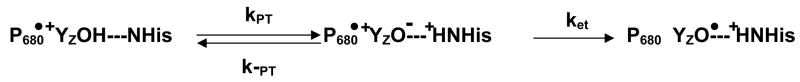Abstract
The catalytic cycle of numerous enzymes involves the coupling between proton transfer and electron transfer. Yet, the understanding of this coordinated transfer in biological systems remains limited, likely because its characterization relies on the controlled but experimentally challenging modifications of the free energy changes associated with either the electron or proton transfer. We have performed such a study here in Photosystem II. The driving force for electron transfer from TyrZ to P680•+ has been decreased by ~ 80 meV by mutating the axial ligand of P680, and that for proton transfer upon oxidation of TyrZ by substituting a 3-fluorotyrosine (3F-TyrZ) for TyrZ. In Mn-depleted Photosystem II, the dependence upon pH of the oxidation rates of TyrZ and 3F-TyrZ were found to be similar. However, in the pH range where the phenolic hydroxyl of TyrZ is involved in a H-bond with a proton acceptor, the activation energy of the oxidation of 3F-TyrZ is decreased by 110 meV, a value which correlates with the in vitro finding of a 90 meV stabilization energy to the phenolate form of 3F-Tyr when compared to Tyr (Seyedsayamdost et al., 2006, JACS 128:1569–79). Thus, when the phenol of YZ acts as a H-bond-donor, its oxidation by P680•+ is controlled by its prior deprotonation. This contrasts with the situation prevailing at lower pH, where the proton acceptor is protonated and therefore unavailable, in which the oxidation-induced proton transfer from the phenolic hydroxyl of TyrZ has been proposed to occur concertedly with the electron transfer to P680•+. This suggests a switch between a concerted proton/electron transfer at pHs < 7.5 to a sequential one at pHs > 7.5 and illustrates the roles of the H-bond and of the likely salt-bridge existing between the phenolate and the nearby proton acceptor in determining the coupling between proton and electron transfer.
Introduction
Many enzymes produce radical intermediates in cycling through their catalytic mechanism. Prominent among these is Photosystem IIa which drives the oxidation of water to dioxygen and supplies our atmosphere with the substrate of aerobic metabolism. In PSII, absorption of a photon leads to the formation of a radical pair, P680•+Phe•−, which traps a major part of the photon energy. P680•+ is then quickly reduced by YZ which is in turn reduced by an electron abstracted from the (Mn)4Ca cluster 1–3. Once this sequence is repeated four times, two water molecules are split into O2 and the enzyme is reset to its lowest oxidation state. Although the overall catalytic mechanism is known at the phenomenological level, the description at the molecular or atomic level still remains a challenge. Beside its academic interest, a molecular description of the sequence of reactions that produce the strongest oxidants known in biological systems 4–6 should help to design artificial compounds that mimic the light-driven splitting of water and which could eventually sustain hydrogen production 7–9. Although the (Mn)4Ca cluster is the catalytic center which drives water splitting, the redox cycling of (YZOH/YZO•) has received considerable attention because both the oxidation and reduction of YZ are coupled to a proton transfer, making YZox a likely player in the chemical scenario of the abstraction of four electrons and four protons from the two water molecules 10–13. Also, as it is amenable to time-resolved study, it serves as a paradigm for understanding the coupling between electron and proton transfer in bio-mimetic systems 14–17.
The strongest evidence for tight coupling between electron and proton transfer from YZ came from the studies of site-directed PSII mutants lacking the His D1-190, in which the oxidation of YZ was markedly slowed down and could be kinetically rescued by addition of exogenous proton acceptor 18–21. This finding, together with YZ being protonated in its reduced form 22–24 and YZox being a neutral radical, raises the question of the reaction pathway followed to abstract both a proton and an electron from the phenol side chain. Efforts to disentangle this pathway have heretofore relied on the study of the dependence of the rate of oxidation of YZ on pH 11,21 or on the driving force of the electron transfer reaction proper 25. The former approach led to the characterization of two different regimes for the oxidation of YZ, depending on the protonation state of the base serving as a proton acceptor from YZox. Below pH 7, the oxidation of YZ in Mn-depleted PSII is slow and has significant activation energy (Ea ≈ 300 meV 11). The rate can be accelerated by the addition of mobile proton carriers 21 indicating that proton release is kinetically limiting the electron transfer reaction from YZ to P680•+. Yet, in this pH range, the rate of oxidation of YZ shows a similar dependence on the driving force for either H+ release or electron transfer indicating that the overall reaction proceeds by a concerted proton-electron transfer 25. Above pH ~ 7, the oxidation rate of YZ is increased about 30-fold but only very weakly depends on the driving force for H+ release and its activation energy is smaller (Ea ≈ 150 meV 11). In an artificial complex, the oxidation of a phenol group bound to a Ru(II)-polypyridine showed similar characteristics, that is a strong dependence on pH and a significant activation energy below the pKa of the phenol group and a weak activation energy and no dependence on pH above the pKa 26. In this case, the dependence upon pH of the oxidation rate of the phenol group and its significant activation energy were accounted for by a concerted electron/proton transfer from the phenol and, by analogy, this mechanism was proposed to apply as well in Mn-depleted PSII 26. Yet, this analogy only holds in the pH range where the oxidation of YZ indeed displays similar characteristics, that is a pH dependent and significantly activated oxidation rate or in other terms below pH ~7. In addition, whereas in the artificial system the transition between the regime where the oxidation rate of the phenol group depends on pH to the regime where it is pH independent likely reflects the transition from the phenol to the phenolate, in Mn-depleted PSII it is interpreted as reflecting the deprotonation of a nearby proton-accepting base rather than the deprotonation of the phenol group of YZ proper.
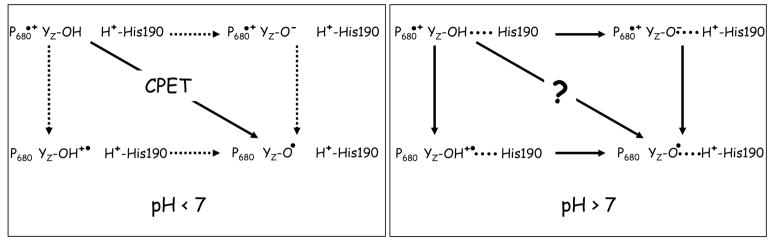
While, as illustrated in Fig. 1, in the absence of this nearby H+ accepting base (i.e. in Mn-depleted PSII and in the low pH range), there is therefore a consensus on the nature of the coupling between the proton and electron transfer from YZ, the mechanism by which oxidation of YZ proceeds when the nearby base is available as a proton-accepting group at high pH in Mn-depleted PSII remains uncharacterized.
Figure 1.
A scheme depicting the reaction pathways for the electron and proton transfer from the phenol group of YZ in Mn-depleted PSII. The left panel illustrates the situation proposed to prevail at low pH, where the nearby proton-accepting base (shown here as being D1-His190, see text for a discussion) is protonated and thus unavailable as a proton acceptor. As discussed in the text, the characteristics of the oxidation rate of YZ are those expected for a concerted electron/proton transfer (CPET). The right panel illustrates the situation thought to prevail above pH ~ 7, where a proton- accepting base in close vicinity to the phenol group of YZ is available. The nature of the coupling between the electron transfer and proton transfer from the phenol group of YZ is the subject of the present study.
This latter case is particularly important since, in these circumstances, the structural and kinetic characteristics which determine the electron and proton transfer from or to YZ more closely correspond to those prevailing when water oxidation is active. Indeed, according to the 3D structure of O2-evolving PSII, the oxygen of the phenol group of YZ is at H-bonding distance (2.7 Å) to the ε-N of the imidazole ring of His-190 27,28. In addition, the time constant of the oxidation of YZ lies in the 50–400 ns time range depending on the oxidation state of the (Mn)4Ca cluster 29, a situation which markedly differs from that observed in Mn-depleted PSII below pH 7 but which is close to that reached in Mn-depleted PSII at alkaline pH (21 and this work). Many studies, including infra-red and magnetic resonance spectroscopy, support a model in which, in Mn-depleted PSII, both the YZOH and YZO• states are involved in a H-bond 22,30,31. These observations, combined with the faster rate for the oxidation of YZ at alkaline pH, call for the characterization of the coupling between proton and electron transfer upon YZ oxidation under conditions where the H-bond is preserved.
In order to better understand the coordination of the proton and electron transfer, we have changed the driving force for the electron and/or proton transfer and assessed the kinetic consequences of such changes. The driving force for electron transfer proper was modified by site-directed mutagenesis to alter the midpoint potential of P680 32 (by 80 mV), and the driving force for proton transfer was tuned by incorporating a fluorinated tyrosine in place of YZ (ΔpKa 1.5 pH units33).
Materials and Methods
Incorporation of 3F-tyrosine and purification of PSII core complexes
The incorporation of 3F-Tyr in Thermosynechococcus elongatus was performed as described in 34. The culture medium was supplemented with 125 μM L-Phe, 125 μM L-Trp and 125 μM L-3F-Tyr purchased from Sigma-Aldrich. PSII core complexes were purified as described in 35. Mn-depletion was performed as in 36
Synechocystis Olive WT cells bearing a hexahistidine C-terminal tail on PsbB 37 were grown photoautotrophically in 10 L of BG-11 minimal medium. Once the OD730 had reached 0.4–0.5/cm, the medium was supplemented with 0.5 mM 3-F-tyrosine and 0.5 mM phenylalanine and 0.25 mM tryptophan added under sterile conditions to bring the volume up to 11 L. The cells were cultivated from 24–72 h under these conditions and harvested in an Amicon DC10 hollow-fiber concentrator. The cells were broken and the PSII core complexes isolated as described in 37 and concentrated in a Centricon 100 concentrator. The core complexes were treated with 5 mM hydroxylamine for 30 min on ice in 50 mM MES-NaOH, pH 6.5, 5 mM MgCl2, 20 mM CaCl2, 25% glycerol + 0.03% dodecyl maltoside to reduce and extract the manganese cluster and then applied to an ECONOPAK 10DG gel filtration column equilibrated with the same buffer to remove free Mn2+ and residual hydroxylamine.
Synechocystis WT and D1-His198Ala core complexes were isolated from strains described previously 32. The core complexes were prepared according to the combined procedures of Tang and Diner 38 and Rögner et al. 39 in that order, and were stored at −80°C until use.
EPR spectroscopy
ESE-field-swept spectra where recorded at 4.2 K with a Bruker Elexsys 580 X-band spectrometer equipped with an Oxford Instruments cryostat for T. elongatus and at 40 K with a home-built 40 spectrometer for Synechocystis. In both cases, a 2-pulse sequence was used with a duration of 48 and 96 ns for the π/2 and π pulses. The τ value was 300 ns. The shot repetition time was 16 ms for T. elongatus and 2 ms for Synechocystis.
In T. elongatus, ferricyanide (5 mM, final concentration) was added to the sample as an artificial electron acceptor. The YZ• radical was induced by illumination of the Mn-depleted PSII at −40°C for 5 s. Then, the samples were immediately transferred in the dark to 198 K and then to 77 K. Illumination of the samples was performed in a non-silvered dewar flask filled with ethanol and cooled to −40 °C with liquid N2. The light source was a 800-W tungsten lamp, light the beam of which was filtered through water and IR filters. The YZ• component of the spectrum recorded after illumination was obtained by subtracting the YD• spectrum recorded after thawing the sample in the dark at which point the YZ• signal rapidly decays.
Synechocystis PSII samples were loaded into EPR tubes with 300μM K3Fe(CN)6. YZ• and YD• were trapped together by freezing under illumination (~5000 W/m2) in ≤ 20 s above a pool of liquid N2 contained in a transparent dewar. YD• alone was recorded after a further dark-adaptation for 15 min on ice. The sample was then frozen in the dark in liquid N2 prior to recording the ESE field-swept spectra.
Time-resolved absorption spectroscopy
Absorption changes were measured using a laboratory-built spectrophotometer described in ref. 41. The PSII core complexes were resuspended at a chlorophyll concentration of 25 μg/mL in a medium containing 10 mM CaCl2, 10 mM MgCl2 and 50 mM of the appropriate buffer; (2-(N-morpholino)ethanesulfonic acid (MES), for the 5.0–6.5 pH range, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), for the 7.0–8.5 pH range, and N-cyclohexyl-2-aminoethanesulfonic acid (CHES), for the 8.5–10.5 pH range). 2,6 Dichloro-p-benzoquinone (100 μM) was added as an electron acceptor. The temperature of the cuvette was controlled by a thermostated circulating bath.
Results
Two different strategies can be followed for substituting an exogenous 3F-Tyr for a Tyr. In the case of the ribonucleotide reductase, the specific substitution of a single Tyr residue in the entire protein was achieved through purification, cleavage and fusion with a synthetic peptide containing a F-Tyr 42. PSII being a membrane protein comprising more than 20 subunits such an elegantly targeted strategy was unrealistic, and we resorted to a global labeling approach as in 43,44, in the thermophilic organism Thermosynechococcus elongatus and mesophilic organism Synechocystis sp. PCC 6803. We thus first assessed the incorporation yield of the 3F-Tyr by EPR, which allows one to probe specifically the redox active tyrosines in the protein.
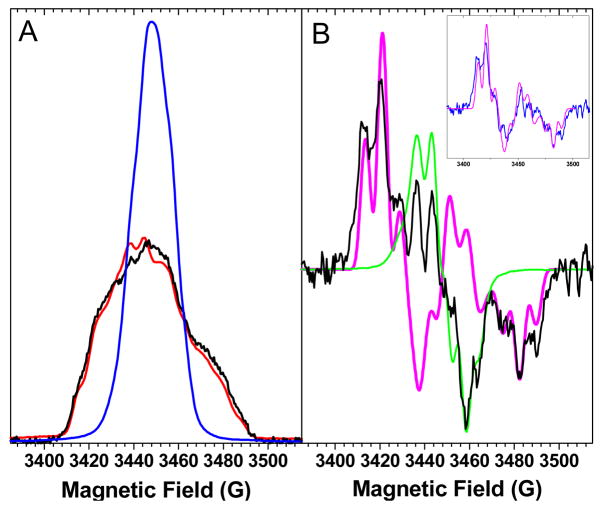
Fig. 2A compares the electron spin echo field swept-EPR difference spectra of Mn-depleted PSII core complexes from 3F-YZ• T. elongatus and Synechocystis with that from unlabeled T. elongatus.
Figure 2.
Panel A: ESE field-swept difference spectra of labeled PSII core complexes from T. elongatus (black) and Synechocystis sp. PCC 6803 (red) and of unlabeled PSII core complexes from T. elongatus (blue). All spectra have been normalized to an area of one spin. Panel B: A comparison between the first derivative of the ESE field-swept spectrum of labeled PSII core complexes from T. elongatus (black) with the simulated spectrum of 3F-YZ• (magenta); shown in green is the spectrum of YZ• scaled to account for 25% of the total spin number in the experimental spectrum. The inset compares the difference between the experimental spectrum and the scaled spectrum of YZ• (blue) to the simulated spectrum (magenta).
These spectra were generated by first recording the ESE-field swept spectra of YZ• and YD• and then subtracting the ESE-field swept spectra of YD•. First it is clear that the incorporation of 3F-Tyr leads to a spectra significant broadening of the spectra from both organisms. Second, it is also clear that the 3F-YZ• are of similar amplitude, indicating an equivalent extent of incorporation of the fluorinated tyrosine into both organisms. Fig. 2B shows the ESE field-swept spectrum of the (3F)-YZ• radicalb in PSII from T. elongatus (black) after a pseudo modulation has been applied. To quantify the yield of the incorporation, the simulated spectrum of 3F-YZ• (magenta, see the supporting information for details) was scaled to match the spectral patterns of the external “wings”, diagnostic of the large hyperfine splitting due to the presence of the 19F nucleus and absent in the unlabeled YZ• radical (shown in green). The comparison of the integrated areas of the scaled 3F-YZ• simulated spectrum with the experimental one sets the percent of 3F-Tyr incorporation at 75 ± 5% for T. elongatus. As shown in the inset of Fig. 2B the difference between the ESE field-swept spectrum of (3F)-YZ• radical and of unlabeled YZ• scaled to an area of 0.25 spin (blue spectrum), nicely matches the simulated spectrum of 3F-YZ• (magenta).
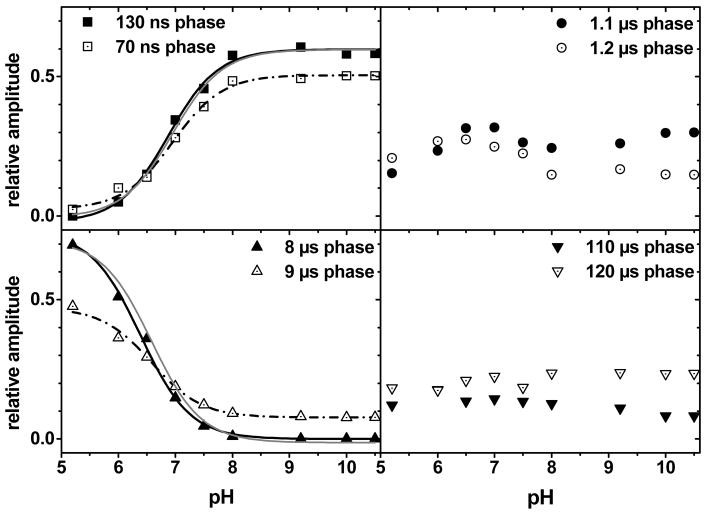
The consequence of the substitution on the rate of oxidation of 3F-YZ was then characterized. We studied the pH dependence of the kinetics of P680•+ reduction in labeled and unlabeled PSII from T. elongatus. These were followed at 432 nm where the oxidation of P680 is associated with a strong bleaching band. The reduction of P680•+ in T. elongatus was well fit with four exponential decays as found earlier by Hays et al. in Synechocystis sp. PCC 680321 (see Fig. S2 in the supplementary materials). The amplitude of the individual decay components varied with pH whereas the individual rate constants were rather pH insensitive as also found earlier in Synechocystis sp. PCC 6803 21. We thus globally fit the various kinetic components obtained at different pH’s using four pH independent rate constants. The amplitude of each of these four components as a function of pH is shown in Figure 3. Two main conclusions can be drawn. The overall pH dependence of P680•+ reduction is similar in T. elongatus and Synechocystis PSII. Indeed, as in the Synechocystis case (see Fig. 4 of Hays et al. 21), the amplitudes of the first and third components are those which more strongly depend on pH and their dependencies are, to a first approximation, complementary. Even more saliently, the substitution of 3F-YZ for YZ had hardly any effect on the pH dependence of the kinetics of reduction of P680•+. Again, the first and third components were those displaying the more pronounced pH dependence and, although the maximum amplitudes were slightly smaller in the labeled cases, the pKa’s characterizing the pH dependencies of each component were similar to those observed with unlabeled PSII.
Figure 3.
Relative amplitudes of the four exponentials components associated with the reduction of P680•+ as a function of pH. The reduction of P680•+ was measured at 432 nm from 5 ns to 10 ms at various pHs and the kinetics were globally fit with four exponentials (the half-times of which are indicated in the different panels). The solid symbols correspond to the unlabeled PSII from T. elongatus. Fitting these data (solid lines) with the Henderson-Haselbach equation (n=1) yielded pKas of 6.85 ± 0.15 and 6.4 ± 0.2 for the 130 ns and 8 μs components, respectively. The open symbols correspond to the labeled PSII from T. elongatus. Fitting these data (dashed lines) with the Henderson-Haselbach equation (n=1) yielded pKas of 6.9 ± 0.2 and 6.55 ± 0.3 for the 70 ns and 9 μs components, respectively. The solid lines in gray are the fit of the data obtained with labeled PSII normalized to the data obtained with the unlabeled PSII to facilitate the comparison.
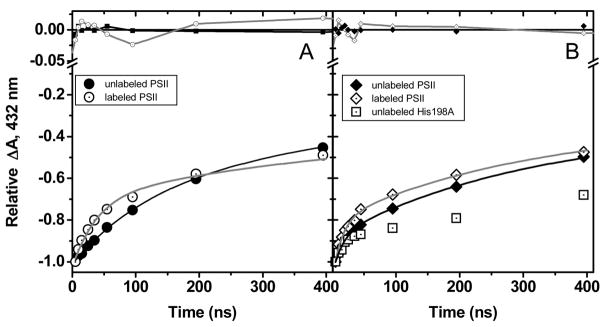
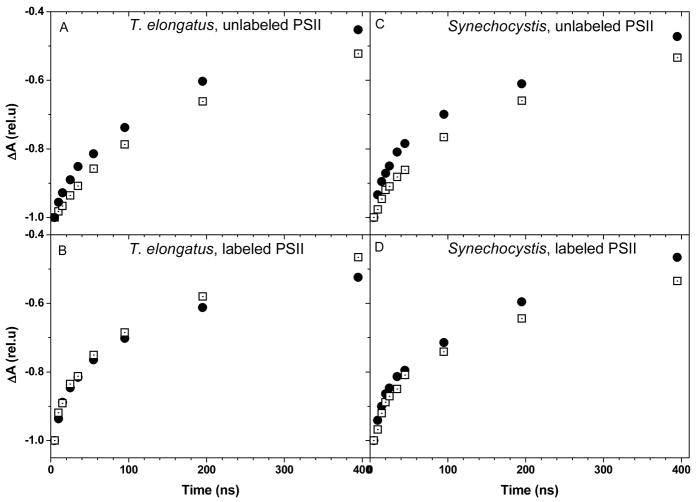
The kinetic complexity found here for the reduction of P680•+ in Mn-depleted PSII core complexes from T. elongatus nicely reproduces previous results from different groups 11,20,21,45 and discussing the assignment of the different components is beyond the scope of the present study. Briefly, whereas the fast component is commonly assigned to the coupled electron/proton transfer from YZOH, the subsequent components are commonly understood as reflecting the relaxation of the protein moiety and as such are kinetically controlled by local structural rearrangement such as proton transfer or formation of H-bonds. Since the aim of this study was to investigate the nature of the coupling between proton and electron transfer from (3F)-YZ we focused on the fast component. In addition, since we were particularly interested in dissecting these processes under conditions which kinetically resemble those prevailing in oxygen-evolving PSII, we focused our analysis on the high pH range where the proton acceptor is deprotonated and thus the phenol hydroxyl group is thought to be a H-bond donor. Fig. 4 shows that in labeled PSII, the fast component of the reduction of P680•+ (in the hundreds of the ns time-range), at pH 9.2, is slightly but significantly faster than in unlabeled PSII. This observation holds true in both the T. elongatus and Synechocystis species. As such a rate increase may stem from a change in the driving force for electron and/or proton transfer, we also compared the rate of reduction of P680•+ in unlabeled but mutated PSII in which the driving force for electron transfer between YZ and P680•+ has been specifically modified.
Figure 4.
Comparison of the reduction kinetics of P680•+ at pH 9.2 in unlabeled (solid symbols) and labeled (open symbols) PSII complexes from T. elongatus (left panel) and Synechocystis sp. PCC 6803 (right panel) at 15°C. The kinetics were normalized to their initial amplitude. The lines show the best fit of the data with four exponentials (the gray and black lines correspond respectively to the labeled and unlabeled PSII complexes). The fit yielded the following time constants for the fast component: T. elongatus: 1.3 107 s−1 ± 1.5 106 (unlabeled), 2.8 107 s−1 ± 2.2 106 (labeled); Synechocystis: 1.9 107 s−1 ± 2.4 106 (unlabeled), 3.7 107 s−1 ± 2.5 106 (labeled), H198A 2.1 107 s−1 ± 3.1 106.
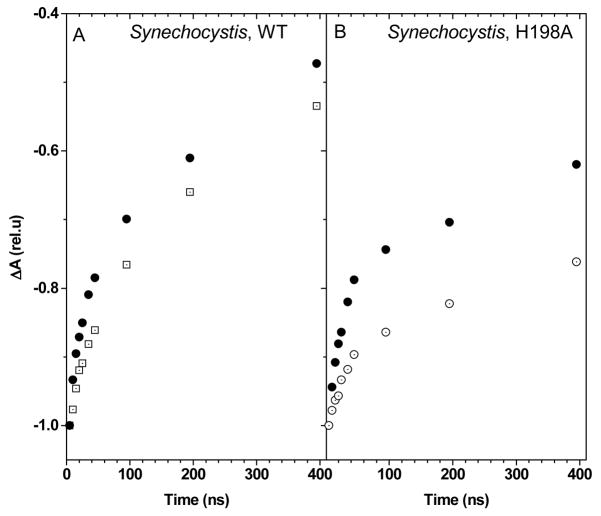
A collection of PSII mutants has been described in which the axial His ligand to the P680 chlorophyll was replaced by other amino acids 32. We chose the D1-His198A mutant, in which the midpoint potential of the P680•+/P680 couple is down-shifted by ≈ 80 mV 32, a change which qualitatively corresponds to that resulting from the 3F-Tyr labeling since, according to in vitro studies, the fluorination induces an up-shift of the (3F)-YO•/(3F)-YO− midpoint potential by ≈ 60 mV 33. Fig. 4B shows the kinetics of reduction of P680•+ in labeled, unlabeled WT and unlabeled D1-His198A PSII from Synechocystis sp. PCC 6803 c. The rate was similar in the mutant and unlabeled PSII (compare the initial slope of the absorption changes in Fig. 4) but slightly faster in the labeled PSII. Thus, a decrease in the midpoint potential of the P680•+/P680 couple does not produce a change in the reduction rate of P680•+. We conclude that a change in the driving force for electron transfer from YZ to P680•+ cannot be the explanation for the acceleration of P680•+ reduction produced by tyrosine fluorination.
However, the decrease in the midpoint potential of the P680•+/P680 couple qualitatively correlates with a decreased equilibrium constant for the YZP680•+ to YZoxP680 reaction, as the amplitude of the fast phase was almost halved in the mutant. In the case of the labeled WT PSII, the amplitude of the fast component was diminished by only ≈ 10% (see Fig. 3 and 4A), indicating that the substitution with 3F-Tyr only modestly affects the driving force of the electron transfer reaction.
We further investigated this difference in the overall oxidation rate of (3F)-YZ by measuring its temperature-dependence in the labeled and unlabeled PSII core complexes from both Synechocystis and T. elongatus. As shown, in Fig. 5, the fast component in the reduction of P680•+ in the labeled samples was more weakly dependent on the temperature than in the unlabeled samples.
Figure 5.
Consequences of the substitution of 3F-YZ for YZ on the dependence upon temperature of the fast component in the reduction of P680•+, at pH 9.2. The left column shows the reduction of P680•+ in the hundreds of ns time range in the unlabeled (panel A) and labeled (panel B) PSII core complexes from T. elongatus. The closed circles and open squares show the kinetics at 25–28°C and 5°C, respectively. The right column shows the reduction of P680•+ in the hundreds of ns time range in the unlabeled (panel C) and labeled (panel D) PSII core complexes from Synechocystis. The closed circles and open squares depict the kinetics at high and low temperature, respectively. Note that the ordinate axis does not go to 0 and that the figure is a focus on the early events. The fit yielded the following time constants for the fast component: T. elongatus: (panel A, unlabeled) 1.75 107 s−1 ± 1.7 106 at 25°C and 1.05 107 s−1 ± 1.8 106 at 5°C; (panel B, labeled) 3.2 107 s−1 ± 4.2 106 at 25°C and 3.1 107 s−1 ± 4.5 106 at 5°C; Synechocystis: (panel C, unlabeled) 3.7 107 s−1 ± 5.7 106 at 28°C and 1.3 107 s−1 ± 5.8 106 at 5°C, (panel D, labeled) 5.2 107 s−1 ± 8.2 106 at 28°C and 3.1 107 s−1 ± 7.5 106 at 5°C.
However, in the labeled PSII, this lesser dependence of the rate of reduction of P680•+ on temperature does not stem from a smaller driving force for electron transfer. Indeed, as shown in Fig. 6, the qualitative comparison of the dependence on the temperature of the kinetics of reduction of P680•+ in PSII core complexes from Synechocystis WT and D1-H198A mutant shows that decreasing the driving force for the electron transfer does not result in a weaker temperature dependence.
Figure 6.
Consequences of the D1-H198A mutation on the dependence upon temperature of the fast component in the reduction of P680•+, at pH 9.2. The left panel shows the reduction of P680•+ in the hundreds of ns time range in the unlabeled PSII core complexes from Synechocystis WT. The right panel shows the reduction of P680•+ in the hundreds of ns time range in the unlabeled PSII core complexes from Synechocystis D1- H198A mutant. Closed circles, 25°C; Open squares, 5°C. Note that the ordinate axis does not go to 0 and that the figure is a focus on the early events.
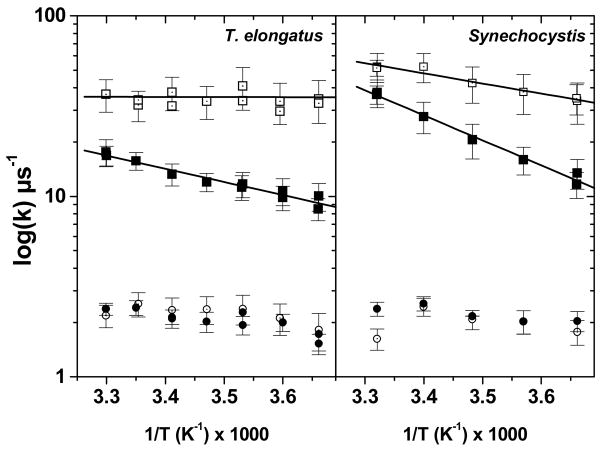
To further characterize the consequences of the substitution of 3F-YZ for YZ the temperature-dependence of each of the four exponentials was determined. We found that the activation enthalpy of the fast component of the oxidation of YZ is larger than that of the oxidation of 3F-YZ in both organisms (Fig. 7). Although in Synechocystis PSII the activation enthalpy of the oxidation of YZ was larger than in T. elongatus PSII (220 ± 35 meV and 120 ± 10 meV, respectively), the changes in activation enthalpy resulting from the fluorination were similar ≈ 110 meV.
Figure 7.
Arrhenius plot of the rates of the first (square) and second component (circle) in the reduction of P680•+ at pH 9.2, in unlabeled (solid symbols) and labeled (open symbols) PSII complexes from T. elongatus (left panel) and Synechocystis sp. PCC 6803 (right panel). The data were linearly fitted to extract the activation energy (the results are shown as the solid lines) and the fit yielded the following values: T. elongatus: 120 ± 30 meV (unlabeled PSII), 0 ± 25 meV (labeled PSII); Synechocystis: 220 ± 35 meV (unlabeled PSII) and 110 ± 25 meV (labeled PSII).
Discussion
1. pH-dependence of the oxidation of YZ and pKa of YZOH
As the substitution of 3F-YZ for YZ is expected to decrease the pKa of the phenol group by 1.5 pH unit 33, the present results provide an unambiguous way to address two correlated issues: the rationale(s) for the pH dependence of the oxidation of YZ in Mn-depleted PSII, as well as the pKa of YZ. The similarity of the dependence on pH of the reduction of P680•+ in labeled and unlabeled PSII shows that they reflect the protonation of an amino-acid other than YZ. The absence of marked differences in the pH-dependence of the reduction of P680•+ in the pH range extending from 5.2 to 10.5 suggests that the pKa of 3F-YZOH is larger than 10.5: thus the apparent pKa of YZOH would be larger by 1.5 pH units, i.e. ≥ 12.0.
The mechanistic model which follows from the present data is consistent with the current views on the role of H+ transfer in determining the overall rate constant for the oxidation of YZ 11,13,21,25,46,47. In the alkaline pH range, the phenol proton of YZ would be involved in a H-bond network, with the ε-N of the imidazole ring of His-190 acting as a direct or indirect H+ acceptor. This network facilitates the deprotonation of YZOH and allows fast electron transfer to P680•+. Below pH ≈ 7 the protonation of the H-bond acceptor (likely the ε-N of the imidazole ring) perturbs this H-bond network and the oxidation of YZ, proceeding via a concerted proton-electron transfer, is significantly slowed down. Although supporting in essence a similar mechanism, the present results provide further insights into the energetics of the H-bond network in which YZOH is involved. Indeed, based upon the acceleration of the oxidation of YZ above pH 9 in a mutant lacking D1-His190, Hays et al. concluded that the pKa of YZ is 10.3 in this mutant21. The difference between this previous estimate and the present one (pKa ≥ 12.0) likely stems from differences in the H-bonded status of the YZ phenol group in the two studies. Indeed, the present data were obtained in the presence of D1-His190 the ε-N of which is likely involved, directly or indirectly, in a H-bond with YZ (in the following we will denote this H-bond network as YZOH---NHis, with no assumption as to whether D1-His190 is directly or indirectly H-bonded). This H-bond should stabilize the singly protonated state of the YZOH---NHis ensemble because its deprotonation requires the prior rupture of the H-bond (e.g. ref. 17), and would thus induce an up-shift of the apparent pKa of the YZOH---NHis ensemble. Accordingly, the H-bond would account for the different pKa for YZ in both studies and this difference would provide an estimate of its strength: EH ≅ (12.0-10.3) × 60 ≥ 105 meV, which is in the range corresponding to a H-bond of moderate to significant strength.
2. Mechanism of (3F)-YZ oxidation
The study of the fastest component in the reduction of P680•+ at pH 9.5 shows the following: (1) it is accelerated by the fluorination of the phenol group of YZ, (2) the fluorination results in a decrease of its activation energy by about 110 meV, a value which nicely matches the difference in free energy of deprotonation between 3F-YZO− and YZO− as derived from the pKa shift of 1.5 pH units (90 meV) induced by the fluorination 33. Although this correlation strongly suggests that the oxidation of YZ requires its prior deprotonation, we will first discuss the possibility that the kinetic effect of the fluorination reflects a change in the driving force for electron transfer rather than for proton transfer.
Dependence of the rate of oxidation of YZ on the driving force for electron transfer
A first argument against a hypothesis which asserts that the kinetic consequences of the fluorination would reflect a change in the driving force for electron transfer comes from the electrochemical properties of 3F-Tyr. Indeed, despite the 63 mV difference between the Em’s of the (3F-YO•/3F-YO−) and (YO•/YO−) couples, the Em’s of the (3F-YO•/3F-YOH) and (YO•/YOH) couples are similar (within 20 mV, at most; see Fig. 4 in 33). This similarity stems from the down-shift of the pKa of the phenol group induced by the fluorination 33 which compensates for the difference in Em. Thus, the substitution of 3F-YZ for YZ is not expected to result in a significant change in the driving force for the oxidation of (3F)-YZ, since as discussed above, (3F)-YZ is protonated when reduced and neutral when oxidized so that, irrespective of the mechanistic pathway, the relevant redox couple is, on thermodynamic grounds, ((3F)-YO•/(3F)-YOH). As regards this issue, the amplitude of the fast component of the reduction of P680•+ proved to be a sensitive measure of the driving force. It was significantly decreased in the D1-His198A mutant, which induces a down-shift by ≈ 80 mV of the midpoint potential of the P680•+/P680 couple 32. The similar amplitudes observed in the labeled and unlabeled PSII thus show that, as expected, the driving force for electron transfer is hardly affected by the fluorination and that, given the respective electrochemical properties of (3F)-YZOH and (3F)-YZO−, it is (3F)-YZOH that is the electron donor to P680•+.
In addition, we found that, in the unlabeled PSII, the oxidation of YZ is slightly activated (ΔHa ≈ 110 meV and ≈ 220 meV in T. elongatus and Synechocystis, respectively), consistent with previous reports of an activation enthalpy of 150 meV for Mn-depleted PSII at pH 9 from pea 11, or of 100–250 meV in O2-evolving PSII 48,49. Yet, a smaller driving force for the electron transfer reaction due to the lowering of the midpoint potential of the electron acceptor in the D1-H198A PSII mutant, did not weaken the dependence upon temperature of the reduction of P680•+ (Fig. 5). Thus, the decrease in the activation enthalpy induced by the substitution of 3F-YZ for YZ cannot result from a change in the driving force for electron transfer for the two following reasons: firstly this substitution is not expected to alter this driving force, and, secondly, were it to do so, it would not decrease the activation energy.
Dependence of the rate of oxidation of YZ on the driving force for proton transfer
The question remains as to whether the kinetic effect of the fluorination on the fast component of P680•+ reduction can be accounted for by a change in pKa of (3F)-YZOH. Since the fluorination induces a decrease of the pKa 33, the transfer of the phenol proton to a nearby acceptor should be more favorable. This increased driving force for proton transfer should facilitate the formation of the (3F)-YO− species, which, in turn, is a more rapid electron donor than (3F)-YOH, and the fluorination would thus be expected to speed up the reduction of P680•+, as observed here. Accordingly, the coupled transfer of a proton and an electron from YZOH would follow:
Yet, the translation of these views into their formal expression has often led to their rebuttal. In such a model, which implicitly assumes that the overall oxidation is kinetically controlled by the deprotonation of the phenol, the apparent rate constant for the oxidation of (3F)-YZ is:
| (1) |
where ket is the intrinsic rate constant of the tyrosinate oxidation and KH is the equilibrium constant between the (3F)-YZOH---NHis and (3F)-YZO−---HN+His states.
This model has often been discounted because KH was assumed to be equal to 10ΔpKa, where ΔpKa is the difference between the pKas of the proton acceptor (here, NHis) and the proton donor (here, YZOH). Were this latter assumption to apply, the estimated pKa of 10.3–12 for YZ and 7–7.5 for His190 would yield kapp ≅10−3 × ket and for kapp to be ≈107 s−1 ket would need to be in the 1010 s−1 range, a value which is hard to reconcile with the ≈ 10 Å separating the electron donor and acceptor (YZ, and P680•+, respectively) 26,50–52. Yet, as argued on thermodynamic grounds46, and on experimental grounds53, the salt bridge between the phenolate and the imidazolium in the (3F)-YZO−---HN+His state could affect the coupling between proton and electron transfer with contributions from the electrostatic potentials or electronic coupling arising from the charge distribution. In other words, KH may differ from 10ΔpKa as soon as the proton transfer between the H+ donor and acceptor results in the possible formation of a salt bridge as in the (3F)-YZO−---HN+His case (we note that this holds as long as the proton acceptor is positively charged when protonated, as would be the case if it were a water molecule or an imidazolium ion). Interestingly, if one assumes that the intrinsic rate constant for electron transfer from (3F)-YZO−---HN+His is temperature independent in the narrow temperature range studied here, the free energy change associated with the transition between (3F)-YZOH---NHis and (3F)-YZO−---HN+His should correspond to the activation energy of the overall reaction, provided the proton transfer along the H-bond is not associated with a significant change in entropy. The above model thus predicts that, provided the strength of the salt bridge is unaltered upon the (3F-Tyr)/Tyr substitution (see the supporting information for an experimental support to this assumption), a variation of the pKa of the proton donor should induce a similar variation of the activation enthalpy. This expectation is satisfyingly met, since in both the T. elongatus and Synechocystis cases the 3F-Tyr/Tyr substitution resulted in a decrease of the activation enthalpy by ≈ 110 meV which compares to the 90 meV change expected from the difference in pKa. We thus conclude that: (1) there is no strong theoretical reason to dismiss the reaction path through which the electron transfer to P680•+ proceeds from the (3F)-YZO−---HN+His state, the relative steady-state concentration of which state is determined by the equilibrium constant, KH, between the (3F)-YZO−---HN+His and (3F)-YZOH---NHis states; (2) the present data support this model.
Equation (1) relies on the assumptions that the steady-state approximation applies and that the rates of proton transfer from, kPT, and to the phenol, k-PT, are larger than the electron transfer rate, ket. As just discussed the satisfying correlation between the change in the activation energy induced by the fluorination and the expected change in KH support both assumptions. In addition, the observation that the rate of the proton-coupled electron transfer through a carboxylate-guanidinium bridge was 3 108 s−1 experimentally ascertains the fact that such high rates are indeed conceivable in a H-bonded salt bridge as the one considered here 53.
The question then arises as to the value of KH. The mere observation that kapp varies with KH shows that it cannot be much larger than 1. Yet, the acceleration, at 25°C which results from the substitution is moderate so that KH cannot be much smaller than 1 either. A more quantitative derivation can be made, based on equation (1). The ratio between the apparent oxidation rates of YZ versus 3F-YZ is:
| (2) |
| (3) |
where, the superscript F denotes the values of KH or pK in the case of the 3F-YZ. By combining equations (2) and (3) one obtains:
| (4) |
With a shift in pKa of 1.5 pH units (β=10−1.5), and a ratio of 0.3 between the apparent rate constant for the unlabeled and labeled PSII at 25°C, one obtains KH(25°C) = 0.4 (ΔG = 25 meV), which corresponds to a probability of ~ 0.25 for the YZO−---HN+His state (we note that this figure is, in actual fact, an overestimate owing to the incomplete incorporation of the fluorinated tyrosine). Although present in a minority of centers, the proportion of tyrosinate state may be high enough to allow its detection by optical spectroscopy. Indeed, the difference between the (YZox-YZ) spectra measured at pH 9.0 and 6.1 displays similarities with the deprotonation spectrum of a tyrosine 30.
3. Relevance to the O2 evolving PSII
Finally, we believe that the above conclusions on the coupling between, proton and electron transfer from YZ are not restricted to Mn-depleted PSII and also apply to O2-evolving PSII. At alkaline pH’s, Mn-depleted PSII shows significant kinetic similarities with O2-evolving PSII. Indeed, the overall oxidation rate of YZ lies in a similar range as the one observed in O2-evolving PSII. Furthermore, as discussed above, the activation energies of the fastest component of P680•+ reduction are also comparable. The main difference thus lies in the relative weight of the components which occur in the 1–100 μs time range. It is commonly accepted that these phases, in Mn-depleted PSII, reflect heterogeneities in the H-bond network in which the phenol side chain of YZ is involved. Such proposals come from kinetic studies 11,21,54,55 and magnetic resonance and FTIR studies directly assessing the strength and order of the H-bonding network to YZO• or YZOH 22,30,56. Signigicantly, the FTIR spectra of YZO•/YZOH in Mn-depleted PSII and in Ca2+ depleted PSII containing the (Mn)4 cluster shows strong similarities 22. In O2-evolving PSII, the reduction of P680•+ is also markedly multiphasic. The fastest component is thought to reflect the electron transfer reaction between P680•+ and YZ and the slow components, developing in the μs time range, would result from the electrostatic relaxation of the surrounding medium due to proton transfers57,58. Thus, although the kinetic complexity which is a common feature to Mn-depleted and O2-evolving PSII likely stems from different causes, it seems reasonable to assume that, in Mn-depleted PSII, the fastest component probes those PSII centers in which the YZOH---NHis/YZO−---HN+His state exists prior to the actinic flash. If indeed O2-evolving PSII behaves, kinetically speaking, similarly to this subpopulation, the mechanism proposed here would apply to the active enzyme as well.
Conclusion
The combined use of site-directed mutants together with the global labeling by 3F-Tyr produced modifications of the driving force for electron transfer and for proton transfer from (3F)-YZ. The finding that the overall oxidation rate was increased with increased probability of (3F)-tyrosinate formation and was insensitive to a change in the driving force for electron transfer, points to the deprotonation of the phenol group as being the determining factor in the process. Although the apparent pKa of (3F)-YOH is significantly higher than that of the proton acceptor, making a priori highly unfavorable the formation of the phenolate, this deprotonation could be kinetically and thermodynamically facilitated by the H-bond network in which (3F)-YZ is involved as well as by the salt bridge between the phenolate and the proton acceptor. These bonds, which form at high pH’s, would promote the switch from the concerted proton-electron transfer proposed to occur at lower pH’s, to the stepwise transfer of the proton and the electron from (3F)-YZ. This sequential transfer contrasts with the case of a phenol-carboxylate compound, which provides a H-bond to the phenol group but no salt bridge, in which a concerted proton-electron transfer was observed 52. Significantly, this overall reaction was slower than the electron transfer from the phenolate. By fulfilling the requirements for a fast proton/electron transfer from the YO−---HN+His ensemble, the combination of the H-bond with the phenolate-imidazolium bridge may prove more efficient as a catalyst in bio-inspired photo-voltaic devices as recently exemplified by a PSII-inspired construct 59.
Supplementary Material
Scheme 1.
The stepwise proton transfer/electron transfer pathway.
Acknowledgments
B.A.D. gratefully acknowledges the support of the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service grant no. 2003-35318-13589. This study was supported in part by the JSPS and CNRS under the Japan-France Research Cooperative Program (F.R., A.B., M.S.) and the Solar H program STRP from the European Union (A.B.). R.D.B. acknowledges the support of NIH, grant no. NIH GM073789.
Footnotes
Abbreviations: 3F-Tyr, fluorinated tyrosine on the carbon 3 of the phenol ring, PS, Photosystem, YZ Tyr-161 of the D1 subunit, ESE, electron spin echo.
When referring specifically to the fluorinated YZ or to the non-fluorinated YZ, we will use the notation 3F-YZ and YZ, respectively. (3F)-YZ will be used when referring without distinction to the fluorinated or non-fluorinated YZ.
We did not perform this experiment with T. elongatus PSII, since, in this species, the D1-His198 mutations hardly affected the redox properties of P680 34.
Supporting information available: Figure S1 and Table S1 describe in more detail how the simulation of the 3F-Tyr EPR spectrum was done and how the incorporation yield was quantified. Figure S2 illustrates the pH dependence of the reduction of P680+• in Mn-depleted PSII from T. elongatus. It also shows the improved quality of the fit when using 4 pH independent rate constants rather than 3. Figure S3 shows the high-field EPR spectra of YZO° and 3F-YZO° in PSII complexes from T. elongatus at pH 9, and Table S2 gives a summary of the geometries and hydrogen bonding energies of the 4-Ethylphenolate/imidazole, 3-Fluoro-4-Ethylphenolate/imidazole hydrogen-bonded supercomplexes and their respective oxidized radical versions as determined from B3LYP/6-31+G(D,P) density functional theory calculations. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Diner BA, Rappaport F. Annu Rev Plant Biol. 2002;53:551–580. doi: 10.1146/annurev.arplant.53.100301.135238. [DOI] [PubMed] [Google Scholar]
- 2.Rappaport F, Diner BA. Coord Chem Rev. 2008;252:259–272. [Google Scholar]
- 3.Dau H, Haumann M. Coord Chem Rev. 2008;252:273–295. [Google Scholar]
- 4.Rappaport F, Guergova-Kuras M, Nixon PJ, Diner BA, Lavergne J. Biochemistry. 2002;41:8518–8527. doi: 10.1021/bi025725p. [DOI] [PubMed] [Google Scholar]
- 5.Cuni A, Xiong L, Sayre RT, Rappaport F, Lavergne J. Phys Chem Chem Phys. 2004;6:4825–4831. [Google Scholar]
- 6.Grabolle M, Dau H. Biochim Biophys Acta. 2005;1708:209–218. doi: 10.1016/j.bbabio.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Esswein AJ, Nocera DG. Chem Rev. 2007;107:4022–4047. doi: 10.1021/cr050193e. [DOI] [PubMed] [Google Scholar]
- 8.Irebo T, Reece SY, Sjödin M, Nocera DG, Hammarström L. J Am Chem Soc. 2007;129:15462–15464. doi: 10.1021/ja073012u. [DOI] [PubMed] [Google Scholar]
- 9.Herrero C, Lassalle-Kaiser B, Leibl W, Rutherford AW, Aukauloo A. Coord Chem Rev. 2008;252:456–468. [Google Scholar]
- 10.Rappaport F, Blanchard-Desce M, Lavergne J. Biochim Biophys Acta. 1994;1184:178–192. [Google Scholar]
- 11.Ahlbrink R, Haumann M, Cherepanov D, Bögershausen O, Mulkidjanian A, Junge W. Biochemistry. 1998;37:1131–1142. doi: 10.1021/bi9719152. [DOI] [PubMed] [Google Scholar]
- 12.Haumann M, Liebisch P, Muller C, Barra M, Grabolle M, Dau H. Science. 2005;310:1019–1021. doi: 10.1126/science.1117551. [DOI] [PubMed] [Google Scholar]
- 13.Renger G. Biochim Biophys Acta. 2004;1655:195–204. doi: 10.1016/j.bbabio.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Mayer JM, Rhile IJ, Larsen FB, Mader EA, Markle TF, DiPasquale AG. Photosynth Res. 2006;87:3–20. doi: 10.1007/s11120-005-8164-3. [DOI] [PubMed] [Google Scholar]
- 15.Cukier RI, Nocera DG. Annu Rev Phys Chem. 1998;49:337–369. doi: 10.1146/annurev.physchem.49.1.337. [DOI] [PubMed] [Google Scholar]
- 16.Lomoth R, Magnuson A, Sjödin M, Huang P, Styring S, Hammarström L. Photosynth Res. 2006;87:25–40. doi: 10.1007/s11120-005-9005-0. [DOI] [PubMed] [Google Scholar]
- 17.Markle TF, Rhile IJ, Dipasquale AG, Mayer JM. Proc Natl Acad Sci U S A. 2008;105:8185–8190. doi: 10.1073/pnas.0708967105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diner BA, Nixon PJ, Farchaus JW. Curr Opin Struct Biol. 1991;1:546–554. [Google Scholar]
- 19.Mamedov F, Sayre RT, Styring S. Biochemistry. 1998;37:14245–14256. doi: 10.1021/bi980194j. [DOI] [PubMed] [Google Scholar]
- 20.Hays AMA, Vassiliev IR, Golbeck JH, Debus RJ. Biochemistry. 1998;37:11352–11365. doi: 10.1021/bi980510u. [DOI] [PubMed] [Google Scholar]
- 21.Hays AMA, Vassiliev IR, Golbeck JH, Debus RJ. Biochemistry. 1999;38:11851–11865. doi: 10.1021/bi990716a. [DOI] [PubMed] [Google Scholar]
- 22.Berthomieu C, Hienerwadel R, Boussac A, Breton J, Diner BA. Biochemistry. 1998;37:10548–10554. doi: 10.1021/bi980788m. [DOI] [PubMed] [Google Scholar]
- 23.Noguchi T, Inoue Y, Tang XS. Biochemistry. 1997;36:14705–14711. doi: 10.1021/bi971760y. [DOI] [PubMed] [Google Scholar]
- 24.Berthomieu C, Hienerwadel R. Biochim Biophys Acta. 2005;1707:51–66. doi: 10.1016/j.bbabio.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Diner BA, Bautista JA, Nixon PJ, Berthomieu C, Hienerwadel R, Britt RD, Vermaas WF, Chisholm DA. Phys Chem Chem Phys. 2004;6:4844–4850. [Google Scholar]
- 26.Sjödin M, Styring S, Åkermark B, Sun L, Hammarström L. J Am Chem Soc. 2000:122. [Google Scholar]
- 27.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 28.Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Nature. 2005;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- 29.Brettel K, Schlodder E, Witt HT. Biochim Biophys Acta. 1984;766:403–415. [Google Scholar]
- 30.Diner BA, Force DA, Randall DW, Britt RD. Biochemistry. 1998;37:17931–17943. doi: 10.1021/bi981894r. [DOI] [PubMed] [Google Scholar]
- 31.Force DA, Randall DW, Britt RD, Tang XS, Diner BA. J Am Chem Soc. 1995;117:12643–12644. [Google Scholar]
- 32.Diner BA, Schlodder E, Nixon PJ, Coleman WJ, Rappaport F, Lavergne J, Vermaas WF, Chisholm DA. Biochemistry. 2001;40:9265–9281. doi: 10.1021/bi010121r. [DOI] [PubMed] [Google Scholar]
- 33.Seyedsayamdost MR, Reece SY, Nocera DG, Stubbe J. J Am Chem Soc. 2006;128:1569–1579. doi: 10.1021/ja055926r. [DOI] [PubMed] [Google Scholar]
- 34.Boussac A, Verbavatz JM, Sugiura M. Photosynth Res. 2008;98:285–292. doi: 10.1007/s11120-008-9305-2. [DOI] [PubMed] [Google Scholar]
- 35.Sugiura M, Boussac A, Noguchi T, Rappaport F. Biochim Biophys Acta. 2008;1777:331–342. doi: 10.1016/j.bbabio.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Un S, Boussac A, Sugiura M. Biochemistry. 2007;46:3138–3150. doi: 10.1021/bi062084f. [DOI] [PubMed] [Google Scholar]
- 37.Lakshmi KV, Reifler MJ, Chisholm DA, Wang JY, Diner BA, Brudvig GW. Photosynth Res. 2002;72:175–189. doi: 10.1023/A:1016140902662. [DOI] [PubMed] [Google Scholar]
- 38.Tang XS, Diner BA. Biochemistry. 1994;33:4594–4603. doi: 10.1021/bi00181a021. [DOI] [PubMed] [Google Scholar]
- 39.Rögner M, Nixon PJ, Diner BA. J Biol Chem. 1990;265:6189–6196. [PubMed] [Google Scholar]
- 40.Sturgeon BE, Ball JA, Randall DW, Britt RD. J Phys Chem. 1994;98:12871–12883. [Google Scholar]
- 41.Beal D, Rappaport F, Joliot P. Rev Sci Instr. 1999;70:202–207. [Google Scholar]
- 42.Seyedsayamdost MR, Stubbe J. J Am Chem Soc. 2006;128:2522–2523. doi: 10.1021/ja057776q. [DOI] [PubMed] [Google Scholar]
- 43.Barry BA, Babcock GT. Proc Natl Acad Sci U S A. 1987;84:7099–7103. doi: 10.1073/pnas.84.20.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ayala I, Perry JJ, Szczepanski J, Tainer JA, Vala MT, Nick HS, Silverman DN. Biophys J. 2005;89:4171–4179. doi: 10.1529/biophysj.105.060616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conjeaud H, Mathis P. Biochim Biophys Acta. 1980;590:353–359. doi: 10.1016/0005-2728(80)90206-6. [DOI] [PubMed] [Google Scholar]
- 46.Rappaport F, Lavergne J. Biochim Biophys Acta. 2001;1503:246–259. doi: 10.1016/s0005-2728(00)00228-0. [DOI] [PubMed] [Google Scholar]
- 47.Diner BA, Britt RD. In: Photosystem II: The Light Driven Water: Plastoquinone Oxidoreductase. Wydrzynski TJ, Satoh K, editors. Springer; Dordrecht: 2005. pp. 207–233. [Google Scholar]
- 48.Eckert HJ, Renger G. FEBS Lett. 1988;236:425–431. [Google Scholar]
- 49.Jeans C, Schilstra MJ, Klug DR. Biochemistry. 2002;41:5015–5023. doi: 10.1021/bi0118862. [DOI] [PubMed] [Google Scholar]
- 50.Renger G, Christen G, Karge M, Eckert HJ, Irrgang KD. J Biol Inorg Chem. 1998;3:360–366. [Google Scholar]
- 51.Tommos C, Babcock GT. Biochim Biophys Acta. 2000;1458:199–219. doi: 10.1016/s0005-2728(00)00069-4. [DOI] [PubMed] [Google Scholar]
- 52.Sjödin M, Irebo T, Utas JE, Lind J, Merenyi G, Åkermark B, Hammarström L. J Am Chem Soc. 2006;128:13076–13083. doi: 10.1021/ja063264f. [DOI] [PubMed] [Google Scholar]
- 53.Kirby JP, Roberts JA, Nocera DG. J Am Chem Soc. 1997;119:9230–9236. [Google Scholar]
- 54.Lavergne J, Rappaport F. Biochemistry. 1998;37:7899–7906. doi: 10.1021/bi9801210. [DOI] [PubMed] [Google Scholar]
- 55.de Wijn R, Schrama T, van Gorkom HJ. Biochemistry. 2001;40:5821–5834. doi: 10.1021/bi002824z. [DOI] [PubMed] [Google Scholar]
- 56.Tommos C, McCracken J, Styring S, Babcock GT. J Am Chem Soc. 1998;120:10441–10452. [Google Scholar]
- 57.Schilstra MJ, Rappaport F, Nugent JHA, Barnett CJ, Klug DR. Biochemistry. 1998;37:3974–3981. doi: 10.1021/bi9713815. [DOI] [PubMed] [Google Scholar]
- 58.Christen G, Renger G. Biochemistry. 1999;38:2068–2077. doi: 10.1021/bi982188t. [DOI] [PubMed] [Google Scholar]
- 59.Moore GF, Hambourger M, Gervaldo M, Poluektov OG, Rajh T, Gust D, Moore TA, Moore AL. J Am Chem Soc. 2008;130:10466–10467. doi: 10.1021/ja803015m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.