Abstract
Phages are promising alternatives to antibodies as the biorecognition element in a variety of biosensing applications. In this study, a monolayer of bacteriophage P22 whose tailspike proteins specifically recognize Salmonella serotypes was covalently bound to glass substrates through a bifunctional cross linker 3-aminopropyltrimethoxysilane. The specific binding of Salmonella typhimurium to the phage monolayer was studied by enzyme-linked immunosorbent assay and atomic force microscopy. Escherichia coli and a Gram-positive bacterium Listeria monocytogenes were also studied as control bacteria. The P22 particles show strong binding affinity to Salmonella typhimurium. In addition, the dried P22 monolayer maintained 50% binding capacity to Salmonella typhimurium after a one-week storage time. This is a promising method to prepare phage monolayer coatings on surface plasmon resonance and acoustic biosensor substrates in order to utilize the nascent phage display technology.
Keywords: bacteriophage, Salmonella typhimurium, lipopolysaccharide membrane
Introduction
The development of accurate and rapid detection methods of microorganisms, particularly those infectious pathogens in soil, food, and water, remains a technical challenge. It becomes more urgent due to the growing threat of bioterrorism. Different types of chemicals, toxins, and biological molecules are either being used or have the potential to be used as warfare agents [1–3]. Biorecognition strategies are being developed to utilize biomolecules such as enzymes, DNA, bacteriophages (or phages), and antibodies to detect complementary molecules through bio-specific interactions. Phages are virus particles that carry their genetic information in the form of DNA or RNA and can attach to specific receptors that are present on a limited range of host bacterial cells [4–7]. Since their discovery a century ago, phages have found new applications including phage therapy [8, 9], water treatment [10], high-throughput screening [11, 12], and biosensing [4, 5, 13–15]. The specific targeting phage probes can be directly attached to the sensor surface in surface plasmon resonance (SPR) and acoustic biosensors [16]. This paper presents a method to chemically immobilize phage particles on glass substrates while maintaining their binding affinity and specificity to bacterial surface receptors. The selectivity and durability of phage are key attributes to its ultimate use as a biosensing element.
Currently, antibodies are the most prevalent biosensing element for bacteria in immunosensors [17–20]. Phage display for bio-detection is a nascent technology suitable for real-time and inexpensive field detection [5]. Antibodies can be immobilized on solid substrates via their Fc fraction leaving the Fab portion free for detection using physical adsorption [21], Langmuir-Blodgett (LB) technique [22, 23], and covalent coupling [24, 25]. Immunosensors have serious limitations including the non-antigenic nature of the analyte, incompatibility with the sample matrix or extraction process, and the time- and labor-intensive process of making the antibodies. In addition to the high cost, antibodies are highly fragile and sensitive to environmental conditions.
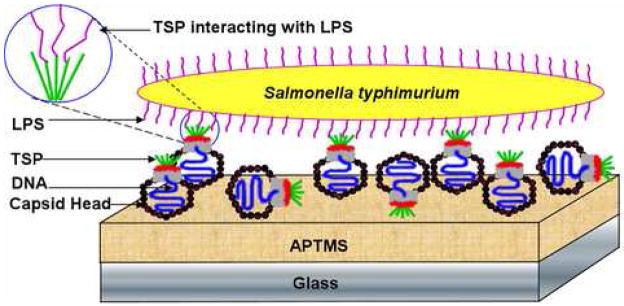
Compared to antibodies, phages are less fragile and less sensitive to environmental stress such as pH and temperature fluctuation [26–28], which give them longer field life for detecting toxins, bacteria, and spores. In addition, the new phage display technology offers billion clone libraries of recombinant phages for high-throughput detection. This paper explores the possibility of using a covalently linked phage monolayer for specific bacterial detection by combining the traditional enzyme-linked immunosorbent assay (ELISA) and molecular-level atomic force microscopy (AFM) characterization techniques. The well-documented P22 phage and its interactions with Salmonella enterica serovar typhimurium (S. typhimurium) [29–34] was chosen as a model system. P22 is known to bind to the repetitive O-antigen part present in the lipopolysaccharides (LPS) of Salmonella outer membrane. P22 consists of double-stranded DNA packaged in an icosahedral capsid head and the O-antigen recognizing tailspike protein (TSP). A total of 6 TSP gp9 (6 × 215.4 kDa) copies are non-covalently attached to the capsid head by the N-terminal domain of the gp9 while the C-terminal domain binds to their cellular LPS receptor. Gp9 exhibits endoglycosidase activity by hydrolyzing the repeating sections of the O-antigen portion of LPS specifically at the Rha–Gal α (1→po 3)-glycosidic linkage.
Here we report a method to chemically immobilize P22 in a monolayer and the study of the interactions of P22 with various bacteria including Gram-negative S. typhimurium (Figure 1) and Escherichia coli, and the Gram-positive Listeria monocytogenes. Previous reports on phage immobilization include physical adsorption [13–15] and Langmuir-Blodgett deposition [5]. Chemical adsorption based immobilization utilizing the versatile silane chemistry may improve the durability of the phage coating. We employed a chemical vapor deposition (CVD) method developed in our group to make a smooth aminosilane monolayer on silicon oxide surface of the glass substrate [35]. The phage P22 was bound to the aminosilane monolayer using the well-established sulfo-NHS and EDC chemistry [36–41]. To the best of our knowledge, our results are among the first to demonstrate successful immobilization of phage by EDC/NHS activation. Other covalent attachment schemes are also available in literature for phage immobilization [42, 43]. Through ELISA and AFM characterization, we showed that the phage-coated substrates are capable of differentiating among different bacterial types in an aqueous environment. Such phage coatings promise to achieve the same degree of sensitivity and selectivity as current immunosensors but with a fraction of the cost and significantly improved durability.
Figure 1.
Scheme of phage P22 immobilization for the detection of Salmonella typhimurium.
Experimental section
Materials
3-Aminopropyltrimethoxysilane (APTMS) and o-phenylenediamine dihydrochloride (OPD) were purchased from Sigma-Aldrich. N-hydroxysulfosuccinimide (Sulfo-NHS) and 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) were purchased from Pierce. HRP-labeled anti Salmonella IgG and anti E coli IgG were purchased from U.S. biological (s0060-20) and HRP-labeled anti Listeria IgG was purchased from Abcam (ab20357). Phage P22 (Salmonella enterica subsp. Enterica serovar typhimurium bacteriophage) ATCC (19585-B1), S. typhimurium ATCC (19585), and E coli strain ATCC (33780) were used. 0.22-μm cellulose acetate filters were purchased from Corning.
Bacteria culture
Three-ml Luria-Bertani (LB Broth) was added to a 15-ml centrifuge tube. The media was inoculated with S. typhimurium. The tube was shaken in water bath at 37 °C overnight. Cells were removed from the bath in log phase at a titer of approximately 1 × 106 – 1 × 107 cfu/ml.
Propagation of phage
One hundred μl P22 (titer approximately 108 pfu/ml) and one hundred μl S. typhimurium (titer approximately 106 cfu/ml) were stirred with 50 ml of LB media overnight in water bath at 37 °C. Uninfected cells and other debris were removed the following day by centrifugation for 20 min at 10,000 rpm (Sorvall GSA rotor). The solution was filtered using 0.22-μm cellulose acetate filters. The filtrate containing P22 was tittered and stored at 4 °C.
Silane monolayer deposition
Chemical vapor deposition (CVD) was used to form the aminosilane layer instead of the usual solution-based dip coating method. We found that CVD provides more consistent and smoother coating [35]. Prior to CVD, glass substrates were immersed in Piranha solution (H2SO4:H2O2 = 3:1) for 10 min followed by 5 rinses in deionized water (Nanopure System, Barnstead) and 1 rinse in ethanol (200 proof, manufactured by Aaper). The CVD deposition was conducted in a clean desiccator. The glass desiccator was silanized before the deposition on glass substrates in order to passivate the desiccator surface. Finally the glass substrates were silanized with APTMS in the desiccator with an active vacuum pumping time of 15 min at 0.67 kPa followed by a standing time of 16 hr under vacuum. The treated substrates were subsequently rinsed with deionized water and ethanol and dried.
Immobilization of P22
Twelve-ml P22 stock solution (ca. 108 pfu/ml) was placed in a 13-ml ultracentrifuge tube. The solution was spun at 35,000 rpm in a Beckman SW41-Ti rotor for 2 hr. After centrifugation a small transparent pellet could be faintly observed on the bottom of the tube. The pellet was then resuspended in 1-ml distilled deionized water with an added 2-mM CaCl2 to stabilize the phage heads. The concentration of the final solution is approximately between 1010 to 1012 pfu/ml for optimal crosslinking to the surface. For crosslinking P22 to the APTMS layer, 2.2 mg of Sulfo-NHS and 0.8 mg of EDC were added to each 1-ml aliquot of concentrated phage solution. The samples were mixed briefly and allowed to react for 30 min at room temperature. 100 μl of phage was then pipetted onto the silanized surface and incubated overnight at 4 °C in microtiter plates.
Binding of S. typhimurium on P22 monolayer
The P22 phage solution was removed from the substrate and 100-μl of freshly prepared overnight-cultured bacteria with a titer value of 106–107 was added to each well of the 96-well plate. The bacteria culture was allowed to react for 15 min at room temperature followed by thorough wash with Phosphate Buffered Saline (PBS, pH = 7.2). As a control, 100 μl of freshly prepared overnight culture was added directly to the wells containing silanized glass without any P22 phage. In addition, E coli strain 33780 and a Gram-positive bacterium L. monocytogenes were used as control bacteria. One hundred μl of freshly prepared overnight culture of E coli and L. monocytogenes (106–107 cfu/ml) were added on P22-coated glass for 15 min at 37 °C and subsequently washed with PBS before imaging.
Enzyme-linked immunosorbent assay
Enzyme-linked immunosorbent assay (ELISA) is based on the principle of antibody and antigen interactions, and is used to provide the value of relative amount of bacteria attached to P22. After the binding of S. typhimurium to P22 the chips were incubated with 100 μl of a 3w/w% bovine serum albumin (BSA) and 0.05v/v% tween-20 in PBS solution for 30 min at 37 °C. This solution acts as a blocking agent to reduce nonspecific binding of the labeled antibody in the next step. After 30-min incubation the chips were washed 40 times with a 0.05% tween-20 PBS solution in order to remove any excess blocking agent and non-phage bound bacteria. Anti-Salmonella or anti-Listeria HRP-labeled antibodies were used depending on the experiment. The antibodies were diluted 103 times with 3% BSA/0.05% Tween-20 solution before use. The chips containing Salmonella or the control bacteria E. coli or Listeria attached to P22 were incubated with 100-μl HRP-labeled antibody for 1 hr at 37 °C. The chips were washed thoroughly with 0.05% Tween-20 and PBS respectively. OPD peroxidase was used as the ELISA substrate. Tablets of the substrate were dissolved in 20-ml deionized water. Two hundred μl of the substrate solution was allowed to react for 30 min at room temperature. Finally, the absorbance was measured at 450 nm using a micro-titer plate spectrophotometer (Bio-Tek instruments EL340).
Atomic force microscopy
Atomic force microscopy (AFM) images were obtained with either an E scanner (maximum scan area = 14.2 × 14.2 μm2) or J scanner (maximum scan area = 125 × 125 μm2) (Nanoscope IIIa, VEECO). Height, amplitude, and phase images were obtained in tapping mode in ambient air with silicon tips (TESP, VEECO). All AFM height images are reported unless specified. Height images have been plane-fit in the fast scan direction with no additional filtering operation. The surface roughness was determined using the root-mean-square surface roughness where zi is the height value and n is n the number of pixels in the image. The scan rate used was in the range of 0.2 to 1 Hz depending on the scan size. Integral and proportional gains were approximately 0.4 and 0.7, respectively.
Contact angle goniometry
The surface hydrophobicity was measured with an NRL contact angle goniometer (Model 100, Rame-Hart) in the laboratory atmosphere. A 20-μl water droplet was placed on the substrate and the static contact angles were measured on both sides of the droplet. Three droplets were placed at various spots on the substrate and the average readings are reported. The typical error is ±3°.
Results and Discussion
In order to covalently bind phage to the solid substrate, we use bifunctional aminosilanes to crosslink phage and the substrate. Surface silanization is a mild chemical reaction suitable for silicon oxide and metal oxide substrates. The silanized surface not only maintains the smoothness and chemical homogeneity of the surface but also improves the immobilization of bacteriophage by establishing strong covalent bonds with the phage molecules.
After piranha cleaning, the glass substrate exposes free hydroxyl groups, which can react with the silane to form the siloxane bond as shown in Figure 2 (Warning: Piranha solution is a strong oxidizing agent, so extreme care is necessary when using it). The surface roughness (Rq) of the silanized glass was found to be 0.4–0.5 nm for 10 × 10 μm2 images as represented by Figure 3A, similar to previous observations [35].
Figure 2.
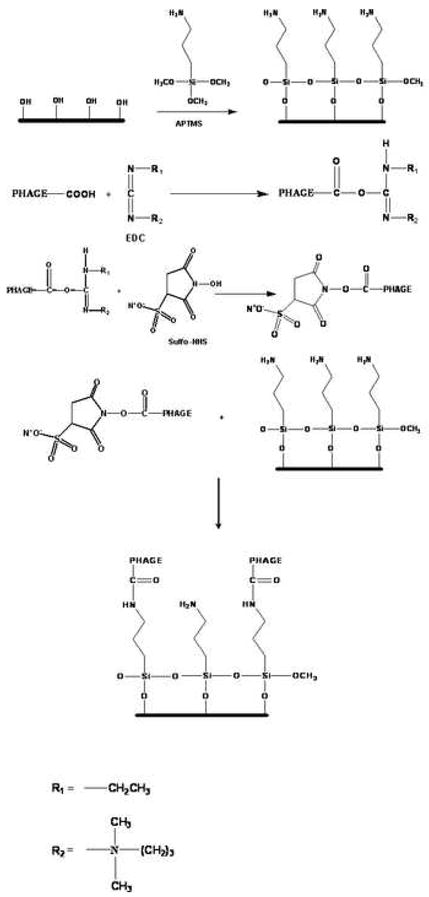
Reaction mechanism of phage P22 with silanized glass. (Note: For simplification only one carboxylic acid group is shown, other carboxylic acid groups on the phage will be activated too.)
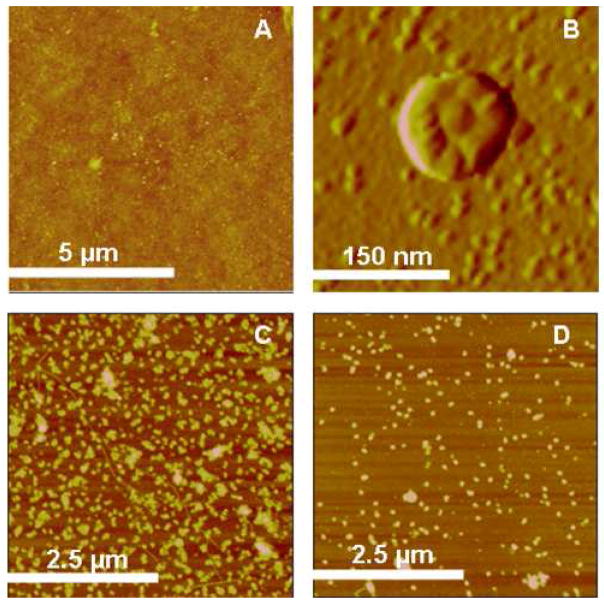
Figure 3.
AFM images of A) Silanized glass (z-range = 15 nm), B) Magnified image of one phage particle on silanized glass (amplitude image, z-range = 135 mV C) P22 on silanized glass from a concentrated solution (z-range = 60 nm), D) P22 on silanized glass from a diluted solution (z-range = 45 nm), and C)).
The amine-terminated glass reacts with the carboxylic groups on P22 to form amide bonds. EDC and Sulfo-NHS have commonly been used to immobilize proteins The covalent nature of EDC/NHS activated bonding of peptides to amine-terminated surfaces, i.e. the amide bond, has been established using X-ray photoelectron spectroscopy (XPS) [44, 45] and Fourier transform infrared spectroscopy (FTIR) [46].EDC reacts with the carboxylic group on P22 and makes an unstable intermediate while Sulfo-NHS reacts further with the intermediate and makes a reactive intermediate that is more vulnerable to attack by the amino groups on the substrate (Figure 2).
Contact angle measurements provide a quick check of surface treatments. Contact angles were measured for the untreated glass, piranha-cleaned glass, and glass after silanization. The contact angle of glass was reduced from 15° to 5–7° after piranha clean. Silanization with APTMS raised the contact angle to 65°, which is in the range of the published contact angle values of amine-terminated surfaces [47–51].
The immobilized phage particles were stable against AFM scanning in tapping mode in air. The surface coverage of P22 was found to vary with solution concentration. For example, when the P22 concentration was 1010–1011 pfu/ml, approximately 67% surface coverage was achieved as shown in Figure 3C. The surface coverage is estimated by the bearing area analysis using Nanoscope software from VEECO (version 5.30). The bearing area gives the percentage of the surface above the chosen reference plane. When the concentration was 108–109 pfu/ml, the surface coverage was reduced to 12% (Figure 3D) while all other conditions were kept the same. The height and diameter of individual P22 particles estimated from AFM height images are 22 ± 7 nm and 130 ± 40 nm respectively. The diameter of P22 head capsid reported in the literature is 70 nm [52]. Phage particles are expected to flatten significantly upon surface adsorption and drying. In addition, the discrepancy may come from the AFM tip convolution. The chemical reaction does not specifically control the orientation since carboxylic groups exist both on the head and TSP domains of P22. However, our results will show that there must be sufficient P22 particles with the TSP part oriented away from the substrate to bind specifically and strongly to Salmonella. It is conceivable that the attachment of the large and smooth capsid head to the glass substrate is favored over the pointy tail spikes of the gp9 proteins. Figure 3B is an AFM height image of one P22 particle. The spherical particle contains one center core surrounded by smaller protrusions toward the periphery. The center core likely corresponds to the portal closure of the capsid and the needle protein gp26 while the smaller protrusions correspond to the collapsed TSPs upon drying. The image suggests the outward orientation of the TSPs. It is possible to resolve more clearly the orientation and morphology of surface-attached phage particles in the future via high-resolution in-situ AFM imaging experiments.
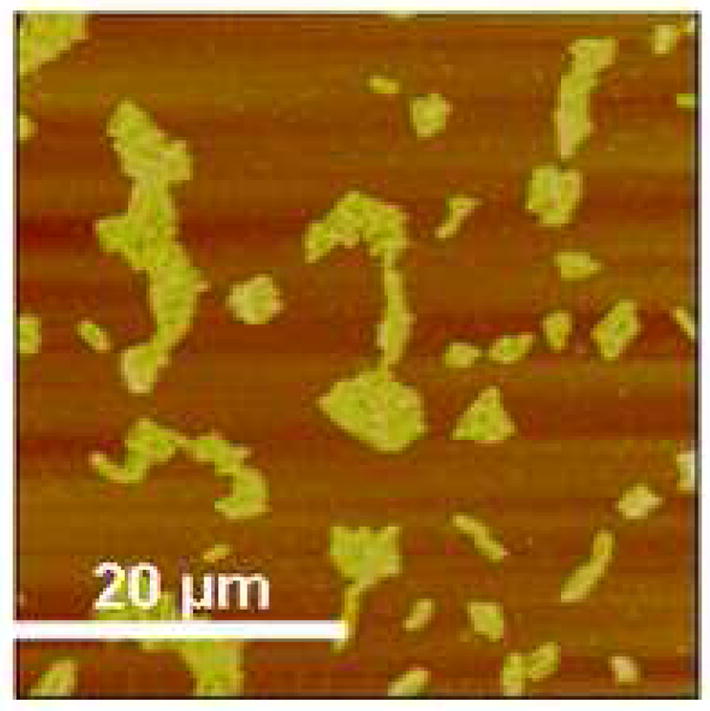
The adsorption of bacteriophage P22 on amine-terminated monolayers without EDC/NHS was studied. The adsorbed phage structure without NHS/EDC is represented by Figure 4, which was prepared under the same conditions as the film in Figure 3C minus the EDC/NHS procedure. It is clear that EDC/NHS provides a higher surface coverage and prevents aggregate formation on the amine-terminated surface.
Figure 4.
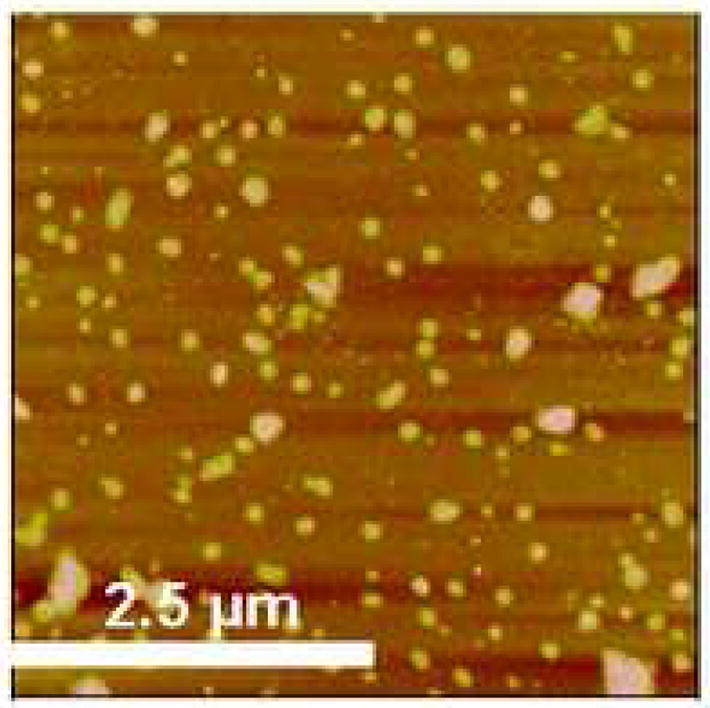
AFM height image of physically adsorbed P22 on silanized surface (z range = 50 nm)
Next the recognition of S. typhimurium by immobilized P22 was studied as a function of P22 monolayer coverage. The S. typhimurium concentration (1 × 106–1 × 107 cfu/ml) was kept constant while the surface coverage of P22 was varied by its solution concentration. One hundred μl of freshly prepared Salmonella culture was applied to each well containing silanized chips. The chips have different monolayer coverage at different concentrations as shown in Figure 5. The concentrations of P22 are 1010–1011 pfu/ml (Figure 5A), 109–1010 pfu/ml (Figure 5B), 2 × 108–109 pfu/ml (Figure 5C), and 108–109 pfu/ml (Figure 5D) respectively. S. typhimurium has a distinctive rodlike shape with length = 2 ± 0.3 μm, width = 790 ± 70 nm, and height = 185 ± 15 nm. Clearly, the bacterial coverage is directly correlated to phage coverage. For example, the percentage of surface coverage of S. typhimurium was 64% when 67% surface contained P22. But when P22 only covered 12% of the amine-terminated substrate, the S. typhimurium coverage was decreased to 11%. In the absence of immobilized P22, few bacterial particles were deposited (Figure 5E) on the APTMS-coated glass substrate. Figure 5F is a close-up image near the edge of a S. typhimurium particle where the smaller phage particles can be clearly seen on the substrate surrounding bacterial particle. Figure 6 summarizes the average number of S. typhimurium particles per 40 × 40 μm2 area (left) and the corresponding ELISA analysis (right). The average was assessed from 2–3 scans on 3 different samples prepared at the same conditions. The error bars correspond to multiple separate experiments. Approximately 250/1600 μm2 S. typhimurium bacteria were captured by the P22 monolayer with 67% P22 coverage. Approximately 60 were found in the case of 12% P22 coverage, and only 10 were found when no P22 was present. Spectral analysis by ELISA showed the same trend. The absorbance related to the amount of S. typhimurium decreased linearly with P22 amount. The results demonstrate that the bio-specific interactions between P22 and Salmonella are necessary for the permanent attachment of Salmonella.
Figure 5.
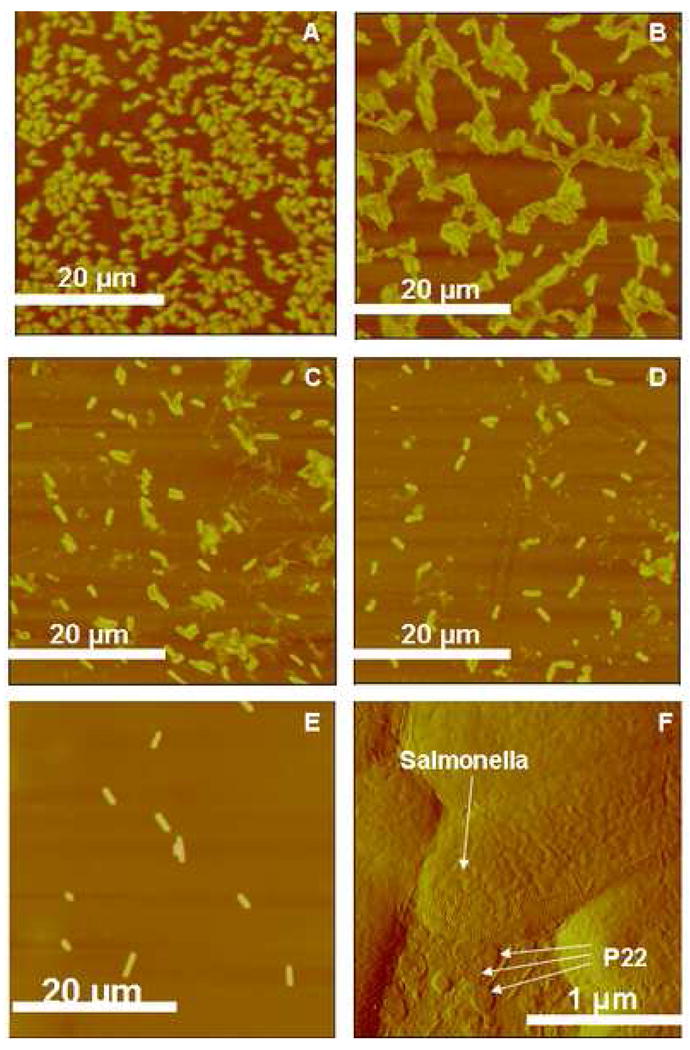
AFM images of S. typhimurium deposited on P22 monolayers of varying surface coverage. The P22 monolayers were made from A) 1010–1011 pfu/ml (z-range = 1000 nm), B) 109–1010 pfu/ml (z-range = 1000 nm), C) 2 × 108–109 pfu/ml (z-range = 1000 nm), D) 108–109 pfu/ml (z-range = 1000 nm), and E) Salmonella attached directly on silanized glass without P22 (z-range = 500 nm). F) AFM amplitude image showing S. typhimurium attached on P22 (z-range = 225 mV).
Figure 6.
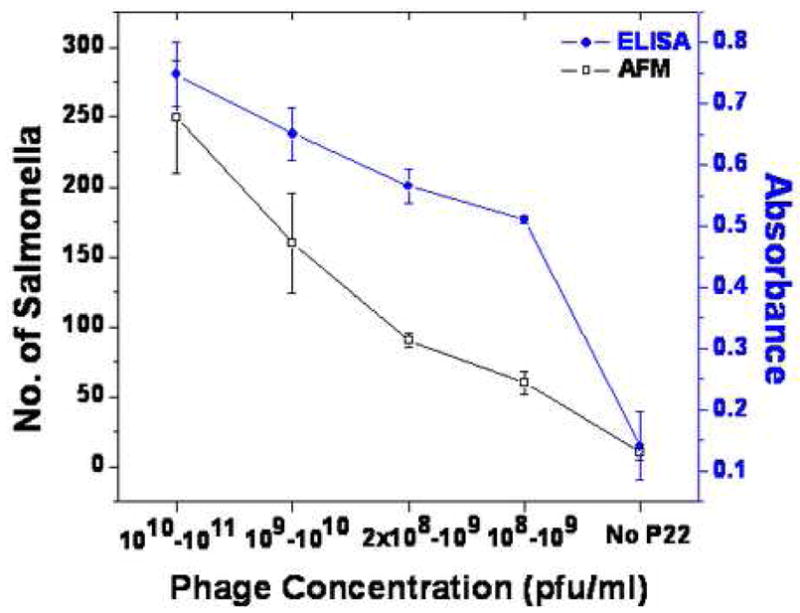
The comparison between AFM and ELISA results of the correlation between phage concentration and the number of surface-bound S. typhimurium.
We also conducted experiments with control bacteria in order to examine the selectivity of the P22 coating. The deposited amount of S. typhimurium is compared to those of E coli (strain 33780), a Gram-negative bacterium with weaker affinity to P22, and L. monocytogenes, a Gram-positive bacterium without the O-antigenic LPS outer layer while all other conditions are the same. Figure 7 shows the ELISA results, which indicate the expected decrease in adsorbed amount with deceasing P22 affinity. ELISA was repeated on three different samples for each of the bacterium. The results show that the immobilized P22 layer is capable of discriminating among different bacterial types according to TSP and bacterial membrane receptor affinity.
Figure 7.
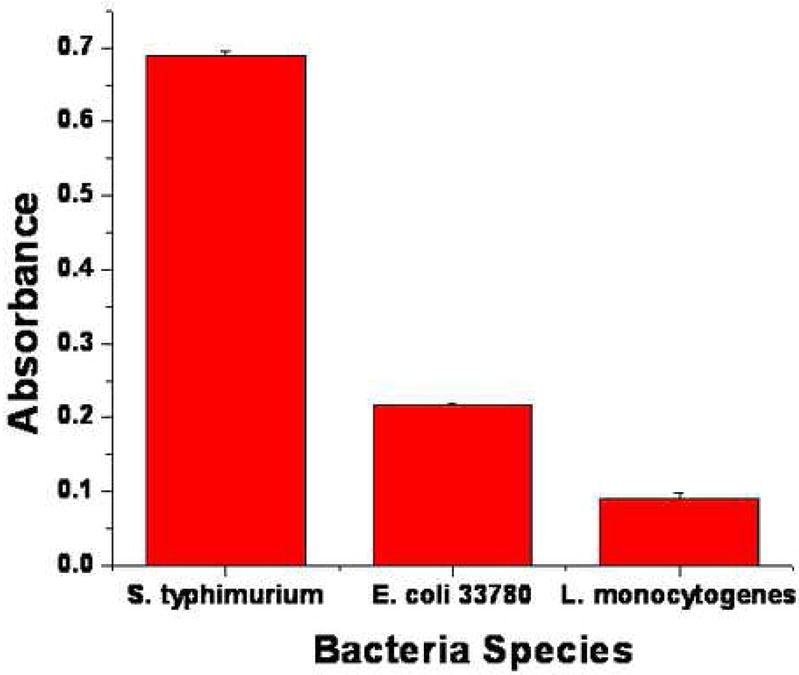
ELISA results showing the difference in binding efficiency of S. typhimurium, E. coli, and L. monocytogenes to P22 monolayer on silanized glass.
We also conducted preliminary tests of the durability of our phage coatings. One of the main advantages of phage over antibody sensors is that phage remains active in adverse environmental conditions such as drying while antibodies are fragile [17]. A P22-coated substrate was left drying in laboratory atmosphere for 7 days before introducing it to the S. typhimurium solution. Figure 8 shows approximately 120 bacterial particles on a40 × 40 μm2 area. This is lower than the bacterial density on freshly prepared P22 samples. The number suggests that approximately 40–60% of phage particles can still actively bind to S. typhimurium after 7 days.
Figure 8.
A durability test of the number of bound S. typhimurium on a one-week old P22 substrate kept in air. The z-range of the AFM image is 500 nm.
CONCLUSIONS
This paper reports a method to prepare monolayers of chemically bound phage P22 on glass substrates. AFM and ELISA were used to characterize the phage monolayer as well as its interactions with bacteria. The results show that the phage particles are permanently attached to the substrate via a combination of bifunctional aminosilane linkers and the sulfo-NHS enhanced surface reaction. The P22 coverage can be varied by solution concentration. The role of bio-specific interactions between immobilized phage and bacteria in solution is demonstrated both by the preference of P22 to S. typhimurium that contains the P22-binding O-antigenic receptors in its outer membrane and by the lack of S. typhimurium binding to areas not covered by P22. The results imply the favorable orientation of the 6 tailspike gp9 proteins toward the solution since they need to be accessible to recognize the O-antigenic repeating units on the cell surface of S. typhimurium. The durability of the P22 coating in ambient air was tested to show that 50% of immobilized phage particles are still capable of recognizing S. typhimurium after left in the dry state for 7 days. It is hopeful that the same phage coatings can be deposited on biosensor substrates such as piezoelectric aluminum nitride in order to test their applicability in real-time field monitoring devices. We are also conducting single molecule force spectroscopy studies to determine the bio-specific force magnitudes between phages and bacteria.
Acknowledgments
We acknowledge the National Science Foundation (G. Mao for CTS-0553533), Department of Defense (G. Auner for U.S. Army TACOM, DAAE07-03-C-L140), and National Institute of Health (G. Auner for 1 R01 EB00741) for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ivnitski D, Abdel-Hamid I, Atanasov P, Wilkins E. Biosensors for detection of pathogenic bacteria. Biosensors & Bioelectronics. 1999;14:599–624. doi: 10.1016/s0956-5663(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 2.Compton JAF. Military Chemical and Biological Agents. 1987. p. 458. [Google Scholar]
- 3.Dando M. Biological Warfare in the 21st Century. 1994. p. 258. [Google Scholar]
- 4.Smith GP, Petrenko VA. Phage Display. Chem Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 5.Petrenko VA, Vodyanoy VJ. Phage display for detection of biological threat agents. Journal of Microbiological Methods. 2003;53:253–262. doi: 10.1016/s0167-7012(03)00029-0. [DOI] [PubMed] [Google Scholar]
- 6.Benhar I. Biotechnological applications of phage and cell display. Biotechnology Advances. 2001;19:1–33. doi: 10.1016/s0734-9750(00)00054-9. [DOI] [PubMed] [Google Scholar]
- 7.Goldman ER, Pazirandeh MP, Mauro JM, King KD, Frey JC, Anderson GP. Phage-displayed peptides as biosensor reagents. Journal of Molecular Recognition. 2000;13:382–387. doi: 10.1002/1099-1352(200011/12)13:6<382::AID-JMR511>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 8.Sulakvelidze A, Alavidze Z, Morris JG. Bacteriophage therapy. Antimicrobial Agents and Chemotherapy. 2001;45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Summers WC. Bacteriophage therapy. Annual Review of Microbiology. 2001;55:437–451. doi: 10.1146/annurev.micro.55.1.437. [DOI] [PubMed] [Google Scholar]
- 10.Withey S, Cartmell E, Avery LM, Stephenson T. Bacteriophages - potential for application in wastewater treatment processes. Science of the Total Environment. 2005;339:1–18. doi: 10.1016/j.scitotenv.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 11.Azzazy HME, Highsmith WE. Phage display technology: clinical applications and recent innovations. Clinical Biochemistry. 2002;35:425–445. doi: 10.1016/s0009-9120(02)00343-0. [DOI] [PubMed] [Google Scholar]
- 12.Yin J, Liu F, Schinke M, Daly C, Walsh CT. Phagemid encoded small molecules for high throughput screening of chemical libraries. Journal of the American Chemical Society. 2004;126:13570–13571. doi: 10.1021/ja045127t. [DOI] [PubMed] [Google Scholar]
- 13.Olsen EV, Sorokulova IB, Petrenko VA, Chen IH, Barbaree JM, Vodyanoy VJ. Affinity-selected filamentous bacteriophage as a probe for acoustic wave biodetectors of Salmonella typhimurium. Biosensors & Bioelectronics. 2006;21:1434–1442. doi: 10.1016/j.bios.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Nanduri V, Sorokulova IB, Samoylov AM, Simonian AL, Petrenko VA, Vodyanoy V. Phage as a molecular recognition element in biosensors immobilized by physical adsorption. Biosensors & Bioelectronics. 2007;22:986–992. doi: 10.1016/j.bios.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 15.Nanduri V, Balasubramaniam S, Sista S, Vodyanoy VJ, Simonian AL. Highly sensitive phage-based biosensor of β-galactosidase. Anal Chim Acta. 2007;589:166–172. doi: 10.1016/j.aca.2007.02.071. [DOI] [PubMed] [Google Scholar]
- 16.Cooper MA. Advances in membrane receptor screening and analysis. Journal of Molecular Recognition. 2004;17:286–315. doi: 10.1002/jmr.675. [DOI] [PubMed] [Google Scholar]
- 17.Babacan S, Pivarnik P, Letcher S, Rand AG. Evaluation of antibody immobilization methods for piezoelectric biosensor application. Biosensors & Bioelectronics. 2000;15:615–621. doi: 10.1016/s0956-5663(00)00115-9. [DOI] [PubMed] [Google Scholar]
- 18.Olsen EV, Pathirana ST, Samoylov AM, Barbaree JM, Chin BA, Neely WC, Vodyanoy V. Specific and selective biosensor for Salmonella and its detection in the environment. Journal of Microbiological Methods. 2003;53:273–285. doi: 10.1016/s0167-7012(03)00031-9. [DOI] [PubMed] [Google Scholar]
- 19.Pathirana ST, Barbaree J, Chin BA, Hartell MG, Neely WC, Vodyanoy V. Rapid and sensitive biosensor for Salmonella. Biosensors & Bioelectronics. 2000;15:135–141. doi: 10.1016/s0956-5663(00)00067-1. [DOI] [PubMed] [Google Scholar]
- 20.Skladal P. Piezoelectric quartz crystal sensors applied for bioanalytical assays and characterization of affinity interactions. Journal of the Brazilian Chemical Society. 2003;14:491–502. [Google Scholar]
- 21.Caruso F, Rodda E, Furlong DN. Orientational aspects of antibody immobilization and immunological activity on quartz crystal microbalance electrodes. Journal of Colloid and Interface Science. 1996;178:104–115. [Google Scholar]
- 22.Vikholm I, Gyorvary E, Peltonen J. Incorporation of lipid-tagged single-chain antibodies into lipid monolayers and the interaction with antigen. Langmuir. 1996;12:3276–3281. [Google Scholar]
- 23.Guntupalli R, Hu J, Lakshmanan RS, Huang TS, Barbaree JM, Chin BA. A magnetoelastic resonance biosensor immobilized with polyclonal antibody for the detection of Salmonella typhimurium. Biosensors & Bioelectronics. 2007;22:1474–1479. doi: 10.1016/j.bios.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 24.Williams RA, Blanch HW. Covalent Immobilization of Protein Monolayers for Biosensor Applications. Biosensors & Bioelectronics. 1994;9:159–167. doi: 10.1016/0956-5663(94)80108-8. [DOI] [PubMed] [Google Scholar]
- 25.Ye JM, Letcher SV, Rand AG. Piezoelectric biosensor for detection of Salmonella typhimurium. Journal of Food Science. 1997;62:1067–1086. [Google Scholar]
- 26.Zillig W, Reiter WD, Palm P, Gropp F, Neumann H, Rettenberger M. Viruses of Archaebacteria. The Bacteriophages Vol 1. Calendar R. 1988:517. [Google Scholar]
- 27.Sharp RJ, Ahmad SI, Munster A, Dowsett B, Atkinson T. The Isolation and Characterization of Bacteriophages Infecting Obligately Thermophilic Strains of Bacillus. Journal of General Microbiology. 1986;132:1709–1722. doi: 10.1099/00221287-132-6-1709. [DOI] [PubMed] [Google Scholar]
- 28.Raven NDH, Munster AP, Sharp RJ. Characterization of thermus bacteriophage YBIO. 1986. [Google Scholar]
- 29.Israel V, Rosen H, Levine M. Binding of bacteriophage P22 tail parts to cells. J Virol. 1972;10:1152–1158. doi: 10.1128/jvi.10.6.1152-1158.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews D, Butler JS, Al-Bassam J, Joss L, Winn-Stapley DA, Casjens S, Cingolani G. Bacteriophage P22 tail accessory factor gp26 is a long triple-stranded coiled-coil. Journal of Biological Chemistry. 2005;280:5929–5933. doi: 10.1074/jbc.C400513200. [DOI] [PubMed] [Google Scholar]
- 31.Baxa U, Steinbacher S, Miller S, Weintraub A, Huber R, Seckler R. Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophysical Journal. 1996;71:2040–2048. doi: 10.1016/S0006-3495(96)79402-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwashita S, Kanegasaki S. Enzymic and molecular properties of base plate parts of bacteriophage P22. European Journal of Biochemistry. 1976;65:87–94. doi: 10.1111/j.1432-1033.1976.tb10392.x. [DOI] [PubMed] [Google Scholar]
- 33.Poteete AR. P22 bacteriophage. In: Webster RG, Granoff A, editors. Encyclopedia of virology. 1994. pp. 1009–1013. [Google Scholar]
- 34.Maurides PA, Schwarz JJ, Berget PB. Intragenic Suppression of a Capsid Assembly-Defective P22 Tailspike Mutation. Genetics. 1990;125:673–681. doi: 10.1093/genetics/125.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong J, Wang A, Ng KYS, Mao G. Self-assembly of octadecyltrichlorosilane monolayers on silicon-based substrates by chemical vapor deposition. Thin Solid Films. 2006;515:2116–2122. [Google Scholar]
- 36.Campbell GA, Mutharasan R. Detection and quantification of proteins using self-excited PZT-glass millimeter-sized cantilever. Biosensors & Bioelectronics. 2005;21:597–607. doi: 10.1016/j.bios.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 37.Fung YS, Wong YY. Self-assembled monolayers as the coating in a quartz piezoelectric crystal immunosensor to detect salmonella in aqueous solution. Anal Chem. 2001;73:5302–5309. doi: 10.1021/ac010655y. [DOI] [PubMed] [Google Scholar]
- 38.Lahiri J, Isaacs L, Tien J, Whitesides GM. A strategy for the generation of surfaces presenting ligands for studies of binding based on an active ester as a common reactive intermediate: A surface plasmon resonance study. Anal Chem. 1999;71:777–790. doi: 10.1021/ac980959t. [DOI] [PubMed] [Google Scholar]
- 39.Patel N, Davies MC, Hartshorne M, Heaton RJ, Roberts CJ, Tendler SJB, Williams PM. Immobilization of protein molecules onto homogeneous and mixed carboxylate-terminated self-assembled monolayers. Langmuir. 1997;13:6485–6490. [Google Scholar]
- 40.Su XL, Li Y. A self-assembled monolayer-based piezoelectric immunosensor for rapid detection of Escherichia coli O157:H7. Biosensors and Bioelectronics. 2004;19:563–574. doi: 10.1016/s0956-5663(03)00254-9. [DOI] [PubMed] [Google Scholar]
- 41.Wong YY, Ng SP, Ng MH, Si SH, Yao SZ, Fung YS. Immunosensor for the differentiation and detection of Salmonella species based on a quartz crystal microbalance. Biosensors & Bioelectronics. 2002;17:676–684. doi: 10.1016/s0956-5663(02)00030-1. [DOI] [PubMed] [Google Scholar]
- 42.Yang LMC, Tam PY, Murray BJ, McIntire TM, Overstreet CM, Weiss GA, Penner RM. Virus electrodes for universal biodetection. Anal Chem. 2006;78:3265–3270. doi: 10.1021/ac052287u. [DOI] [PubMed] [Google Scholar]
- 43.Gervals L, Gel M, Allain B, Tolba M, Brovko L, Zourob M, Mandeville R, Griffiths M, Evoy S. Immobilization of biotinylated bacteriophages on biosensor surfaces. Sensor Actuat B-Chem. 2007;125:615–621. [Google Scholar]
- 44.Nam K, Kimura T, Kishida A. Prepaaration and characterization of cross-linked collagen-phospholipid polymer hybrid gels. Biomaterials. 2007;28:1–8. doi: 10.1016/j.biomaterials.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 45.Wang XH, Li DP, Wang WJ, Feng QL, Cui FZ, Xu YX, Song XH. Covalent immobilization of chitosan and heparin on PLGA surface. Int J Biol Macromol. 2003;33:95–100. doi: 10.1016/s0141-8130(03)00072-2. [DOI] [PubMed] [Google Scholar]
- 46.Frey BL, Corn RM. Covalent attachment and derivatization of poly(L-lysine) monolayers on gold surfaces as characterized by polarization-modulation FT-IR spectroscopy. Anal Chem. 1996;68:3187–3193. [Google Scholar]
- 47.Lee I, Wool RP. Controlling amine receptor group density on aluminum oxide surfaces by mixed silane self assembly. Thin Solid Films. 2000;379:94–100. [Google Scholar]
- 48.Wei ZQ, Wang C, Zhu CF, Zhou CQ, Xu B, Bai CL. Study on single-bond interaction between amino-terminated organosilane self-assembled monolayers by atomic force microscopy. Surface Science. 2000;459:401–412. [Google Scholar]
- 49.Wu B, Mao GZ, Ng KYS. Stepwise adsorption of a long trichlorosilane and a short aminosilane. Colloids and Surfaces a-Physicochemical and Engineering Aspects. 2000;162:203–213. [Google Scholar]
- 50.Faucheux N, Schweiss R, Lutzow K, Werner C, Groth T. Self-assembled monolayers with different terminating groups as model substrates for cell adhesion studies. Biomaterials. 2004;25:2721–2730. doi: 10.1016/j.biomaterials.2003.09.069. [DOI] [PubMed] [Google Scholar]
- 51.Heise A, Stamm M, Rauscher M, Duschner H, Menzel H. Mixed silane self assembled monolayers and their in situ modification. Thin Solid Films. 1998;329:199–203. [Google Scholar]
- 52.Jiang W, Li ZL, Zhang ZX, Baker ML, Prevelige PE, Chiu W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nature Structural Biology. 2003;10:131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]