Abstract
Poly-N-acetyllactosamine (polyLacNAc) is a linear carbohydrate polymer composed of alternating N-acetylglucosamine and galactose residues involved in cellular functions ranging from differentiation to metastasis. PolyLacNAc also serves as a scaffold on which other oligosaccharides such as sialyl Lewis X are displayed. The polymerization of the alternating N-acetylglucosamine and galactose residues is catalyzed by the successive action of UDP-GlcNAc:βGal β-1,3-N-acetylglucosaminyltransferase 1 (B3GNT1) and UDP-Gal:βGlcNAc β-1,4-galactosyltransferase, polypeptide 1 (B4GALT1), respectively. The functional association between these two glycosyltransferases led us to investigate whether the enzymes also associate physically. We show that B3GNT1 and B4GALT1 colocalize by immunofluorescence microscopy, interact by coimmunoprecipitation, and affect each other's subcellular localization when one of the two proteins is artificially retained in the endoplasmic reticulum. These results demonstrate that B3GNT1 and B4GALT1 physically associate in vitro and in cultured cells, providing insight into possible mechanisms for regulation of polyLacNAc production.
Keywords: endoplasmic reticulum, enzyme complexes, glycosyltransferase, Golgi complex, poly-N-acetyllactosamine
Introduction
A fundamental question in oligosaccharide assembly is how cells template the synthesis of specific carbohydrate structures. As proteins and lipids pass through each cisterna of the Golgi complex, they encounter distinct glycosyltransferases that generate substrates for modification by subsequently encountered enzymes (Munro 1998; de Graffenried and Bertozzi 2004; Maccioni 2007). Despite an enormous amount of work by many laboratories, how glycosyltransferases achieve their unique localizations is still poorly understood (reviewed by Colley (1997); Opat et al. (2001); and de Graffenried and Bertozzi (2004)).
Two nonexclusive mechanisms have been proposed to explain how the type II single transmembrane-spanning glycosyltransferases are properly localized. According to the “bilayer thickness” model, enzymes sort according to the size match between the lengths of their transmembrane domains and the local membrane thickness (Bretscher and Munro 1993; Mitra et al. 2004). In the “kin recognition” model, glycosyltransferases that are localized in a common compartment interact to form complexes that are excluded from inclusion in forward moving cargo (Machamer 1991; Nilsson et al. 1993; Opat et al. 2000). These models are not mutually exclusive: membrane thickness could contribute to the localization of enzyme complexes while enzyme complexes could establish domains of particular membrane thickness.
Importantly, the known hetero-oligomeric kin recognition complexes are composed of glycosyltransferases that catalyze successive processing reactions (de Graffenried and Bertozzi 2004) as proposed by Roseman (1970). The first reported case of association was between α-1,3-1,6-mannosidase II and β-1,2-N-acetylglucosaminyltransferase I; in this case, the action of β-1,2-N-acetylglucosaminyltransferase I is a necessary precondition for α-1,3-1,6-mannosidase-II-catalyzed mannose trimming (Nilsson et al. 1994; Moremen 2002). Associations have also been reported in glycolipid synthesis: β-1,4-N-acetylgalactosaminyltransferase 1 (B4GALNT1) and the UDP-Gal:GA2/GM2/GD2 β-1,3-galactosyltransferase physically associate (Giraudo et al. 2001), and UDP-Gal:glucosylceramide galactosyltransferase, CMP-NeuAc:lactosylceramidesialyltransferase, and ST8 α-N-acetylneuraminide α-2,8-sialyltransferase 1 (ST8SIA1) have been reported to form a complex (Giraudo and Maccioni 2003a). ST8SIA1 can also associate with B4GALNT1 in a different cell line (Bieberich et al. 2002). Glycosaminoglycan biosynthetic enzymes exostosin 1 and 2 associate with one another as do uronosyl 5-epimerase and iduronic acid 2-O-sulfotransferase (McCormick et al. 2000; Pinhal et al. 2001). Hetero-oligomeric glycosyltransferase complexes have been identified in organisms from Saccharomyces cerevisiae to humans (Jungmann and Munro 1998; Jungmann et al. 1999). Thus, kin recognition may represent an important strategy to direct substrate traffic and prevent off-target glycosylation events.
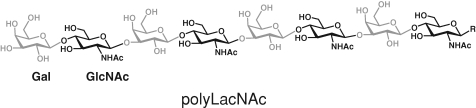
Poly-N-acetyllactosamine (polyLacNAc) is a linear carbohydrate polymer composed of alternating galactose (Gal) and N-acetylglucosamine (GlcNAc) residues (Figure 1). This polysaccharide can be incorporated into either N-linked or mucin-type O-linked glycans and can act as a marker for development, apoptosis, and metastasis (Kasai and Hirabayashi 1996; Elola et al. 2005). PolyLacNAc polymers can be further modified by various glycosyltransferases to create branched structures and display terminal epitopes such as the sialyl Lewis X modification, an important adhesion marker (Hakomori 1999; Dall’Olio 2000; Zhou 2003).
Fig. 1.
Structure of poly-N-acetyllactosamine. Poly-N-acetyllactosamine is composed of alternating residues of galactose-linked β1→4 and N-acetylglucosamine-linked β1→3. Sugars are color-coded in gray and black, respectively.
PolyLacNAc is biosynthesized by the alternating addition of GlcNAc by a UDP-GlcNAc:βGal β-1,3-N-acetylglucosaminyltransferase (B3GNT) and Gal by a UDP-Gal:βGlcNAc β-1,4-galactosyltransferase (B4GALT). Two of the primary glycosyltransferases within the B3GNT and B4GALT families are UDP-GlcNAc:βGal β-1,3-N-acetylglucosaminyltransferase 1 (B3GNT1) and UDP-Gal:βGlcNAc β-1,4-galactosyltransferase, polypeptide 1 (B4GALT1), respectively (Trayer and Hill 1971; Narimatsu et al. 1986; Shaper et al. 1986; Sasaki et al. 1997). While little is known about the localization mechanism for B3GNT1, several studies have suggested that proper B4GALT1 localization requires all or part of its cytoplasmic and transmembrane domains (Nilsson et al. 1991; Aoki et al. 1992; Russo et al. 1992; Teasdale et al. 1992; Evans et al. 1993; Masibay et al. 1993; Yamaguchi and Fukuda 1995). The linear and repetitive characteristics of the polyLacNAc structure have led to the hypothesis that the glycosyltransferases involved may form complexes to aid in polyLacNAc assembly (de Graffenried and Bertozzi 2004). Indeed, Seko and Yamashita (2008) recently demonstrated the interaction of two glycosyltransferases, B3GNT2 and B3GNT8, from the B3GNT family and showed that the presence of B3GNT8 can stimulate the activity of B3GNT2. It has been observed that B3GNT1 displays an in vitro preference for the GlcNAcβ1→2Man branch while B4GALT1 shows a complementary preference for the GlcNAcβ1→6Man branch. The equal prevalence of polyLacNAc on both branches therefore suggests tight functional association between the two enzymes (Ujita et al. 1999). Thus, we sought to determine whether these enzymes, from the complementary families of polyLacNAc catalyzing glycosyltransferases, also physically associate. Using coimmunoprecipitation and an endoplasmic reticulum (ER) retention assay adapted from an approach developed by Nilsson et al. (1994), we show that B3GNT1 and B4GALT1 interact with each other in the trans-Golgi.
Results
B3GNT1 and B4GALT1 colocalize
To investigate the mechanism of polyLacNAc synthesis, we explored the potential association of two of the primary glycosyltransferases, B3GNT1 and B4GALT1, involved in polyLacNAc production. To track subcellular localization of these enzymes, we employed plasmids encoding B3GNT1 with a myc epitope tag at its C-terminus and B4GALT1 with an HA epitope tag at its C-terminus. The C-termini of these type II membrane proteins are expected to reside in the lumen of the secretory pathway. HeLa cells were utilized for microscopy experiments because of their generally flat morphology; COS-1 cells were used in coimmunoprecipitation experiments because of their high transfection efficiency and good protein expression.
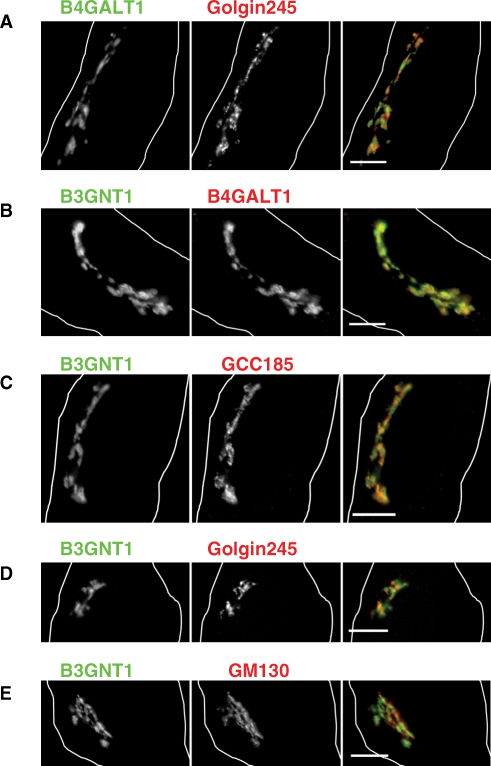
In order to associate in a physiologically relevant manner, B3GNT1 and B4GALT1 must localize within the same compartment of the Golgi complex. Control experiments (Figure 2A) demonstrated that exogenously expressed B4GALT1-HA localized near the trans-Golgi network (TGN) marker Golgin245 supporting its well-established trans-Golgi localization (Roth and Berger 1982; Gleeson et al. 1996; Llopis et al. 1998). To determine the localization of B3GNT1, we compared the colocalization of B3GNT1 with several different markers: B4GALT1, GCC185, Golgin245, and GM130. As shown in Figure 2B, B3GNT1-myc and B4GALT1-HA colocalized strongly with Pearson's coefficient of 0.87 ± 0.02. Moreover, B3GNT1-myc colocalized to some extent with the TGN marker GCC185 (Pearson's coefficient of 0.85 ± 0.02) followed by Golgin245 with Pearson's coefficient of 0.83 ± 0.03 (Figure 2C and D) (Luke et al 2003). B3GNT1-myc colocalized least well with the cis-Golgi marker GM130 (Pearson's coefficient of 0.82 ± 0.03) (Figure 2E) (Nakamura et al. 1995). The trend in colocalization among the TGN and cis-Golgi markers is consistent with B3GNT1 being localized with B4GALT1 at the trans-Golgi near the TGN and further from the cis-Golgi complex.
Fig. 2.
Immunofluorescence microscopy of B4GALT1 and B3GNT1 in HeLa cells. (A) B4GALT1 transfected cell (left panel, green in merge) with endogenous Golgin245 (middle panel, red in merge). (B) B3GNT1 (left panel, green in merge) and B4GALT1 (middle panel, red in merge) cotransfected cell. (C–E) B3GNT1 transfected cells (left panels, green in merge) with endogenous GCC185 (C), Golgin245 (D), or GM130 (E) (middle panels, red in merge). White outlines show cell boundaries. Scale bar is 10 μm.
B3GNT1 and B4GALT1 coimmunoprecipitate
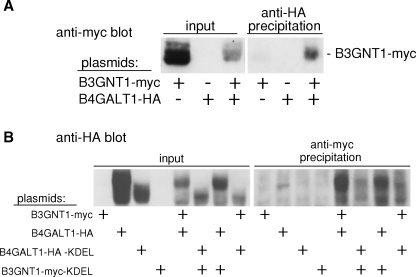
Next, we conducted coimmunoprecipitation experiments to test whether B3GNT1 and B4GALT1 associate with each other. To test whether B4GALT1 could precipitate B3GNT1, the epitope-tagged forms of the constructs were transfected into COS-1 cells and lysates were incubated with an anti-HA antibody to precipitate B4GALT1-HA and any associated proteins. A subsequent immunoblot was probed with an anti-myc antibody to detect immunoprecipitated B3GNT1-myc. As shown in Figure 3A, lane 6, B3GNT1-myc was coimmunoprecipitated with B4GALT1-HA only when both proteins were present in the extract.
Fig. 3.
Reciprocal physical association of B4GALT1 and B3GNT1. (A) Indicated COS-1 cell extracts were subjected to immunoprecipitation with an anti-HA antibody. Input (2%) and immunoprecipitated fractions were immunoblotted with an anti-myc antibody. (B) COS-1 cells transfected as indicated were immunoprecipitated using an anti-myc antibody. Input (1%) and immunoprecipitated fractions were immunoblotted with an anti-HA antibody.
We also tested whether B3GNT1 could conversely precipitate B4GALT1. Transfected COS-1 cell lysates were incubated with an anti-myc antibody to precipitate B3GNT1-myc and its associated proteins. Immunoblot analysis of the precipitated proteins with an anti-HA antibody revealed that B4GALT1-HA was coimmunoprecipitated by the anti-myc antibody only in the presence of B3GNT1-myc (Figure 3B, lane 13). It is interesting to note that two B4GALT1-HA bands were coimmunoprecipitated. Furthermore, there was a larger amount of the higher molecular weight species (lanes 2 and 5), and the B3GNT1 precipitation seems enriched for the higher molecular weight B4GALT1 band (lane 13). As discussed below, the larger forms may represent more highly glycosylated forms of B4GALT1. These experiments demonstrate a reciprocal physical interaction between at least a fraction of B3GNT1-myc and B4GALT1-HA, two enzymes that function together in polyLacNAc synthesis.
KDEL sequences cause glycosyltransferases to localize to the ER
To determine if the physical interaction between B3GNT1 and B4GALT1 could be demonstrated in cultured cells, we utilized a cell-based relocalization assay. Previously, Warren and colleagues pioneered an approach for demonstrating Golgi-resident glycosyltransferase interactions using an ER retention assay (Nilsson et al. 1994). Specifically, one enzyme is retained in the ER through an N-terminal fusion of a cytoplasmically oriented retention signal provided by the ER-localized human invariant chain p33. Changes in localization of putative binding partners were then examined for complementary relocalization to the ER. A change in localization, from the Golgi to the ER, was interpreted as being indicative of an interaction between the two proteins.
Rather than modifying the native cytoplasmic domain sequences that may include localization information (Russo et al. 1992; Evans et al. 1993), we used a lumenally oriented C-terminal KDEL retention signal to relocalize Golgi-resident glycosyltransferases to the ER. A similar approach was used by Munro (1995) to investigate the retention determinants of medial and trans-Golgi glycosyltransferases. KDEL sequences are typically found at the C-termini of soluble, ER-resident proteins (Pelham 1990). However, the unusual type II transmembrane topology of glycosyltransferases permits their lumenal C-termini to interact with the KDEL receptor, thereby making it possible to use a C-terminal KDEL sequence to recruit glycosyltransferases to the ER. Indeed, KDEL-related sequences may also contribute to the natural ER localization of certain ER-resident glycosyltransferases (Heinonen et al. 2003; Okajima et al. 2005; Heinonen et al. 2006 but see also Moloney and Haltiwanger (1999); Kozma et al. (2006) and Sato et al. (2006)).
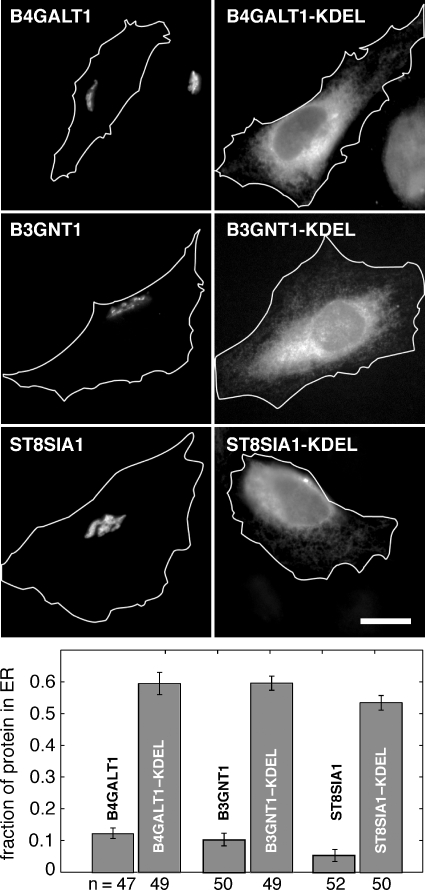
Expression plasmids were generated that encode the amino acid sequence SEKDEL at the C-termini of epitope-tagged B3GNT1, B4GALT1, and ST8SIA1 constructs. ST8SIA1, another trans-Golgi resident glycosyltransferase, participates in the orthologous glycolipid biosynthesis pathway (Daniotti et al. 2000). Immunofluorescence microscopy was used to determine the capacity of the SEKDEL sequence to relocalize the normally Golgi-associated glycosyltransferases to the ER. As shown in Figure 4 (top-right column), B4GALT1-HA-KDEL, B3GNT1-myc-KDEL, and ST8SIA1-myc-KDEL were all efficiently relocalized to the ER compared with the corresponding non-KDEL-tagged constructs (top-left column). Relocalization was still observed after 2 h of cycloheximide treatment (100 μg/mL) prior to fixation, suggesting that the localization was stable and did not reflect proteins in the process of folding prior to ER export. The cycloheximide incubation did not perturb the normal Golgi localization of the non-KDEL-tagged constructs.
Fig. 4.
KDEL relocalizes glycosyltransferases. Representative images show the localization of non-KDEL-tagged and KDEL-tagged B4GALT1, B3GNT1, and ST8SIA1 (top). White outlines show cell boundaries as determined by phase contrast microscopy. Scale bar, 20 μm. The percent ER localization was quantified (bottom). Error bars represent 95% confidence intervals from a sampling of ≥47 cells per condition (number of cells analyzed is shown below each bar).
Quantitative analysis of protein localization
A metric was established to permit quantitative analysis of ER versus Golgi localization for each glycosyltransferase. First, we approximated the cell as a flat two-dimensional structure and defined three compartments: nucleus, Golgi, and ER (bulk cytoplasm). Total cell intensity was determined by summing the signal within a mask drawn from the phase contrast image of the cell edge; background staining and cell autofluorescence, calculated from cells that had been immunostained but not transfected, was subtracted. The intensity within the Golgi and nuclear areas was determined using masks derived from a Golgi marker protein, GM130, and a nuclear marker, DAPI, respectively. Any signal not localized to the nuclear or Golgi area was defined as ER. During instances where the Golgi and nucleus slightly overlapped, the signal was attributed to the Golgi. Golgi and nuclear areas were defined liberally to decrease the possibility of attributing signal derived from these areas to the ER. It is important to note that ER intensity will be underestimated by this method because some ER intensity overlaps with the nuclear envelope and Golgi areas.
As quantified at the bottom of Figure 4, the addition of a KDEL tag to the C-termini of B4GALT1, B3GNT1, or ST8SIA1 shifted their distributions to the ER compared with their untagged counterparts even after 2 h of cycloheximide treatment. Golgi-localized B4GALT1-HA showed a mean ER fraction of 0.12; KDEL tagging this glycosyltransferase increased the mean ER fraction to 0.59, a 4.8-fold increase. Similarly, the addition of KDEL to B3GNT1-myc shifted the mean ER fraction from 0.10 to 0.60, a 6-fold enhancement. The mean fraction of ST8SIA1-myc intensity localized to the ER was 0.05. KDEL-tagging ST8SIA1-myc raised this value to 0.53, a 10.6-fold difference. These data demonstrate that the addition of KDEL can efficiently relocalize these type II Golgi resident glycosyltransferases to the ER.
KDEL-tagged B3GNT1 and B4GALT1 can also coimmunoprecipitate
Coimmunoprecipitation experiments were carried out as an independent confirmation that KDEL-tagged versions of B3GNT1 and B4GALT1 can also associate in vitro. B3GNT1-myc or B3GNT1-myc-KDEL and their associated proteins were precipitated from transfected COS-1 cell lysates with an anti-myc antibody. Subsequent probing of an immunoblot with an anti-HA antibody revealed that B4GALT1-HA was efficiently precipitated by B3GNT1-myc-KDEL, and B4GALT1-HA-KDEL was precipitated by both KDEL-tagged and non-KDEL-tagged B3GNT1-myc (Figure 3B, lanes 14–16). Interestingly, the KDEL-tagged B4GALT1-HA was predominantly present as the lower molecular weight form (lane 3). Conversely, B3GNT1-myc- and B3GNT1-myc-KDEL-precipitated B4GALT1-HA-KDEL was enriched in the higher molecular weight form of B4GALT1 (lane 14 and 16). While one molecular weight form of B4GALT1-HA-KDEL is enriched, the KDEL-tagged constructs can efficiently associate with one another as well as their non-KDEL-tagged partners.
B3GNT1 and B4GALT1 can recruit one another to the ER
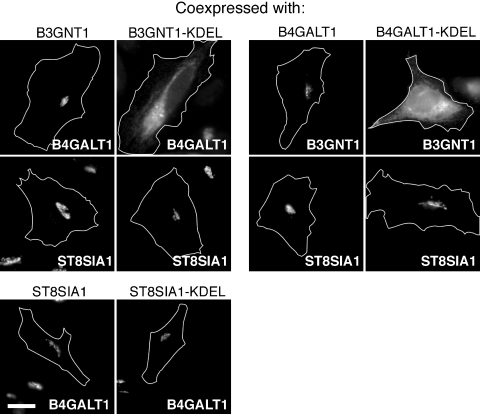
Possible interactions between glycosyltransferases were tested in our cell-based assay by cotransfecting a non-KDEL-tagged glycosyltransferase with a KDEL-tagged glycosyltransferase and monitoring the localization of the non-KDEL-tagged glycosyltransferase after 2 h of cycloheximide treatment. When non-KDEL-tagged B4GALT1 and B3GNT1 were coexpressed, B4GALT1 was localized to the Golgi complex (Figure 5, upper left). In contrast, cotransfection of B4GALT1 with B3GNT1-KDEL readily shifted B4GALT1's localization to the ER (Figure 5, upper left). This effect was specific, as cotransfection of the orthologous ST8SIA1 with B3GNT1-KDEL failed to relocalize ST8SIA1 (Figure 5, upper left). Similarly, coexpression of B3GNT1 with B4GALT1 did not perturb B3GNT1's normal Golgi localization (Figure 5, upper right), while coexpression of B3GNT1 and B4GALT1-KDEL resulted in the relocalization of B3GNT1 to the ER (Figure 5, upper right). Again, cotransfection of ST8SIA1 with B4GALT1-KDEL did not relocalize ST8SIA1 to the ER (Figure 5, upper right). As expected, ST8SIA1-KDEL was not capable of relocalizing B4GALT1 to the ER (Figure 5, bottom row).
Fig. 5.
B4GALT1 and B3GNT1 affect each other's subcellular localization. The localization of the one glycosyltransferase (within image) was examined to determine the ability of a partner glycosyltransferase (indicated above image) to recruit the shown glycosyltransferase to the ER. White outlines show the cell boundaries as determined by phase contrast microscopy. Scale bar is 20 μm.
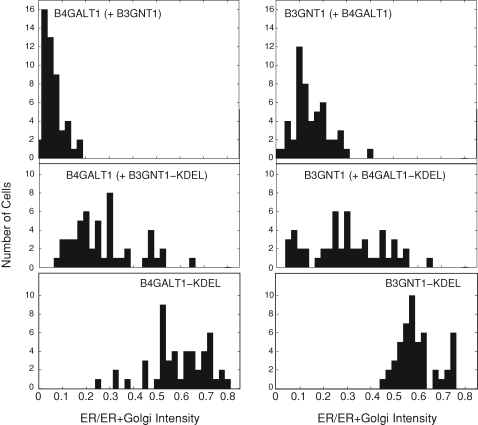
The ability of two proteins to alter each others’ localizations is likely to be dependent upon their relative levels of expression. Because this aspect of our experiment is difficult to control, we carried out careful quantitation of at least 49 cells for each cotransfected pair to ensure reliability of our observations. Figure 6 (top-left column) shows the fraction of the ER-localized B4GALT1 signal for individual HeLa cells cotransfected with B4GALT1 and B3GNT1. The peak of the distribution was centered at 0.06, consistent with the Golgi localization of B4GALT1. When B4GALT1 was cotransfected with B3GNT1-KDEL, the fraction of B4GALT1 found in the ER tended to increase: the mean of the distribution of ER fraction shifted to 0.27 and the distribution overall broadened (Figure 6, middle-left column). The distribution of ER fraction of B4GALT1-KDEL alone is provided as a positive control and was centered about 0.59 (bottom panel). This represents the maximum shift that could have been observed in these experiments.
Fig. 6.
Fraction of ER localization of glycosyltransferases in individual cells. HeLa cells were transiently transfected and imaged as in Figure 4 and the fraction of the glycosyltransferase residing in the ER was quantified and plotted on a histogram. In each plot, the signal from the indicated glycosyltransferase was quantified under the cotransfection conditions indicated in parenthesis. More than 48 cells were quantified per condition.
Similarly, when B3GNT1 was cotransfected with B4GALT1 (Figure 6, top-right column), the fraction of B3GNT1 localized to the ER was distributed around a peak centered at 0.15. Cotransfection with B4GALT1-KDEL resulted in a broadened ER-fraction distribution with a mean of 0.30 (Figure 6, middle-right column). Again, the maximum shift that could have been observed was provided by the B3GNT1-KDEL-alone control, which showed a mean ER fraction of 0.60 (bottom panel). These data show that the presence of the KDEL-tagged partner causes a normally Golgi-localized protein to be retained in the ER in a large representative sampling of transfected cells.
One limitation of our approach is the fact that KDEL-terminating proteins often leave the ER and are normally retrieved by the KDEL receptor. In the course of retrieval, an artificially KDEL-tagged glycosyltransferase would be expected to traverse the Golgi compartments where it might interact preferentially with untagged counterparts. Thus, we also analyzed our data by normalizing for this possibility. Specifically, for each non-KDEL-tagged glycosyltransferase, we calculated the fraction of that marker in the ER as a percentage of the KDEL-tagged partner protein's ER fraction. This would enable a direct comparison of different pairs of glycosyltransferases and account for any variation in ER retention between different KDEL-tagged proteins.
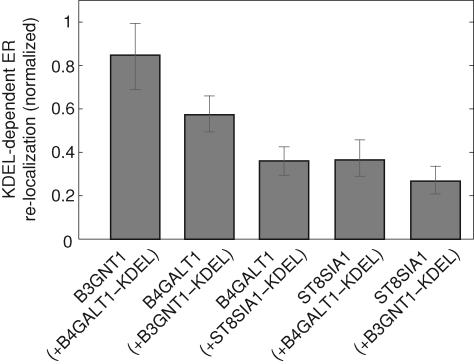
When B3GNT1 was transfected with B4GALT1-KDEL, we detected an average normalized, ER localization value of 0.85 for B3GNT1. Similarly, when B4GALT1 was transfected with B3GNT1-KDEL, we observed a mean normalized ER localization value of 0.57 for B4GALT1. In contrast, this value was only 0.36 when ST8SIA1-KDEL was cotransfected with B4GALT1 or B4GALT1-KDEL was cotransfected with ST8SIA1. Similarly, ST8SIA1 was not relocalized to the ER by B3GNT1-KDEL, as a value of 0.27 was obtained for ST8SIA1 under these conditions. These data suggest that B3GNT1 and B4GALT1 can associate with one another within the secretory pathway and that this association is stronger than that of ST8SIA1 with either enzyme (Figure 7).
Fig. 7.
B4GALT1–B3GNT1 interaction results in a higher percentage of relocalization to the ER when compared to interactions with non-related glycosyltransferases. Shown is the mean ER relocalization of the non-KDEL-tagged partner (indicated below each bar) normalized by the efficiency with which the KDEL-tagged protein (indicated in parenthesis) is itself ER retained. Error bars represent 95% confidence intervals from a sampling of n = 51, 50, 52, 50, and 50, respectively.
Discussion
We have reported here the use of two independent methods to demonstrate the association of B3GNT1 and B4GALT1 within the secretory pathway. Both coimmunoprecipitation and cellular relocalization assays provide evidence for an interaction between these two enzymes. The physical association between B3GNT1 and B4GALT1 likely contributes to the functional coupling of these two enzymes in producing the polyLacNAc glycan structure.
The distinctive structures of glycosyltransferases, namely their common type II transmembrane orientation, made it possible to adapt the ER retention assay (Nilsson et al. 1994) by employing a C-terminal KDEL tag for recruitment. C-terminal KDEL tags are well-established cellular signals for soluble protein retention to the ER (Pelham 1990) and have been exploited previously to relocalize type II transmembrane proteins to the ER (Munro 1995). Our data show that the KDEL tag can be applied to other Golgi glycosyltransferases and used to retain trans-Golgi glycosyltransferases and their partners in the ER.
Recent reports have suggested that Golgi localization of glycosyltransferases can depend on native cytoplasmic signals (Grabenhorst and Conradt 1999; Milland et al. 2001, 2002; Giraudo and Maccioni 2003b; Schmitz et al. 2008; Tu et al. 2008). While the original p33-mediated ER retention assay was able to detect interactions between medial glycosyltransferases (Nilsson et al. 1994), the use of a KDEL motif has advantages over the original system. First, the KDEL fusion requires a smaller overall change to the chimeric protein: fusion of 6 amino acids (SEKDEL), compared to the addition of the 47 amino acids that comprise the cytoplasmic domain of the p33 protein. Second, the addition of a KDEL motif does not involve replacement of any amino acids. Lastly, the KDEL motif does not necessitate the removal of glycosyltransferase cytoplasmic domains that may contribute to protein localization. The efficiency of partner protein ER-relocalization is likely to depend on the individual glycosyltransferases studied, and the p33 system may be more effective than the KDEL motif for certain enzyme pairs.
It is worth noting that the relative efficiency of relocalization for trans-Golgi partners versus medial Golgi partners cannot be compared directly since the different native pH environments may influence the ability of such partners to achieve ER-relocalization. Indeed, Colley and co-workers demonstrated that β-galactoside α-2,6-sialyltransferase 1, a trans-Golgi resident enzyme, formed insoluble oligomers only when harvested under pH conditions similar to those found at the trans-Golgi (Chen et al. 2000). The same pH gradient may explain why the KDEL-mediated ER retention assay reported here did not result in the relocalization of 100% of the examined trans-Golgi glycosyltransferases.
Probing lysates of cells transfected with B4GALT1-HA using an anti-HA antibody revealed two isoforms: one at the expected molecular weight for unmodified B4GALT1-HA (45 kDa) and another at a higher molecular weight (approximately 53 kDa) (Figure 3B, lanes 2, 5, 7). We hypothesize that this higher molecular weight band represents the presence of an extended glycan modification added during passage through the Golgi. This hypothesis is consistent with the observation that lysates from cells transfected with B4GALT1-HA-KDEL contain predominantly the lower molecular weight form (Figure 3B, lanes 3, 6, 8), since the KDEL-tagged isoform is less likely to traffic through the Golgi and be modified by glycan extending enzymes. We were intrigued to observe that B3GNT1-myc and B3GNT1-myc-KDEL both preferentially precipitated the higher molecular weight isoform of B4GALT1-HA-KDEL (Figure 3B, lanes 14 and 16). This finding suggests that an extended glycan on B4GALT1 may enhance binding to B3GNT1.
B4GALT1 transmembrane residues Cys29 and His32 are required for Golgi retention and appear to also contribute to B4GALT1 homodimerization (Aoki et al. 1992; Yamaguchi and Fukuda 1995). An intriguing possibility is that these same residues might also mediate association between B4GALT1 and B3GNT1. Also noteworthy is B3GNT1's unusually long, 28-amino-acid transmembrane domain, which is particularly unexpected in a trans-Golgi-resident glycosyltransferase. Perhaps an interaction between B3GNT1 and B4GALT1 allows B3GNT1 to take advantage of the more canonical B4GALT1 transmembrane domain to stabilize B3GNT1 localization to the trans-Golgi. Future experiments will be needed to test these possibilities directly. Furthermore, experiments using purified components will help to address whether the interaction observed is direct.
The present study has focused on two of the main enzymes that cooperate in polyLacNAc synthesis. However, other galactosyltransferases also participate in this biosynthetic process (Lo et al. 1998; Hennet 2002). Preliminary results using the ER recruitment assay described here provide evidence for an association between B3GNT1 and UDP-Gal:βGlcNAc β-1,4-galactosyltransferase, polypeptide 4, B4GALT4, in cotransfected cells (data not shown). In vitro, B4GALT1 has been shown to display substrate preference for N-linked and core 4 O-linked glycans while B4GALT4 has substrate preference for core 2 O-linked glycans (Ujita et al. 2000). B4GALT1 and B4GALT4 might therefore compete for association with B3GNT1 and bias B3GNT1 substrate preference in vivo. More detailed information about the molecular interfaces of B3GNT1: B4GALT complexes will aid in investigating this hypothesis. Alternatively, B3GNT1, B4GALT1, and B4GALT4 may associate with each other within a single, large heterocomplex that allows B3GNT1 to process both N-linked and O-linked glycans. By disrupting or strengthening associations between B3GNT1 and B4GALT1 or between B3GNT1 and B4GALT4, it may be possible to alter the glycan synthetic capacity in cells. Furthermore, engineering an enzyme containing both B3GNT1 and B4GALT1 catalytic domains may generate increased polyLacNAc length by increasing the processivity of the reaction and allow further studies on how polyLacNAc length affects its biological activities.
In conclusion, we have shown the first example of enzyme association among trans-Golgi-localized glycosyltransferases using an ER retention assay that does not alter the N-terminal cytoplasmic, transmembrane, and stem domains of these type II transmembrane proteins. The physical association between these two glycosyltransferases likely contributes to the localization of both B3GNT1 and B4GALT1 to the trans-Golgi and, importantly, to regulating the production of polyLacNAc from these enzymes.
Material and methods
Cell culture and transfections
HeLa and African green monkey kidney fibroblast (COS-1) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 7.5% fetal calf serum, penicillin, and streptomycin. HeLa and COS-1 cells were transfected using FuGENE 6 (Roche Applied Science, Indianapolis, IN).
Plasmids
Standard recombinant DNA technology was used to construct all plasmids. All regions that were amplified by PCR were analyzed by DNA sequencing. Plasmid DNA was prepared from large-scale bacterial cultures and purified by a Midiprep kit (QIAGEN, Valencia, CA). Restriction enzymes were purchased from New England Biolabs (Ipswich, MA).
B4GALT1-HA was amplified from pBS-β1,4 GT-1 kindly provided by Dr. Michiko Fukuda (The Burnham Institute, La Jolla, CA) by PCR with a 3′ primer containing the HA epitope tag, digested, and ligated into pcDNA 3.1 Zeo (+) (Invitrogen, Carlsbad, CA) for mammalian expression. B4GALT1-HA-KDEL was amplified from B4GALT1-HA using a 3′ primer containing codons for the amino acid sequence SEKDEL and ligated into pcDNA 3.1. B3GNT1-myc was amplified from pcDNA3.1-iGnT (B3GNT1) kindly provided by Dr. Minoru Fukuda (The Burnham Institute, La Jolla, CA) by PCR using a 3′ primer containing the myc epitope tag, digested, and ligated into pcDNA 3.1 (+). B3GNT1-myc-KDEL was amplified from B3GNT1-myc in an analogous manner as described for B4GALT1-HA-KDEL. Similar to the method used to produce B4GALT1 and B3GNT1 constructs above, ST8SIA1-HA, ST8SIA1-myc, and ST8SIA1-myc-KDEL were made by PCR amplifying the respective genes with primers encoding the appropriate tags. ST8SIA1 constructs were kindly provided by Chad Whitman (UT Southwestern, Dallas, TX).
Immunofluorescence
For colocalization studies, HeLa cells were grown on coverslips and transfected with B3GNT1-myc and/or B4GALT1-HA for 24 h. Cells were fixed using 3.7% formaldehyde and permeabilized with 0.1% Triton X-100. Cells were stained with combinations of chicken anti-Myc antibodies (1:750, Bethyl Laboratories, Montgomery, TX), rabbit anti-HA antibodies (1:1000, Abcam, Cambridge, MA), mouse anti-GM130 antibodies (1:500, BD Biosciences, San Jose, CA), mouse anti-Golgin245 antibodies (p230 trans-Golgi, BD Biosciences, 1:400), and rabbit anti-GCC185 antibodies (Reddy et al. 2006). Secondary immunostaining was conducted with goat anti-chicken antibodies conjugated to Alexa 488, goat anti-mouse antibodies conjugated to Alexa 488 or Alexa 546, and goat anti-rabbit antibodies conjugated to Alexa 633 (Invitrogen). Coverslips were mounted with a VectaShield mounting medium with DAPI (Vector Laboratories, Burlingame, CA). Images were captured with an Eclipse 80i microscope (Nikon, Tokyo, Japan) equipped with epifluorescence optics using a 100× numerical aperture 1.40 plan apochromat oil immersion objective and a charge-coupled device camera (CoolSnapHQ; Photometrics, Tucson, AZ) and MetaMorph image acquisition software (Molecular Devices, Sunnyvale, CA). Pearson's coefficients were calculated using the Manders’ Coefficients plugin (Tony Collins, Wayne Rasband) for ImageJ (NIH, Bethesda, MD).
To quantify KDEL-induced relocalization, HeLa cells were grown on coverslips and 22 hours after transfection were treated with 100 μg/mL cycloheximide for 2 h to clear the ER of newly synthesized proteins. Cells were subsequently fixed 24 h posttransfection, stained and mounted as above with the exception of different secondary antibodies: goat anti-chicken antibodies conjugated to Alexa 633, goat anti-mouse antibodies conjugated to Alexa 488 or Alexa 610, and goat anti-rabbit antibodies conjugated to Alexa 488 or Alexa 610 (Invitrogen). Fluorescence images were captured with a Zeiss Axioplan 2 microscope (Carl Zeiss, Thornwood, NY) equipped with phase contrast and epifluorescence optics by using a Fluor 40×, numerical aperture 1.3 oil immersion objective and a back-thinned cooled charge-coupled device camera (Princeton Instruments, Trenton, NJ) and MetaMorph image acquisition software (Universal Imaging, Downingtown, PA). Image masks were defined using Photoshop (Adobe Systems, Mountain View, CA) and ImageJ (NIH). Cell masks were defined by manually outlining the phase contrast image of the cell. Nucleus masks were defined by thresholding on the DAPI image of the cell. Golgi masks were defined by thresholding on the GM130 image or the Golgi-localized signal from the glycosyltransferase. MATLAB (The MathWorks, Natick, MA) was used for image quantitation and automated mask joining algorithms.
Immunoprecipitation
COS-1 cells grown on 10 cm plates were transiently transfected with combinations of B3GNT1-myc, B4GALT1-HA, and their KDEL-tagged plasmids for 24 h. After two washes with phosphate-buffered saline, cells were harvested in 1% CHAPS, 30 mM Tris-HCl, pH 7.5, 150 mM NaCl supplemented with protease inhibitors (complete, EDTA free, Roche Applied Science). Protein extracts were centrifuged at 55,000 × g for 15 min, and supernatants collected. Bovine serum albumin (0.1%) was added to the supernatants, which were then precleared with 30 μL Protein A-agarose (Roche Applied Science) for 20 min at 4°C. A rabbit anti-HA antibody (Abcam) or rabbit anti-myc antibody (Bethyl Laboratories) was added to a final concentration of 10 μg/mL and incubated for 2 h at 22°C. A 1:1 slurry of Protein A-agarose (30 μL) was added for 20 min at 4°C. The beads were washed once in 30 mM Tris-HCl, pH 7.5, 150 mM NaCl and boiled in a 20 μL SDS–PAGE sample buffer. Samples were then resolved by SDS–PAGE and transferred onto nitrocellulose. The blot was probed using either a mouse anti-myc antibody conjugated to horseradish peroxidase (Invitrogen) or a rabbit anti-HA antibody conjugated to horseradish peroxidase (Bethyl Laboratories) and detected using the ECL Plus Western blotting detection kit (GE Healthcare Bio-Sciences, Piscataway, NJ) and HyBlot CL autoradiography film (Denville Scientific Inc., Metuchen, NJ).
Funding
The National Institutes of Health (R37 DK37332 to S.R.P.); Stanford University; the Camille and Henry Dreyfus Foundation (New Faculty Award to J.J.K.); and the National Science Foundation (to P.L.L.).
Acknowledgments
We thank Dr. Minoru Fukuda for plasmids and reagents, Dr. Michiko Fukuda for plasmids, and Chad Whitman for the ST8SIA1 constructs. PLL is grateful to Dr. J. Theriot for continued support and advice.
Conflict of interest statement
None declared.
Abbreviations
- B3GNT
UDP-GlcNAc:βGal β-1,3-N-acetylglucosaminyl transferase
- B3GNT1
UDP-GlcNAc:βGal β-1,3-N-acetylgluco saminyltransferase 1
- B4GALT
UDP-Gal:βGlcNAc β-1,4- galactosyltransferase
- B4GALT1
UDP-Gal:βGlcNAc β-1,4- galactosyltransferase, polypeptide 1
- B4GALT4
UDP- Gal:βGlcNAc β-1,4-galactosyltransferase, polypeptide 4
- B4GALNT1
β-1,4-N-acetylgalactosaminyltransferase 1
- ER
endoplasmic reticulum
- Gal
galactose
- GlcNAc
N-acetylglucosamine
- polyLacNAc
poly-N-acetyllactosamine
- ST8SIA1
ST8 α-N-acetylneuraminide α-2,8-sialyltransferase 1
- TGN
trans-Golgi network
References
- Aoki D, Lee N, Yamaguchi N, Dubois C, Fukuda MN. Golgi retention of a trans-Golgi membrane protein, galactosyltransferase, requires cysteine and histidine residues within the membrane-anchoring domain. Proc Natl Acad Sci USA. 1992;89(10):4319–4323. doi: 10.1073/pnas.89.10.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberich E, MacKinnon S, Silva J, Li DD, Tencomnao T, Irwin L, Kapitonov D, Yu R. Regulation of ganglioside biosynthesis by enzyme complex formation of glycosyltransferases. Biochemistry. 2002;41(38):11479–11487. doi: 10.1021/bi0259958. [DOI] [PubMed] [Google Scholar]
- Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261(5126):1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- Chen C, Ma J, Lazic A, Backovic M, Colley KJ. Formation of insoluble oligomers correlates with ST6Gal I stable localization in the Golgi. J Biol Chem. 2000;275(18):13819–13826. doi: 10.1074/jbc.275.18.13819. [DOI] [PubMed] [Google Scholar]
- Colley KJ. Golgi localization of glycosyltransferases: More questions than answers. Glycobiology. 1997;7(1):1–13. doi: 10.1093/glycob/7.1.1-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Olio F. The sialyl-alpha2,6-lactosaminyl-structure: Biosynthesis and functional role. Glycoconj J. 2000;17(10):669–676. doi: 10.1023/a:1011077000164. [DOI] [PubMed] [Google Scholar]
- Daniotti JL, Martina JA, Giraudo CG, Zurita AR, Maccioni HJF. GM3 alpha2,8-sialyltransferase (GD3 synthase): Protein characterization and sub-golgi location in CHO-K1 cells. J Neurochem. 2000;74(4):1711–1720. doi: 10.1046/j.1471-4159.2000.0741711.x. [DOI] [PubMed] [Google Scholar]
- de Graffenried CL, Bertozzi CR. The roles of enzyme localisation and complex formation in glycan assembly within the Golgi apparatus. Curr Opin Cell Biol. 2004;16(4):356–363. doi: 10.1016/j.ceb.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Elola MT, Chiesa ME, Alberti AF, Mordoh J, Fink NE. Galectin-1 receptors in different cell types. J Biomed Sci. 2005;12(1):13–29. doi: 10.1007/s11373-004-8169-5. [DOI] [PubMed] [Google Scholar]
- Evans S, Lopez L, Shur B. Dominant negative mutation in cell surface beta 1,4- galactosyltransferase inhibits cell–cell and cell–matrix interactions. J Cell Biol. 1993;120(4):1045–1057. doi: 10.1083/jcb.120.4.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo CG, Daniotti JL, Maccioni HJF. Physical and functional association of glycolipid N-acetyl-galactosaminyl and galactosyl transferases in the Golgi apparatus. Proc Natl Acad Sci USA. 2001;98(4):1625–1630. doi: 10.1073/pnas.031458398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo CG, Maccioni HJF. Ganglioside glycosyltransferases organize in distinct multienzyme complexes in CHO-K1 cells. J Biol Chem. 2003a;278(41):40262–40271. doi: 10.1074/jbc.M305455200. [DOI] [PubMed] [Google Scholar]
- Giraudo CG, Maccioni HJF. Endoplasmic reticulum export of glycosyltransferases depends on interaction of a cytoplasmic dibasic motif with Sar1. Mol Biol Cell. 2003b;14(9):3753–3766. doi: 10.1091/mbc.E03-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson PA, Anderson TJ, Stow JL, Griffiths G, Toh BH, Matheson F. p230 is associated with vesicles budding from the trans-Golgi network. J Cell Sci. 1996;109(12):2811–2821. doi: 10.1242/jcs.109.12.2811. [DOI] [PubMed] [Google Scholar]
- Grabenhorst E, Conradt HS. The cytoplasmic, transmembrane, and stem regions of glycosyltransferases specify their in vivo functional sublocalization and stability in the Golgi. J Biol Chem. 1999;274(51):36107–36116. doi: 10.1074/jbc.274.51.36107. [DOI] [PubMed] [Google Scholar]
- Hakomori S. Antigen structure and genetic basis of histo-blood groups A, B and O: Their changes associated with human cancer. Biochim Biophys Acta. 1999;1473(1):247–266. doi: 10.1016/s0304-4165(99)00183-x. [DOI] [PubMed] [Google Scholar]
- Heinonen TYK, Pasternack L, Lindfors K, Breton C, Gastinel LN, Mäki M, Kainulainen H. A novel human glycosyltransferase: Primary structure and characterization of the gene and transcripts. Biochem Biophys Res Commun. 2003;309(1):166–174. doi: 10.1016/s0006-291x(03)01540-7. [DOI] [PubMed] [Google Scholar]
- Heinonen TYK, Pelto-Huikko M, Pasternack L, Mäki M, Kainulainen H. Murine ortholog of the novel glycosyltransferase, B3GTL: Primary structure, characterization of the gene and transcripts, and expression in tissues. DNA Cell Biol. 2006;25(8):465–474. doi: 10.1089/dna.2006.25.465. [DOI] [PubMed] [Google Scholar]
- Hennet T. The galactosyltransferase family. Cell Mol Life Sci. 2002;59(7):1081–1095. doi: 10.1007/s00018-002-8489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Munro S. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with alpha-1,6-mannosyltransferase activity. EMBO J. 1998;17(2):423–434. doi: 10.1093/emboj/17.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Rayner JC, Munro S. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J Biol Chem. 1999;274(10):6579–6585. doi: 10.1074/jbc.274.10.6579. [DOI] [PubMed] [Google Scholar]
- Kasai K, Hirabayashi J. Galectins: A family of animal lectins that decipher glycocodes. J Biochem. 1996;119(1):1–8. doi: 10.1093/oxfordjournals.jbchem.a021192. [DOI] [PubMed] [Google Scholar]
- Kozma K, Keusch JJ, Hegemann B, Luther KB, Klein D, Hess D, Haltiwanger RS, Hofsteenge J. Identification and characterization of a beta1,3-glucosyltransferase that synthesizes the Glc-beta1,3-Fuc disaccharide on thrombospondin type 1 repeats. J Biol Chem. 2006;281(48):36742–36751. doi: 10.1074/jbc.M605912200. [DOI] [PubMed] [Google Scholar]
- Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci USA. 1998;95(12):6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo NW, Shaper JH, Pevsner J, Shaper NL. The expanding beta 4-galactosyltransferase gene family: Messages from the databanks. Glycobiology. 1998;8(5):517–526. doi: 10.1093/glycob/8.5.517. [DOI] [PubMed] [Google Scholar]
- Luke MR, Kjer-Nielsen L, Brown DL, Stow SL, Gleeson PA. GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J Biol Chem. 2003;278(6):4216–4226. doi: 10.1074/jbc.M210387200. [DOI] [PubMed] [Google Scholar]
- Maccioni HJF. Glycosylation of glycolipids in the Golgi complex. J Neurochem. 2007;103(Suppl 1):81–90. doi: 10.1111/j.1471-4159.2007.04717.x. [DOI] [PubMed] [Google Scholar]
- Machamer CE. Golgi retention signals: Do membranes hold the key? Trends Cell Biol. 1991;1(6):141–144. doi: 10.1016/0962-8924(91)90001-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masibay A, Balaji P, Boeggeman E, Qasba P. Mutational analysis of the Golgi retention signal of bovine beta-1,4- galactosyltransferase. J Biol Chem. 1993;268(13):9908–9916. [PubMed] [Google Scholar]
- McCormick C, Duncan G, Goutsos KT, Tufaro F. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc Natl Acad Sci USA. 2000;97(2):668–673. doi: 10.1073/pnas.97.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milland J, Russell SM, Dodson HC, McKenzie IFC, Sandrin MS. The cytoplasmic tail of alpha 1,3-galactosyltransferase inhibits Golgi localization of the full-length enzyme. J Biol Chem. 2002;277(12):10374–10378. doi: 10.1074/jbc.M111799200. [DOI] [PubMed] [Google Scholar]
- Milland J, Taylor SG, Dodson HC, McKenzie IF, Sandrin MS. The cytoplasmic tail of alpha 1,2-fucosyltransferase contains a sequence for golgi localization. J Biol Chem. 2001;276(15):12012–12018. doi: 10.1074/jbc.M010018200. [DOI] [PubMed] [Google Scholar]
- Mitra K, Ubarretxena-Belandia I, Taguchi T, Warren G, Engelman DM. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc Natl Acad Sci USA. 2004;101(12):4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney D, Haltiwanger R. The O-linked fucose glycosylation pathway: Identification and characterization of a uridine diphosphoglucose: Fucose-beta1,3- glucosyltransferase activity from Chinese hamster ovary cells. Glycobiology. 1999;9(7):679–687. doi: 10.1093/glycob/9.7.679. [DOI] [PubMed] [Google Scholar]
- Moremen KW. Golgi alpha-mannosidase II deficiency in vertebrate systems: Implications for asparagine-linked oligosaccharide processing in mammals. Biochim Biophys Acta. 2002;1573(3):225–235. doi: 10.1016/s0304-4165(02)00388-4. [DOI] [PubMed] [Google Scholar]
- Munro S. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 1995;14(19):4695–4704. doi: 10.1002/j.1460-2075.1995.tb00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. Localization of proteins to the Golgi apparatus. Trends Cell Biol. 1998;8(1):11–15. doi: 10.1016/S0962-8924(97)01197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131(6):1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narimatsu H, Sinha S, Brew K, Okayama H, Qasba PK. Cloning and sequencing of cDNA of bovine N-acetylglucosamine (beta 1–4)galactosyltransferase. Proc Natl Acad Sci USA. 1986;83(13):4720–4724. doi: 10.1073/pnas.83.13.4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Hoe MH, Slusarewicz P, Rabouille C, Watson R, Hunte F, Watzele G, Berger EG, Warren G. Kin recognition between medial Golgi enzymes in HeLa cells. EMBO J. 1994;13(3):562–574. doi: 10.1002/j.1460-2075.1994.tb06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Lucocq JM, Mackay D, Warren G. The membrane spanning domain of beta-1,4-galactosyltransferase specifies trans Golgi localization. EMBO J. 1991;10(12):3567–3575. doi: 10.1002/j.1460-2075.1991.tb04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Slusarewicz P, Hoe MH, Warren G. Kin recognition. A model for the retention of Golgi enzymes. FEBS Lett. 1993;330(1):1–4. doi: 10.1016/0014-5793(93)80906-b. [DOI] [PubMed] [Google Scholar]
- Okajima T, Xu A, Lei L, Irvine KD. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science. 2005;307(5715):1599–1603. doi: 10.1126/science.1108995. [DOI] [PubMed] [Google Scholar]
- Opat AS, Houghton F, Gleeson PA. Medial Golgi but not late Golgi glycosyltransferases exist as high molecular weight complexes. Role of luminal domain in complex formation and localization. J Biol Chem. 2000;275(16):11836–11845. doi: 10.1074/jbc.275.16.11836. [DOI] [PubMed] [Google Scholar]
- Opat AS, van Vliet C, Gleeson PA. Trafficking and localisation of resident Golgi glycosylation enzymes. Biochimie. 2001;83(8):763–773. doi: 10.1016/s0300-9084(01)01312-8. [DOI] [PubMed] [Google Scholar]
- Pelham HR. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem Sci. 1990;15(12):483–486. doi: 10.1016/0968-0004(90)90303-s. [DOI] [PubMed] [Google Scholar]
- Pinhal MA, Smith B, Olson S, Aikawa J, Kimata K, Esko JD. Enzyme interactions in heparan sulfate biosynthesis: Uronosyl 5-epimerase and 2-O-sulfotransferase interact in vivo. Proc Natl Acad Sci USA. 2001;98(23):12984–12989. doi: 10.1073/pnas.241175798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy JV, Burguete AS, Sridevi K, Ganley IG, Nottingham RM, Pfeffer SR. A functional role for the GCC185 Golgin in mannose 6-phosphate receptor trafficking. Mol Biol Cell. 2006;17(10):4353–4363. doi: 10.1091/mbc.E06-02-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman S. The synthesis of complex carbohydrates by multiglycosyltransferase systems and their potential function in intercellular adhesion. Chem Phys Lipids. 1970;5(1):270–297. doi: 10.1016/0009-3084(70)90024-1. [DOI] [PubMed] [Google Scholar]
- Roth J, Berger EG. Immunocytochemical localization of galactosyltransferase in HeLa cells: Codistribution with thiamine pyrophosphatase in trans-Golgi cisternae. J Cell Biol. 1982;93(1):223–229. doi: 10.1083/jcb.93.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo R, Shaper N, Taatjes D, Shaper J. Beta 1,4-galactosyltransferase: A short NH2-terminal fragment that includes the cytoplasmic and transmembrane domain is sufficient for Golgi retention. J Biol Chem. 1992;267(13):9241–9247. [PubMed] [Google Scholar]
- Sasaki K, Kurata-Miura K, Ujita M, Angata K, Nakagawa S, Sekine S, Nishi T, Fukuda M. Expression cloning of cDNA encoding a human beta-1,3-N-acetylglucosaminyltransferase that is essential for poly-N-acetyllactosamine synthesis. Proc Natl Acad Sci USA. 1997;94(26):14294–14299. doi: 10.1073/pnas.94.26.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Sato M, Kiyohara K, Sogabe M, Shikanai T, Kikuchi N, Togayachi A, Ishida H, Ito H, Kameyama A, et al. Molecular cloning and characterization of a novel human beta1,3-glucosyltransferase, which is localized at the endoplasmic reticulum and glucosylates O-linked fucosylglycan on thrombospondin type 1 repeat domain. Glycobiology. 2006;16(12):1194–1206. doi: 10.1093/glycob/cwl035. [DOI] [PubMed] [Google Scholar]
- Schmitz KR, Liu J, Li S, Setty TG, Wood CS, Burd CG, Ferguson KM. Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev Cell. 2008;14(4):523–534. doi: 10.1016/j.devcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seko A, Yamashita K. Activation of beta1,3-N-acetyl-glucosaminyltransferase-2 (beta3Gn-T2) by beta3Gn-T8. Possible involvement of beta3Gn-T8 in increasing poly-N-acetyllactosamine chains in differentiated HL-60 cells. J Biol Chem. 2008;283(48):33094–33100. doi: 10.1074/jbc.M806933200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaper NL, Shaper JH, Meuth JL, Fox JL, Chang H, Kirsch IR, Hollis GF. Bovine galactosyltransferase: Identification of a clone by direct immunological screening of a cDNA expression library. Proc Natl Acad Sci USA. 1986;83(6):1573–1577. doi: 10.1073/pnas.83.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale R, D’Agostaro G, Gleeson P. The signal for Golgi retention of bovine beta 1,4-galactosyltransferase is in the transmembrane domain. J Biol Chem. 1992;267(6):4084–4096. [PubMed] [Google Scholar]
- Trayer IP, Hill RL. The purification and properties of the A protein of lactose synthetase. J Biol Chem. 1971;246(21):6666–6675. [PubMed] [Google Scholar]
- Tu L, Tai WCS, Chen L, Banfield DK. Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science. 2008;321(5887):404–407. doi: 10.1126/science.1159411. [DOI] [PubMed] [Google Scholar]
- Ujita M, McAuliffe J, Hindsgaul O, Sasaki K, Fukuda MN, Fukuda M. Poly-N-acetyllactosamine synthesis in branched N-glycans is controlled by complemental branch specificity of I-extension enzyme and beta1,4-galactosyltransferase I. J Biol Chem. 1999;274(24):16717–16726. doi: 10.1074/jbc.274.24.16717. [DOI] [PubMed] [Google Scholar]
- Ujita M, Misra AK, McAuliffe J, Hindsgaul O, Fukuda M. Poly-N-acetyllactosamine extension in N-glycans and core 2- and core 4-branched O-glycans is differentially controlled by i-extension enzyme and different members of the beta 1,4-galactosyltransferase gene family. J Biol Chem. 2000;275(21):15868–15875. doi: 10.1074/jbc.M001034200. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Fukuda MN. Golgi retention mechanism of beta-1,4-galactosyltransferase. J Biol Chem. 1995;270(20):12170–12176. doi: 10.1074/jbc.270.20.12170. [DOI] [PubMed] [Google Scholar]
- Zhou D. Why are glycoproteins modified by poly-N-acetyllactosamine glyco-conjugates? Curr Protein Pept Sci. 2003;4(1):1–9. doi: 10.2174/1389203033380304. [DOI] [PubMed] [Google Scholar]