Abstract
We have demonstrated that nanostructures, and in particular nanorings incorporating a homodimeric enzyme, can be prepared by chemically induced self-assembly of dihydrofolate reductase (DHFR)-histidine triad nucleotide binding 1(Hint1) fusion proteins. The dimensions of the nanorings were found by static light scattering and atomic force microscopy studies to be dependent on the length and composition of the peptide linking the fusion proteins, ranging in size from 10 to 70 nm in diameter and 64 to 740 kDa. The catalytic efficiency of the nanorings was found to be dependent on ring size, thus suggesting that the arrangement of supermolecular assemblies of enzymes maybe be used to control their catalytic parameters.
Keywords: Nanoring, Histidine triad nucleotide binding proteins (Hints), Phosphoramidase, human Hint1, Dihydrofolate Reductase (DHFR), self-assembly, chemical dimerization
Introduction
Designing and producing biological based assemblies that can be used for the fabrication of advanced materials is a rapidly developing area of research.1 Self-assembling DNA and protein biomolecular building blocks have been used to produce a number of novel nanomaterials that may be applied to microelectronics, tissue engineering and drug delivery.2, 3 Because of the well-understood set of rules governing nucleic acid duplex assembly, substantial advances have been made toward the development of DNA based nanostructures.4 Although the potential for a greater variety of structural and, therefore, functional uses can be envisioned for protein based materials, the development of a general set of rules for the de novo design of protein-protein interfaces has been challenging due to the highly idiosyncratic nature of protein surfaces. 3, 5, 6
Recently, we have discovered a method for the preparation of stable and homogeneous protein polygons, or nanorings. In particular, dihydrofolate reductase (DHFR) molecules when fused together by a peptide chain of variable length were found to spontaneously self-assemble in to protein macrocycles after treatment with a dimeric enzyme inhibitor, Bis-MTX-C9, (Figure 5S), that efficiently dimerizes DHFR.7 The size of the nanoring (8–30 nm diameter) was found to be dependent on the length of the linker peptide between the two DHFRs. To expand the potential size of protein nanoring self-assembly, we hypothesized that replacement of a portion of the amino acid linker by a rigid protein might favor formation of larger nanostructures. We chose to incorporate the enzyme human Hint1 between the two DHFRs for three reasons. First, hHint1 is a highly stable homodimer with an estimated monomer dissociation constant of less than 1.0 nM. 8 Secondly, fully enzymatically active DHFR-hHint1 fusion proteins can be expressed and purified as exclusively homodimers.8, 9 Lastly, the incorporation of hHint1, which is a phosphoramidate10 and acyl-adenylate hydrolase11, would allow the effect of protein oligomerization and, in particular, macrocycliztion on enzymatic activity to be determined.
Results & Discussion
We prepared expression plasmids for DHFR-hHint1 fusion proteins as described in the supplemental material and methods. The proteins were fused together through a one amino acid (Glycine or Threonine), three amino acid (GLG or GLE), seven amino acid (GLGGGLE) or fifteen amino acid (GLGGGGGLVPRGTLE) linker and isolated by AMP-agarose affinity chromatography. To assess the effect of the fusion protein on catalytic activity, the turnover number (kcat), Michaelis-Menten constant (Km) and efficiency (kcat/Km) for hydrolysis of the fluorogenic substrate, tryptamine adenosine phosphoramidate monoester (TpAd) were determined.10 (Table 1) The kcat values for all but one of the six fusion proteins were either identical (1DHG, 1DHT, 3DH-GLG) or slightly greater (7DH, 15DH) than those for hHint1. The Km values for 3DH-GLG, 7DH and 15DH are nearly identical to the value for hHint1, while the Km values for the two one amino acid linker fusion proteins, 1DHG and 1DHT, were found to be significantly lower. Surprisingly, the kcat for the 3DH-GLE was found to be approximately three to four fold lower than the value for either hHint1 or 3DH-GLG, while the Km was reduced by 5- fold. Given the close proximity of the C-terminus to the active site, these differences are likely a reflection of the ability of the glutamate side chain to interact with either the substrate or active site residues. Nevertheless, the catalytic efficiency of each of the fusion proteins was found to be similar to that of native hHint1.
Table 1.
Comparison of steady-state kinetic parameters of hydrolysis of a fluorogenic substrate, tryptamine adenosine phosphoramidate (TpAd) monoester by DHFR-hHint1 fusion proteins with different length of peptide linkers. Data was obtained at 25 °C in HEPES buffer (20 mM HEPES pH 7.2, 1 mM MgCl2) as previously described a.10
| Protein | kcat (s−1) |
Km (nM) |
kcat/Km (s−1M−1)×10−7 |
|---|---|---|---|
| hHint1* | 2.1 ± 0.1 | 130 ± 20 | 1.5 ± 0.3 |
| 1DHG | 2.1 ± 0.02 | 69 ± 5 | 3.1 ± 0.4 |
| 1DHT | 2.1 ± 0.01 | 93 ± 10 | 2.3 ± 0.6 |
| 3DH-GLG | 2.9 ± 0.09 | 150 ± 20 | 2.0 ± 0.5 |
| 3DH-GLE | 0.73 ± 0.01 | 28 ± 1 | 2.6 ± 0.2 |
| 7DH | 3.2 ± 0.01 | 130 ± 10 | 2.4 ± 0.1 |
| 15DH | 3.3 ± 0.01 | 120 ± 10 | 2.7 ± 0.2 |
Measurements were carried out in duplicate and variants are given as standard deviations.
Data for hHint1 were adapted from ref 10.
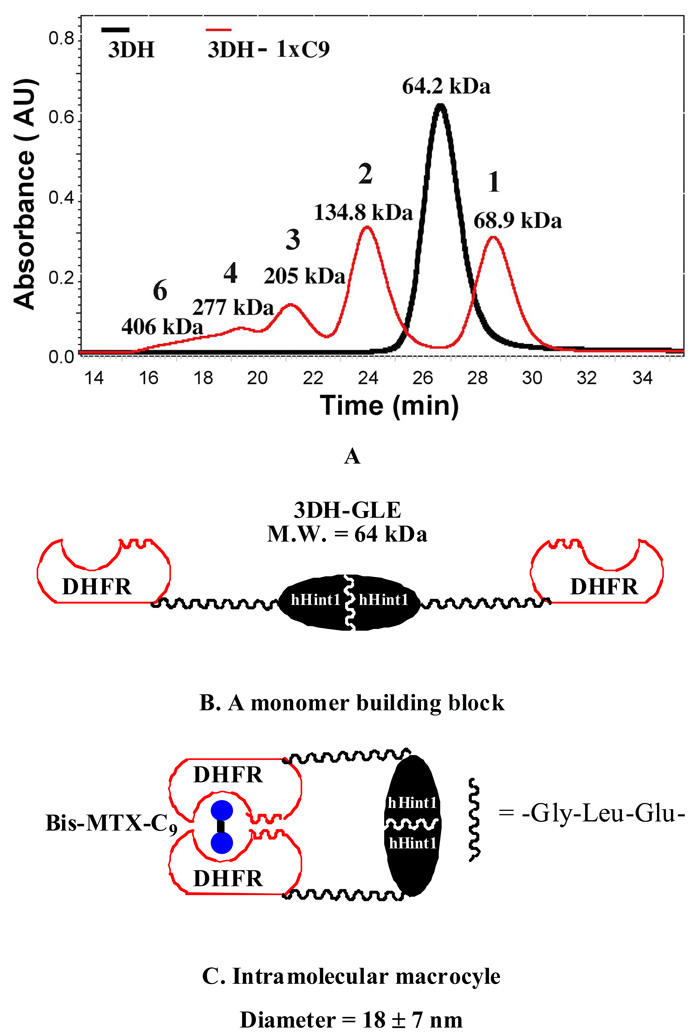
Since we had previously found that DHFR-DHFR fusion proteins containing linkers with at least three amino acids (including sterically constraining amino acids) were able to oligomerize into a range of nanorings, we chose to first examine nanoring formation by oligomerization of 3DH-GLE.12 The addition of one equivalent of Bis-MTX-C9 to 3DH-GLE (20 µM, 64 kDa, 27 min) resulted in formation of predominantly a dimeric oligomer (134.8, 43%, 24 min), followed by trimer (205 kDa, 12%, 21 min), tetramer (277 kDa, 4%, 19.5 min) and a small amount of higher molecular weight species (5%). (Figure 1 and Figure 1S) Surprisingly, a significant amount (36.8 %) of the oligomerized protein was found to exist as a smaller species, eluting at 29 min. Given the size and flexibility of the linker, it was assumed that this species represented that portion of fusion protein that under went intramolecular cyclization, thus converting the monomer into a species with a smaller hydrodynamic radius. (Figures 1B & C) Consistent with this conclusion, when oligmerization experiments at a lower 3DH-GLE concentration (2.5 µM) and one equivalent of Bis-MTX-C9 were carried out, the intramolecular macrocyle was found to be the predominant species (83%), with only a minor amount of dimeric (17%) oligomer was observable. (Figure 2S)
Figure 1.
Overlaid size exclusion profiles for 3DH-GLE and 3DH-GLE with 1 equiv of added Bis-MTX-C9. Samples were eluted at a flow rate of 0.5 mL/min on Superdex G200 column in P500 buffer monitored by an in-line multiangle static light scattering detector a refractive index detector and a UV detector. (A) 3DH-GLE (20 µM, 500 µL, black) was eluted at 27 min and estimated molecular mass is 64.2 kDa consistent with linear dimeric DHFR-hHint1 as shown in (B); assembly of 3DH-GLE (20 µM) with Bis-MTX-C9 (20 µM, 1 equiv) was shown in red trace and appearance of various sizes of oligomeric rings (16~24 min) and a stable intramolecular macrocyle as shown in (C) was observed.
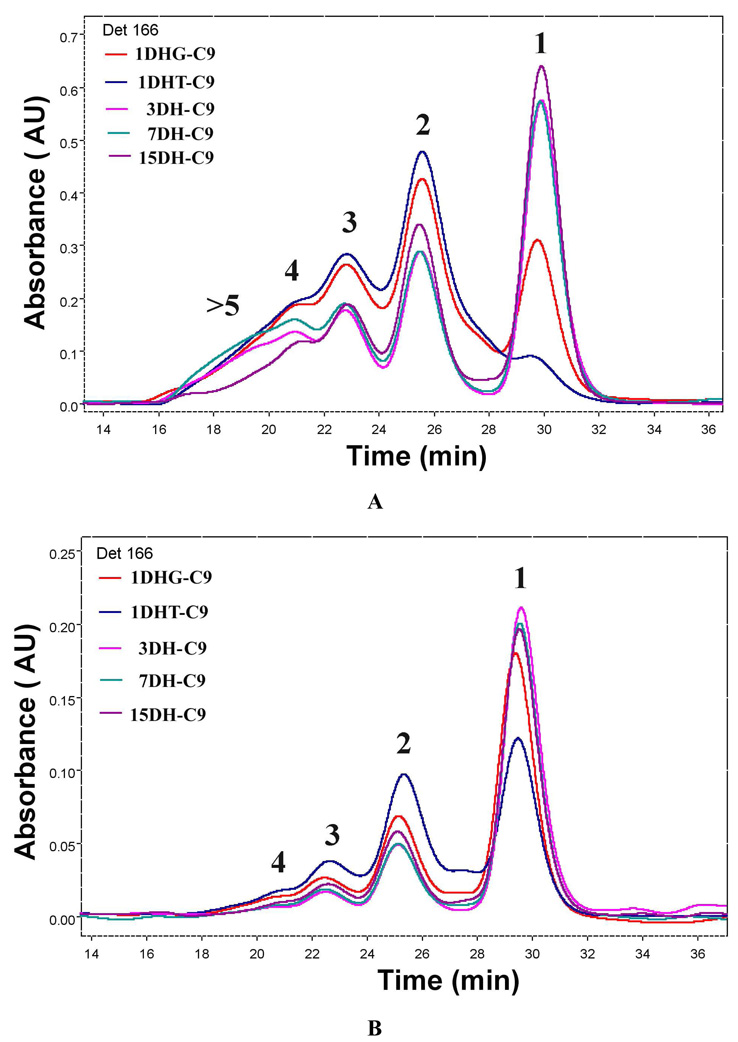
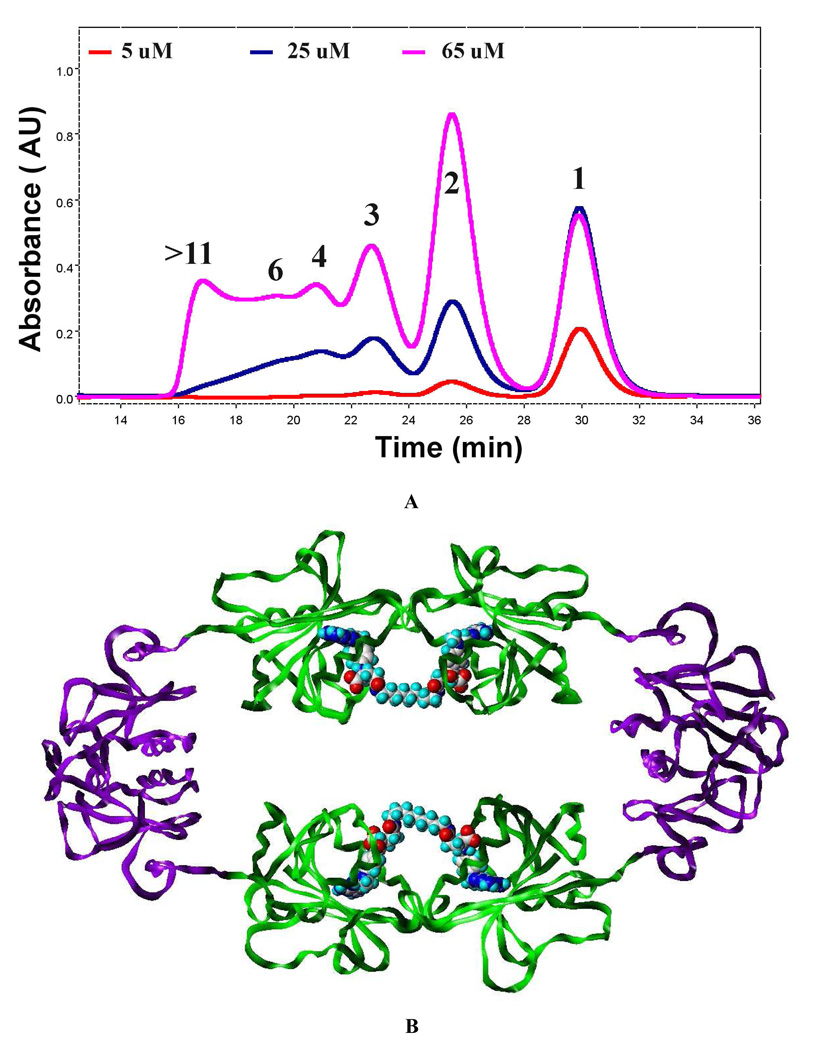
The effects of both longer and shorter peptide-linkers on oligomerization were evaluated at a concentration favoring intramolecular oligomerization (25 µM). (Figure 2A) Not surprisingly, given that 15DH has the longest peptide linking DHFR and hHint1, the greatest amount of intramolecular cyclization was observed. Nevertheless, significant amounts of dimer, trimer, tetramer and higher molecular weight species were noticeable. Reducing the length of the linker to seven (7DH) and then three (3DH-GLE) amino acids did slightly reduce the amount of intramolecular macrocyclization with a comparable decrease in the amount of dimer. However, the amount of larger molecular weight oligomers (4 to >6) did increase. In contrast, a 54% reduction in the amount of intramolecular macrocyclization was observed during oligomerization of 1DHG. The amount of dimer and higher molecular weight species increased by 35%, relative to 15DH. When the one amino acid glycine linker was replaced with threonine, intramolecular cyclization was decreased by 65% with an even greater amount of dimer and trimer observed. Consequently, shortening the linker to one amino acid substantially reduced the effective molarity necessary for intramolecular macrocyclization.13 Nevertheless, regardless of the linker length, the ratio of intramolecular to intermolecular cyclization should be dependent on the fusion protein concentration. As can be seen in Figure 2A, at a 5 µM concentration, intramolecular cyclization predominates, even for the one amino acid linker, 1DH-T. When we examined the effect of concentration on just 3DH-GLE, we found that, while little intermolecular nanoring formation was observed at 5 uM (Fig. 3a), when the concentration was increased to 65 µM, oligomers consisting of 2 to 12 monomers with molecular weights ranging from 130 to 740 kDa, respectively, not only were observable, but predominate. A molecular model of the dimeric species was built (Fig. 3b) and revealed that when fully extended, the three amino acid linker was fully capable of facilitating ring formation.
Figure 2.
Comparison of Bis-MTX-C9 polymerized DHFR-hHint1 fusion proteins with different lengths of peptide linkers. Overlaid elution profiles for (A) 25 µM of protein mixed with 1 equiv Bis-MTX-C9; (B) 5 µM of proteins mixed 1 equiv Bis-MTX-C9.
Figure 3.
(A) Concentration dependent formation of macro-nanorings. Overlaid elution profiles of Bis-MTX-C9 (1 equiv) oligomerized 3DH-GLE (red : 5 µM, blue: 25 µM, pink : 65 µM). (B) Model of 3DH-GLE dimer ; DHFR (Green) and Hint1 (Purple).
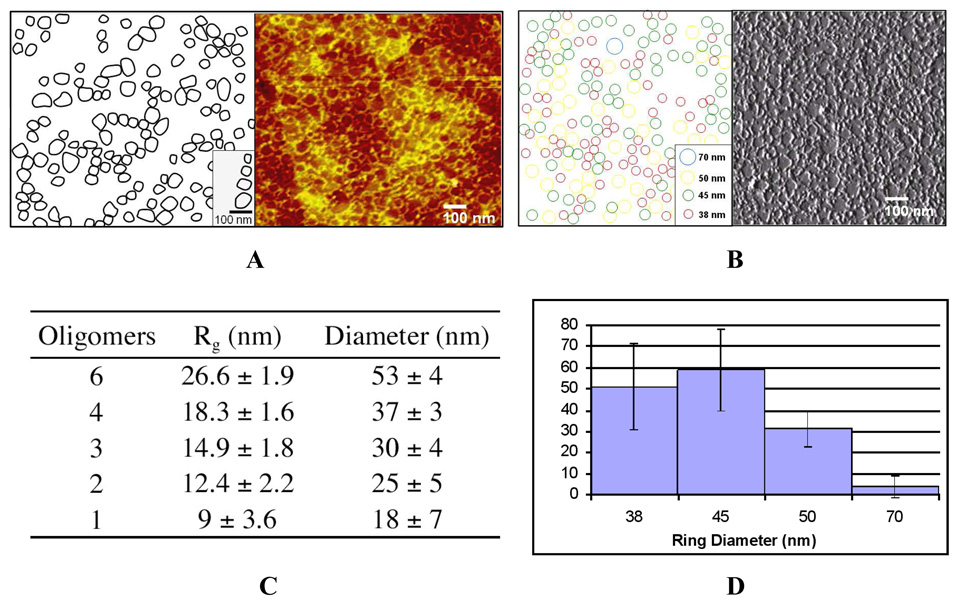
The cyclic structure of the nanorings was confirmed by AFM analysis (supplemental material and methods) of a sample isolated from the oligomerizaton of 3DH-GLE-C9 (Table 2, fraction 2 and Figure 3, pink trace). To our surprise, the expected ring sizes did not exhibit sufficient affinity towards mica. (see supplemental Fig. 6S). Interestingly, one particular ring size seemed to dominantly adhere to mica with dimensions above 200 nm in diameter, possibly the largest species in the solution. As an alternative, ultra-flat gold and silicon wafer were tested under the same preparation conditions as mica for protein ring adhesion. Like mica, silicon showed poor binding affinity (Data not shown), while ultra-flat template-stripped gold (TS-Au)14 surfaces showed high surface coverage of the protein nanorings with mono-molecular thickness. (Figure 4A) TS-Au was used to perform large area ring size analysis for statistical relevance and because it is not limited by traditional grain-to-grain roughness exhibited by most gold surfaces (see supplemental for preparation).
Table 2.
Six eluted fractions of the 65 µM 3DH-GLE-C9 sample (Figure 3) were collected and the specific activity of Hint1 hydrolase was determined with TpAd substrate (1.5 µM).
| Fraction | RT (min) |
Oligomer (subunit) |
M.W. (kDa, SLS) |
Specific Activity (s-1) |
|---|---|---|---|---|
| 1 | 16.5–18.5 | >11 | 737.3 | 1.49 |
| 2 | 18.5–20.5 | 6 | 390.7 | 0.78 |
| 3 | 20.5–22.9 | 4 | 246.6 | 0.89 |
| 4 | 22.9–24.8 | 3 | 186.4 | 0.72 |
| 5 | 24.8–27.2 | 2 | 133.2 | 0.86 |
| 6 | 29.2–32.3 | 1 | 66.4 | 0.45 |
Figure 4.
AFM ring size quantification of protein nanorings on TS-Au. (A, right) Height data showing apparent rings dispersed across a TS-Au surface (A, left) Schematic representation of rings from masking and their four representative diameters (B) Binned amplitude dataset to the four observed sizes in 4A with schematic (C) Nanoring sizes as from SLS of collected SEC fraction used for AFM study and (D) Quantification of AFM images gathered from 5 samplings of 3.292 um2 total surface area showing predominantly 4–6 oligomer sized nanorings on the TS-Au surface.
Ring size analysis was carried out over 5 separate images sampled randomly from the surface with a total surface area of 3.292 um2. This was done manually using a digital masking technique where the representative rings were fitted to the actual AFM image for analysis in Figure 4A. Four representative ring sizes were found to be prevalent: 38, 45, 50 and 70 nm, shown in the schematic scale-bar on the left of 4A. For statistical quantification analysis, the amplitude data output was used to eliminate background topography and rings were binned to the previously established prevalent diameters shown in schematic 4B and graphed in figure 4D. Static light scattering (shown in Fig. 4C) and SEC studies indicated that nanorings consisting of 4 to 6 monomers are formed in significant abundance. With diameters from ~37–53 nm, they are measurable (Fig. 4A–D) and toroidal in morphology with AFM analysis. Although observed, species below 4 monomers form crowded populations (Fig 4B, interstitial spaces) and are indiscernible due to tip convolution and are omitted from the analysis.
The experimentally determined ring dimensions were supported by modeling of nanoring hydrodynamic parameters using the HYDRO++ program. Toroids containing 6 and 4 DHFR-HINT-DHFR subunits (N = 12 and 8 in Table 3S) were predicted to have radii of gyration of 20.0 and 13.5 nm, respectively, which are in good agreement with the SLS results of 26.6 and 18.3 nm in Fig. 4C. Additionally, diameters for N = 12 and 8 toroids were calculated to be 45.5 and 32.2 nm, respectively, also in good agreement with the experimental values of 53 and 37 nm. Discrepancies in these data can likely be attributed to a combination of solvent and amino acid linker effects that contribute to physical nanoring behavior that differs from the rigid bead model.
Once formed and purified, the 3DH-GLE dimeric nanorings and intramolecular macrocycle are stable and do not re-sort into a mixture of species after 3 months at 4°C. (Fig. 3S, A). In contrast, redistribution of the higher molecular weight fraction 2 into a range of smaller and more thermodynamic stable nanorings was observed after 3 month at 4°C (Fig. 3S, B). Thus, as observed previously for DHFR-DHFR fusion proteins, DHFR-hHint1 fusion proteins are fully capable of oligomerizing to form stable nanorings in which ring size is highly dependent on the effective molarity enforced by the composition and length of the linker peptide.13
To assess the effect of oligomer size on enzymatic activity, peaks corresponding to selected oligomers were purified by SEC after oligomerization of 3DH-GLE. (Table 2) The greatest specific activity was found for the largest oligomers (> 11 monomers). When the ring size was reduced by approximately 50% to 75%, a reduction of 50% to 60% in activity was observed. The greatest reduction in activity (70%) was found for the intramolecular macrocycle. To quantitatively assess the effect of embedding hHint1 within the nanoring assembly, the Michaelis-Menten parameters for the intramolecular macrocycle and dimeric nanorings were determined and compared to the constituent monomer. (Table 3) Both the monomer and the dimeric nanoring exhibited identical kcat values, while the kcat for the intramolecular macrocycle was reduced by nearly 50%. Surprisingly, the Km value for the dimeric nanoring was 1.6- fold greater than the value for the monomer, while the lowest Km value was found for the intramolecular macrocyle. Consequently, the enzymatic efficiency (kcat/Km) for the monomer and intramolecular macrocycle were found to be nearly identical, while the larger dimeric nanoring was found to have a modestly lower kcat/Km value. In either case, the efficiencies of the three complexes were found to either exceed or to be equivalent to the efficiency found for wild-type hHint1. The retention of enzymatic efficiency, even in the apparently highly constrained intramolecular macrocyle is largely due to the unexpected 7- fold reduction in the Km value, when compared to hHint1, despite a 3- fold reduction the kcat value.
Table 3.
Comparison of steady-state kinetic parameters of hydrolysis of TpAd by 3DHGLE and 3DH-GLE-C9 nanorings isolated from SEC fraction 5, and 6 shown in table 2.
| Fraction | Protein | Oligomer (Subunit) |
kcat (s−1) |
Km (nM) |
kcat/Km (s−1 M−1)×10−7 |
|---|---|---|---|---|---|
| NA | 3DH-GLE* Linear monomer |
1 | 0.73±0.01 | 28 ± 1 | 2.6 ± 0.2 |
| 5 | 3DH-GLE-C9 (24.8~27.2 min) Dimeric Nanoring |
2 | 0.73±0.01 | 44 ± 4 | 1.7 ± 0.2 |
| 6 | 3DH-GLE-C9 (31~32.3 min) Intramolecular Macrocyle |
1 | 0.39±0.003 | 19 ± 1 | 2.1 ± 0.2 |
Data for 3DH-GLE were adapted from table 1.
Collectively, our results demonstrate that nanostructures, and in particular nanorings incorporating a homodimeric enzyme, can be prepared by chemically induced self-assembly. As observed for previous nanoring assemblies, ring size is dependent on the length and composition of the peptide linking the fusion proteins. The relatively large size of the internal hHint1 protein resulted in a significant amount of intramolecular cyclization. Not surprisingly, we observed that shorter linkers led to formation of the largest unnatural protein assemblies ever reported, to our knowledge. While at the moment we are unable to tune the dimensions of the nanorings with precision, future studies examining the role of the linker length and composition on nanoring assembly may allow greater control over there size to be exerted.12 In addition, we observed for the first time the effect of induced conformational constrain on enzyme catalysis, suggesting that control of enzymic parameters maybe engineered into enzyme based supermolecular assemblies and arrays.
Materials & Methods
Protein Expression and Purification
The expression plasmids for 15 and 7 amino acids linker, pTFC15DH and pTFC7DH, were constructed previously.8 Plasmids for 3DH-GLE (DHFR and Hint linked with 3 amino acids – GLE) and 3DH-GLG, 1DH-G and 1DH-T were generated using the QuickChange site-directed mutagenesis kit with primers listed in Table 1S. DHFR-hHint1 fusion proteins were purified to homogeneity as shown in Fig. 4S as previously described.8
Size Exclusion Chromatography and Static Light Scattering (SLS)
The molar concentration of DHFR-hHint1 was calculated based on the theoretical molecular weight of a monomer shown in Table 2S. Protein samples mixed with Bis-MTX-C9 (structure shown in Fig. 5S) were prepared in P500 buffer (0.5 M NaCl, 50 mM potassium phosphate, 1 mM EDTA, pH 7.0, filtered through a 0.02 µm filter) with 5 % (v/v) glycerol. Samples were subjected to size exclusion chromatography (Superdex G200 or G75, GE Healthcare) after incubation at 23 °C for at least 1h, eluted at a flow rate of 0.5 mL/min with P500 buffer, and monitored by an in-line multiangle light scattering detector with a power 690 nm argon laser light source (DAWN EOS, Wyatt Technology), a refractive index detector (Altex) and a UV detector (Beckman Gold 166). Data were collected across a range of angles from 14.5 – 163.3 degrees. The instrument was calibrated with a bovine serum albumin (BSA) molecular standard (1 mg/mL, 500 µL). All samples were filtered with a 0.02 µm filter just prior to analysis. Molecular masses and radius of gyration were calculated with the ASTRA software.15
Atomic Force Microscopy of Nanorings on Surfaces
Mica surfaces (Ted Pella, Redding, CA) are used as-cleaved and untreated prior to protein solution exposure. Surfaces are cleaved with normal Scotch-brand tape. Ultra-flat gold surfaces were prepared via template-stripping (TS-Au) as detailed by Hegner and coworkers.14 Briefly, bare ruby muscovite mica surfaces are heated at 300°C for 20 hrs in a metal evaporation system (BOC Edwards Auto 306, West Sussex, UK) equipped with a sample heating stage, then gold is evaporated at 1 Å/sec for a final thickness of ~200 nm. Gold substrates are then epoxied face down onto precut silicon wafers using epo-tek no. 377 (Epoxy Technology, Billerica, MA) and cured for 1 hr at 150°C. TS-Au-Si sandwiches are then placed in tetrahydrofuran (THF) for 10 minutes and mica backings are removed to reveal the first-deposited layer of gold. The nascent gold film is exposed to protein solution immediately upon THF treatment to prevent contamination. Verification of the exposed surface is firstly from EDAX measurements (Fig 7S, though not highly surface sensitive) and secondly through the observation of significantly different reactivity towards the nanorings between mica and TS-Au substrates.
A Digital Instruments Nanoscope IIIa Multimode AFM (Santa Barbara, CA) was used in tapping-mode under acoustic and mechanical isolation at RTP conditions. Nanosensor (Neuchatel, Switzerland) silicon nitride tapping-mode tips were used at ~300kHz resonance frequency. Images were scanned at an average tip velocity of 5 um/sec and processed via Nanoscope v5.30r1 standard software.
For AFM analysis of nanoring sizes, 2 µL of SEC elution is pipetted onto freshly cleaved muscovite mica surfaces and allowed to completely dry under a clean-hood. 100 µL of DI H2O is then placed over the mica surface for 10 min. to dissociate salt crystals left from the dried protein buffer solution. Surfaces are blown dry with argon and scanned in air at RT.
Toroid Hydrodynamic Parameter Modeling
For the calculation of hydrodynamic parameters on 3DH toroid bead models, we utilized the HYDRO++ program developed by Garcia de la Torre, et al.16 A model of 3DH was constructed in Maestro (Schrodinger, Inc.) by linking the C-terminus of E. coli DHFR (pdb 4DFR) to the N-terminus of the rabbit Hint monomer (pdb 6RHN). The newly constructed fusion protein was relaxed via minimization and a short molecular dynamics simulation using the OPLS2005 force field. Measurements were taken from several different points on the exterior of the two proteins to yield average diameters of ~5.0 and 4.0 nm for DHFR and Hint, respectively. These measurements were used to establish bead radii for HYDRO++ input files of 2.5 and 2.0 nm for DHFR and Hint. A Fortran95 script was written to automatically generate HYDRO++ structural input files containing Cartesian coordinates for bead aggregates of n = 2 to n = 30 fusion proteins based on trigonometric calculations of bead spacing in an appropriate polygon of n sides. The ICASE value for the HYDRO++ calculation was 20 for n ≤ 10 and 21 for n ≥ 12. The temperature was set to 296 K for all calculations. All other input variables were unchanged from the suggested values in the HYDRO++ examples found at the program website. To verify the structure of the bead model used in the calculations, the results were loaded into the CORTONA brower plug-in for visual inspection.
Supplementary Material
The Fortran script for generating HYDRO++ input parameters; static light scattering, size exclusion, and SDS-PAGE of fusion proteins and generated nanorings; atomic force microscopy images of nanorings; the structure of bis-MTX; EDAX spectra of TS-Au surface; primers used in genetic engineering; and calculated hydrodynamic parameters of nanorings. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgement
We thank Dr. Terry W. Steele for his assistance with the SLS experiments and Dr. R. A. Siegel for the use of the SLS instrument. We wish to thank the University of Minnesota Nanobiotechnology Initiative (CRW), NIH-NCI (CA120116, CRW), the Leukemia Research Foundation, and NSF-MRSEC (CS and MS) for partial support of this study (DMR-0520567).
References
- 1.Service RF. Nanotechnology Takes Aim at Cancer. Science. 2005;310:1132–1134. doi: 10.1126/science.310.5751.1132. [DOI] [PubMed] [Google Scholar]
- 2.Sarikaya M, Tamerler C, Jen A, Shulten K, Baneyx F. Molecular Biomimetics: Nanotechnology Through Biology. Nature-Mater. 2003;2:577–585. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- 3.Zhang S, Marini DM, Hwang W, Santoso S. Design of Nanostructured Biological Materials Through Self-Assembly of Peptides and Proteins. Cur. Opin. Chem. Biol. 2002;6:865–871. doi: 10.1016/s1367-5931(02)00391-5. [DOI] [PubMed] [Google Scholar]
- 4.Seeman NC. From Genes to Machines: DNA Nanomechanical Devices. Trends Biochem. Sci. 2005;30:119–125. doi: 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grueninger DTN, Ziegler MOP, Koetter JWA, Schulze MS, Schulz GE. Designed Protein-Protein Association. Science. 2008;319:206–209. doi: 10.1126/science.1150421. [DOI] [PubMed] [Google Scholar]
- 6.Yeates TO, Padilla JE. Designing Supramolecular Protein Assemblies. Cur. Opin. Struc. Biol. 2002;12:464–470. doi: 10.1016/s0959-440x(02)00350-0. [DOI] [PubMed] [Google Scholar]
- 7.Carlson JCT, Kanter A, Thuduppathy GR, Cody V, McIvor RS, Wagner CR. Designing Protein Dimerizers: the Importance of Ligand Conformational Equilibria. J. Amer. Chem. Soc. 2003;125:1501–1507. doi: 10.1021/ja026264y. [DOI] [PubMed] [Google Scholar]
- 8.Chou T-F, Tikh IB, Horta BA, Ghosh B, de Alencastro RB, Wagner CR. Engineered Monomeric Human Histidine Triad Nucleotide Binding Protein 1 Hydrolyzes Fluorogenic Acyl –Adenylate And Lysyl-tRNA Synthetase-Generated Lysyl-Adenylate. J. Bio. Chem. 2007;282:15137–15147. doi: 10.1074/jbc.M606972200. [DOI] [PubMed] [Google Scholar]
- 9.Chou T-F, Bieganowski P, Shilinski K, Cheng J, Brenner C, Wagner CR. 31P NMR and Genetic Analysis Establish hinT as the Only Escherchia Coli Purine Nucleoside Phosphoramidase and as Essential for Growth Under High Salt Conditions. J. Biol. Chem. 2005;280:15356–15361. doi: 10.1074/jbc.M500434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou T-F, Baraniak J, Kaczmarek R, Zhou X, Cheng J, Ghosh B, Wagner CR. Phosphoramidate Pronucleotides: a Comparison of the Phosphoramidase Substrate Specificity of Human and E. Coli Histidine Triad Nucleotide Binding Proteins (Hint1) Mol. Pharmaceutics. 2007;4:208–217. doi: 10.1021/mp060070y. [DOI] [PubMed] [Google Scholar]
- 11.Chou T-F, Wagner CR. Lysyl-tRNA Synthetase-Generated Lysyl-Adenylate is a Substrate for Histidine Triad Nucleotide Binding Proteins. J. Biol. Chem. 2007;282:4719–4727. doi: 10.1074/jbc.M610530200. [DOI] [PubMed] [Google Scholar]
- 12.Carlson JCT, Sidhartha SJ, Flenniken M, Chou T-F, Siegel RA, Wagner CR. Chemically Controlled Self-Assembly of Protein Nanorings. J. Amer. Chem. Soc. 2006;128:7630–7638. doi: 10.1021/ja060631e. [DOI] [PubMed] [Google Scholar]
- 13.Ercolani G. A Model for Self-Assembly in Solution. J. Phys. Chem. B. 2003;107:5052–5057. [Google Scholar]
- 14.Hegner M, Wagner P, Semenza G. Ultralarge Atomically Flat Template-Stripped Au Surfaces for Scanning Probe Microscopy. Surface Science. 1993;291:39–46. [Google Scholar]
- 15.Wyatt JP. Light Scattering and the Absolute Characterization of Macromolecules. Anal. Chim. Acta. 1993;272:1–40. [Google Scholar]
- 16.Garcia de la Torre J, del Rio G, Ortega A. Improved Calculation of Rotational Diffusion and Intrinsic Viscosity of Bead Models for Macromolecules and Nanoparticles. J. Phys. Chem. B. 2007;111:955–961. doi: 10.1021/jp0647941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Fortran script for generating HYDRO++ input parameters; static light scattering, size exclusion, and SDS-PAGE of fusion proteins and generated nanorings; atomic force microscopy images of nanorings; the structure of bis-MTX; EDAX spectra of TS-Au surface; primers used in genetic engineering; and calculated hydrodynamic parameters of nanorings. This material is available free of charge via the Internet at http://pubs.acs.org.