Abstract
Synthetic siRNA duplexes are used widely as reagents for silencing of mRNA targets in cells and are being developed for in vivo use. Serum stability is a major concern if siRNA is to be used for therapeutic delivery within blood circulation. We have developed the use of MALDI-TOF mass spectrometry as a rapid and convenient analytical tool to identify the most vulnerable sites within siRNA to serum degradation. Using this approach, we found that one siRNA duplex (Dh3) with UpA sequences close to one end was particularly vulnerable to rapid cleavage. This produced a fragment of mass consistent with the presence of a 2′,3′-cyclic phosphate that was slowly hydrolysed to a 2′-(3′-)phosphate on extended incubation. Substitution of these sites with 2′-O-methyl U residues prevented cleavage and confirmed that the major pathway for initial degradation is via cleavage by an RNAse A-like activity. Mass spectral analysis was used to follow the serum degradation of siRNA over more prolonged periods to show the accumulation of many fragments, almost all showing cleavage following pyrimidine nucleoside residues. Overall, the MALDI-TOF mass spectral analysis technique should prove useful for preliminary screening of the serum stability of siRNA duplexes and for identification of the most vulnerable cleavage sites.
Introduction
Small interfering RNAs (siRNAs) are short double-stranded nucleic acids, commonly containing 19–21 residues and 3′-dinucleotide overhangs, that are used widely as synthetic reagents to reduce gene expression of target RNA in cells.1 When introduced into cells, one strand of the siRNA (the antisense or guide strand) becomes incorporated into the RNA-induced silencing complex (RISC), which directs cleavage of the complementary target mRNA. A substantial reduction in gene expression activity is often achieved at low siRNA concentration (nM or less), and the effect can last for several days. Thus siRNAs are being developed to target therapeutically important genes in cancer, viral infections and other indications.2
It is well-known that RNAs, particularly single-stranded RNAs, are unstable within cells and biological media such as serum, and are degraded rapidly by ribonucleases (RNAses), such as those in the RNAse A family, originally known as pancreatic RNAses.3 However, relatively little has been published concerning the serum RNAse cleavage profile of double-stranded synthetic siRNA, despite a number of studies showing that introduction of synthetic analogues into one or both strands often increases the serum stability.4,5 Very recently, strong evidence was obtained of an RNAse A-like activity within serum that preferentially attacks U/A-rich sequences in siRNAs.6 Such cleavages were characterised by a difference in mobility of the siRNA when subjected to polyacrylamide gel electrophoresis (PAGE). Degradation of the RNA was inhibited when an RNAse A inhibitor was added, or when human serum was immunodepleted by pre-treatment with an antibody to bovine RNAse A.6
During a recent study of the delivery into mouse lungs of synthetic conjugates of siRNA targeted to p38 MAP kinase (Moschos et al., manuscript in preparation), we observed by PAGE analysis a rapid cleavage of the siRNA when this was incubated in mouse serum, but not in bronchiolar lavage (BAL). This observation raised the more general question of how to rapidly assess the stability of siRNA in biological fluids, such as serum, and to reveal the locations of the most vulnerable cleavage sites. We have devised an informative and convenient MALDI-TOF mass spectrometric technique that can be used with serum-treated siRNA samples to identify the major cleavage products. The results confirm the vulnerability to very rapid cleavage of UpA sequences close to the end of an siRNA, to give products consistent with an RNAse A cleavage mechanism. 2′-O-Methyl U substitution at such UpA sites blocks this cleavage and stabilises the siRNA significantly. Further RNAse A-type cleavages are significantly slower and are seen within a few hours at many other sites within siRNAs, particularly following pyrimidine nucleoside residues. This technique may help to screen siRNA sequences for serum sensitivity and inform as to where chemical modification of siRNA might be best directed.
Results
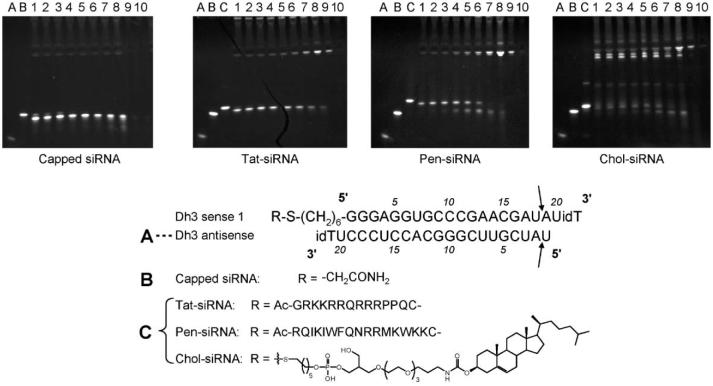
PAGE analysis is commonly used to separate fragments of nucleic acids by molecular size and to demonstrate cleavage of siRNA in serum.4,6 We initially used this method to observe the serum stability of certain synthetic conjugates of siRNA, targeted to mouse p38 MAP kinase, which were designed for mouse lung knockdown studies. In these siRNA constructs (Fig. 1), the Dh3 antisense strand (A) contains a 3′-inverted dT residue, whilst the sense strand (Dh3 sense-1) also contains a 3′-inverted dT residue as well as a 5′-C6-thiol linker. In the siRNA duplexes, the sense strand linker is either acetamide-capped (B), or conjugated with either the cell penetrating peptide Tat (residues 48–60),7 Penetratin,7 or cholesterol8 (duplexes C) in order to improve cell delivery. After treatment with 90% mouse serum for just 1 minute at 37 °C, PAGE analysis showed the degradation of the unconjugated and peptide-conjugated siRNA duplexes into a single smaller product (compare lanes B and 1 for capped-linker siRNA as well as lanes C and 1 for Tat–siRNA and Pen–siRNA). The uniform nature of the migration change suggested that cleavage occurred in the siRNA rather than the peptide parts.
Fig. 1.
Stability of capped siRNA and siRNA conjugates in 90% mouse serum at 37 °C. Capped siRNA (B) and siRNA conjugates (C) were incubated with 90% mouse serum at 37 °C at various time intervals and then separated on a 20% PAGE gel with siRNA products detected using a SYBR Gold nucleic acid stain. Lanes A, B and C refer to untreated controls of Dh3 antisense, capped siRNA, and corresponding conjugate, respectively. Lanes 1–10 = 1 min, 2.5 min, 5 min, 15 min, 30 min, 1 h, 3 h, 6 h, 12 h, 24 h serum incubation time. Arrows refer to clip sites as determined in Fig. 2, 3′ inverse deoxythymidine is denoted by idT.
For the cholesterol conjugate (Fig. 1, Chol–siRNA), most of the duplexes appeared to be sequestered in lower mobility bands, presumably serum protein-bound products. However even in these circumstances, a degradation (higher mobility) product is seen within 1 minute of serum treatment (compare lanes C and 1). With each siRNA construct, further degradation products were only seen after 1–3 h of serum treatment. Overall, these results showed that a rapid cleavage of the siRNA had taken place to form a single product. However, it is not possible to infer from the PAGE technique anything about the nature of the cleavage(s) or their locations on the siRNA.
Capillary gel electrophoresis has been employed to separate serum cleavage products of siRNA,5 but this also does not allow identification of the siRNA cleavage fragments. Reversed-phase liquid chromatography coupled with electrospray ionization mass spectrometry (ESI-MS) has also been used to separate and identify degradation products of siRNA,9 but sample pre-treatments are often needed to remove contaminating salts that interfere with ESI mass analysis. To our knowledge, the relatively accessible technique of MALDI-TOF mass spectrometry has not been described previously for the identification of siRNA degradation products. Such mass spectrometry instrumentation is often readily available for bench-top use in laboratories involved in oligonucleotide and siRNA chemical synthesis, and could be used to determine the masses of siRNA cleavage products without the need for a pre-treatment to separate salts and serum.
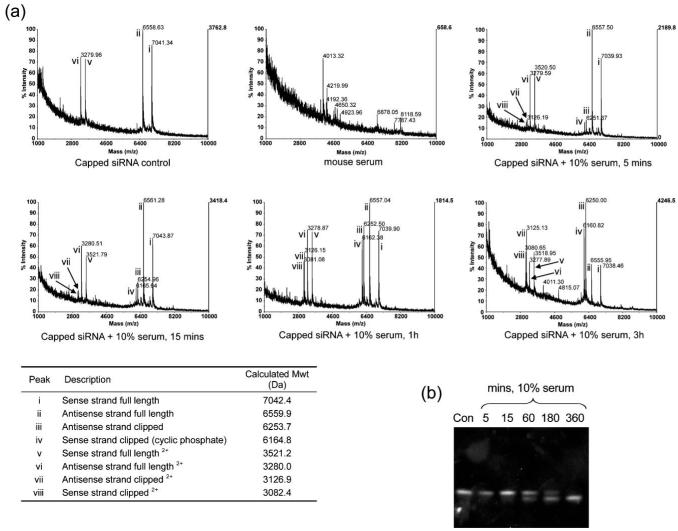
Therefore we treated duplex siRNA (capped Dh3 sense-1/Dh3-antisense, B, 100 μM) with 10% mouse serum at room temperature for various times and examined the cleavage patterns by direct MALDI-TOF analysis. A lower temperature and concentration of serum was used in order to slow down the cleavage rate to a suitable time scale for mass spectral analysis. Conditions were found with matrix (a) (see the Experimental section and the ESI†) under which it was possible to suppress the intensities of peaks that were due to the presence of serum (presumably protein or peptide), whilst maintaining good peak intensities of the siRNA strands and their cleavage products (Fig. 2a). Control Dh3 siRNA duplex showed peaks with masses corresponding to both sense and antisense full length strands (top left, peaks i and ii) and doubly charged (half values for m/z) full length strands (v and vi). No peak corresponding to duplex siRNA is observed, since the strands are expected to separate during analysis. Mouse serum alone showed peaks in this same mass range, but they are much smaller in intensity and can only be seen clearly when the sensitivity scale is boosted several fold (top centre). Upon incubation of the siRNA with 10% serum for 5 min (top right), 15 min (bottom left), 1 h (bottom centre) or 3 h (bottom right), two new peaks emerged (iii and iv) corresponding respectively in mass to antisense strand with residue 1 and its 3′-phosphate removed, and sense strand with residues 19–21 removed and leaving a 2′,3′-cyclic phosphate (Fig. 1, arrowed). Doubly charged peaks were also seen (vii and viii). On both strands, cleavage appeared to have occurred between UpA sequences on opposite sides of the siRNA close to one of the duplex ends (after U18 and U1, respectively). The products are consistent with an RNAse A-type cleavage mechanism, particularly the mass of the sense strand fragment which could only correspond to a 2′,3′-cyclic phosphate and not for example a 2′- or 3′-phosphate. The rate of emergence of the two peaks iii and iv appeared to match the rate of formation of the higher mobility product by PAGE analysis for the 10% serum-treated sample (Fig. 2b).
Fig. 2.
Stability of 100 μM capped siRNA in 10% mouse serum at room temperature. (a) MALDI-TOF mass spectra acquired using matrix a. Capped siRNA and mouse serum controls, capped siRNA incubated with 10% serum at room temperature for 5 min, 15 min, 1 h and 3 h, showing decomposition of the full length strands into shorter fragments (refer to the table). The same laser power was used for all spectra shown (b) 20% PAGE gel of capped siRNA incubated with 10% mouse serum at room temperature at 5–360 min time intervals. Con = untreated capped siRNA control.
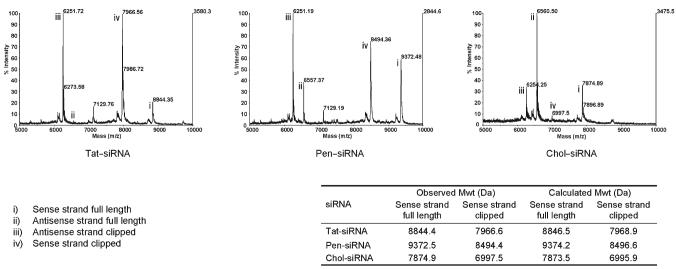
siRNA conjugates C (Tat–siRNA, Pen–siRNA and Chol–siRNA, Fig. 1) treated under the same conditions with 10% serum also showed the emergence of two peaks (Fig. 3, peaks iii and iv in each case, 3 h time point shown) with masses corresponding to the same cleavage positions in the siRNA and the same 2′,3′-cyclic phosphate cleavage product on the sense strand. Interestingly, no obvious degradation of the peptides was seen in either case under these conditions.
Fig. 3.
MALDI-TOF mass spectra of Tat–, Penetratin– and cholesterol–siRNA conjugates (100 μM) after 3 h incubation with 10% mouse serum at room temperature using matrix a. Spectra show the same mass difference between parent and clipped species (refer to the table), and therefore the same clips as capped siRNA (see Fig. 2).
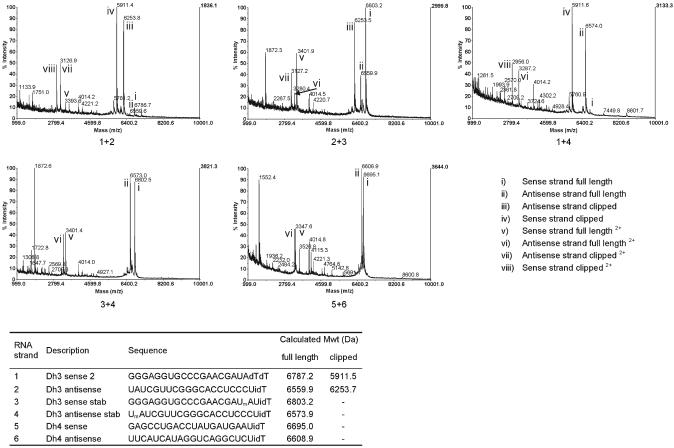
Soutschek et al had suggested previously that the predominant mechanism for siRNA degradation in serum is by exonucleolytic activity.8 Thus, they used a number of chemical modifications at the 3′-end of each siRNA strand in order to stabilise the siRNA in vivo, but no proof of such an exonucleolytic cleavage mechanism in serum was provided. Since we had also used an inverted residue (idT) at each end of our siRNA strands, we investigated the effect of serum exonucleases by use of a Dh3 sense strand with a simple dTdT 3′-dinucleotide overhang and without the 5′-thiol linker, thus leaving a free 5′-hydroxyl end (Fig. 4, Dh3 sense-2, strand no. 1) After annealing with Dh3-antisense (strand no. 2) the siRNA duplex (50 μM) was treated at room temperature with 10% mouse serum for 1 h, and the products observed by MALDI-TOF mass spectrometry using matrices (b) or (c), which improved the signal to noise ratio further. At this lower siRNA concentration the cleavage rate was faster than before. The major peaks (Fig. 4, 1 + 2, iii and iv with m/z 6254 and 5911, respectively) correspond to sense strand cleaved once again after U18 to leave a 2′,3′-cyclic phosphate and antisense strand cleaved after U1. The doubly charged species were also visible. No evidence was seen for exonuclease activity under these conditions, i.e. removal of a single pdT from the 3′-end of the sense strand or a single rGp residue from the 5′-end, which would have been evident by peaks at approximately 6483 and 6442 m/z, respectively.
Fig. 4.
Comparison of MALDI-TOF mass spectra of Dh3 and corresponding stabilised duplexes, and Dh4 at 1 h time point with 10% mouse serum at room temperature using matrix b. 50 μM solutions of Dh3 (1 + 2), Dh3 stabilised on one strand (using 2′-OMe modification to block RNAse A-like clipping at cleavage site; 2 + 3 and 1 + 4), Dh3 stabilised on both strands (3 + 4) and Dh4 (5 + 6) siRNA duplexes were incubated with 10% mouse serum for 1 h. Unstabilised strands 1 and 2 were rapidly degraded whereas the full length stabilised RNA strands 3 and 4, as well as strands 5 and 6, which do not have terminal UA sequences did not show extensive degradation at this time point. Note that a higher guide wire voltage was used in these experiments, which has the result of enhancing the intensities of lower molecular mass fragments.
If the siRNA duplex 1 + 2 was incubated with 10% serum for a longer period (26 h), it was possible to see that the peak at 5911.4 had been partially converted into a second peak with mass of m/z 5930.2 (see the ESI, Fig. S1†). This second peak corresponds in mass to the 2′- (or 3′-) phosphate of Dh3 sense strand residues 1–18, which would be expected to form by slow hydrolysis of the 2′,3′-cyclic phosphate.
We also examined Dh3 siRNA sense and antisense strands containing single 2′-O-methyl U (OMe) residues at positions U18 (strand no. 3) and U1 (strand no. 4) respectively (in these cases the 3′-ends contained UidT overhangs). An OMe residue would be unable to be cleaved by an RNAse A-type mechanism, owing to the lack of a proximal 2′-hydroxyl group. Treatment of siRNA composed of strands 2 + 3 with 10% serum for 1 h showed the cleavage product of the antisense strand (Fig. 4, 2 + 3, peak iii, m/z 6253) whilst the sense strand appeared intact (peak i, m/z 6803). Similarly with serum treatment of siRNA duplex composed of strands 1 + 4, only the cleavage product of the sense strand was seen (Fig. 4, 1 + 4, peak iv, m/z 5911), whilst the antisense strand remained intact (peak ii, m/z 6574). When both strands where OMe-substituted (strands 3 + 4), the predominant peaks were the intact siRNA strands (Fig. 4, 3 + 4, peaks i and ii, m/z 6803 and 6573, respectively).
We decided also to check the serum stability of a completely different siRNA duplex targeted to mouse p38 mRNA, Dh4. This sequence has one UpA sequence in the central part of each strand. Under the same incubation conditions as for Dh3, the majority of the two strands of duplex Dh4 were still intact (Fig. 4, 5 + 6, peaks i and ii). However, a small amount of cleavage of Dh4 sense strand was seen after U11 to give a 5′-fragment with a terminal 2′,3′-cyclic phosphate (m/z 3527) and on the antisense strand after U8 (3′-fragment m/z 4115) but these peaks were amongst several others seen.
PAGE analysis of Dh3 siRNA duplex 1 + 2 following 90% serum treatment showed almost complete conversion within 5 min into a higher mobility band (see the ESI, Fig. S2†), just as for the analogous Dh3 siRNA duplex (Fig. 1), but neither doubly OMe-stabilised Dh3 duplex 3 + 4, nor Dh4 duplex 5 + 6 showed any higher mobility band for short serum incubation periods (see the ESI, Fig. S2†). It should also be noted that doubly OMe stabilised Dh3 duplex 3 + 4 did not affect the biological activity compared to the unstabilised Dh3 siRNA duplex 1 + 2 when we examined siRNA knockdown of mouse p38 mRNA, delivered by cationic lipids into mouse L929 cells, as judged by an RT-PCR assay (see the ESI, Fig. S3†).
Overall, these results indicate that the rapid cleavage of siRNA in Dh3 occurs at the UpA sequences by an RNAse A-type mechanism, as previously inferred from the indirect studies of Haupenthal et al.,7 that protection by 2′-OMe substitution prevents such cleavage, and that such protection, at least in the case examined, did not affect the biological activity.
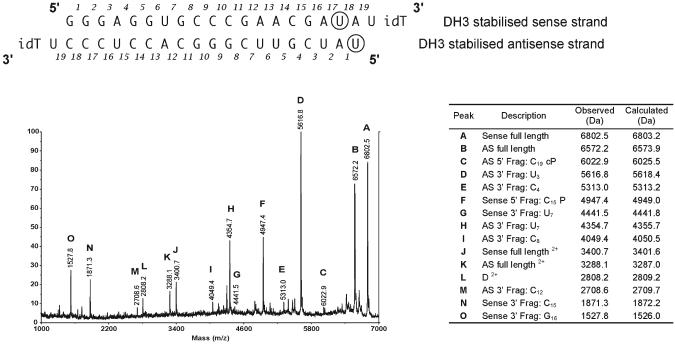
It was clear from the MALDI-TOF mass spectral analysis of the 1 h serum treatment of the OMe-stabilised DH3 siRNA (Fig. 4) that small amounts of degradation products are visible even though the intact siRNA strands are the major components. It was of interest to see if the mass spectral technique could be used to obtain additional data to identify some of these cleavage products. Accordingly, we incubated doubly OMe-stabilised Dh3 siRNA duplex 3 + 4 with 10% mouse serum for a very extended period (120 h) and subjected the product to direct MALDI-TOF mass spectral analysis using matrix (c) (Fig. 5). In addition to the full length strands 3 and 4 (peaks A and B) and doubly charged species (peaks J and K), a series of other fragments could be identified from their mass values. For the sense strand, cleavages were identified following residues U7 (peak G), C15 (both 5′-fragment with terminal 3′-phosphate, peak F, and 3′-fragment, peak N), and G16 (3′-fragment, peak O). For the antisense strand, cleavages were identified after U3 (3′-fragment, peak D), C4 (3′-fragment, peak E), U7 (3′-fragment, peak H), C8 (3′-fragment, peak I), C12 (3′-fragment, peak M) and C19 (5′-fragment 2′,3′-cyclic phosphate, peak C). Thus in all but one case, the cleavages were found after pyrimidine residues, consistent with an RNAse A-type mechanism. In one case (the G16, sense strand) cleavage is more consistent with an RNAse T1-like activity, but the 5′-fragment corresponding with this cleavage could unfortunately not be identified and either must have been degraded further, or fragment O is derived from further degradation of the small fragment N.
Fig. 5.
MALDI-TOF mass spectrum of siRNA duplex consisting of RNAs 3 + 4 (50 μM) incubated with 10% mouse serum for 5 days using matrix c. Evidence of cleavage at several other pyrimidine nucleoside sites at long time periods. The circled U in both strands of RNA 3 and 4 denote that these are 2′-OMeU derivatives; 3′ inverse deoxythymidine is denoted with idT; cP denotes 2′,3′-cyclic phosphate; P denotes 2′- (3′-) phosphate.
In the case of extended 10% serum treatment of Dh4–siRNA duplex 5 + 6, at least seven large fragments were identified with cleavage following pyrimidine nucleoside residues, some containing 2′,3′-cyclic phosphate or the hydrolysis product, 2′- (or 3′-) phosphate (see the ESI, Fig. S4†). There was also evidence for two smaller fragments terminating in A or G residues but without 3′-phosphate (see the ESI, Fig. S4†), which accumulated at even longer incubation times (data not shown). These are likely to have arisen by exonuclease degradation of fragments previously formed by initial RNase A-like degradation. It is of interest to note also that extended 10% serum incubation of duplex Dh4 showed more overall fragmentation and less intact strands than for Dh3 that had been doubly OMe-stabilised at U18 and U1 on sense and antisense strands respectively (data not shown).
Discussion and conclusions
The vulnerability of synthetic siRNA duplexes to cleavage by serum nucleases has been known for some time, but the susceptibility of particular sequences and the mode of degradation has been less clear. This has been due partly to the lack of a rapid and convenient method for siRNA analysis that can be used following serum treatment. We have shown that MALDI-TOF mass spectrometry is a convenient method for such analysis and provides a significant amount of information about the vulnerability of particular siRNA sequences to serum-induced cleavage.
In particular, we have obtained direct analytical evidence for an RNAse A-type cleavage mechanism by identification of peaks that in practically all cases can be attributed to fragments arising as a result of cleavage after pyrimidine nucleoside residues. Some of these fragments are consistent only with oligonucleotides carrying 2′,3′-cyclic phosphates. Such fragments, upon extended incubation, are gradually converted into peaks with mass ca. +18, consistent with hydrolysis to 2′- (or 3′-) phosphates, another known property of 2′,3′-cyclic phosphates. No evidence was seen for exonuclease degradation of either end of a strand of an siRNA, at least under the initial serum conditions used. The results are consistent with the recent indirect data implicating RNAse A-like activity as the primary degradation mechanism for siRNA.6 We observed no significant differences in the fragmentation patterns of siRNAs treated with 10% human serum as compared to mouse serum (data not shown), suggesting that RNAse A-like degradation is common to mammalian serum samples, in line with a previous report.6 We found evidence for exonucleolytic degradation only with extended 10% serum incubation and such fragments appeared to be most likely derived following generation of an initial fragment by RNAse A-like degradation.
It is unlikely that an RNAse A-like enzyme can recognise and cleave a double stranded RNA, and it is therefore probable that cleavage only takes place when the siRNA duplex “breathes” (i.e. during transient breaking of hydrogen bonds between the two strands). The amount of breathing is expected to be highest at A/U-rich sequences. Our observation is that Dh3 contains a particularly vulnerable sequence at one end of the duplex such that both strands are cleaved very rapidly at UpA by the RNAse A-like activity. By contrast UpA sequences near the centre of Dh4 were found to be cleaved at a much slower rate and did not appear to be significantly more vulnerable than other pyrimidine nucleoside residues in the Dh4 sequence. It has been common practice for siRNA duplexes to be designed with one end more A/U rich than the other, in order to facilitate the desired orientation of the siRNA duplex to be loaded into the RISC complex and thus present the correct antisense (guide) strand ready for cleavage of the desired mRNA. Such practices clearly risk serum instability of this end and we have shown that UpA sequences appear particularly vulnerable in this respect.
With regards to their potential use as therapeutics in blood or other biological fluids, the practice of chemical modification of siRNA sequences to increase their stability towards nucleases has become popular in the last few years.4,10 Common modifications include sugar or phosphate alterations at the 3′-end designed for protection against exonucleases as well as the inclusion of 2′-protected nucleosides at internal sites (particularly 2′-O-methyl, 2′-O-methoxyethyl, 2′-fluoro or locked nucleic acids, LNA). In favourable cases even a complete strand of an siRNA duplex has been 2′-modified without substantial loss of RNA silencing activity.5 Our results showing protection from RNAse A-like degradation by OMe U substitution at the two most vulnerable UpA sites in Dh3 are consistent with the many other reports of enhancement of serum stability by such substitution.4
Extensive degradation was seen at numerous other pyrimidine nucleoside sites with 10% serum treatment over extended periods (Fig. 5 and the ESI, Fig. S4†), and such degradation becomes particularly significant within 1–3 hours in the presence of 90% serum (Fig. 1). It thus seems likely that most or all pyrimidine nucleoside sites may need chemical modification for siRNA therapeutic use, although whether such substitution allows full RNA silencing activity to be maintained has to be determined by case-by-case experimentation. Significant additional protection appears to be achieved in vivo when siRNA is packaged by purpose-designed cationic liposomes.11
Overall, MALDI-TOF mass spectrometry has good potential for rapid identification of the most vulnerable sites to serum cleavage in siRNA as well as for determining many of the more slowly obtained cleavage products, and the technique does not require sample pre-treatment. However, we have been careful not to draw too many conclusions from the peak heights or areas of different fragments from MALDI-TOF mass spectral analysis, and hence we have avoided attempting to compare cleavage rates which appear rather similar, since the MALDI ionization efficiency may depend on the specific analyte composition. Nevertheless, in conclusion, this mass spectral technique may become a first line choice for those wishing to establish the most suitable siRNA sequences for potential therapeutic use in regard to serum stability.
Experimental
RNAs and annealing
siRNA containing Dh3 antisense and Dh3 sense-1 strands (Fig. 1) and corresponding conjugates were obtained from RNA-Tec (Leuven, Belgium). For annealing, the strands were dissolved in RNAse free water (Fluka) to give 200 μM solutions. Equimolar amounts of sense and antisense strands were added to a microfuge tube and this was heated to 95 °C for 5 min using a PCR block, followed by slow cooling to 37 °C over a period of 1 h and then cooled to 4 °C. The final concentration of siRNA was 100 μM. RNAs were stored at −20 °C.
RNA strands Dh3 sense-2, Dh3 antisense, corresponding stabilised strands and Dh4 sense and antisense strands (Fig. 4 and 5) were purchased from Dharmacon (Lafayette, USA) as crude material which was 2′-ACE deprotected according to manufacturer's instructions, purified by ion-exchange chromatography and desalted by dialysis.12 Sense and antisense complementary siRNA strands were annealed using the same protocol as above except that an annealing buffer was used and the final concentration of siRNA was 50 μM. Annealing buffer is 5 mM Tris·HCl (pH 7.5), 2.5 mM NaCl, 0.5 μM EDTA. All newly formed duplexes were examined by electrophoresis using a 5% MetaPhor® agarose (Cambrex Bio Science, Rockland, USA) gel in Tris borate-EDTA buffer (TBE) with visualisation of the bands using SYBR-gold dye (Molecular Probes) in combination with a transilluminator.
MM2 siRNA, used as a control in the cell assay (see the ESI, Fig. 3†), was obtained from Dharmacon (Lafayette, USA), pre-annealed and PAGE purified. Sequence of MM2: sense: 5′ GCG AGC UGC GCG AAG GAU AUU 3′; antisense: 5′- UAU CCU UCG CGC AGC UCG CUU-3′ (bold residues in the antisense strand show the sites of mismatch).
Serum stability experiments
For experiments using 90% serum, to 2 μl of a 50 μM solution of siRNA was added 18 μl of mouse serum (Sigma). The resulting solution was vortexed, centrifuged and placed in a PCR block and heated to 37 °C for the desired length of time. The solution was rapidly frozen using liquid nitrogen and stored in a freezer (−20 °C) until all time points were collected.
For experiments using 10% serum, 2.7 μl of the requisite siRNA solution was added to a microfuge tube and 0.3 μl of either mouse (Sigma) or human (Sigma) serum was added. The solution was vortexed and spun down using a microcentrifuge and allowed to stand at room temperature.
Polyacrylamide gel electrophoresis (PAGE) analysis
Samples were analysed on a 20% PAGE-TBE gel (Invitrogen, Carlsbad, USA) and RNAs were detected using a SYBR Gold dye (Molecular Probes) with visualisation on a transilluminator.
MALDI-TOF mass spectrometry
MALDI-TOF mass spectra were recorded on a Voyager-DE™ PRO Biospectrometry™ Workstation (Applied Biosystems) using a flat sample, 100-well, laser-etched, stainless steel plate. Matrices used for preparing the samples were as follows: (a) a saturated solution of 2,4,6-trihydroxyacetophenone (Fluka) in 50 mg ml−1 diammonium hydrogen citrate (Fluka)–50% acetonitrile, (b) a 20 mg ml−1 solution of 2,4,6-trihydroxyacetophenone in acetonitrile diluted with an equal volume of 50 mg ml−1 diammonium hydrogen citrate, or (c) a 20 mg ml−1 solution of 2,4,6-trihydroxyacetophenone in acetonitrile diluted with an equal volume of 80 mg ml−1 diammonium hydrogen citrate. Matrix (a) was used for initial studies with conjugates and control (Fig. 2 and 3). Matrices (b) and (c) were used for all other studies interchangeably.
Sample preparation
Using matrix (a), a 0.1 μl sample to be analysed was taken and spotted on a well on the MALDI plate. Before this dried, 1 μl of matrix (a) was mixed with it by quickly withdrawing and expelling the solution 5 times with a pipette and allowing it to crystallise with the aid of air flow from a fan (in this case at the back of the MALDI machine). If allowed to crystallise on the bench, the quality of the MALDI acquisition was found to be diminished.
Using matrix (b) or (c), a 0.1 μl sample to be analysed was taken and spotted on a well on the MALDI plate. Before this dried, 1 μl matrix (b) or (c) was mixed with it by quickly withdrawing and expelling the solution 5 times with a pipette and allowing to crystallise. Alternatively, a 0.1 μl sample was taken and spotted on a well on the MALDI plate. Before this dried, 3 μl matrix (b) or (c) was mixed with it as above and then approximately 75% of the solution was immediately removed. The remainder of the solution was equally distributed to other wells, quickly withdrawing and expelling the solution 5 times with a pipette before removing the excess solution and applying it to another well. The solutions were allowed to crystallise on the bench before mass measurement.
Mass spectra acquisition
The parameters used for the mass acquisition were: accelerating voltage, 20000 V; grid voltage, 95%; guide wire, 0.01% or 0.05%; extraction delay time, 400 nsec; laser repetition rate, 20.0 Hz; low mass gate, 500 Da. MALDI spectra were acquired in 200 shot segments that were accumulated until background noise had receded to an acceptable level. Typically this was 1–5 segments. MALDI spectra were calibrated using the external calibration of parent siRNA controls. The MALDI spectra were often best obtained using the edges of the well (see the ESI†). All measurements were carried out in linear mode, since in reflector mode such high mass analytes do not give good spectra. Note that the accuracy of MALDI-TOF measurements in linear mode is quoted by the manufacturers of the instrumentation to be ±0.05% (i.e. 3–4 mass units for an siRNA strand of ca. 6000 Da) and that identification of a fragment is based on this allowance from the calculated values.
SiRNA activity studies
Lyophilised sense and antisense siRNA strands were resuspended at 400 μM in annealing buffer (10 mM Tris pH 7.5, 50 mM NaCl, 1 mM EDTA) and then mixed in equimolar amount (50 μl). The mixture was heated at 95 °C for 5 min using a PCR block, followed by slow cooling over 1 h. Mouse L929 cells were grown to 70% confluency in DMEM (Sigma) supplemented with 10% foetal calf serum (FCS), 1% penicillin–streptomycin–glutamine and 5 ml non-essential amino acids (Invitrogen). On the day of transfection, cells were trypsinised and resuspended at 2 × 105 in media. SiRNAs were mixed at 2 × final concentration with OptiMEM media and Lipofectamine2000 (Invitrogen), following manufacturer's procedures. After 20 min incubation at room temperature, the cell–lipid suspension was mixed 1 : 1 with the cell suspension to give a final serum concentration of 5%. The cells were transferred on to a 96-well plate in triplicate in a volume of 100 μl (1 × 104 cells per well) and left at 37 °C in an incubator for 24 h.
Following transfection, the supernatant was removed and RNA extracted using the RNeasy 96 kit (Qiagen). For the determination of p38 expression, Taqman primers and probes were designed using Primer Express version 2.0 (Perkin-Elmer). The Taqman probe was labelled with a reporter dye, FAM (6-carboxyfluorescein), at the 5′ end, and a fluorescent quencher dye TAMRA (6-carboxytetramethylrhodamine) at the 3′ end. Real-time PCR was carried out using the one-step Quanti-Tect RT-PCR Kit (Qiagen) in a MicroAmp 96-well Reaction Plate. Each well contained 25 ng total RNA, 300 nM (forward and reverse) primers and 125 nM Taqman probe in a reaction volume of 25 μl. All sample and non-template control reactions were carried out in the ABI Prism 7700 Sequence Detection System (Applied Biosystems) in triplicate. Comparative slopes of the relationship between log [RNA] and Ct (gradient = −3.3) for both the gene of interest and 18 S rRNA control indicated that the comparative Ct method could be used for the relative quantification of gene expression.
Supplementary Material
Acknowledgements
This work was supported in part by a grant from the EC Framework 5 (contract QLK3-CT-2002-01989). We thank Martin Fabani for helpful discussions.
Footnotes
Electronic supplementary information (ESI) available: Technical comments about obtaining good MALDI-TOF mass spectra, mass spectrum showing conversion of 2′,3′-cyclic phosphate into 2′- (3′-) phosphate during siRNA degradation by serum, PAGE analyses showing effect of 2′-O-methyl U substitution to stabilised siRNA, siRNA biological activity results, and mass spectrum of extended serum cleavage of Dh4 siRNA. See DOI: 10.1039/b611612d
References
- 1.Jones SW, de Souza PM, Lindsay MA. Curr. Opinion Pharmacol. 2004;4:522–527. doi: 10.1016/j.coph.2004.06.003. [DOI] [PubMed] [Google Scholar]; Gilmour IR, Fox SP, Hollins AJ, Sohail M, Akhtar S. J. Drug Targeting. 2004;12:315–340. doi: 10.1080/10611860400006257. [DOI] [PubMed] [Google Scholar]; Morris KV. BioTechniques. 2006;40:S7–S13. [Google Scholar]
- 2.Fabani MM, Turner JJ, Gait MJ. Curr. Opin. Mol. Ther. 2006;8:108–114. [PubMed] [Google Scholar]; Morris K, Rossi J. Curr. Opin. Mol. Ther. 2006;8:115–121. [PubMed] [Google Scholar]
- 3.Blackburn P, Moore S. In: The Enzymes. part B. Boyer PD, editor. XV. Academic Press; New York: 1982. pp. 317–433. [Google Scholar]
- 4.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]; Czauderna F, Fechtner M, Dames S, Aygün H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]; Elmén J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Ørum H, Koch T, Wahlestedt C. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]; Choung S, Kim YJ, Park H-O, Choi Y-C. Biochem. Biophys. Res. Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 5.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Swayze EE, Bhat B. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 6.Haupenthal J, Baehr C, Kiermayer S, Zeuzem S, Piiper A. Biochem. Pharmacol. 2006;71:702–710. doi: 10.1016/j.bcp.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 7.Turner JJ, Arzumanov AA, Gait MJ. Nucleic Acids Res. 2005;33:27–42. doi: 10.1093/nar/gki142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soutschek J, Akinc A, Bramlage B, Charisse K, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Röhl I, Toudjarska I, Wang W, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher H-P. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 9.Rose SD, Kim D-H, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, Rossi JJ, Behlke MA. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]; Beverly M, Hartsough K, Machemer L, Pavco P, Lockridge J. J. Chromatogr., B. 2006;835:62–70. doi: 10.1016/j.jchromb.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Manoharan M. Curr. Opin. Chem. Biol. 2004;8:1–10. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]; Nawrot B, Sipa K. Curr. Top. Med. Chem. 2006;6:913–925. doi: 10.2174/156802606777303658. [DOI] [PubMed] [Google Scholar]
- 11.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, Vaish N, Zinnen S, Vargeese C, Bowman K, Shaffer CS, Jeffs L, Judge A, MacLachlan I, Polisky B. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 12.Mestre B, Arzumanov A, Singh M, Boulmé F, Litvak S, Gait MJ. Biochim. Biophys. Acta. 1999;1445:86–98. doi: 10.1016/s0167-4781(99)00019-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.