Abstract
We present a technique for labeling the contents of acidic organelles and rapidly releasing, separating, and detecting their labeled contents with laser-induced fluorescence. We have performed solution-phase separation of the contents of single mitochondria and single 100 nm vesicles, which represents a demonstration of an analyzed volume of ~1 attoliter. Our strategy to label the acidic contents of the mitochondrion relies on the use of the membrane-permeable dye, Oregon Green diacetate succinimidyl ester, and a membrane-permeable base to raise intra-mitochondrial pH. In order to measure the contents, we utilized a glass microfluidic chip and high voltage gradient for millisecond capillary-electrophoresis separation after single-mitochondrion photolysis. We observed heterogeneity among a population of mitochondria with respect to a constituent chemical component.
Keywords: microfluidics, mitochondria, capillary electrophoresis, vesicles, photolysis, laser-induced fluorescence, heterogeneity, single mitochondria, single organelle
Just as there are unique cells within a population, the heterogeneity among organelles within a cell has also been implicated as an important aspect of their function. A number of studies have characterized the inhomogeneity of mitochondria at the single-mitochondrion level. The Arriaga group has pioneered studies in this field1-4. In their experiments, fluorescent mitochondria were separated under the influence of an electric field in a capillary vaguely analogous to flow cytometry. The mitochondria crossed a detector, where parameters like peak width and peak height served to give one or more scalar values of information about each mitochondrion. Unlike flow cytometry, retention time is related to surface charge density and mobility of the mitochondrion, which may be relevant to the biological activity of the organelle. For instance, free-flow electrophoresis has been used to isolate permeable mitochondria from non-permeable mitochondria5. The permeability of mitochondria is intimately related to apoptosis6, 7.
These single-mitochondrion studies yield information about the distribution of properties within the population of mitochondria and even reveal such basic facts as the number of mitochondria per cell and the relative enzyme activity among mitochondria4. These studies on intact mitochondria are not small-scale analogues to the analysis of the contents of single cells8, 9 or single secretory vesicles10. Such solution-phase separation experiments provide an electropherogram of the contents of a single biological capsule. As such, they contain more molecular information than can be obtained by the interrogation of a transiting whole mitochondrion. Mass-spectrometry is another method to gather such chemical data, and it offers high-content information on the single cells and vesicles being analyzed11, 12, but the current sensitivity offered by mass spectrometry is still orders of magnitude lower than fluorescence-based detection13.
Here, we describe the use of capillary-electrophoresis (CE) and laser induced fluorescence (LIF) detection to analyze the dye-tagged contents of single mitochondria. In principle, interrelated values can be extracted using such analysis, such as the concentration ratios of metabolites. The ratio of oxidized to reduced mitochondrial glutathione in the liver, for instance, can indicate a starved organism14. How that property might vary among mitochondria is unknown. Our work builds upon a rich literature of experiments that employ CE for the analysis of single-cell contents in the past decades15-22. Decreasing the sample volume from cellular to subcellular, however, brings a number of challenges. The subcellular volume contains fewer molecules - 1 μM of analyte in a 10 μm cell corresponds to the presence of 3×105 molecules but to only 40 molecules in a 0.5 μm mitochondrion - and so obviously the detection system must be sufficiently sensitive. Sensitive detection can be performed with electrochemistry23, 24 or native fluorescence22. Many chemical species within the mitochondria, however, have no native fluorescence or are not electro-active. Therefore, we have elected to label the amine contents of the mitochondria with a fluorescent dye; proteins, peptides, and many small molecules all have free amine groups.
In previous single-cell work, the cell was often lysed prior to derivatization so the cellular contents could mix and react with the dye. This approach is effective for single-cell analysis because a typical cell is still relatively large and diffusive dilution is still manageable. At the level of single subcellular organelles, however, this approach becomes highly inefficient. For example, after one second, 1 mM of a small-molecule (diffusion coefficient of 4.9×10-6 cm2 sec-1) analyte is diluted to 30 μM after lysis of a 10μm cell but to 10 nM after lysis of a 0.5 μm organelle. As a result, it is imperative to spatially confine the dye-tagging reaction. In other reports, we have presented a platform that employs femtoliter-volume droplets as nanolabs for controlling single-molecule reactions25, 26. This paper explores an alternative strategy in which we derivatize the contents of single mitochondria prior to their lysis, thus confining the dye-tagging reaction within the intact mitochondria.
Our strategy relies on the use of the membrane-permeable dye, Oregon Green diacetate succinimidyl ester (OGDA-SE). Oregon Green (OG) is a derivative of fluorescein, which is an excellent dye for sensitive detection27. The diacetate group makes OG and similar dyes membrane-permeant. Diacetate derivatives of Oregon Green, fluorescein28 and Calcein AM20 have all been added to cells for subsequent analysis by single-cell CE. These dyes pass through the cell membrane and are cleaved by intracellular esterases. Once cleaved, the dyes are fluorescent and membrane impermeable, and thus trapped within the cell. Inside the cell, reactive groups on the dye molecule can tag targets of interest for subsequent analysis. Reactive versions of these dyes can be made to tag amines or thiol groups. It is worth noting for our purposes that these intracellular esterases are also known to be present in the mitochondrion29. This strategy is superficially similar to the technique that Presley et. al. demonstrated for labeling intra-mitochondrial protein for analysis by bulk CE30. In that case, a thiol reactive version of Mito Tracker Green enters the mitochondria and labels mitochondrial proteins with thiol groups.
Reaction of free amines with the succinimidyl ester (SE) group is facilitated with basic pH. Depending on the pKa of the amine, neutral to basic pH is needed to deprotonate the quaternary amine to react with the SE group. Unlike the cell cytoplasm where pH is ~7.6, mitochondrial intermembrane pH is acidic31. As a result, OGDS-SE will not react with mitochondrial amine groups as it would with many cytoplasmic amine groups. To overcome this issue, we used benzylethanolamine (BEA) to raise the pH of mitochondria once OGDA-SE had permeated and become trapped within the mitochondria. BEA, like ammonia, is membrane permeant and thus enters intact mitochondria with ease. Unlike ammonia, however, BEA is very bulky and thus cannot react with the SE group. This fact allows us to raise the pH of intact mitochondria without competing with and adversely affecting the derivatization reaction.
Once the contents of single mitochondria have been dye-tagged, each mitochondrion needs to be lysed and analyzed by CE. We chose to use a single nanosecond laser pulse to lyse individual mitochondria32-34 because of the laser-photolysis technique's robustness, speed, and ease of interfacing with our microfluidic system. A wide range of methods exist to lyse single cells, including electrical lysis28, detergents35, shear flow15, and even the use of a sharpened capillary tip for insertion into a cell and direct sampling of the cytoplasm of the cell36. For the analysis of single subcellular organelles, we believe single-pulse laser photolysis offers the best result: it is much faster than most other techniques in disrupting the membrane34, 37, does not require microfabrication of electrodes or other elements, has high spatial resolution, and offers high throughput and duty cycle.
The next challenge in CE analysis of single mitochondria is the speed of separation. Again, because of the minute volume involved and the rapidity of diffusion in diluting the dye-tagged analyte, the separation must be sufficiently fast. The 10 attoliter (aL) volume of a 0.5 μm mitochondrion will be diffusionally broadened to 6 pL within 1 s, resulting in a ten thousand fold dilution of the analyte. To minimize diffusive dilution and to maintain concentrated and separated bands, we carried out ultra-fast (a few ms) CE separation following single-pulse photolysis of individual mitochondria. Because ultra-fast separations require intense voltage conditions, our experiments preclude the use of PDMS (polydimethylsiloxane) despite the ease of fabricating microchannels in PDMS. Glass is ideal for our experiments, as it gives superior performance in CE and can withstand much higher voltages. To simplify the fabrication of glass-based microfluidic systems, we have developed a low-temperature calcium-assisted bonding scheme that allows us to rapidly prototype glass microchannels38, 39.
Approaching the problem in this manner allows for rapid profiling of many mitochondria. The separation itself takes only a few milliseconds, and lining up each mitochondrion in the focus region of the pulsed UV laser takes only a few seconds.
Materials and Methods
Chip fabrication
We etched 1 or 0.5 cm long, 50 μm wide separation channels into D263T glass (Precision Glass and Optics, Santa Ana, CA) using a PDMS etch guide and hydrofluoric acid (HF)40. Briefly, the PDMS guide was produced by replica molding from a SU-8-2050 photoresist (Microchem, Newton, MA) on a silicon master produced by standard photolithography. The pattern included four straight channels approximately 3 cm long. This guide was punched with fluidic access holes and conformally sealed to glass. A solution of 5% HF was then introduced to the channels by negative pressure. Etching was allowed to continue with periodic reapplication of vacuum for 1 min.
We machined wide, perpendicular inlet and outlet access channels to these small separation channels with a diamond bit and Dremel rotary tool. We then added inlet and outlet ports with the same. The spacing between the two large channels defines the length of the high-resistance region for voltage gradient calculation. We then sealed a glass coverslip to the substrate with calcium-assisted low temperature glass-glass bonding as described in detail elsewhere40. Briefly, the etched glass substrate and cover were washed with Alconox detergent (Alconox, Inc. White Plains, NY) then with a 0.5% Alconox/0.5% calcium acetate slurry. The panes were then pressed together under a flow of deionized water and allowed to dry. Finally they were treated with heat at 115 °C for 2 hours or more. Following low-temperature bonding, we applied high-temperature fusion bonding using a programmable furnace (Vulcan 3-550, ESP Chemicals, Tucson, AZ) with a top temperature of 625 °C. We used epoxy to attach cut Eppindorf tubes to the inlets and outlets of the glass chip for buffer and sample wells.
Preparation of 100 nm labeled glutamate vesicles
We prepared a solution of 10 mM glutamic acid. The final pH of the solution was adjusted to approximately pH 8 with NaOH to facilitate labeling. We then reacted the glutamate primary amine with Oregon Green 488 succinimidyl ester (OG-SE, Invitrogen, Carlsbad, CA) per manufacturer's instructions. After the labeling reaction, we prepared vesicles by first drying 0.6 mg 1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphocholine (POPC, Avanti Polar Lipids, Alabaster, AL) from chloroform with nitrogen for 3 hours. We then vortexed the solution to suspend the lipid in 300 μl of the labeled 10 mM amino acid mixture. Later we extruded by 50 passes through a 100 nm pore filter. The extruded vesicles were then frozen in liquid nitrogen and thawed three times. After the last freeze cycle, they were stored at -20 °C until needed. Just prior to the experiment, the vesicles were thawed then purified of extra-vesicular dye using a size exclusion column packed with Sephacryl S-200 HR media (Sigma, St. Louis, MO).
Preparation and labeling of the contents of acidic vesicles
We prepared an unlabeled amino acid buffer containing 10 mM glutamic acid. The final pH of the solution was adjusted to approximately pH 6 with HCl. This solution was encapsulated into vesicles as described above except that in this case we used egg-derived phosphatidyl choline (Egg PC, Avanti). We then exchanged the vesicles into a basic buffer (10 mM PBS adjusted to pH 8) using a size exclusion column packed with Sephacryl S-200 HR media (Sigma, St. Louis, MO). These vesicles were then treated with benzylethanolamine (BEA, Sigma, St. Louis, MO) at a final concentration of 1 mM and labeled with Oregon Green diacetate succinimidyl ester (OGDA-SE, Invitrogen, Carlsbad, CA) per manufacturer's specifications. A control sample was labeled without the addition of BEA. After labeling for 2 hours and just prior to the experiment, the vesicles were purified of extra-vesicular dye using a size-exclusion column packed with Sephacryl S-200 HR media (Sigma, St. Louis, MO). A control portion of the non-basified vesicles was not purified in this manner. The results were analyzed by bulk CE (discussed below).
Preparation of mitochondria
We prepared mitochondria from B cells using the Pierce Mitochondria Isolation kit (Pierce, Rockford, IL) using the Dounce homogenization method according to manufacturer's instructions. We resuspended the final mitochondrial pellet in 150 mM total salt concentration phosphate buffered saline (PBS, from 10x concentrate; Fisher Scientific, Fairlawn, NJ). We then dissolved 50 μg of OGDA-SE in 1 μl DMSO and added 50 μl of the mitochondrial isolate. OGDA-SE is membrane permeant until cleaved by esterases within the mitochondrion, and it is reactive with free amines. The OGDA-SE and mitochondria were allowed to react at 4 °C for 3 hours. This is similar to the technique developed by Presley et. al.30 to label cysteine residues on mitochondrial proteins with a thiol-reactive version of the fluorescent dye Mito Tracker Green.
Use of CE chip
The chip was prepared before use by cleaning with 0.1 M NaOH for 20 minutes, then running buffer (10 mM total salt concentration PBS; Fisher brand 10x concentrate PBS diluted 150x) for 2 hours. Vesicles in 10 mM total salt concentration PBS (or mitochondria in 150 mM total salt concentration PBS) were loaded into the waste access channel and reverse voltage was applied to introduce them into the separation channels. The surface density was then checked. When a reasonable density was obtained, electrodes were then placed in the wells in the forward voltage orientation as shown in Figure 1A. The surface density was chosen to be approximately 5-10 mitochondria per viewable frame for convenience in choosing a target. Forward voltage was then applied for several minutes to pull clear buffer into the separation channel. The wells were then rinsed and filled to roughly equal levels.
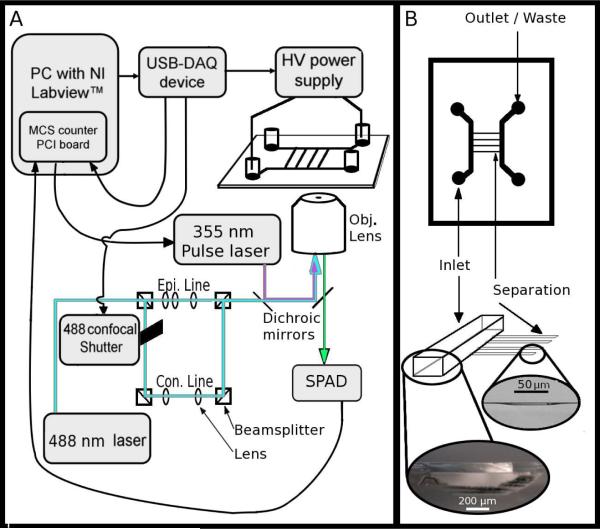
Figure 1.
(A) A schematic diagram of the instrument, including the connections among the modules and essential optics. The USB-DAQ triggers the opening of a mechanical shutter for confocal illumination then activates the HV power supply. Once the power supply has reached full voltage, the USB-DAQ board triggers the MCS card (which is located in the PC). The MCS card records the photon counts from the single photon avalanche diode (SPAD); at 50% time of the full data collection run, the MCS card sends a TTL pulse to the nanosecond pulsed UV laser to initiate photolysis. The 488 nm laser is shuttered such that while aiming, it is configured for epi-fluorescence (to show the location of all mitochondria within the field of view), and during data acquisition it is configured for line confocal detection with the SPAD. Epi. indicates epifluorescence optics, Con. indicates confocal optics, Obj. indicates objective lens. (B) Schematic and images showing the essential elements of the microfluidic chip design including the 750 μm wide, 120 μm deep inlet and outlet channels and the 50 μm wide, ~1 μm deep separation channels. Light micrographs of the two respective side profiles are shown at bottom.
A target (vesicle or mitochondrion) was selected from the surface and centered in the UV focus region. The USB-6008 DAQ module programmed in Labview (National Instruments, Austin, TX) initiated the high voltage for 300 msec (in order to give the power supply sufficient time to come to steady state) then opened the shutter for confocal illumination and initiated data collection using a MCS card (Multi Channel Scaler, Ortec, Oak Ridge, TN). The MCS card triggered a single nanosecond laser pulse to photolyze the selected mitochondrion. The UV source was a frequency tripled YAG laser at 355 nm with a pulse width of 3-5 nsec (Minilite, Continuum, Santa Clara, CA). The time resolution of the MCS trigger to initiate UV laser pulse is very high (within 10 μsec) unlike the reproducibility of the USB-6008 used to initiate other events, which is accurate only to within about 10 msec. As a result, we used the initiation of the photolysis laser pulse to define time zero in our electropherogram. Figure 1A shows schematically the connections among the experimental modules. All data collection and imaging was performed through a SuperFluor 100x objective (Nikon Instruments, Melville, NY).
Bulk CE analysis
To interpret the CE electropherograms of single mitochondria and vesicles, we performed bulk CE of the contents of the mitochondria and vesicles. The results of the bulk studies serve as expectation results for the single-mitochondrial separations. In the case of the synthetic vesicles, we performed bulk CE on the labeled amino acids used to fill the 100 nm vesicles. In the case of the mitochondria, we performed CE analysis on a mitochondrial lysate. Mitochondria were prepared as above for single-mitochondrion CE and lysed with 4% Triton X-100 (Fisher Biotech, Fairlawn, NJ).
For all mitochondrial and vesicle runs, a 30 cm capillary with an inner diameter of 50 μm was rinsed for 20 min with 0.1 M NaOH, then the capillary was equilibrated with 10 mM borate buffer for at least 2 hours. Amino acid standard samples were diluted into the same 10 mM borate by 1000x; mitochondrial and vesicle lysates were not diluted. Injection was made by dipping the capillary end into the diluted sample and raising the capillary inlet approximately 5 cm above the outlet for 10 sec. The inlet was then placed into an Eppendorf tube that was connected to ground by a platinum electrode. High voltage (8 kV) was then applied to the outlet of the capillary with a second platinum electrode. A 20x objective was focused on the capillary 1 cm from the outlet and detection was performed with a line-focused 488 nm laser and a CCD camera.
Acidic bulk vesicle CE was performed similarly with the following exceptions: the location of the detection window was 10 cm from the exit of the capillary; separation voltage was 6 kV; and data was collected with a PMT and National Instruments DAQ board.
Results and Discussion
Chip design
The chip design consisted of very short and shallow separation channels (1-2 μm deep, etched with 50 μm wide guide) sandwiched between two deep and wide access channels (~100 μm deep, 750 μm wide) (Figure 1B). We arrived at this design because of the following considerations: (1) shallow separation channels result in reduced room for diffusion in the Z direction; (2) the use of separation channels with small cross sections in comparison with the large access channels causes most of the voltage drop to occur across the separation channels; and (3) shallow channels are difficult to fill and access with electrodes, so large, deep channels led up to the small separation channels to facilitate electrical and fluidic access. Figure 1A shows the schematic of the experimental setup with the electrical contacts shown at opposite ends of the chip. Electroosmotic flow (EOF) is directed toward the negative high voltage, and so the high voltage electrode is placed in the outlet channel (Figure 1B).
The total length of the separation channel and its effect on the voltage gradient is critical as there exists a practical limit on the total applied voltage. Coverslips for use with oil-immersion high-numerical-aperture objectives are ~ 150 μm thick. The microscope stage on which the coverslip rests is grounded for safety reasons, which means the voltage gradient across the glass coverslip can easily exceed the breakdown potential for glass. Thus, the total voltage is generally kept below 12 kV. Given this constraint, short and shallow separation channels are the best way to increase the voltage gradient needed for fast separations. Additionally, the electrodes must be placed far enough from each other such that the applied voltage does not exceed the breakdown limit for air. The micrograph in Figure 1B shows the cross section of the large and deep machined channels for the inlets and outlets contrasted with the shallow HF-etched separation channels. Within the separation channels, membrane-bounded samples will settle on the surface, ready for analysis by CE.
Diffusion after photolysis
Our general experimental protocol was to position a target (vesicle or mitochondrion) (Figure 2A) and photolyze it with a single tightly-focused nanosecond UV laser pulse (Figure 2B). The contents of the target diffuse out (Figure 2C) and are separated according to their electrophoretic mobilities by a strong applied electric field (Figure 2D). In this experiment, there is a competition between diffusion, which rapidly dilutes the released contents, and CE separation. To understand this competition, we carried out both simulations and experiments to characterize diffusion after photolysis in our separation channels.
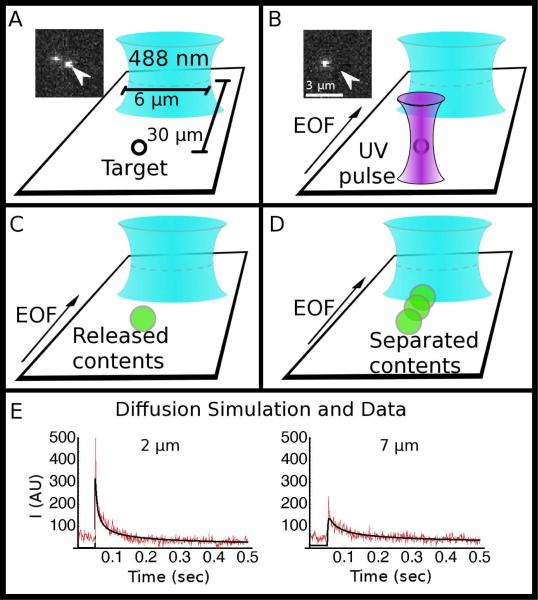
Figure 2.
(A-D) Schematics showing the sequence of events in a capillary-electrophoresis experiment, starting with (A) a target (vesicle or mitochondrion) aligned in the UV laser focus, which caused the target to be lysed with a single nanosecond UV laser pulse (B); the insets show the target vesicle (arrow) before and after lysis. The released components (C) are separated as they travel toward and across the probe volume (blue elliptical focus) (D). (E) Simulation of 1-D free diffusion (black) overlaid on data collected (red) with the line confocal probe volume parked at 2 μm from the release point, and 2-D free diffusion (black) overlaid on data collected (red) at 7 μm from the release point.
To enhance detection, we used the line-confocal geometry where the probe volume is shaped by cylindrical lenses into a line, and the collected fluorescence passes through a slit placed at the primary image plane before being recorded by the detector27. For shallow channels (on the order of 1 μm), the diffusion in the Z direction is restricted for points greater than 1 μm away. It would be possible to reduce diffusional dilution further by making the channels more narrow or to reduce wall effects with a deeper channel. Choosing wide, shallow channels restricts diffusion in only one dimension (Z) but introduces only one wall into the vicinity. This is a compromise between detrimental wall effects and sample dilution. While the analyte diffuses in two dimensions, we integrate across the second dimension with our line confocal detector to reduce dilution effect still farther. By integrating optically in one dimension and restricting physically in the second dimension, we reduce the diffusion to 1-D equivalent while introducing wall effects on only 2 sides41.
This condition breaks down if the sample diffuses to a diameter beyond the width of the line confocal. Our line confocal is approximately 6 μm wide and so we anticipated that diffusion at a radius of less 3 μm will give 1-D diffusion profiles, and at 3 μm and greater will give results consistent with higher dimension diffusion.
To test this theory, we photolyzed 100 nm-diameter synthetic vesicles prepared in a buffer containing OG-labeled glutamate and observed the encapsulated fluorophores diffuse across the detector. We simulated the process using both a 1-D and 2-D diffusion model. We found the 1-D model fits well with the experimental data when the line-confocal probe volume was placed at 2 μm from the release point, while the 2-D model fits well the data collected when the probe volume was placed at 7 μm away from the release point. In Figure 2E we show the best fit 1-D model overlaid upon the free diffusion data for the 2 μm; we show the 2-D diffusion overlaid upon the data collected at 7 μm. In Figure 2E we show the best fit 1-D model overlaid upon the free diffusion data for the 2 μm; we show the 2-D diffusion overlaid upon the data collected at 7 μm. In order to maintain the 1-D case during the electrophoresis, we keep the separation time sufficiently low that the diffusive radius is less than 3μm. The decrease in peak signal as the distance increases is also evident. We carried out our experimental CE separations at distances greater than 24 μm; based on this, we can presume that, had electrophoresis/EOF not carried the fluorophores across the detector, we would have seen no signal.
CE separation of the contents of a 100 nm vesicle
To test our experimental design and protocol, we first analyzed the contents of single 100 nm-diameter synthetic vesicles containing glutamate pre-labeled with Oregon Green succinimidyl ester (OG-SE). To our knowledge, this is the smallest volume analyzed thus far with CE. These vesicles were made by extrusion through a polycarbonate filter with 100 nm-diameter pores34. We have characterized previously that at this small size, virtually all vesicles are unilamellar42. The vesicles were prepared in the presence of 4 mM total dye (OG-SE) concentration. Assuming an encapsulation efficiency of ~ 10%41, 43 and a vesicle volume of 1 aL, we estimate each vesicle contains ~ 250 fluorescent molecules. This number is an estimate and can change depending on the exact volume and the encapsulation efficiency of each vesicle. Nevertheless, the number of contained molecules likely range below a thousand.
Figure 3 shows the results of the photolysis and electrophoretic separation of the contents of a single 100 nm vesicle; the insets to the right show a vesicle before and after photolysis. The reaction of glutamate with OG-SE was not complete, and there is a substantial proportion of free OG remaining in solution despite the use of excess glutamate during labeling. The single-vesicle electropherogram thus shows two peaks as does the bulk electropherogram shown in the inset.
Figure 3.
Electropherogram of a single 100 nm vesicle (1 attoliter in volume) containing Oregon Green (OG) and OG labeled glutamate (OG-Glu) after single-pulse photolysis. The data shows resolved glutamate and free dye peaks. The inset above shows a bulk electropherogram of the glutamate labeling products prior to encapsulation; the images to the right shows the vesicle prior to (top image) and after (bottom image) photolysis; the targeted vesicle is indicated with an arrow.
It is interesting to note how the relative size of the two peaks changes from the bulk to the single-vesicle scenarios, although in both cases we used the same amounts of OG-SE and glutamate and conducted the labeling under identical conditions. Additionally, bulk studies of purified and then lysed vesicles show a significant change in the proportions of the components from the original, extra-vesicular concentrations in which the vesicles were made (see supplementary data). We have insufficient data to draw conclusions about this phenomenon at this time. It is possible that the two species encapsulate into the vesicles with different efficiencies or, conceivably, the altered proportions could also be caused by an enhanced labeling reaction efficiency within the vesicle.
Verification of succinimidyl ester labeling strategy for acidic organelles
After we successfully tested our experimental design, we needed to verify our intra-organelle labeling strategy. Unlike the cytoplasm of cells, mitochondria are known to maintain an acidic pH within their intermembrane space relative to the cytosol31. This would be expected to inhibit labeling by OGDA-SE. To overcome this issue, we chose to use a membrane permeant base, benzylethanolamine (BEA), to raise the mitochondrial pH. Because BEA is very bulky, it should not react with OGDA-SE. To verify the efficacy of this strategy, we characterized it with vesicle-encapsulated glutamate.
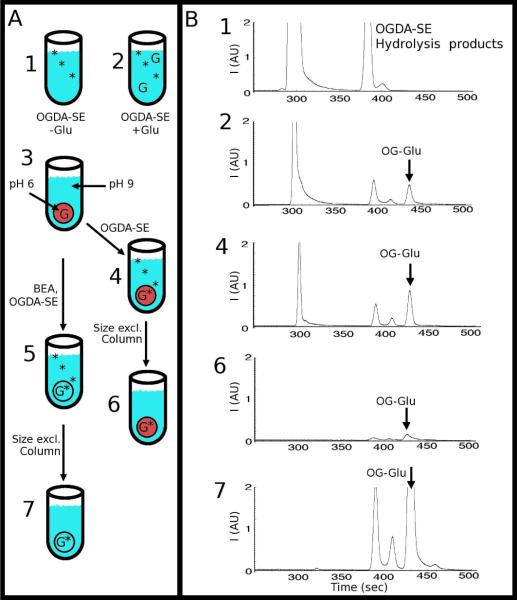
Figure 4A outlines our experiment. We prepared synthetic vesicles at an acidic pH to inhibit the dye-labeling reaction. We exchanged these acidic vesicles into a basic buffer with a size-exclusion column, after which the intra-vesicular solution was acidic while the extra-vesicular solution was basic (solution #3 in Figure 4A). This scenario is similar to the conditions encountered in the analysis of actual acidic organelles, where the organelles are present in a slightly basic physiological buffer. Next, we split the above solution (solution #3) in two. For the first solution (right set of procedure in Figure 4A), we introduced OGDA-SE but without BEA (solutions #4 and #6); for the other solution (left branch of procedure in Figure 4A), we introduced both OGDA-SE and BEA (solutions #5 and #7). We then analyzed these solutions using CE.
Figure 4.
(A) Schematic showing the labeling scheme for encapsulated contents. Solutions 1 and 2 show the Control Samples containing Oregon Green Diacetate Succinimidyl Ester (OGDA-SE) without (1) and with (2) Glutamate (Glu). The flow chart shows acidic, glutamate-containing vesicles (red circles) are suspended in a basic buffer (3). To this solution (3), OGDA-SE was added without (4) and with (5) benzylethanolamine (BEA), which raised the intra-vesicular pH to basic. The red vesicle in (4) represents acidic intra-vesicular pH, while the clear vesicle in (5) represents basic intra-vesicular pH (same pH as the extra-vesicular solution). Both solutions (4) and (5) were purified with a size-exclusion column to remove extra-vesicular OGDA-SE, Glu, and OG-Glu (solutions 6 and 7). (B) Electropherograms of the corresponding numbered solutions in the labeling scheme. The vesicles were lysed with Triton X-100 and analyzed with CE. A small amount of OG-Glu was seen in (4) and (5) because of the presence of a small amount of Glu in the extra-vesicular solution that reacted with OGDA-SE, because the size-exclusion column might not be 100% efficient in removing all extra-vesicular Glu and because a small amount of lysis of the vesicles might have released some Glu into the extra-vesicular solution.
Figure 4B shows our results. Electropherograms of solutions #1 and #2 are control experiments without vesicles; these experiments show the location of the OGDA-SE related peaks and the OG-Glu peak. In the absence of BEA (solution #4), some OG-Glu can still be detected. We attribute this presence of OG-Glu to residual glutamate in the extra-vesicular solution, which was caused by either incomplete removal of glutamate during size-exclusion chromatography or from leakage of glutamate from the vesicles during or after size-exclusion chromatography. We ran the sample through a second size-exclusion column, just prior to CE. This removed any free glutamate and OG-Glu in the extra-vesicular solution and caused a dramatic reduction in the OG-Glu peak (solution #6). The electropherogram of solution #6 (which represents the true acidic intra-vesicular reaction products) contrasts with the results we obtained for the solution treated with both OGDA-SE and BEA (solution #7). As is evident by comparing the electropherograms of solutions #6 and #7, there is a large increase in the OG-Glu signal when BEA was used. And as anticipated, BEA did not react with OGDA-SE. We took this result as confirmation that our strategy works in raising the intra-vesicular pH with BEA while avoiding any adverse effects on the labeling reaction itself.
CE analysis of single mitochondria
We next applied our strategy for the CE analysis of single mitochondria. We isolated mitochondria from B-cells using a commercial kit. We then labeled the mitochondrial contents with OGDA-SE. Again, we tested the labeling with and without BEA treatment. In our experiments, we loaded the mitochondria using pressure into the outlet channels then applied a reverse field to use EOF to draw the mitochondria into the separation channels. Once a reasonable surface density of mitochondria was obtained, forward voltage was applied to sweep out any free-floating mitochondria from the separation channels and to introduce the low-molarity running buffer. We avoided high-molarity running buffer because it is much more conductive and thus increases the Joule heating within the channel.
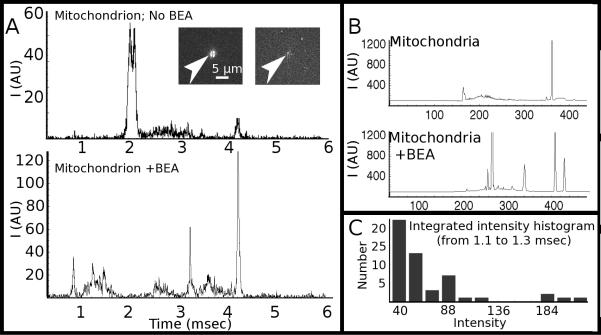
Figure 5A shows electropherograms of two single mitochondria, one labeled without BEA (top) and the other with BEA (bottom). Figure 5B shows electropherograms of bulk mitochondria, again without (top) and with (bottom) BEA present. For bulk analysis, we lysed the mitochondria using 4% Triton-X100. For both the single-mitochondria and bulk experiments, the use of BEA resulted in the presence of more detected peaks because of more efficient labeling. The two visible peaks in the electropherograms without BEA are OG and OG-SE (the acetate groups are cleaved by intra-mitochondrial esterases). With BEA, many more peaks are detected. The results, however, do not lend themselves easily to analysis, which is not surprising given the complexity of the mitochondrial contents. As a result, we decided to compare the variations in a small constituent range of the amine contents of single mitochondria, which we approximated by integrating the photon counts around 1.2 msec (Figure 5C). Our results indicate the presence of heterogeneity, in which we observed what appear to be three populations. The histogram shows the majority of cases with low amounts of analyte (intensities from 40-72), a significant population with medium analytes (intensities 88-120) and a third population with relatively large amounts of analytes (intensities above 184).
Figure 5.
(A) Single-mitochondrion electropherograms obtained without the use of benzylethanolamine (BEA) (top trace) and with BEA (bottom trace). Inset shows before and after photolysis of a mitochondrion prior to capillary-electrophoresis separation. (B) Bulk electropherograms of mitochondrial lysate without (top) and with (bottom) BEA in the labeling step. (C) Histogram of integrated photon counts from 1.1 to 1.3 msec, which shows a multi-modal distribution among those mitochondria displaying a peak at this retention time.
Conclusions
This paper describes a technique for analyzing the contents of single mitochondria with CE. The principle of this approach is simple: label the contents of mitochondria with a membrane-permeable dye by raising the intra-mitochondrial pH with BEA, then photolyze individual mitochondria followed by millisecond CE separation to minimize diffusive dilution. Using synthetic vesicles, we have characterized this approach and demonstrated the ability to use CE for analyzing attoliter-volume samples. When applied to single mitochondria, this technique is able to separate and detect the contents of individual mitochondria within seconds, with a current duty cycle of about one minute, which is limited by our ability to locate and position manually each mitochondrion for photolysis. With automation, the current duty cycle can be improved to a few seconds. Our preliminary results suggest the presence of variability in the contents of mitochondria; a more biologically meaningful study will require a detailed assignment of the detected peaks. Besides mitochondria, we believe our strategy is generalizable to other acidic organelles.
Supplementary Material
ACKNOWLEDGEMNTS
The authors gratefully acknowledge funding provided through NIH grant EB005197.
Footnotes
Supporting Information Available: Electropherograms of labeled amino acids prior to and after encapsulation inside lipid vesicles, which show changes in relative concentrations.
REFERENCES
- (1).Duffy CF, MacCraith B, Diamond D, O'Kennedy R, Arriaga EA. Lab Chip. 2006;6:1007–1011. doi: 10.1039/b601896c. [DOI] [PubMed] [Google Scholar]
- (2).Fuller KM, Arriaga EA. Curr. Opin. Biotech. 2003;14:35–41. doi: 10.1016/s0958-1669(02)00008-3. [DOI] [PubMed] [Google Scholar]
- (3).Fuller KM, Duffy CF, Arriaga EA. Electrophoresis. 2002;23:1571–1576. doi: 10.1002/1522-2683(200206)23:11<1571::AID-ELPS1571>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- (4).Strack A, Duffy C. n. F., Malvey M, Arriaga EA. Analytical Biochemistry. 2001;294:141–147. doi: 10.1006/abio.2001.5148. [DOI] [PubMed] [Google Scholar]
- (5).Zischka H, Larochette N, Hoffmann F, Hamoller D, Jagemann N, Lichtmannegger J, Jennen L, Muller-Hocker J, Roggel F, Gottlicher M, Vollmar AM, Kroemer G. Anal. Chem. 2008;80:5051–5058. doi: 10.1021/ac800173r. [DOI] [PubMed] [Google Scholar]
- (6).Bernardi P, Scorrano L, Colonna R, Petronilli V, Lisa FD. Eur. J. Biochem. 2001;264:687–701. doi: 10.1046/j.1432-1327.1999.00725.x. [DOI] [PubMed] [Google Scholar]
- (7).Grimm S, Brdiczka D. Apoptosis. 2007;12:841–855. doi: 10.1007/s10495-007-0747-3. [DOI] [PubMed] [Google Scholar]
- (8).Hu S, Zhang L, Newitt R, Aebersold R, Kraly JR, Jones M, Dovichi NJ. Anal. Chem. 2003;75:3502–3505. doi: 10.1021/ac034154j. [DOI] [PubMed] [Google Scholar]
- (9).Olson KJ, Ahmadzadeh H, Arriaga EA. Anal Bioanal Chem. 2005;382:906–917. doi: 10.1007/s00216-005-3135-2. [DOI] [PubMed] [Google Scholar]
- (10).Chiu DT, Lillard SJ, Scheller RH, Zare RN, Rodriguez-Cruz SE, Williams ER, Orwar O, Sandberg M, Lundqvist JA. Science. 1998;279:1190–1193. doi: 10.1126/science.279.5354.1190. [DOI] [PubMed] [Google Scholar]
- (11).Gutstein HB, Morris JS, Annangudi SP, Sweedler JV. Mass Spectrom Rev. 2008;27:316–330. doi: 10.1002/mas.20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Li L, Sweedler JV. Ann. Rev. Anal. Chem. 2008;1:451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- (13).Amantonico A, Oh Joo Y., Sobek J, Heinemann M, Zenobi R. Angewandte Chemie International Edition. 2008;47:5382–5385. doi: 10.1002/anie.200705923. [DOI] [PubMed] [Google Scholar]
- (14).Muscari C, Pappagallo M, Ferrari D, Giordano E, Capanni C, Caldarera CM, Guarnieri C. J. Chrom. B. 1998;707:301–307. doi: 10.1016/s0378-4347(97)00595-1. [DOI] [PubMed] [Google Scholar]
- (15).Chen S, Lillard SJ. Anal. Chem. 2001;73:111–118. doi: 10.1021/ac0009088. [DOI] [PubMed] [Google Scholar]
- (16).Gao J, Yin XF, Fang ZL. Lab Chip. 2004;4:47–52. doi: 10.1039/b310552k. [DOI] [PubMed] [Google Scholar]
- (17).Hu S, Michels DA, Fazal MA, Ratisoontorn C, Cunningham ML, Dovichi NJ. Anal. Chem. 2004;76:4044–4049. doi: 10.1021/ac0498314. [DOI] [PubMed] [Google Scholar]
- (18).Jankowski JA, Tracht S, Sweedler JV. Trends Anal. Chem. 1995;14:170–177. [Google Scholar]
- (19).Klein CA, Seidl S, Petat-Dutter K, Offner S, Geigl JB, Schmidt-Kittler O, Wendler N, Passlick B, Huber RM, Schlimok G, Baeuerle PA, Riethmuller G. Nature Biotechnology. 2002;20:387–392. doi: 10.1038/nbt0402-387. [DOI] [PubMed] [Google Scholar]
- (20).Munce NR, Li J, Herman PR, Lilge L. Anal. Chem. 2004;76:4983–4989. doi: 10.1021/ac0496906. [DOI] [PubMed] [Google Scholar]
- (21).Oilman SD, Ewing A. Anal. Chem. 1995;67:58–64. doi: 10.1021/ac00097a010. [DOI] [PubMed] [Google Scholar]
- (22).Yeung ES. J. Chrom. A. 1999;830:243–262. doi: 10.1016/s0021-9673(98)00920-0. [DOI] [PubMed] [Google Scholar]
- (23).Zhang L, Qv S, Wang Z, Cheng J. J. Chrom. B. 2003;792:381–385. doi: 10.1016/s1570-0232(03)00323-4. [DOI] [PubMed] [Google Scholar]
- (24).Troyer KP, Heien MLAV, Venton BJ, Wightman RM. Curr. Opin. Chem. Bio. 2002;6:696–703. doi: 10.1016/s1367-5931(02)00374-5. [DOI] [PubMed] [Google Scholar]
- (25).Edgar JS, Pabbati CP, Lorenz RM, He M, Fiorini GS, Chiu DT. Anal. Chem. 2006;78:6948–6954. doi: 10.1021/ac0613131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lorenz RM, Edgar JS, Jeffries GD, Chiu DT. Anal. Chem. 2006;78:6433–6439. doi: 10.1021/ac060748l. [DOI] [PubMed] [Google Scholar]
- (27).Schiro PG, Kuyper CL, Chiu DT. Electrophoresis. 2007;28:2430–2438. doi: 10.1002/elps.200600730. [DOI] [PubMed] [Google Scholar]
- (28).Han F, Wang Y, Sims CE, Bachman M, Chang R, Li GP, Allbritton NL. Anal. Chem. 2003;75:3688–3696. doi: 10.1021/ac0341970. [DOI] [PubMed] [Google Scholar]
- (29).Kirby S, Moe D, Zelander T. Histochem. J. 1990;22:95–101. doi: 10.1007/BF01885787. [DOI] [PubMed] [Google Scholar]
- (30).Presley AD, Fuller KM, Arriaga EA. J. Chrom. B. 2003;793:141–150. doi: 10.1016/s1570-0232(03)00371-4. [DOI] [PubMed] [Google Scholar]
- (31).Porcelli AM, Ghelli A, Zanna C, Pinton P, Rizzuto R, Rugolo M. Biochem. Biophys. Res. Comm. 2005;326:799–804. doi: 10.1016/j.bbrc.2004.11.105. [DOI] [PubMed] [Google Scholar]
- (32).Dendramis KA, Allen PB, Reid PJ, Chiu DT. Chem Commun (Camb) 2008:4795–4797. doi: 10.1039/b806685j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Sun B, Chiu DT. Langmuir. 2004;20:4614–4620. doi: 10.1021/la0364340. [DOI] [PubMed] [Google Scholar]
- (34).Sun B, Lim DS, Kuo JS, Kuyper CL, Chiu DT. Langmuir. 2004;20:9437–9440. doi: 10.1021/la048444m. [DOI] [PubMed] [Google Scholar]
- (35).Shoemaker GK, Lorieau J, Lau LH, Gillmor CS, Palcic MM. Anal Chem. 2005;77:3132–3137. doi: 10.1021/ac0481304. [DOI] [PubMed] [Google Scholar]
- (36).Olefirowicz TM, Ewing AG. Anal. Chem. 1990;62:1872–1876. doi: 10.1021/ac00216a026. [DOI] [PubMed] [Google Scholar]
- (37).Sun B, Chiu DT. J. Am. Chem. Soc. 2003;125:3702–3703. doi: 10.1021/ja029942p. [DOI] [PubMed] [Google Scholar]
- (38).Allen PB, Rodriguez I, Kuyper CL, Lorenz RM, Spicar-Mihalic P, Kuo JS, Chiu DT. Anal. Chem. 2003;75:1578–1583. doi: 10.1021/ac020498i. [DOI] [PubMed] [Google Scholar]
- (39).Rodriguez I, Spicar-Mihalic P, Kuyper CL, Fiorini GS, Chiu DT. Analytica Chimica Acta. 2003;496:205–215. [Google Scholar]
- (40).Allen PB, Chiu DT. Anal. Chem. 2008;80:7153–7157. doi: 10.1021/ac801059h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Sun B, Chiu DT. Anal. Chem. 2005;77:2770–2776. doi: 10.1021/ac048439n. [DOI] [PubMed] [Google Scholar]
- (42).Kuyper CL, Kuo JS, Mutch SA, Chiu DT. J. Am. Chem. Soc. 2006;128:3233–3240. doi: 10.1021/ja057349c. [DOI] [PubMed] [Google Scholar]
- (43).Lohse B, Bolinger PY, Stamou D. J Am Chem Soc. 2008;130:14372–14373. doi: 10.1021/ja805030w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.