10 gene result in complete loss of flagella, giving the cells a “bald” appearance, due to a failure to assemble centrioles (Matsuura et al., 2004; Hiraki et al., 2007). Bld10p functions in the formation of the cartwheel, a ninefold symmetrical scaffold structure essential for an early step of centriole/basal body assembly (Hiraki et al., 2007). From RNA interference (RNAi) studies in cell culture, the human ortholog of Bld10p, Cep135, was found to be required for PCM integrity (Ohta et al., 2002; Uetake et al., 2004), for cohesion of centrosomes through recruitment of the centriolar C-NAP1 protein (Kim et al., 2008), and for assembly of excess procentrioles in Plk4-overexpressing cells (Kleylein-Sohn et al., 2007). With these fundamental roles for Bld10 orthologs in centriole biogenesis in Chlamydomonas and in mammalian cell culture, we sought to investigate the function of Bld10 in an animal model.
In Drosophila, the ortholog of Chlamydomonas Bld10p and human Cep135 is encoded by the uncharacterized CG17081 gene, which we hereafter refer to as bld10. Here we show that Bld10 is a ubiquitous centriolar protein that also localizes to spermatid basal bodies. We have characterized two loss-of-function bld10 mutants and show that centrioles and basal bodies are shorter compared with the wild-type, suggesting a defect in the assembly of these organelles. bld10 mutant flies are viable but male sterile, producing immotile sperm whose axonemes are deficient in the central pair of microtubules. Therefore, Bld10 functions in centriole and sperm basal body assembly, being essential for axoneme central pair assembly and flagellum motility.
MATERIALS AND METHODS
Fly Strains and Genetics
The PBac(PB)c04199 (bld10c04199) and PBac(WH)f01951 (bld10f01951) stocks were obtained from the Exelixis Drosophila Stock Collection at Harvard Medical School, and the Df(3L)Brd15, Mi(ET1)MB04996 (bld10MB04996), and P(EPgy2)EY05589 (bld10EY05589) stocks from the Bloomington Drosophila Stock Center. Piggybac transposon mobilization and reversion of bld10c04199 was accomplished with Piggybac Transposase [CyO, P(Tub-PBac™)2, from Bloomington] and selection for loss of the w+ marker on the transposon. The lethal mutation on the original bld10c04199 chromosome was removed by meiotic recombination with a wild-type third chromosome followed by selection for the w+-marked bld10c04199 allele. A “clean” stock was obtained that was homozygous viable but male sterile.
Plasmids and Rescue
To construct the pUASp-bld10-GFP plasmid, full-length bld10-coding sequence was amplified by PCR from the LD35990 clone (Berkeley Drosophila Genome Project), cloned into pENTR/D-TOPO (Invitrogen, Carlsbad, CA), and then recombined into pPWG (Terence Murphy, The Drosophila Gateway Vector Collection, Carnegie Institution of Washington, Baltimore, MD). The plasmid was injected into w1118 embryos by BestGene. Rescue of bld10 mutants was performed by driving Bld10-GFP in testes with nos-GAL4VP16.
Production of Antibodies and Western Blotting
Two DNA sequences encoding amino acids 1-255 and 1247-1059 of Bld10 were cloned into the pET100/DTOPO vector (Invitrogen) for expression of 6XHis-tagged Bld10 protein fragments in Escherichia coli BL21(DE3)pLysE. The two 6XHis-tagged proteins were then purified by Ni2+-Immobilized Metal Affinity Chromatography and used to immunize rabbits (Cocalico Biologicals, Reamstown, PA). The rabbit sera were affinity-purified against the 6XHis-tagged proteins coupled to Affigel-10 (Bio-Rad, Hercules, CA). For Western blotting, affinity-purified rabbit anti-Bld10 (UT530) was diluted 1:10,000 and mouse anti-α-tubulin DM1A (Sigma, St. Louis, MO) was used at 1:15,000.
Immunostaining
For immunostaining, brains from third instar larvae were dissected, fixed, and stained according to Bonaccorsi et al. (2000), embryos according to Megraw et al. (1999), testes according to Li et al. (1998), and Kc cells according to Kao and Megraw (2004). Slides were incubated overnight at 4°C with the following primary antibodies diluted in PBS: mouse anti-γ-tubulin clone GTU88 (Sigma; 1:1000), mouse anti-α-tubulin antibody (clone DM1A, Sigma, 1:1000), rabbit affinity-purified anti-Bld10 UT530 or UT530 antibodies (1:1000), anti-Cnn (1:1000), rabbit anti-SPD-2 (1:1000; Giansanti et al., 2008); mouse anti-green fluorescent protein (GFP; Invitrogen, Carlsbad, CA, 1:250). Secondary antibody conjugates to Alexa 488 or 546 (Invitrogen) were used at 1:400 dilution. DNA was stained with DRAQ5 (Axxora, San Diego, CA) at 1:400 dilution. Images were captured on a Leica TCS SP2 confocal microscope (Deerfield, IL) using a 63×/NA1.4 oil immersion objective. Time-lapse live imaging of Drosophila embryos was performed at room temp (23–24°C), and frames were captured every 10 s. Movies were compressed using ImageJ software (NIH; http://rsb.info.nih.gov/ij/) at 15 frames per second.
Electron Microscopy
Testes were dissected in PBS, fixed in 2.5% glutaraldehyde for 1 h, and postfixed with 1% osmium tetroxide. Tissues were then incubated in 1% aqueous uranyl acetate, dehydrated in a graded ethanol series, and embedded in Embed-812 (Electron Microscopy Sciences, Fort Washington, PA; 14120). For immunogold labeling, testes were fixed in 4% paraformaldehyde for 1 h, dehydrated in a graded ethanol series, and embedded in Full LR White resin (Electron Microscopy Sciences; 14381). Affinity-purified Bld10 antibody UT530 was applied at a 1:50 dilution for 3 h, and goat anti-rabbit/12-nm gold (Jackson ImmunoResearch Laboratories, West Grove, PA) secondary antibody was applied at a 1:40 dilution for 90 min. Grids were poststained with 2.5% glutaraldehyde followed by an incubation in 2% aqueous uranyl acetate.
Statistical Methods
Statistical analysis was done with Prism4 software (GraphPad, San Diego, CA). Because the data met the assumptions of the t tests, we used unpaired Student's t tests (two-tailed, with samples that do not have equal variances) to assess differences between the lengths of centrioles and basal bodies in the wild-type and bld10 mutant groups.
RESULTS
Drosophila Bld10 Is the Ortholog of Chlamydomonas Bld10p and Human Cep135
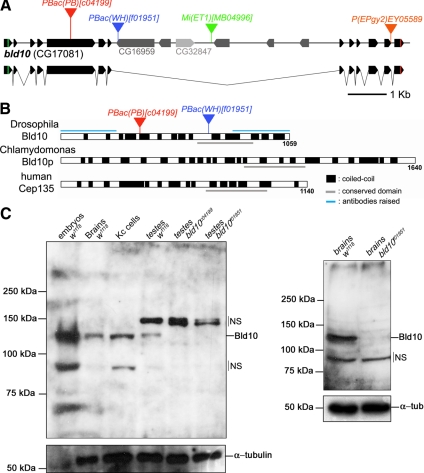
BLAST queries of the Drosophila genome with Chlamydomonas Bld10p and human Cep135 sequences revealed only one ortholog gene in Drosophila, the uncharacterized CG17081 gene. CG17081 encodes a protein of 1059 amino acids (Figure 1, A and B) with significant similarity to Chlamydomonas Bld10p and to human Cep135 (Table 1 and Supplemental Figure S1). We refer to CG17081 hereafter as bld10. Like its orthologs, Bld10 is predicted to contain many coiled-coils (Figure 1B) and contains a highly conserved domain that in Chlamydomonas was shown to be required for Bld10p function in vivo (Hiraki et al., 2007; Supplemental Figure S1).
Figure 1.
Drosophila bld10 encodes a coiled-coil protein with homology to Chlamydomonas Bld10p and human Cep135. (A) A schematic representation of the bld10 locus showing the sites of four transposon insertions and the two enclaved genes, CG16959 and CG32847. The start and stop codons are indicated by green and red lines, respectively. (B) Schematic diagram of Drosophila Bld10, C. reinhardtii Bld10p, and human Cep135. Regions predicted to have a high percentage (>50%) of forming coiled-coils are shown in black (nCOILS program, window 21, matrix MTIDK; Lupas et al., 1991). A conserved domain among these three proteins is underlined in gray. Polypeptide regions used to raise polyclonal antibodies against Bld10 are indicated with a blue line. (C) Western blots of extracts from wild-type embryos, third instar larval brains, Kc cells, and adult testes and from bld10 mutant testes and brains. The anti-Bld10 antibody (C-terminus) recognized a specific 123-kDa protein, corresponding to the expected molecular weight of Bld10. This band was absent in extracts of hemizygous bld10c04199 and homozygous bld10f01951 adults testes. In adult testis extracts, the antibody also recognized apparent nonspecific (NS) bands around 150 kDa and, in embryos, third instar larval brains and Kc cells a band around 80 kDa. α-Tubulin was probed as a loading control. All samples were prepared from specimens acquired from a cross between heterozygous males and females (heterozygous maternal contribution was intact).
Table 1.
Sequence similarities among Bld10 orthologs
| Chlamydomonas Bld10p | Human Cep135 | |
|---|---|---|
| Full-length Drosophila Bld10 | 15% identical | 22% identical |
| 28% similar | 43% similar | |
| Bld10 conserved domain | 19% identical | 24% identical |
| 41% similar | 48% similar |
Protein sequences were aligned using the ClustalW program in MacVector 8.0 using the default parameters. The conserved domain corresponds to the regions boxed in green in Supplementary Figure S1.
To investigate the function of Bld10 in Drosophila, we examined four uncharacterized transposon insertion mutations that mapped to the bld10 locus: bld10c04199, bld10f01951, bld10MB04996, and bld10EY05589 (Figure 1A). The bld10c04199 allele contains a PiggyBac element inserted within the fifth exon of bld10, predicted to produce a truncation product of 369 amino acids. The PiggyBac insertion associated with bld10f01951 lies within the seventh intron of bld10 and at the 3′ end of the enclaved CG16959 gene. The bld10MB04996 and bld10EY05589 alleles are Minos and P element insertions within the seventh and twelve introns of bld10, respectively. Only the bld10c04199 and bld10f01951 alleles presented an obvious phenotype (see below).
To examine the expression of Bld10, we raised antibodies against the N- and C-terminal regions of the protein. On Western blots, the antibody against the C-terminal region recognized a band of the predicted molecular weight, 123 kDa, from wild-type embryos, larval brains, Kc cells, and adult testis extracts (Figure 1C).
Bld10 Is a Centriolar and Sperm Basal Body Protein
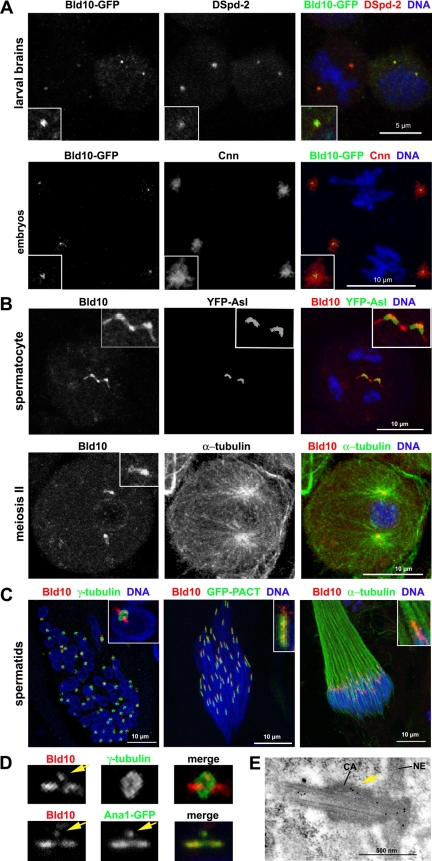
Affinity-purified antibodies directed against either the C- or N-terminal regions of Bld10 immunostained the centrosomes of all tissues examined. In addition, we constructed transgenic flies that express a Bld10-GFP fusion protein, which also localized to centrosomes. Bld10 colocalized with the centriole marker Spd-2 in interphase neuroblasts (Figure 2A) and to small dots at the center of the PCM in mitotic neuroblasts, embryos, and Kc cells (Figure 2A; Supplemental Figure S2 and Movie S1). No significant increase of Bld10 signal was observed at the onset of mitosis, indicating that Bld10 is primarily associated with centrioles rather than with the PCM. Moreover, Bld10 remained localized after depolymerization of the microtubules with colcemid in Kc cells, showing that Bld10 is a core component of the centrosomes (Supplemental Figure S2B). These data indicate that Bld10 is likely a ubiquitous component of centrioles throughout development. Two recent reports corroborated the centriole localization of Bld10 (Blachon et al., 2008; Dobbelaere et al., 2008).
Figure 2.
Bld10 is a centriolar and sperm basal body protein. (A) Bld10-GFP colocalized with DSpd-2 in interphase larval neuroblasts (top panels) and localized to small dots in the middle of the PCM, labeled with Cnn staining, in early embryos (bottom panels). (B) Bld10 is a centriolar protein in spermatocytes. Top panels, wild-type primary spermatocyte expressing YFP-Asl immunostained for Bld10. Bottom panels, wild-type primary spermatocyte in meiosis II immunostained for α-tubulin (green) and Bld10 (red). Note that Bld10 is enriched at one end of the centrioles during meiotic division. (C) Bld10 localized to sperm basal bodies. Cysts of wild-type elongating spermatids stained for Bld10 and γ-tubulin (left panel), for Bld10 and GFP in GFP-PACT spermatids (middle panel), and for Bld10 and α-tubulin (right panel). These three panels represent a chronological series from early to late spermatid development from left to right. (D) Localization of Bld10 and Ana1 to the basal body and to a dot (yellow arrow) within the centriolar adjunct. Centrioles from early elongating spermatids stained for Bld10 and γ-tubulin (top panels) and for Bld10 and GFP in Ana1-GFP expressing flies (bottom panels). (E) Basal body localization of Bld10 by immunoelectron microscopy. Bld10 localized within the lumen of the basal body and was concentrated at the distal end near the site of central pair assembly and to the proximal end at the junction between the basal body and nuclear envelope (NE). Bld10 is also localized to the centriolar adjunct (CA, yellow arrow).
In Drosophila spermatocytes, where the centrioles are ∼10-fold longer than in somatic cells (Gonzalez et al., 1998), the centriole-specific marker YFP-Asl (Varmark et al., 2007) colocalized with Bld10 at centrioles throughout spermatocyte development. In mature spermatocytes and throughout the two meiotic divisions, Bld10 was concentrated at the proximal end of the centrioles, a subregion of the centriole where YFP-Asl and Bld10 localization overlapped (Figure 2B).
In spermatid stages, the centriole transforms into a basal body and templates assembly of the microtubule-based flagellum axoneme. We counterstained spermatids with GFP-pericentrin-AKAP450 centrosomal targeting (PACT; a centriole-targeting domain found in D-PLP; Gillingham and Munro, 2000; Martinez-Campos et al., 2004) to label basal bodies and found that Bld10 localized to the rod-shaped basal body, which is surrounded by the centriolar adjunct (Tates, 1971), a collar-like PCM structure that contains γ-tubulin (Figure 2, C and D; Sunkel et al., 1995; Wilson et al., 1997). Bld10 was concentrated at the proximal and distal ends of the basal body, with lower expression at the region encircled by the centriolar adjunct (Figure 2, C and D). In addition, Bld10 consistently localized to a small discrete dot within the centriolar adjunct of elongating spermatids (Figure 2D). This singular signal was observed with both anti-Bld10 antibodies and the Bld10-GFP fusion protein (not shown). We examined the colocalization of Bld10 with other centriolar proteins at early spermatids including GFP-PACT, D-PLP, YFP-Asl, Sas6-GFP, UNC-GFP, and Ana1-GFP. Expression of GFP-PACT, D-PLP, and Sas6-GFP were restricted to the basal body (Sas6-GFP was concentrated at the distal tip), whereas YFP-Asl and UNC-GFP localized throughout the centriolar adjunct at this stage. Only Ana1-GFP replicated the localization pattern of Bld10 to the basal body and to the dot within the centriolar adjunct, where Ana1-GFP and Bld10 were colocalized (Figure 2D). Bld10 persists at the basal body late into spermiogenesis, but is no longer detected after the onset of individualization (Supplemental Figure S3).
To localize Bld10 to the basal body at the ultrastructural level, we used immunoelectron microscopy using anti-Bld10 antibodies. High-resolution imaging showed that Bld10 localized within the lumen of the basal body and was enriched at its distal and proximal ends and also to a region within the centriolar adjunct (Figure 2E), a pattern consistent with immunofluorescence staining (Figure 2D).
Bld10 Is Not Required for Centriole/Centrosome Assembly or for Mechanosensory Neuron Function
Mutations in genes encoding centriolar and basal body proteins often lead to defects in the biogenesis of centrioles and in the assembly of functional centrosomes at mitosis, loss of locomotion due to impaired basal bodies of ciliated neurons, and immotile sperm due to axoneme dysfunction (Baker et al., 2004; Martinez-Campos et al., 2004; Bettencourt-Dias et al., 2005; Basto et al., 2006; Rodrigues-Martins et al., 2007; Varmark et al., 2007; Blachon et al., 2008; Giansanti et al., 2008).
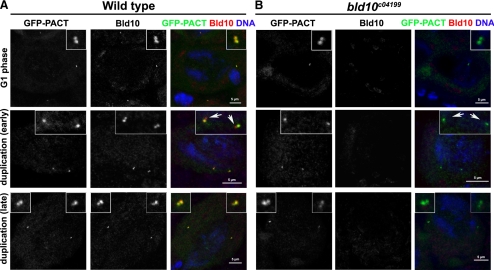
To determine whether bld10 mutations affect centrosome duplication, we examined adult testes from homozygous bld10c04199 males that were derived from a cross with homozygous bld10c04199 females and therefore lack any maternal or zygotic Bld10 and that express GFP-PACT as a centriolar marker. Because the bld10c04199 allele can conceptually produce a small product, which our N-terminal antibody should detect, we examined procentriole assembly in bld10c04199 testes to test the possibility that a mutant product may participate in early stages of procentriole biogenesis and be detected by immunofluorescent imaging. Wild-type spermatocytes in G1 phase display one centrosome that contains two centrioles, each marked by GFP-PACT and Bld10 (Figure 3A, top panels). Bld10 was present on nascent procentrioles at an early stage of centriole duplication before the incorporation of GFP-PACT (Figure 3A, middle panels). At a later stage of centrosome duplication, GFP-PACT was recruited to daughter centrioles where Bld10 persists (Figure 3A, bottom panels). Bld10 was not detected at bld10c04199 centrioles or basal bodies yet, despite the lack of Bld10 protein, centriole duplication appeared to proceed normally in bld10c04199 mutant spermatocytes (Figure 3B). Moreover, no signal for Bld10 could be detected at bld10c04199 nascent centrioles (Figure 3B, middle panels).
Figure 3.
Bld10 is not required for centrosome duplication. Wild-type (A) and bld10 mutant (B) early spermatocytes expressing GFP-PACT were counterstained for with antibodies raised against the N-terminal region of Bld10. For this experiment, homozygous bld10c04199 females were crossed to bld10c04199/TM6B males carrying the GFP-PACT transgene. The resulting homozygous bld10c04199 males lack maternal and zygotic Bld10 and express GFP-PACT. Testes were stained with anti-GFP and anti-Bld10 antibodies and spermatocytes in early and later stages of centriole duplication, judged by the recruitment of GFP-PACT to the daughter centrioles, were imaged. Arrows in the middle row indicate the recruitment of Bld10 to early procentrioles in A and the lack of this signal in B.
We then examined third instar bld10c04199 mutant neuroblasts at mitosis to determine whether bld10 mutations affect mitotic centrosome function. In bld10c04199 brains, lacking zygotic or zygotic plus maternal bld10, Bld10 expression was undetectable (Figure 1C), yet bld10c04199 centrosomes recruited PCM components such as γ-tubulin and Cnn (Megraw et al., 1999), produced robust microtubule asters, and assembled apparently normal mitotic spindles (Supplemental Figure S4, A and B). Therefore, mitotic centrosomes appeared normal in the absence of detectable Bld10. Moreover, the distribution of γ-tubulin at interphase centrioles was indistinguishable between wild-type and bld10c04199 brains (Supplemental Figure S4C). Taken together, these data indicate that centriole biogenesis and assembly of mitotic centrosomes were not dependent on Bld10. Furthermore, bld10 mutant adults displayed no obvious uncoordinated movement, indicating that bld10 is not essential for the function of mechanosensory neurons.
bld10 Mutant Males Are Infertile and Produce Immotile Sperm
The original chromosome harboring bld10c04199 was recessive lethal, however, when heterozygous with Df(3L)Brd15, a deficiency that deletes the entire bld10 gene, the resulting bld10c04199 hemizygous mutant flies were viable. Thus, there is at least one lethal mutation on the bld10c04199 chromosome that is not associated with the PiggyBac[c04199] insertion at bld10. This lethal mutation was removed by recombination, and the “cleaned up” chromosome containing bld10c04199 was homozygous viable and male sterile. Hemizygous bld10c04199, homozygous bld10c04199, and homozygous bld10f01951 and bld10c04199/bld10f01951 females were fertile, but males of these genotypes were completely infertile. Males from all four of these bld10 genotypes developed mature spermatozoa, yet sperm showed no motile flagella. To eliminate the possibility of a persistent maternal contribution of Bld10 to viability or adult motor activity, we crossed homozygous bld10c04199 females to bld10c04199/TM6B males and found that the resulting homozygous bld10c04199 progeny were viable and had normal locomotion, yet males were sterile.
To determine whether the PiggyBac insertion associated with bld10c04199 was responsible for the infertility phenotype, we mobilized the transposon with transposase, achieving precise excision of PBac(PB)[c04199]. This reverted chromosome was fertile when heterozygous with Df(3L)Brd15 or bld10f01951. These results show that the male infertility is associated with the transposon insertion in bld10c04199. In addition, expression of the bld10-GFP transgene in testes rescued bld10c04199 and bld10f01951 male sterility. Thus, bld10c04199 and bld10f01951 are two mutations in bld10, resulting in sterile males with immotile sperm.
To examine Bld10 expression from bld10 mutant alleles, we performed Western blot analysis on testes and brains and found that Bld10 was undetectable from bld10c04199 and bld10f01951 samples (Figure 1C). In addition, no signal was detected at bld10c04199 or bld10f01951 centrioles or basal bodies with the antibody directed against the C-terminus (see Figure 4C). Because bld10f01951 and bld10c04199 are both predicted to produce C-terminal truncations, we also examined whether the antibody directed against the N-terminus of Bld10 (see Figure 1B) could detect a mutant protein localized at the basal bodies. No signal at centrioles or basal bodies was observed in bld10c04199 mutant testes (Figures 3 and 4). However, a weak Bld10 signal was detected at bld10f01951 mutant centrioles and basal bodies (data not shown). Therefore, a low level of Bld10 is expressed from bld10f01951 and is likely a truncated protein that retains centriolar localization, but is not functional to confer motile sperm. On the other hand, bld10c04199 appears to be a strong loss of function allele, with no protein detected by Western blotting or by immunofluorescence staining using antibodies directed against either the N- or C-terminus. For all subsequent analyses we focused on hemizygous bld10c04199 mutant males derived from a cross between Df(3L)Brd15/TM6B females and bld10c04199/TM6B males.
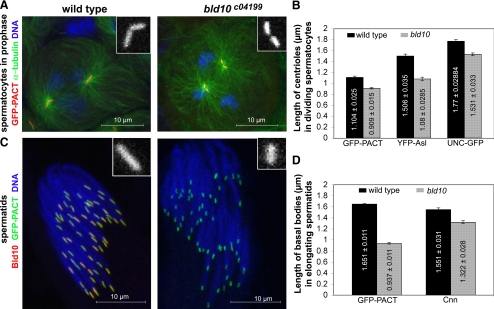
Figure 4.
bld10 mutant centrioles and basal bodies are short. Wild-type and hemizygous bld10c04199 spermatocytes (A) and elongating spermatids (C) expressing GFP-PACT were counterstained for Bld10. Note the bld10c04199 basal bodies are shorter than wild type. The GFP-PACT signal appeared slightly wider on bld10c04199 basal bodies than that of wild type, but we did not observed this with YFP-Asl or UNC-GFP. Bld10 was not detected at bld10c04199 centrioles or basal bodies. (B) Measure of centriole lengths using GFP-PACT, YFP-Asl, and UNC-GFP staining in wild-type and bld10c04199 meiosis I spermatocytes. Approximately 20 centrioles were counted with each marker in two independent experiments. (D) Measure of basal body lengths using GFP-PACT and Cnn signals in wild-type and bld10c04199 spermatids. Approximately 50 basal bodies were counted in at least three independent cysts. Errors bars, SEM. The lengths of centrioles and basal bodies are indicated in the bars in μm. The shorter length of centrioles and basal bodies was highly significant in the bld10 mutant compared with wild type with each of the markers used (unpaired Student's t tests, two-tailed, p < 0.0001).
bld10 Mutants Have Short Centrioles and Basal Bodies
To discern the cause of male sterility in bld10 mutants, we turned our attention to characterization of meiosis and sperm development in mutant testes. We examined the localization of three centriole markers to bld10c04199 centrioles: GFP-PACT, YFP-Asl, and UNC-GFP and found that all three were recruited. However, these markers revealed that bld10c04199 centrioles have a significantly shorter length (Figure 4, A and B). Because centrioles increase in length during the prolonged G2 phase of spermatocyte development, we measured centriole lengths exclusively in dividing spermatocytes (Figure 4A). bld10c04199 centrioles were 18, 28, and 14% shorter than wild-type centrioles with respect to GFP-PACT, YFP-Asl, and UNC-GFP markers (Figure 4B). Despite their altered assembly, centrioles in bld10c04199 spermatocytes recruited the centriolar and PCM proteins Asl, Spd-2, Cnn, γ-tubulin, and D-PLP and appeared to assemble astral microtubules normally (Figure 4A). Moreover, spermatids at the “onion stage,” at the end of the two meiotic divisions, displayed a single round nucleus associated with a nebenkern (mitochondrial derivative) of similar shape and size as in the wild type, indicative of normal completion of meiosis by bld10c04199 spermatocytes (data not shown).
Basal body length was also dependent on bld10 function. We found that bld10c04199 basal bodies were 44% shorter than wild-type basal bodies using GFP-PACT localization and 14% shorter by measuring Cnn localization (Figure 4, C and D). Although the localization of each centriole and basal body marker used in these analyses was impacted to different degrees, together these results indicate that Bld10 is required for the elongation of centrioles and basal bodies or the maintenance of their length.
The requirement for Bld10 in basal body assembly might account for the lack of motility found in mutant sperm. However, sperm tails assembled in the mutant, indicating that the axoneme-templating function of the basal body persists in bld10 mutant testes. This prompted us to examine the structure of the axoneme in bld10c04199 spermatids in order to discern a cause for the lack of flagellum motility.
Bld10 Is Required for the Assembly of the Axoneme Central Pair of Microtubules
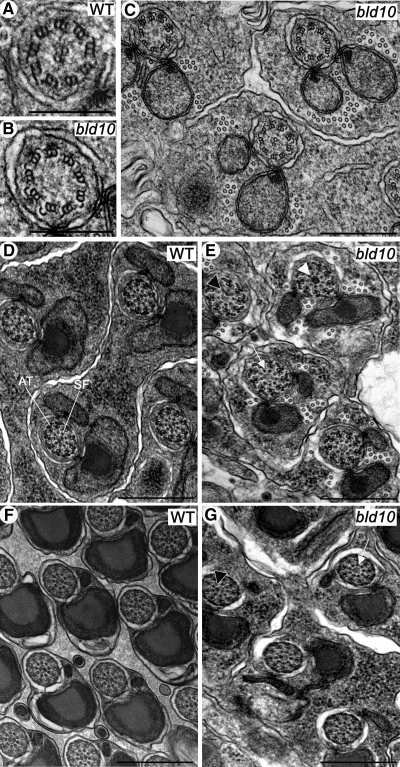
The structural components of the axoneme are highly conserved and include nine outer doublet microtubules with attached dynein arms, nine outer accessory tubules, and two central singlet microtubules (Figure 5A). Electron microscopy of testes revealed that bld10c04199 flagellar axonemes appeared normal in their ultrastructure in all respects except for the formation of the central pair of microtubules (Figure 5). Transverse sections through the tails of elongating spermatids showed that 48% (n = 143) of bld10c04199 mutant axonemes lack the two central tubules (Figure 5, B and C).
Figure 5.
Axoneme defects in bld10c04199 mutant spermatids. (A–C) Transverse sections through the tails of wild-type (A) and mutant (B and C) elongating spermatids. Wild-type axonemes (A) consist of nine microtubule doublets plus two central tubules (9 + 2). Approximately 48% of bld10c04199 axonemes lack the two central tubules. (B) High magnification of one bld10c04199 axoneme in C, showing the loss of the central pair. (D and E) At a later stage of spermatid development, the axoneme assembles nine supplementary accessory tubules (AT) and nine spokes with secondary fibrils (SF). Like the wild-type (D), bld10c04199 axonemes (E) form these structures, but some axonemes exhibit no central tubules (white arrow), some only one central tubule (black arrowhead), and some exhibit a central electron-dense material (white arrowhead). In contrast to mature wild-type spermatozoa (F), bld10c04199 axonemes (G) have one (black arrowhead) or no central tubules (white arrow). Scale bars, (A and B) 250 nm; (C–G) 500 nm.
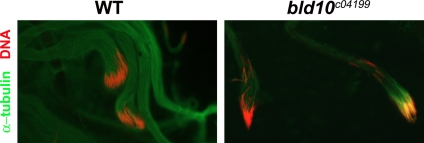
At later stages of spermatid elongation the ultrastructure of the axoneme approaches that of mature spermatozoa, with the development of nine supplementary accessory tubules and nine radial spokes with secondary fibrils (Figure 5, D and F). Like the wild type, bld10c04199 axonemes developed these structures, but 56% of axonemes assembled no central tubules, 27% formed only one central tubule, 15% had a central amorphous electron-dense mass, and only 2% had the normal complement of two central tubules (n = 84; Figure 5, E and G). Note that the association of the axoneme with the nebenkern is maintained in the mutant, but the orientations of spermatid tails within the cyst were irregular (cf. Figure 5, F and G). This is consistent with another observation that nuclei were moderately dispersed along the spermatid bundles in the bld10c04199 mutant instead of being packed at one end of the bundle as in the wild type (Figure 6). The dispersion of nuclei in bld10c04199 cysts was not due to their loss of attachment to the axoneme, as the nuclei were still connected to the axoneme by the basal bodies (Figure 4C).
Figure 6.
Nuclei are detached from bld10c04199 sperm bundles. Wild-type and hemizygous bld10c04199 elongating spermatids were stained with α-tubulin (green) and DAPI (red) for DNA.
DISCUSSION
Here we show that Bld10, the Drosophila ortholog of Chlamydomonas Bld10p and human Cep135, is a centriolar protein. Strong loss-of-function mutations in bld10 disrupt centriole and basal body assembly, producing short centrioles and basal bodies. In addition, Bld10 regulates the assembly of the axoneme central pair of microtubules, a requirement that is the likely cause of sperm immotility and infertility among bld10 mutant males.
Bld10 Is a Conserved Centriolar Protein
We found that Bld10, like its orthologs, is associated with the centrioles in all tissues examined and throughout Drosophila development. In addition, we found that Bld10 is accumulated at the proximal ends of the centrioles and resides within the lumen of spermatid basal bodies, being enriched at its distal and proximal ends. The localization of Bld10 to the lumen of the basal body is consistent with the localization of Bld10p in Chlamydomonas and of Cep135 in human cells (Ohta et al., 2002; Matsuura et al., 2004; Kleylein-Sohn et al., 2007), except that Bld10p was reported only at the proximal aspect of the lumen in Chlamydomonas (Matsuura et al., 2004), whereas Cep135 resided at both ends of the centriole (Kleylein-Sohn et al., 2007).
In addition to its localization to the sperm basal body, Bld10 resides at a distinct spot within the centriolar adjunct in elongating spermatids. The function of the centriolar adjunct is unknown, but appears to be a specialized PCM structure based on the localization of several centrosome proteins including γ-tubulin, Asl, and UNC. However, what is the significance of Bld10 accumulation at a precise single dot within the centriolar adjunct? According to Anderson (1967), the early spermatid contains only one centriole, and the formation of a second centriole occurs in late spermatids within the centriolar adjunct. However, the evidence for two versus one basal body in the mature Drosophila sperm is lacking so far (Tates, 1971). The recruitment of Bld10 orthologs at an early stage of centriole assembly led us to speculate that the Bld10 focus within the centriolar adjunct could correspond to a nascent procentriole. Indeed, the colocalization of Ana1, which is also required for centriole biogenesis (Goshima et al., 2007), within this Bld10 spot lends support for this hypothesis. Therefore, another function of the centriolar adjunct could be to promote the formation of a second centriole in the vicinity of the centriole/basal body. In support of this idea, recent studies have shown that PCM promotes new centriole assembly, perhaps by concentrating γ-tubulin (Dammermann et al., 2008; Loncarek et al., 2008).
Proper Assembly of Centrioles and Basal Bodies Require Bld10, But Not at an Early Step
The earliest centriole precursor with ninefold symmetry is the cartwheel, an intermediate in centriole biogenesis isolated in Chlamydomonas and found to require Bld10p for its assembly (Hiraki et al., 2007). Consistent with an early role for Bld10p in centriole assembly, the human homolog, Cep135, was shown to be required for ectopic procentriole assembly in Plk4-overexpressing cells (Kleylein-Sohn et al., 2007). In Drosophila and mammals, an equivalent to the cartwheel intermediate has not been identified, yet the cartwheel structure is found in the centrioles of Drosophila, mammals, and many other organisms (Gonzalez et al., 1998; Preble et al., 2000). In Tetrahymena basal bodies, Sas6a was identified as a component of the cartwheel (Kilburn et al., 2007) and in Chlamydomonas, Sas-6 is a cartwheel component required to establish ninefold symmetry (Nakazawa et al., 2007). In Caenorhabditis elegans there appears to be no bld10 ortholog (Dutcher, 2007), and centriole biogenesis involves assembly of a precursor tube-like structure that requires sas-6 (Pelletier et al., 2006). Examination of centrioles from Drosophila sas-6 mutants has led to a model whereby centriole assembly involves incorporation of “enatosomes,” modules that comprise one-ninth of the centriole's rotational architecture (Rodrigues-Martins et al., 2007). These findings have led to the hypothesis that the precursor tube that Sas-6 regulates provides ninefold assembly instruction similar to the cartwheel in Chlamydomonas (Pelletier et al., 2006; Rodrigues-Martins et al., 2008). These models are not exclusive and a unified model for the establishment of the elegant ninefold rotational architecture of the centriole may eventually be reconciled. In contrast to Sas-6, which appears to have a conserved function in establishing ninefold symmetry and early steps of centriole biogenesis, our results indicate that Bld10 is not necessary for early steps of centriole assembly in Drosophila.
We show that bld10c04199 mutant flies have no severe defects in centrosome duplication. In Drosophila S2R+ cells, RNAi knockdown of Bld10 gave an intermediate centriole replication phenotype (Dobbelaere et al., 2008). This contrast with our in vivo results, likely reflects a peculiarity of S2R+ cells or possible off-target effects of RNAi. The bld10c04199 allele appears to be a strong, and perhaps null, mutation in bld10 because no protein is detected by Western blotting or by immunostaining at procentrioles or mature centrioles. Bld10 is required for centriole elongation and/or the maintenance of proper centriole length. Despite their shorter length, bld10c04199 centrioles recruit centrosomal proteins and support MTOC activity. However, despite no strict Bld10 requirement for gross centriole biogenesis in flies, its localization to nascent centrioles implicates a conserved role for Bld10 in early centriole assembly between Drosophila and its orthologs in Chlamydomonas and human.
The Role of Bld10 in the Assembly of the Central Apparatus of the Axoneme
We found that bld10c04199 spermatids are defective in the assembly of the central pair microtubules without other discernable axoneme defects. The central pair is required for the motility of 9 + 2 cilia and flagella and is proposed to transmit chemical and/or mechanical signals to the radial spokes in order to regulate flagellar waveform movement (Smith and Yang, 2004; Wirschell et al., 2007). Nonmotile cilia generally lack the central pair (9 + 0 cilia). Therefore, the requirement for Bld10 in central pair formation provides a mechanistic explanation for the impaired motility and lack of fertility in bld10 mutants.
Interestingly, the central pair is lost gradually during progression of spermatogenesis in bld10c04199 mutant. In early stages the central microtubule pair was missing in 48% of spermatids, rising to 71% in later stage spermatids that display no central pair or a central amorphous electron-dense material. An additional 27% have only one central pair in these later stages. A specific disruption of the central pair microtubules which becomes progressively more pronounced as spermatid differentiation proceeds was also reported in a mutant of the Drosophila fragile X mental retardation gene (fxr, also called fmr1; Zhang et al., 2004). fxr encodes an RNA-binding translational regulator that was proposed to regulate microtubule stability in the testes by controlling the translation of specific proteins. This suggests that in many cases the central pair is initially formed in fxr and bld10 mutants, but then is lost due to a lack of stability. Alternatively, it is possible that the prevalence of the central pair in bld10c04199 spermatids depends on its proximity to the basal body.
Unlike the nine doublet microtubules of the axoneme, whose assembly and ninefold rotational symmetry is templated by the corresponding triplet microtubules on the basal body, the central pair is not initiated by a corresponding structure at the basal body, which does not contain central microtubules (Smith and Lefebvre, 1997). Thus, the genesis of the axoneme central pair occurs by unknown mechanisms. However, bld10 joins a few other genes, in addition to fxr mentioned above, that promote assembly of the axoneme central pair of microtubules. In Drosophila, the β2-tubulin isoform is responsible for central pair assembly, requiring the carboxyl terminal polyglycylation domain of β2-tubulin (Nielsen et al., 2001; Hoyle et al., 2008). Mutations in cnn also disrupt assembly of the central pair and, like Bld10, Cnn is a highly coiled-coil protein that localizes to the centrioles in spermatocytes and to the basal body in Drosophila spermatids (Li et al., 1998). It is conceivable that these coiled-coil proteins form a scaffold, providing attachment sites for molecules required for the nucleation and/or stability of central tubules. The localization of Bld10 within the basal body lumen at the distal tip (Figure 2E) positions it to function near the site of nucleation of the central pair at the basal body/axoneme transition zone.
In summary, we show that Drosophila Bld10 is required for proper assembly of centrioles and basal bodies to achieve their normal length. Moreover, Bld10 impacts the centriole-to-basal body transformation in spermatids, being essential for the axoneme central pair assembly and therefore for flagellum motility. With its specialized role in the assembly of the central pair to confer cilium motility, it will be interesting to determine whether Bld10 is regulated to discriminate the assembly of motile 9 + 2 versus nonmotile 9 + 0 cilia to specialize these different cilium types in different tissues.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elisabetta Bucciarelli and Maurizio Gatti (Università di Roma La Sapienza, Rome, Italy) for the anti-DSpd-2 antibody, Jordan Raff (The Gurdon Institute, Cambridge, United Kingdom) for the GFP-PACT stock, Cayetano Gonzalez (Institute for Research in Biomedicine, Barcelona, Spain) for the YFP-Asl stock, Tomer Avidor-Reiss (Harvard Medical School, Boston, MA) for the Ana1-GFP stock, and Tom Januszewski (Green Center, University of Texas Southwestern Medical Center, Dallas, TX) for excellent assistance with electron microscopy. This work was supported by National Institutes of Health Grant GM068756 to T.L.M.
Abbreviations used:
- Asl
Asterless
- Cnn
centrosomin
- DSpd-2
Drosophila spindle defective 2
- D-PLP
Drosophila pericentrin-like protein
- GFP
green fluorescent protein
- PACT
pericentrin-AKAP450 centrosomal targeting
- PCM
pericentriolar material
- Plk-4
Polo-like kinase 4
- UNC
uncoordinated
- WT
wild type
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-11-1115) on March 25, 2009.
REFERENCES
- Anderson W. A. Cytodifferentiation of spermatozoa in Drosophila melanogaster: the effect of elevated temperature on spermiogenesis. Mol. Gen. Genet. 1967;99:257–273. doi: 10.1007/BF01797731. [DOI] [PubMed] [Google Scholar]
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genom. Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Baker J. D., Adhikarakunnathu S., Kernan M. J. Mechanosensory-defective, male-sterile unc mutants identify a novel basal body protein required for ciliogenesis in Drosophila. Development. 2004;131:3411–3422. doi: 10.1242/dev.01229. [DOI] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C. G., Khodjakov A., Raff J. W. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Glover D. M. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M. K., Carmo N., Balloux F., Callaini G., Glover D. M. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Bisgrove B. W., Yost H. J. The roles of cilia in developmental disorders and disease. Development. 2006;133:4131–4143. doi: 10.1242/dev.02595. [DOI] [PubMed] [Google Scholar]
- Blachon S., Gopalakrishnan J., Omori Y., Polyanovsky A., Church A., Nicastro D., Malicki J., Avidor-Reiss T. Drosophila Asterless the ortholog of vertebrate Cep152 is essential for centriole duplication. Genetics. 2008;180:2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi S., Giansanti M. G., Gatti M. Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat. Cell Biol. 2000;2:54–56. doi: 10.1038/71378. [DOI] [PubMed] [Google Scholar]
- Dammermann A., Maddox P. S., Desai A., Oegema K. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J. Cell Biol. 2008;180:771–785. doi: 10.1083/jcb.200709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix C. I., Raff J. W. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 2007;17:1759–1764. doi: 10.1016/j.cub.2007.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J., Josue F., Suijkerbuijk S., Baum B., Tapon N., Raff J. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S. K. Finding treasures in frozen cells: new centriole intermediates. Bioessays. 2007;29:630–634. doi: 10.1002/bies.20594. [DOI] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- Giansanti M. G., Bucciarelli E., Bonaccorsi S., Gatti M. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 2008;18:303–309. doi: 10.1016/j.cub.2008.01.058. [DOI] [PubMed] [Google Scholar]
- Gillingham A. K., Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1:524–529. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C., Tavosanis G., Mollinari C. Centrosomes and microtubule organisation during Drosophila development. J. Cell Sci. 1998;111(Pt 18):2697–2706. doi: 10.1242/jcs.111.18.2697. [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., Vale R. D., Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraki M., Nakazawa Y., Kamiya R., Hirono M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr. Biol. 2007;17:1778–1783. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Hoyle H. D., Turner F. R., Raff E. C. Axoneme-dependent tubulin modifications in singlet microtubules of the Drosophila sperm tail. Cell Motil. Cytoskelet. 2008;65:295–313. doi: 10.1002/cm.20261. [DOI] [PubMed] [Google Scholar]
- Kao L. R., Megraw T. L. RNAi in cultured Drosophila cells. Methods Mol. Biol. 2004;247:443–457. doi: 10.1385/1-59259-665-7:443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi S., Zheng Y. Characterization of a Drosophila centrosome protein CP309 that shares homology with Kendrin and CG-NAP. Mol. Biol. Cell. 2004;15:37–45. doi: 10.1091/mbc.E03-03-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn C. L., Pearson C. G., Romijn E. P., Meehl J. B., Giddings T. H., Jr, Culver B. P., Yates J. R., 3rd, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Lee S., Chang J., Rhee K. A novel function of CEP135 as a platform protein of C-NAP1 for its centriolar localization. Exp. Cell Res. 2008;314:3692–3700. doi: 10.1016/j.yexcr.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y. D., Nigg E. A. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Li K., Xu E. Y., Cecil J. K., Turner F. R., Megraw T. L., Kaufman T. C. Drosophila centrosomin protein is required for male meiosis and assembly of the flagellar axoneme. J. Cell Biol. 1998;141:455–467. doi: 10.1083/jcb.141.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J., Hergert P., Magidson V., Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat. Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Marshall W. F. The cell biological basis of ciliary disease. J. Cell Biol. 2008;180:17–21. doi: 10.1083/jcb.200710085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Campos M., Basto R., Baker J., Kernan M., Raff J. W. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 2004;165:673–683. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura K., Lefebvre P. A., Kamiya R., Hirono M. Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J. Cell Biol. 2004;165:663–671. doi: 10.1083/jcb.200402022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw T. L., Li K., Kao L. R., Kaufman T. C. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 1999;126:2829–2839. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- Nakazawa Y., Hiraki M., Kamiya R., Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Nielsen M. G., Turner F. R., Hutchens J. A., Raff E. C. Axoneme-specific beta-tubulin specialization: a conserved C-terminal motif specifies the central pair. Curr. Biol. 2001;11:529–533. doi: 10.1016/s0960-9822(01)00150-6. [DOI] [PubMed] [Google Scholar]
- Ohta T., Essner R., Ryu J. H., Palazzo R. E., Uetake Y., Kuriyama R. Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. J. Cell Biol. 2002;156:87–99. doi: 10.1083/jcb.200108088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L., O'Toole E., Schwager A., Hyman A. A., Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Preble A. M., Giddings T. M., Jr, Dutcher S. K. Basal bodies and centrioles: their function and structure. Curr. Top. Dev. Biol. 2000;49:207–233. doi: 10.1016/s0070-2153(99)49010-6. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Bettencourt-Dias M., Riparbelli M., Ferreira C., Ferreira I., Callaini G., Glover D. M. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 2007;17:1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D. M., Bettencourt-Dias M. From centriole biogenesis to cellular function: centrioles are essential for cell division at critical developmental stages. Cell Cycle. 2008;7:11–16. doi: 10.4161/cc.7.1.5226. [DOI] [PubMed] [Google Scholar]
- Smith E. F., Lefebvre P. A. The role of central apparatus components in flagellar motility and microtubule assembly. Cell Motil. Cytoskelet. 1997;38:1–8. doi: 10.1002/(SICI)1097-0169(1997)38:1<1::AID-CM1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Smith E. F., Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil. Cytoskelet. 2004;57:8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel C. E., Gomes R., Sampaio P., Perdigao J., Gonzalez C. Gamma-tubulin is required for the structure and function of the microtubule organizing centre in Drosophila neuroblasts. EMBO J. 1995;14:28–36. doi: 10.1002/j.1460-2075.1995.tb06972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tates A.D. Netherlands: Rijksunivrsiteit de Leiden; 1971. Cytodiferentiation during spermatogenesis in Drosophila melanogaster: an electron microscopy study. PhD thesis. [Google Scholar]
- Uetake Y., Terada Y., Matuliene J., Kuriyama R. Interaction of Cep135 with a p50 dynactin subunit in mammalian centrosomes. Cell Motil. Cytoskelet. 2004;58:53–66. doi: 10.1002/cm.10175. [DOI] [PubMed] [Google Scholar]
- Varmark H., Llamazares S., Rebollo E., Lange B., Reina J., Schwarz H., Gonzalez C. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 2007;17:1735–1745. doi: 10.1016/j.cub.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Wilson P. G., Zheng Y., Oakley C. E., Oakley B. R., Borisy G. G., Fuller M. T. Differential expression of two gamma-tubulin isoforms during gametogenesis and development in Drosophila. Dev. Biol. 1997;184:207–221. doi: 10.1006/dbio.1997.8545. [DOI] [PubMed] [Google Scholar]
- Wirschell M., Hendrickson T., Sale W. S. Keeping an eye on I 1, I1 dynein as a model for flagellar dynein assembly and regulation. Cell Motil. Cytoskelet. 2007;64:569–579. doi: 10.1002/cm.20211. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Q., Matthies H. J., Mancuso J., Andrews H. K., Woodruff E., 3rd, Friedman D., Broadie K. The Drosophila fragile X-related gene regulates axoneme differentiation during spermatogenesis. Dev. Biol. 2004;270:290–307. doi: 10.1016/j.ydbio.2004.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.