Abstract
OBJECTIVE
This study evaluated the influence of somatostatin receptor type 2 (SSTR2) polymorphisms on measures of glucose homeostasis in the Insulin Resistance Atherosclerosis Family Study (IRASFS). SSTR2 is a G-protein–coupled receptor that, in response to somatostatin, mediates inhibition of insulin, glucagon, and growth hormone release and thus may affect glucose homeostasis.
RESEARCH DESIGN AND METHODS
Ten single nucleotide polymorphisms (SNPs) spanning the gene were chosen using a SNP density selection algorithm and genotyped on 1,425 Hispanic-American individuals from 90 families in the IRASFS. These families comprised two samples (set 1 and set 2), which were analyzed individually and as a combined set. Single SNP tests of association were performed for four glucose homeostasis measures—insulin sensitivity (SI), acute insulin response (AIR), disposition index (DI), and fasting blood glucose (FBG)—using generalized estimating equations.
RESULTS
The SSTR2 locus was encompassed by a single linkage disequilibrium (LD) block (D′ = 0.91–1.00; r2 = 0.09–0.97) that contained four of the ten SNPs evaluated. Within the SSTR2-containing LD block, evidence of association was observed in each of the two sets and in a combined analysis with decreased SI(βhomozygous = −0.16; Pmeta-analysis = 0.0024–0.0030), decreased DI (βhomozygous = −0.35 to −5.16; Pmeta-analysis = 0.0075–0.027), and increased FBG (βhomozygous = 2.30; Pmeta-analysis = 0.045). SNPs outside the SSTR2-containing LD block were not associated with measures of glucose homeostasis.
CONCLUSIONS
We observed evidence for association of SSTR2 polymorphisms with measures of glucose homeostasis. Thus, variants in SSTR2 may influence pathways of SIto modulate glucose homeostasis.
Somatostatin (SRIF) is a hormone found throughout the body that inhibits the action or expression of several hormones including insulin, glucagon, and growth hormone (1). SRIF is an important regulator of the endocrine system, exerting its effects through interactions with pituitary growth hormone, thyroid-stimulating hormone, and several hormones of the gastrointestinal tract. Five G-protein–coupled receptors (SRIF receptor type [SSTR] 1–5) that bind SRIF exist, each exerting differential effects based on the receptor bound and localization. SSTR2 is found primarily in the pancreas, pituitary, and stomach (2). Several studies have evaluated the effects of SRIF on the respective receptors. Rat studies have found that SSTR2 ligands mediate inhibition of glucagon release, whereas SSTR5 mediates inhibition of insulin release (3). In addition, a study of pancreatic islets from SSTR2 knockout mice supported the finding in rats, concluding that SRIF inhibition of glucagon release is primarily regulated through interaction with SSTR2, whereas insulin secretion is primarily regulated by SSTR5 (4). Human studies of the pancreas revealed slightly different results, suggesting that SRIF can inhibit insulin release through the SSTR2 receptor (5). In contrast, Cheng et al. (6) observed that SSTR2 mediates the SRIF-induced increase in insulin secretion, although the increase was detected only in the presence of arginine vasopressin. Based on these studies, it is apparent that whereas SSTR2 does appear to mediate the regulation of insulin and glucagon, the exact mechanism remains obscure.
To date, no studies have assessed the effects of SSTR2 polymorphisms on glucose homeostasis traits in population-based studies. The Insulin Resistance Atherosclerosis Family Study (IRASFS) is a multicenter study investigating the genetic contributions to glucose homeostasis and adiposity and includes a population suitable for studying such effects. Previous studies in the IRASFS of SSTR2 as a positional candidate gene for alterations in the amount of visceral adipose tissue present in study participants (7,8) revealed no association; however, the role in glucose homeostasis, as suggested by the literature, was not assessed. The IRASFS is an extensively phenotyped population with measures of glucose homeostasis derived from minimal model (MINMOD) analysis of frequently sampled intravenous glucose tolerance tests, which include insulin sensitivity (SI), acute insulin response (AIR), and disposition index (DI) as well as fasting blood glucose (FBG). In this study, we have evaluated the influence of SSTR2 polymorphisms on measures of glucose homeostasis with the goal of defining the biological pathways through which this receptor exerts its effects.
RESEARCH DESIGN AND METHODS
The IRASFS was designed to investigate the genetic determinants of insulin resistance and adiposity that are important risk factors in the development of type 2 diabetes and atherosclerosis. Using well-developed technical methods to accurately assess quantitative measures of glucose homeostasis and adiposity, the IRASFS examines the familial aggregation and genetic contributions for those phenotypes. Hispanic Americans from the IRASFS were recruited from two centers: San Antonio, Texas, and San Luis Valley, Colorado. The study design, recruitment, and phenotyping have previously been described in detail (9). Briefly, 90 multigenerational Hispanic-American families (1,425 individuals) were recruited in two phases denoted set 1 (45 families: 33 from San Antonio, Texas, and 12 from San Luis Valley, Colorado; 827 individuals) and set 2 (45 families: 27 from San Antonio, Texas, and 18 from San Luis Valley, Colorado; 592 individuals). Recruitment was independent of type 2 diabetes or cardiovascular disease status, although 14.2% of individuals were diagnosed with type 2 diabetes.Table 1 summarizes the primary phenotypes of study participants.
TABLE 1.
Demographic summary of IRASFS Hispanic-American participants
| Set 1 | Set 2 | Combined sets 1 and 2 | |
|---|---|---|---|
| Age (years) | 42.2 ± 14.5 (40.9) | 43.5 ± 14.7 (41.6) | 42.8 ± 14.6 (41.3) |
| Female sex (%) | 57 | 60 | 58 |
| Glucose homeostasis | |||
| SI(×10−5min−1/[pmol/l]) | 2.0 ± 2.0 (1.5) | 2.1 ± 1.8 (1.7) | 2.0 ± 1.9 (1.5) |
| AIR (pmol/l) | 685 ± 631 (537) | 709 ± 687 (529) | 695 ± 655 (534) |
| DI (10−5min−1) | 1,193 ± 1,266 (887) | 1,222 ± 1,201 (933) | 1,205 ± 1,238 (919) |
| FBG (mg/dl) | 103.5 ± 33.6 (93.5) | 102.7 ± 36.5 (93.5) | 103.2 ± 34.8 (93.5) |
Data are means ± SD (median) unless otherwise indicated.
Measures of glucose homoeostasis were assessed by frequently sampled intravenous glucose tolerance tests (10) with MINMOD analyses (11). SI was calculated using MINMOD software. AIR was calculated as the mean insulin increment in plasma insulin concentration above basal in the first 8 min after glucose administration. DI was calculated as DI = SI × AIR. FBG values were obtained using standard methods. Individuals with type 2 diabetes were excluded from analysis.
Singlue nucleotide polymorphisms (SNPs) were selected using the SNPbrowser 3.0 (Applied Biosystems) SNP density selection algorithm. The genomic interval assessed included the SSTR2 gene ±15 kb, which extended coverage to two adjacent linkage disequilibrium (LD) blocks, as assessed by evaluation of the HapMap CEU population. Density was selected at 2 kb with a minor allele frequency (MAF) >5%. Supplementary Table A1, available in an online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db08-0189/DC1, contains the characteristics of SNPs selected for genotyping. Primers for PCR amplification and extension were designed using SpectroDESIGNER software, and genotyping was performed on the Sequenom MassARRAY system (12).
SNPs were examined for Mendelian inconsistencies in their genotypes using PedCheck (13). Inconsistent genotypes (n = 43) were converted to missing (0.002% error rate). Maximum likelihood estimates of allele frequencies were computed using the largest set of unrelated individuals and genotypes tested for departures from Hardy-Weinberg equilibrium proportions. To validate SNP coverage, the aggressive tagging option of Tagger (14) implemented in the program Haploview (15) was used to assess coverage in the HapMap CEU population.
To test for association among each SNP-trait combination by set and in the combined analysis, a series of generalized estimating equations (16) was computed. When necessary, quantitative traits were transformed to approximate the distributional assumptions of conditional normality (conditional on covariates) and homogeneity of variance. Familial correlations within a pedigree were adjusted for by assuming exchangeable correlation and computing the sandwich estimator of the variance (17). The sandwich estimator is also denoted the robust or empirical estimator of the variance and is a consistent estimator even under misspecification of the correlation matrix. For each SNP and phenotype, the 2 d.f. overall test of genotypic association was performed and reported. When evidence of association was observed, three individual contrasts defined by the genetic models (dominant, additive, and recessive) were computed. In addition, a meta-analysis was computed treating set 1 and set 2 as different populations. We modeled set 1 or set 2 affiliation as a random effect in a variance component–measured genotype model, as implemented in the sequential oligogenic linkage analysis (18). This standard approach to meta-analysis accounts for unknown sources of confounding affiliated with the subsamples of set 1 and set 2 (19). To test for whether the effect was consistent across the two subsamples, a test of SNP by set (dummy variable parameterization) interaction (effect heterogeneity) was computed as the centered cross product. Absence of an interaction suggests there is no detected evidence of heterogeneity of effect across the subsamples. Tests reported here were computed adjusting for age, sex, recruitment center, and BMI. Association results reported herein were not adjusted for multiple comparisons due to inter-SNP and inter-trait correlations.
RESULTS
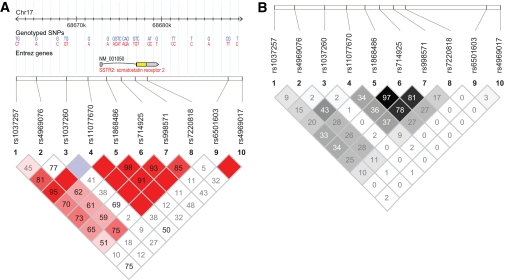
A total of 10 SNPs at the SSTR2 locus were genotyped on 1,425 Hispanic individuals. The majority of SNPs in this study had an MAF >0.15. Using the Haploview Tagger program, a subset of the genotyped SNPs (six SNPs genotyped in the HapMap Build 36 dataset) captured common variation (HapMap CEU, MAF >0.05, aggressive tagging algorithm) with a mean r2> 0.70. D′ values were high between SNPs within the SSTR2 gene (rs11077670, rs1868486, rs714925, rs998571, and rs7220818; D′ ≥0.8) and declined 5′ and 3′ to these markers (D′ <0.6;Fig. 1).
FIG. 1.
Haploview-generated LD map of the 10 SNPs at the SSTR2 locus in unrelated Hispanic Americans from the IRASFS. A: Regions of high LD (D′ = 1 and logarithm of odds [LOD] >2) are shown in the darkest shade. Markers with lower LD (0.45 < D′ < 1 and LOD >2) are shown in dark through light shades, with the color intensity decreasing with decreasing D′ value. Regions of low LD and low LOD scores (LOD <2) are shown in white. The number within each box indicates the D′ statistic value between the corresponding two SNPs. B: Regions of high correlation (r2 = 1) are shown in black. Markers with lower correlations (0 < r2 < 1) are shown in shades of gray with the color intensity decreasing with decreasing r2 value. Regions of low correlation (r2 = 0) are shown in white.
In set 1 (Table 2), the majority of the SNPs in the SSTR2-containing LD block (D′ >0.91; r2> 0.10) were associated (or trending toward association) with SI (P < 0.011), DI (P < 0.036), and FBG (P < 0.019). Association with AIR was not detected. Similar association was detected with individuals in set 2, although many of the SNPs showed a decrease in the magnitude of association. SI and FBG were the most consistently associated glucose homeostasis traits, with three of five SNPs in high LD (D′ >0.77; r2> 0.08) showing association or trending toward association (P < 0.039 and P < 0.030, respectively; Table 3). In the combined analysis (Table 4), the magnitude of association increased within the LD block (D′ >0.85, r2> 0.09;Fig. 1) from that seen in either set independently. SI was the most consistently associated trait within the LD block (Pmeta-analysis < 0.0030). DI was also associated, with four of the five SNPs in high LD showing association (Pmeta-analysis < 0.027). Association with FBG decreased in the combined analysis, and only one SNP remained significant (rs714925, Pmeta-analysis = 0.045).
TABLE 2.
Genotypic means for quantitative measures of glucose homeostasis calculated in set 1 of the IRASFS Hispanic-American population
| Phenotype | SNP | Alleles† | MAF | Genotypic means ± SD (n)* |
P‡ | ||
|---|---|---|---|---|---|---|---|
| 1/1 | 1/2 | 2/2 | |||||
| SI | rs1037257 | C/T | 0.37 | 2.37 ± 2.21 (207) | 1.96 ± 1.59 (280) | 2.13 ± 2.17 (85) | 0.027 |
| rs4969076 | C/G | 0.22 | 2.13 ± 2.01 (315) | 2.05 ± 1.69 (198) | 2.24 ± 2.21 (30) | 0.98 | |
| rs1037260 | T/G | 0.13 | 2.09 ± 1.90 (470) | 2.25 ± 2.05 (94) | 2.18 ± 1.74 (5) | 0.40 | |
| rs11077670 | G/A | 0.14 | 2.09 ± 1.98 (398) | 2.22 ± 1.86 (139) | 2.01 ± 2.12 (8) | 0.91 | |
| rs1868486 | G/T | 0.30 | 2.23 ± 2.06 (254) | 2.10 ± 1.92 (232) | 1.81 ± 1.56 (73) | 0.0040 | |
| rs714925 | A/G | 0.30 | 2.19 ± 2.03 (244) | 2.07 ± 1.89 (243) | 1.76 ± 1.45 (71) | 0.011 | |
| rs998571 | A/G | 0.29 | 2.24 ± 2.01 (261) | 2.14 ± 1.97 (219) | 1.71 ± 1.43 (73) | 0.0040 | |
| rs7220818 | A/G | 0.37 | 1.93 ± 1.67 (216) | 2.11 ± 1.94 (268) | 2.71 ± 2.33 (91) | 0.0020 | |
| rs6501603 | T/G | 0.48 | 2.27 ± 2.16 (152) | 2.03 ± 1.86 (276) | 2.15 ± 1.72 (128) | 0.51 | |
| rs4969017 | T/C | 0.03 | 2.14 ± 1.97 (529) | 1.96 ± 1.13 (20) | 1.01 ± 0.00 (1) | 0.18 | |
| AIR | rs1037257 | C/T | 0.37 | 713 ± 543 (207) | 743 ± 619 (280) | 845 ± 721 (85) | 0.54 |
| rs4969076 | C/G | 0.22 | 788 ± 623 (315) | 698 ± 585 (198) | 820 ± 627 (30) | 0.29 | |
| rs1037260 | T/G | 0.13 | 763 ± 644 (470) | 761 ± 566 (94) | 513 ± 177 (5) | 0.69 | |
| rs11077670 | G/A | 0.14 | 770 ± 620 (398) | 676 ± 698 (139) | 920 ± 737 (8) | 0.17 | |
| rs1868486 | G/T | 0.30 | 727 ± 532 (254) | 801 ± 706 (232) | 755 ± 696 (73) | 0.80 | |
| rs714925 | A/G | 0.30 | 728 ± 532 (244) | 800 ± 699 (243) | 751 ± 708 (71) | 0.81 | |
| rs998571 | A/G | 0.29 | 722 ± 525 (261) | 812 ± 724 (219) | 774 ± 699 (73) | 0.66 | |
| rs7220818 | A/G | 0.37 | 788 ± 643 (216) | 739 ± 637 (268) | 736 ± 567 (91) | 0.66 | |
| rs6501603 | T/G | 0.48 | 759 ± 584 (152) | 764 ± 650 (276) | 702 ± 581 (128) | 0.72 | |
| rs4969017 | T/C | 0.03 | 765 ± 626 (529) | 579 ± 297 (20) | 195 ± 0 (1) | 0.14 | |
| DI | rs1037257 | C/T | 0.37 | 1,375 ± 1,279 (207) | 1,225 ± 1,201 (280) | 1,390 ± 1,345 (85) | 0.053 |
| rs4969076 | C/G | 0.22 | 1,397 ± 1,414 (315) | 1,130 ± 893 (198) | 1,553 ± 1,539 (30) | 0.34 | |
| rs1037260 | T/G | 0.13 | 1,273 ± 1,229 (470) | 1,485 ± 1,409 (94) | 1,207 ± 1,052 (5) | 0.62 | |
| rs11077670 | G/A | 0.14 | 1,344 ± 1,309 (398) | 1,133 ± 1,027 (139) | 1,655 ± 1,780 (8) | 0.036 | |
| rs1868486 | G/T | 0.30 | 1,360 ± 1,349 (254) | 1,342 ± 1,283 (232) | 1,027 ± 939 (73) | 0.0020 | |
| rs714925 | A/G | 0.30 | 1,348 ± 1,337 (244) | 1,327 ± 1,202 (243) | 979 ± 838 (71) | 0.0010 | |
| rs998571 | A/G | 0.29 | 1,352 ± 1,292 (261) | 1,388 ± 1,366 (219) | 994 ± 837 (73) | 0.0010 | |
| rs7220818 | A/G | 0.37 | 1,254 ± 1,176 (216) | 1,262 ± 1,280 (268) | 1,597 ± 1,404 (91) | 0.052 | |
| rs6501603 | T/G | 0.48 | 1,463 ± 1,437 (152) | 1,230 ± 1,176 (276) | 1,227 ± 1,004 (128) | 0.68 | |
| rs4969017 | T/C | 0.03 | 1,328 ± 1,293 (529) | 1,100 ± 975 (20) | 197 ± 0 (1) | 0.94 | |
| FBG | rs1037257 | C/T | 0.37 | 93.28 ± 9.35 (220) | 93.85 ± 10.25 (298) | 92.88 ± 9.68 (88) | 0.62 |
| rs4969076 | C/G | 0.22 | 93.57 ± 10.24 (343) | 93.69 ± 9.01 (202) | 91.16 ± 8.15 (32) | 0.043 | |
| rs1037260 | T/G | 0.13 | 93.26 ± 9.55 (498) | 94.35 ± 10.48 (99) | 94.07 ± 11.80 (7) | 0.14 | |
| rs11077670 | G/A | 0.14 | 93.57 ± 9.99 (427) | 93.38 ± 9.49 (144) | 96.31 ± 11.29 (8) | 0.41 | |
| rs1868486 | G/T | 0.30 | 92.49 ± 9.70 (272) | 94.62 ± 10.02 (243) | 94.24 ± 9.35 (77) | 0.019 | |
| rs714925 | A/G | 0.30 | 92.34 ± 9.52 (264) | 94.48 ± 10.00 (253) | 94.05 ± 9.58 (75) | 0.005 | |
| rs998571 | A/G | 0.29 | 92.41 ± 9.48 (280) | 94.24 ± 10.27 (230) | 94.23 ± 9.32 (77) | 0.014 | |
| rs7220818 | A/G | 0.37 | 93.86 ± 9.46 (222) | 93.88 ± 10.00 (286) | 90.96 ± 9.23 (101) | 0.014 | |
| rs6501603 | T/G | 0.48 | 93.30 ± 9.76 (166) | 94.12 ± 10.37 (293) | 92.52 ± 8.86 (132) | 0.25 | |
| rs4969017 | T/C | 0.03 | 93.50 ± 9.69 (562) | 90.59 ± 9.68 (22) | 93.00 ± 0.00 (1) | 0.19 | |
Boldface data indicate P values <0.05.
*Genotypic means ± SD (n) for homozygotes of the major allele (1/1) as well as heterozygotes (1/2) and homozygotes of the minor allele (2/2).
†Major/minor alleles.
‡Test for association among each SNP-trait combination using a series of generalized estimating equations.
TABLE 3.
Genotypic means for quantitative measures of glucose homeostasis calculated in set 2 of the IRASFS Hispanic-American population
| Phenotype | SNP | Alleles† | MAF | Genotypic means ± SD (n)* |
P‡ | ||
|---|---|---|---|---|---|---|---|
| 1/1 | 1/2 | 2/2 | |||||
| SI | rs1037257 | C/T | 0.40 | 2.35 ± 1.78 (143) | 2.08 ± 1.78 (218) | 2.34 ± 1.91 (61) | 0.60 |
| rs4969076 | C/G | 0.22 | 2.31 ± 1.85 (34) | 2.15 ± 1.75 (162) | 1.82 ± 1.53 (25) | 0.15 | |
| rs1037260 | T/G | 0.15 | 2.19 ± 1.72 (330) | 2.28 ± 1.99 (92) | 3.19 ± 1.55 (2) | 0.83 | |
| rs11077670 | G/A | 0.10 | 2.29 ± 1.84 (318) | 1.96 ± 1.69 (88) | 1.81 ± 1.57 (6) | 0.039 | |
| rs1868486 | G/T | 0.28 | 2.24 ± 1.78 (199) | 2.23 ± 1.79 (191) | 1.68 ± 1.72 (36) | 0.038 | |
| rs714925 | A/G | 0.28 | 2.27 ± 1.81 (196) | 2.25 ± 1.79 (191) | 1.69 ± 1.75 (35) | 0.097 | |
| rs998571 | A/G | 0.25 | 2.26 ± 1.79 (214) | 2.21 ± 1.79 (178) | 1.70 ± 1.64 (36) | 0.11 | |
| rs7220818 | A/G | 0.41 | 2.17 ± 1.72 (132) | 2.20 ± 1.76 (219) | 2.18 ± 1.85 (72) | 0.98 | |
| rs6501603 | T/G | 0.45 | 2.35 ± 1.79 (141) | 2.11 ± 1.77 (203) | 2.22 ± 1.92 (73) | 0.61 | |
| rs4969017 | T/C | 0.02 | 2.21 ± 1.79 (417) | 2.00 ± 1.66 (16) | 0.15 | ||
| AIR | rs1037257 | C/T | 0.40 | 697 ± 629 (143) | 820 ± 739 (218) | 841 ± 695 (61) | 0.32 |
| rs4969076 | C/G | 0.22 | 773 ± 664 (234) | 748 ± 692 (162) | 776 ± 829 (25) | 0.80 | |
| rs1037260 | T/G | 0.15 | 740 ± 681 (330) | 867 ± 712 (92) | 455 ± 6 (2) | 0.34 | |
| rs11077670 | G/A | 0.10 | 773 ± 641 (318) | 815 ± 886 (88) | 831 ± 358 (6) | 0.84 | |
| rs1868486 | G/T | 0.28 | 773 ± 676 (199) | 797 ± 727 (191) | 618 ± 593 (36) | 0.021 | |
| rs714925 | A/G | 0.28 | 777 ± 671 (196) | 794 ± 727 (191) | 629 ± 605 (35) | 0.089 | |
| rs998571 | A/G | 0.25 | 786 ± 660 (214) | 777 ± 740 (178) | 643 ± 596 (36) | 0.26 | |
| rs7220818 | A/G | 0.41 | 757 ± 626 (132) | 789 ± 744 (219) | 747 ± 646 (72) | 0.86 | |
| rs6501603 | T/G | 0.45 | 709 ± 602 (141) | 781 ± 708 (203) | 933 ± 837 (73) | 0.66 | |
| rs4969017 | T/C | 0.02 | 744 ± 661 (417) | 1,236 ± 1,130 (16) | 0.12 | ||
| DI | rs1037257 | C/T | 0.40 | 1,270 ± 1,017 (151) | 1,375 ± 1,245 (218) | 1,520 ± 1,499 (61) | 0.56 |
| rs4969076 | C/G | 0.22 | 1,426 ± 1,247 (234) | 1,255 ± 1,152 (162) | 1,122 ± 1,149 (25) | 0.0050 | |
| rs1037260 | T/G | 0.15 | 1,307 ± 1,163 (330) | 1,462 ± 1,321 (92) | 1,445 ± 685 (2) | 0.53 | |
| rs11077670 | G/A | 0.10 | 1,415 ± 1,236 (318) | 1,143 ± 1,106 (88) | 1,468 ± 1,524 (6) | 0.0010 | |
| rs1868486 | G/T | 0.28 | 1,305 ± 1,049 (199) | 1,436 ± 1,311 (191) | 1,035 ± 1,402 (36) | 0.088 | |
| rs714925 | A/G | 0.28 | 1,315 ± 1,032 (196) | 1,430 ± 1,309 (191) | 1,072 ± 1,457 (35) | 0.21 | |
| rs998571 | A/G | 0.25 | 1,380 ± 1,158 (214) | 1,353 ± 1,216 (178) | 1,096 ± 1,430 (36) | 0.20 | |
| rs7220818 | A/G | 0.41 | 1,443 ± 1,339 (132) | 1,294 ± 1,106 (219) | 1,265 ± 1,098 (72) | 0.63 | |
| rs6501603 | T/G | 0.45 | 1,312 ± 1,180 (141) | 1,313 ± 1,161 (203) | 1,578 ± 1,409 (73) | 0.32 | |
| rs4969017 | T/C | 0.02 | 1,323 ± 1,200 (417) | 1,540 ± 1,199 (16) | 0.59 | ||
| FBG | rs1037257 | C/T | 0.40 | 92.68 ± 9.98 (151) | 93.40 ± 9.01 (231) | 93.93 ± 7.27 (63) | 0.21 |
| rs4969076 | C/G | 0.22 | 93.31 ± 9.04 (246) | 92.47 ± 9.20 (172) | 97.52 ± 10.32 (26) | 0.18 | |
| rs1037260 | T/G | 0.15 | 93.43 ± 9.46 (350) | 92.38 ± 7.98 (95) | 90.33 ± 7.94 (3) | 0.53 | |
| rs11077670 | G/A | 0.10 | 92.75 ± 8.99 (336) | 94.68 ± 9.83 (92) | 97.75 ± 5.40 (6) | 0.099 | |
| rs1868486 | G/T | 0.28 | 93.43 ± 9.53 (212) | 92.18 ± 8.51 (199) | 97.41 ± 9.63 (39) | 0.024 | |
| rs714925 | A/G | 0.28 | 93.40 ± 9.59 (209) | 92.24 ± 8.46 (199) | 97.87 ± 9.44 (38) | 0.030 | |
| rs998571 | A/G | 0.25 | 93.34 ± 9.47 (229) | 92.59 ± 8.55 (185) | 96.25 ± 10.56 (38) | 0.51 | |
| rs7220818 | A/G | 0.41 | 93.89 ± 8.88 (143) | 92.88 ± 9.09 (226) | 93.29 ± 10.29 (77) | 0.64 | |
| rs6501603 | T/G | 0.45 | 93.02 ± 9.20 (147) | 93.15 ± 9.01 (217) | 93.91 ± 9.27 (76) | 0.60 | |
| rs4969017 | T/C | 0.02 | 93.28 ± 9.24 (438) | 92.92 ± 8.70 (18) | 0.92 | ||
Boldface data indicate P values <0.05.
*Genotypic means ± SD (n) for homozygotes of the major allele (1/1) as well as heterozygotes (1/2) and homozygotes of the minor allele (2/2).
†Major/minor alleles.
‡Test for association among each SNP-trait combination using a series of generalized estimating equations.
TABLE 4.
Genotypic means for quantitative measures of glucose homeostasis calculated in the combined IRASFS Hispanic-American population
| Phenotype | SNP | Alleles† | MAF | Genotypic means ± SD (n)* |
P‡ | P§ | ||
|---|---|---|---|---|---|---|---|---|
| 1/1 | 1/2 | 2/2 | ||||||
| SI | rs1037257 | C/T | 0.38 | 2.36 ± 2.04 (350) | 2.01 ± 1.68 (498) | 2.22 ± 2.057 (146) | 0.60 | 0.16 |
| rs4969076 | C/G | 0.22 | 2.21 ± 1.95 (549) | 2.09 ± 1.72 (360) | 2.05 ± 1.93 (55) | 0.43 | 0.41 | |
| rs1037260 | T/G | 0.14 | 2.13 ± 1.83 (800) | 2.26 ± 2.02 (186) | 2.47 ± 1.63 (7) | 0.83 | 0.75 | |
| rs11077670 | G/A | 0.12 | 2.18 ± 1.92 (716) | 2.11 ± 1.80 (227) | 1.92 ± 1.84 (14) | 0.24 | 0.62 | |
| rs1868486 | G/T | 0.29 | 2.23 ± 1.94 (453) | 2.16 ± 1.86 (423) | 1.76 ± 1.61 (109) | 0.000071 | 0.0025 | |
| rs714925 | A/G | 0.29 | 2.23 ± 1.94 (440) | 2.15 ± 1.85 (434) | 1.74 ± 1.55 (106) | 0.0010 | 0.0030 | |
| rs998571 | A/G | 0.27 | 2.25 ± 1.91 (475) | 2.17 ± 1.89 (397) | 1.71 ± 1.50 (109) | 0.00018 | 0.0024 | |
| rs7220818 | A/G | 0.39 | 2.02 ± 1.69 (348) | 2.15 ± 1.86 (487) | 2.48 ± 2.14 (163) | 0.60 | 0.16 | |
| rs6501603 | T/G | 0.47 | 2.31 ± 1.99 (293) | 2.07 ± 1.82 (479) | 2.17 ± 1.79 (201) | 0.29 | 0.13 | |
| rs4969017 | T/C | 0.03 | 2.17 ± 1.89 (946) | 1.98 ± 1.37 (36) | 1.01 ± 0.00 (1) | 0.83 | 0.94 | |
| AIR | rs1037257 | C/T | 0.38 | 706 ± 579 (350) | 777 ± 675 (498) | 844 ± 708 (146) | 0.32 | 0.16 |
| rs4969076 | C/G | 0.22 | 781 ± 640 (549) | 721 ± 635 (360) | 800 ± 719 (55) | 0.48 | 0.66 | |
| rs1037260 | T/G | 0.14 | 754 ± 659 (800) | 814 ± 643 (186) | 496 ± 147 (7) | 0.50 | 0.78 | |
| rs11077670 | G/A | 0.12 | 772 ± 629 (716) | 730 ± 725 (227) | 882 ± 586 (14) | 0.37 | 0.55 | |
| rs1868486 | G/T | 0.29 | 747 ± 599 (453) | 799 ± 715 (423) | 710 ± 664 (109) | 0.43 | 0.58 | |
| rs714925 | A/G | 0.29 | 750 ± 598 (440) | 798 ± 711 (434) | 711 ± 675 (106) | 0.48 | 0.56 | |
| rs998571 | A/G | 0.27 | 751 ± 590 (475) | 796 ± 730 (397) | 731 ± 667 (109) | 0.99 | 0.99 | |
| rs7220818 | A/G | 0.39 | 776 ± 636 (348) | 762 ± 687 (487) | 741 ± 601 (163) | 0.90 | 0.96 | |
| rs6501603 | T/G | 0.47 | 735 ± 592 (293) | 771 ± 674 (479) | 786 ± 692 (201) | 0.46 | 0.47 | |
| rs4969017 | T/C | 0.03 | 756 ± 641 (946) | 871 ± 839 (36) | 195 ± 0 (1) | 0.75 | 0.25 | |
| DI | rs1037257 | C/T | 0.38 | 1,332 ± 1,179 (350) | 1,290 ± 1,221 (498) | 1,444 ± 1,408 (146) | 0.56 | 0.92 |
| rs4969076 | C/G | 0.22 | 1,409 ± 1,344 (549) | 1,186 ± 1,018 (360) | 1,357 ± 1,380 (55) | 0.051 | 0.036 | |
| rs1037260 | T/G | 0.14 | 1,287 ± 1,201 (800) | 1,474 ± 1,362 (186) | 1,275 ± 910 (7) | 0.82 | 0.90 | |
| rs11077670 | G/A | 0.12 | 1,376 ± 1,277 (716) | 1,137 ± 1,056 (227) | 1,575 ± 1,615 (14) | 0.00025 | 0.015 | |
| rs1868486 | G/T | 0.29 | 1,335 ± 1,225 (453) | 1,385 ± 1,295 (423) | 1,030 ± 1,107 (109) | 0.00037 | 0.0075 | |
| rs714925 | A/G | 0.29 | 1,333 ± 1,209 (440) | 1,373 ± 1,250 (434) | 1,009 ± 1,076 (106) | 0.0010 | 0.0061 | |
| rs998571 | A/G | 0.27 | 1,365 ± 1,233 (475) | 1,373 ± 1,299 (397) | 1,028 ± 1,064 (109) | 0.0030 | 0.027 | |
| rs7220818 | A/G | 0.39 | 1,326 ± 1,242 (348) | 1,277 ± 1,203 (487) | 1,450 ± 1,285 (163) | 0.070 | 0.14 | |
| rs6501603 | T/G | 0.47 | 1,391 ± 1,320 (293) | 1,265 ± 1,169 (479) | 1,355 ± 1,177 (201) | 0.30 | 0.24 | |
| rs4969017 | T/C | 0.03 | 1,326 ± 1,252 (946) | 1,295 ± 1,087 (36) | 197 ± 0 (1) | 0.66 | 0.47 | |
| FBG | rs1037257 | C/T | 0.38 | 93.03 ± 9.60 (371) | 93.65 ± 9.72 (529) | 93.31 ± 8.74 (151) | 0.21 | 0.87 |
| rs4969076 | C/G | 0.22 | 93.46 ± 9.75 (589) | 93.13 ± 9.10 (374) | 94.01 ± 9.65 (58) | 0.47 | 0.54 | |
| rs1037260 | T/G | 0.14 | 93.33 ± 9.51 (848) | 93.38 ± 9.37 (194) | 92.95 ± 10.49 (10) | 0.91 | 0.74 | |
| rs11077670 | G/A | 0.12 | 93.21 ± 9.56 (763) | 93.89 ± 9.63 (236) | 96.93 ± 8.97 (14) | 0.056 | 0.11 | |
| rs1868486 | G/T | 0.29 | 92.90 ± 9.63 (484) | 93.52 ± 9.44 (442) | 95.31 ± 9.52 (116) | 0.010 | 0.083 | |
| rs714925 | A/G | 0.29 | 92.81 ± 9.56 (473) | 93.49 ± 9.41 (452) | 95.34 ± 9.66 (113) | 0.010 | 0.045 | |
| rs998571 | A/G | 0.27 | 92.83 ± 9.48 (509) | 93.51 ± 9.57 (415) | 94.90 ± 9.75 (115) | 0.010 | 0.079 | |
| rs7220818 | A/G | 0.39 | 93.87 ± 11.96 (365) | 93.44 ± 1097 (513) | 91.96 ± 9.74 (178) | 0.10 | 0.25 | |
| rs6501603 | T/G | 0.47 | 93.17 ± 9.49 (313) | 93.71 ± 9.82 (510) | 93.03 ± 9.02 (208) | 0.76 | 0.72 | |
| rs4969017 | T/C | 0.03 | 93.40 ± 9.49 (1000) | 91.64 ± 9.21 (40) | 93.00 ± 0.00 (1) | 0.41 | 0.81 | |
Boldface data indicate P values <0.05.
*Genotypic means ± SD (n) for homozygotes of the major allele (1/1) as well as heterozygotes (1/2) and homozygotes of the minor allele (2/2).
†Major/minor alleles.
‡Test for association among each SNP-trait combination using a series of generalized estimating equations.
§Meta-analysis P value computed by sequential oligogenic linkage analysis with the modeling of set 1 or set 2 affiliation as a random effect in a variance component–measured genotype model.
DISCUSSION
This study of SSTR2 gene polymorphisms represents the first detailed population genetic analysis of this gene in relation to glucose homeostasis phenotypes. Whereas the current study design is limited by the SNP selection method, i.e., density versus tagging algorithms, the high LD in the region facilitated sufficient coverage of genetic variation. Strengths of this study include the use of a large Hispanic-American cohort and analysis of quantitative measures of glucose homeostasis. Moreover, since the Hispanic-American subjects were divided into two independent sets with identical recruitment criteria and similar demographic data, results can be compared between the two populations, resulting in an ideal replication population, and can be combined to give maximal power for detecting association.
In total, 10 SNPs located at the SSTR2 locus were genotyped on 1,425 Hispanic Americans. The most consistent region of association fell within the LD block encompassing the SSTR2 gene, with three of the five SNPs highly correlated (r2> 0.78;Fig. 1) and driving the association. IRASFS Hispanic Americans were separated into two distinct groups for analysis, and results were compared between groups. Although set 1 individuals exhibited stronger association between polymorphisms in SSTR2 and SI as well as DI and FBG, similar evidence of association or trends was detected in set 2, albeit of lesser magnitude. Thus, association was detected in two independent sets of Hispanic Americans.
The magnitude of association increased when the two sets of Hispanic Americans were combined. Tests of between-population heterogeneity were nonsignificant for the most significantly associated SNPs and traits (Table A2 in the online appendix). Within the LD block, associations were observed between SSTR2 SNPs and SI (Pmeta-analysis = 0.0024–0.0030) and DI (Pmeta-analysis = 0.0075–0.027). Examination of the genotypic means revealed a consistent pattern of decreased SI (βhomozygous = −0.16) and DI (βhomozygous = −0.35 to −5.16) associated with the minor allele (Table 4; online appendix Table A2). In addition, evidence of association was observed with FBG (Pmeta-analysis = 0.045), with increased values (βhomozygous = 2.30) associated with the minor allele (Table 4; online appendix Table A2). Taken together, these findings are consistent with genetic variants at the SSTR2 locus contributing to insulin resistance, which results in higher circulating blood glucose levels. Evaluation of SSTR2 SNPs for association with type 2 diabetes resulted in an overall lack of association, with only a single SNP showing nominal association (rs7220818, P = 0.040; data not shown). Lack of association should be viewed cautiously, as the result could be attributed to modest sample size for type 2 diabetes (n = 181).
In addition, we evaluated SSTR2 in a collection of 605 African Americans from the IRASFS who were recruited and phenotyped in a similar manner. No evidence of association was observed (data not shown). This absence of association could reflect observable differences in LD patterns in the Hispanic- and African-American populations as reflected in the HapMap reference populations (CEU and YRI, respectively; online appendix Figs. A1 and A2), which results in less efficient tagging of variation in African populations (r2 = 0.47, MAF >0.05, aggressive tagging algorithm). In addition, lack of association could be attributed to reduced power due to smaller sample size or ethnicity-specific genetic differences.
SSTR2 polymorphisms appear to play a role in the modulation of glucose homeostasis through mechanisms consistent with the known biological function of the protein. SRIF inhibits growth hormone and glucagon secretion in a subtype-selective process through SSTR2 (1). Both growth hormone and glucagon exhibit potent antagonistic effects on insulin action by increasing hepatic gluconeogenesis and glycogenolysis. In addition, growth hormone decreases peripheral glucose utilization (20). Decreased utilization of peripheral glucose would directly affect SI as measured herein. Evidence of association with DI is likely to be attributed to associations with SI due to the mathematical relationship (DI = SI × AIR). The physiological effect of decreased glucose utilization coupled with increased glucose production would result in elevated blood glucose levels, as observed through the association with FBG. Lack of association with AIR could be attributed to the subtype-selective actions of SRIF to inhibit insulin secretion, predominantly through SSTR5 (1).
Based on these findings, additional research is warranted to identify a true functional variant(s), as it is unlikely the causal variant was genotyped on these individuals; i.e., SNPs were chosen based on position. Although evidence of association was detected between SNPs in the SSTR2 coding region and glucose homeostasis phenotypes, magnitude of the association was not completely consistent throughout all analyses, suggesting that the association is due to LD with the true functional variant. Additionally, a more thorough evaluation of the proximal 5′ genomic region may provide insight into a causal mechanism, possibly through interaction with the proposed promoter(s) (21–23). In conclusion, SSTR2 polymorphisms are associated with glucose homeostasis and appear to be involved through modulation of SI.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by NIH grants HL060894, HL060931, HL060944, HL061019, and HL061210.
No potential conflicts of interest relevant to this article were reported.
The authors acknowledge the anonymous reviewers for their thoughtful insight during the manuscript review process.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Patel YC: Somatostatin and its receptor family. Front Neuroendocrinol 1999; 20: 157– 198 [DOI] [PubMed] [Google Scholar]
- 2.Yamada Y, Post SR, Wang K, Tager HS, Bell GI, Seino S: Cloning and functional characterization of a family of human and mouse somatostatin receptors expressed in brain, gastrointestinal tract, and kidney. Proc Natl Acad Sci U S A 1992; 89: 251– 255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossowski WJ, Coy DH: Potent inhibitory effects of a type four receptor-selective somatostatin analog on rat insulin release. Biochem Biophys Res Commun 1993; 197: 366– 371 [DOI] [PubMed] [Google Scholar]
- 4.Strowski MZ, Parmar RM, Blake AD, Schaeffer JM: Somatostatin inhibits insulin and glucagon secretion via two receptors subtypes: an in vitro study of pancreatic islets from somatostatin receptor 2 knockout mice. Endocrinology 2000; 141: 111– 117 [DOI] [PubMed] [Google Scholar]
- 5.Moldovan S, Atiya A, Adrian TE, Kleinman RM, Lloyd K, Olthoff K, Imagawa D, Shevlin L, Coy D, Walsh J, et al. : Somatostatin inhibits B-cell secretion via a subtype-2 somatostatin receptor in the isolated perfused human pancreas. J Surg Res 1995; 59: 85– 90 [DOI] [PubMed] [Google Scholar]
- 6.Cheng H, Yibchok-anun S, Coy DH, Hsu WH: SSTR2 mediates the somatostatin-induced increase in intracellular Ca(2+) concentration and insulin secretion in the presence of arginine vasopressin in clonal beta-cell HIT-T15. Life Sci 2002; 71: 927– 936 [DOI] [PubMed] [Google Scholar]
- 7.Sutton BS, Langefeld CD, Campbell JK, Haffner SM, Norris JM, Scherzinger AL, Wagenknecht LE, Bowden DW: Genetic mapping of a 17q chromosomal region linked to obesity phenotypes in the IRAS family study. Int J Obes (Lond) 2006; 30: 1433– 1441 [DOI] [PubMed] [Google Scholar]
- 8.Norris JM, Langefeld CD, Scherzinger AL, Rich SS, Bookman E, Beck SR, Saad MF, Haffner SM, Bergman RN, Bowden DW, Wagenknecht LE: Quantitative trait loci for abdominal fat and BMI in Hispanic-Americans and African-Americans: the IRAS Family study. Int J Obes Relat Metab Disord 2005; 29: 67– 77 [DOI] [PubMed] [Google Scholar]
- 9.Henkin L, Bergman RN, Bowden DW, Ellsworth DL, Haffner SM, Langefeld CD, Mitchell BD, Norris JM, Rewers M, Saad MF, Stamm E, Wagenknecht LE, Rich SS: Genetic epidemiology of insulin resistance and visceral adiposity: the IRAS Family Study design and methods. Ann Epidemiol 2003; 13: 211– 217 [DOI] [PubMed] [Google Scholar]
- 10.Bergman RN, Finegood DT, Ader M: Assessment of insulin sensitivity in vivo. Endocr Rev 1985; 6: 45– 86 [DOI] [PubMed] [Google Scholar]
- 11.Bergman RN: Lilly lecture 1989: toward physiological understanding of glucose tolerance: minimal-model approach. Diabetes 1989; 38: 1512– 1527 [DOI] [PubMed] [Google Scholar]
- 12.Buetow KH, Edmonson M, MacDonald R, Clifford R, Yip P, Kelley J, Little DP, Strausberg R, Koester H, Cantor CR, Braun A: High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc Natl Acad Sci U S A 2001; 98: 581– 584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Connell JR, Weeks DE: PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998; 63: 259– 266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D: Efficiency and power in genetic association studies. Nat Genet 2005; 37: 1217– 1223 [DOI] [PubMed] [Google Scholar]
- 15.Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263– 265 [DOI] [PubMed] [Google Scholar]
- 16.Zeger SL, Liang KY: Longitudinal data analysis for discrete and continuous outcomes. Biometrics 1986; 42: 121– 130 [PubMed] [Google Scholar]
- 17.Hardin JW: Generalized Estimating Equations New York, Chapman & Hall/CRC, 2003 [Google Scholar]
- 18.Almasy L, Blangero J: Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998; 62: 1198– 1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutton AJ, Abrams KR, Sheldon TA, Song F: Methods for Meta-Analysis in Medical Research West Sussex, U.K., John Wiley & Sons, 2000 [Google Scholar]
- 20.Yuen KC, Dunger DB: Therapeutic aspects of growth hormone and insulin-like growth factor-I treatment on visceral fat and insulin sensitivity in adults. Diabetes Obes Metab 2007; 9: 11– 22 [DOI] [PubMed] [Google Scholar]
- 21.Petersenn S, Rasch AC, Presch S, Beil FU, Schulte HM: Genomic structure and transcriptional regulation of the human somatostatin receptor type 2. Mol Cell Endocrinol 1999; 157: 75– 85 [DOI] [PubMed] [Google Scholar]
- 22.Pscherer A, Dorflinger U, Kirfel J, Gawlas K, Ruschoff J, Buettner R, Schule R: The helix-loop-helix transcription factor SEF-2 regulates the activity of a novel initiator element in the promoter of the human somatostatin receptor II gene. Embo J 1996; 15: 6680– 6690 [PMC free article] [PubMed] [Google Scholar]
- 23.Torrisani J, Hanoun N, Laurell H, Lopez F, Maoret J, Souque A, Susini C, Cordelier P, Buscail L: Identification of an upstream promoter of the human somatostatin receptor hSSTR2, that is controlled by epigenetic modifications. Endocrinology 2008; 149: 3137– 3147 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.