Abstract
Urea transporters facilitate urea permeation across cell membranes in prokaryotes and eukaryotes. Bacteria use urea either as a means to survive in acidic environments and/or as a nitrogen source. The urea transporter ApUT from Actinobacillus pleuropneumoniae, the pathogen that causes porcine pleurisy and pneumonia, was expressed in E. coli and purified. Analysis of the recombinant protein using cross-linking and blue-native gel electrophoresis established that ApUT is a dimer in detergent solution. To determine the urea transport kinetics of ApUT, purified protein was reconstituted into proteoliposomes, and urea efflux was measured by stopped-flow fluorometry. The measured urea flux was saturable, could be inhibited by phloretin, and was not affected by pH. Two-dimensional crystals of the biologically active ApUT show that it is also dimeric in a lipid membrane and provide the first structural information on a member of the urea transporter family.
Keywords: Urea transporter, oligomeric state, ApUT, Channel, 2D crystallization
Introduction
Urea is the main catabolite in mammals, which excrete it as waste product in their urine and feces. In the mammalian kidney, urea plays an important role in urine formation. Meanwhile, the ubiquitous urea molecule represents an important nitrogen source for many microorganisms. Urea is thus transported in and out of a variety of cells. Although biological membranes are permeable to urea, a number of mechanisms have evolved that facilitate urea transport across membranes.
Besides porins 1, which form rather unspecific membrane pores, and secondary 2; 3 and ATP binding cassette transporters 4, which transport urea actively across membranes, there are three membrane protein families whose members facilitate urea transport in a channel-like manner. These are the urea/amide channel (UAC) family 5, the urea transporter (UT) family 6, and the aquaporin (AQP) family 7.
Channels of the AQP family conduct mainly water; but a subfamily, the aquaglyceroporins, are also permeable to small, uncharged solutes such as urea and glycerol 8.
The proteins of the UAC family facilitate transport of urea and short-chain aliphatic amides across membranes 9. Channels of the UAC family are only found in bacteria. The best-studied member of the UAC family is UreI from Helicobacter pylori, a 21.7-kDa protein with six putative transmembrane segments. Urea uptake through UreI and subsequent conversion of urea into NH3 and CO2 by urease allow H. pylori to buffer its periplasmic pH and hence enable it to survive in highly acidic environments such as the stomach 10.
The UT family is divided into three classes: the renal tubular type of urea transporters UT-A1-6, which in humans are expressed by alternative splicing of the Slc14A2 gene11; the erythrocyte urea transporter UT-B1, which in humans is encoded by the Slc14A1 gene12; and the bacterial urea transporters. All members of the UT family are found only in animals (only vertebrates) or pathogenic bacteria, with no representation in the other eukaryotic kingdoms or in the archaeal domain 13. Hydropathy plots predict 10 transmembrane segments for UT-A2-6, UT-B1 and bacterial UTs including ApUT, which range in molecular weight from 35 to 55 kDa. UT-A1, a ~110-kDa protein, has 20 predicted transmembrane segments, suggesting that it resulted from a gene duplication 13.
The role of bacterial UACs such as UreI from H. pylori has been established, but the role of UTs, which are expressed by bacteria residing in non-acidic environments, remains to be elucidated. All bacteria that express UTs are pathogens that first have to pass through the highly acidic environment of the stomach to reach their final destination in the intestine or other organs. It has therefore been proposed that the bacterial UTs may serve a similar function in these pathogenic bacteria as UreI in H. pylori 14.
Here, we report the structural and functional characterization of the urea transporter ApUT from Actinobacillus pleuropneumoniae, the pathogen that causes porcine pleurisy and pneumonia. Recombinant ApUT was purified and reconstituted into proteoliposomes, which were used to determine the urea transport kinetics of ApUT by stopped-flow fluorometry. The measured urea flux was saturable, could be inhibited by phloretin, and was not affected by pH. Several methods established that ApUT is a dimer in detergent solution, and two-dimensional crystals of the transporter showed that it also exists as a dimer when embedded in a lipid bilayer.
Results
Purification and oligomeric state of ApUT in detergent solution
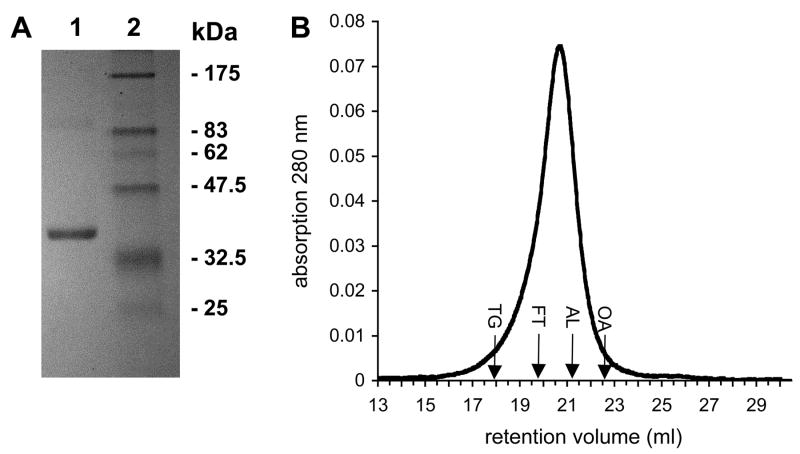
Several detergents were tested for solubilization and purification of recombinant ApUT. Dodecyl-ß,D-maltoside (DDM) proved to be the best detergent in keeping the protein stable in solution and was used for all further experiments. After Ni-NTA affinity and gel filtration chromatography, the protein was >95% pure as judged by SDS-PAGE and Coomassie blue staining (Fig. 1A). ApUT appeared as a strong band at ~34 kDa, close to its calculated molecular weight of ~36.7 kDa. The detergent-solubilized protein was analyzed by gel filtration chromatography on a Superose-12 column calibrated with soluble proteins. ApUT in DDM eluted from the column as a single peak with a retention volume of 20.7 ml (Fig. 1B), corresponding to a molecular mass of ~280 kDa. Mass spectrometry confirmed that the purified protein was indeed ApUT (data not shown).
Fig. 1. SDS-PAGE and gel filtration chromatography of purified ApUT.
(A) ApUT was purified in DDM by Nickel affinity and gel filtration chromatography and resolved on a 12.5% SDS-PAGE gel stained with Coomassie Brilliant Blue. Lane 1: 6 μg purified ApUT, lane 2: molecular weight markers. (B) Elution profile of ApUT in DDM (400 μl of 1 mg/ml) from a Superose-12 gel filtration column. Retention volumes of standard proteins of known molecular weight are indicated: TG, thyroglobulin (669 kDa); FT, ferritin (440 kDa); AL, aldolase (158 kDa); ovalbumin (43 kDa).
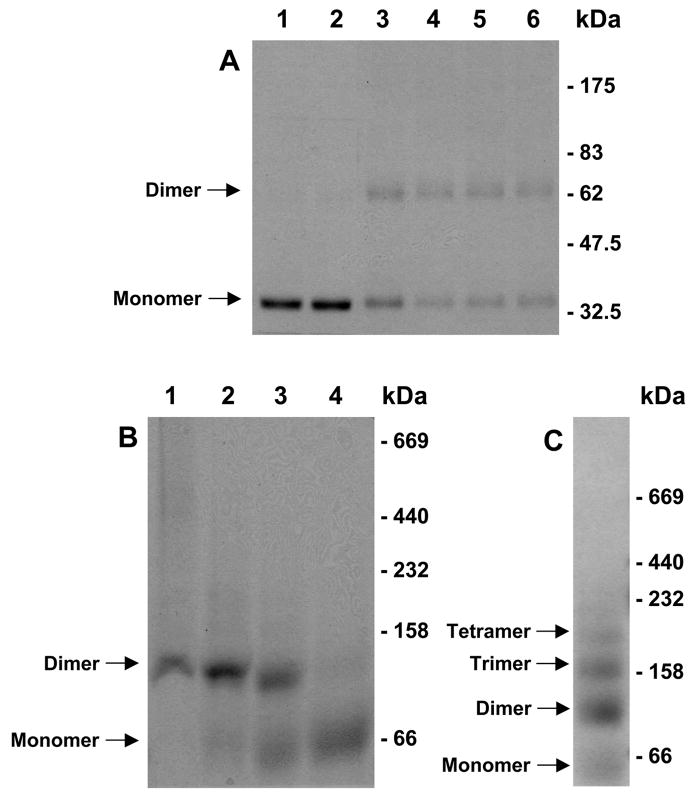
SDS-PAGE analysis of purified ApUT showed a strong monomer band at ~34 kDa (Fig. 1A and Fig. 2A, lane 1). To analyze the oligomeric state of ApUT in DDM, we performed cross-linking studies with glutardialdehyde. A glutardialdehyde concentration of 0.25 mM had little effect on the oligomeric state of ApUT as seen on the SDS-PAGE gel (Fig. 2A, lane 2), but at higher glutardialdehyde concentrations a second band appeared at ~65 kDa (Fig. 2A, lanes 3-6), indicating an ApUT dimer.
Fig. 2. Cross-linking and blue-native gel electrophoresis of ApUT.
(A) ApUT was incubated with glutardialdehyde in varying amounts for 30 min at room temperature. The cross-linking products were resolved on a 10% SDS-PAGE gel and detected by Coomassie blue staining. Lane 1: 6 μg ApUT without cross-linker, lane 2-6: 6 μg ApUT in the presence of 0.25, 1, 2.5, 5, and 25 mM glutardialdehyde. (B) 10 μg of purified ApUT in DDM was mixed with sample buffer and separated on a blue native gradient gel (lane 1). Increasing concentrations of SDS were added to the sample prior to loading to achieve a partial dissociation of the ApUT oligomers (lane 2: 0.05%, lane 3: 0.25%, lane 4: 1% SDS). (C) Oligomeric ladder of unspecific ApUT aggregates formed after denaturation with SDS and heating for 1 h at 40°C.
To confirm that the ApUT dimer seen on the SDS-PAGE gel after cross-linking was not an artifact and that the dimer is the highest oligomer formed by ApUT in DDM, we performed blue-native gel electrophoresis, which showed a strong band at ~120 kDa (Fig. 2B, lane 1). Incubation with SDS prior to blue-native gel electrophoresis caused the oligomer to dissociate, resulting in the appearance of a second band at ~60 kDa (Fig. 2B, lanes 2 and 3). At higher SDS concentrations, the ~120 kDa band disappeared and only the ~60 kDa band remained visible on the gel (Fig. 2B, lane 4). It has been shown that membrane proteins have a larger apparent mass on blue-native gels. To compensate for this fact, the apparent molecular masses can be divided by a factor of 1.8 15. The resulting values of ~66.7 kDa and 33.3 kDa are close to the expected values of an ApUT dimer (73.4 kDa) and monomer (36.7 kDa). To further verify the deduced oligomeric state of ApUT, an SDS-treated sample was heated for 1 h at 40°C to generate a ladder of unspecific ApUT aggregates (Fig. 2C). The lowest band in the ladder appeared again at ~60 kDa and the next band at ~120 kDa, confirming that the two bands seen on the blue-native gel correspond to the ApUT monomer and dimer.
Two-dimensional (2D) crystals of ApUT
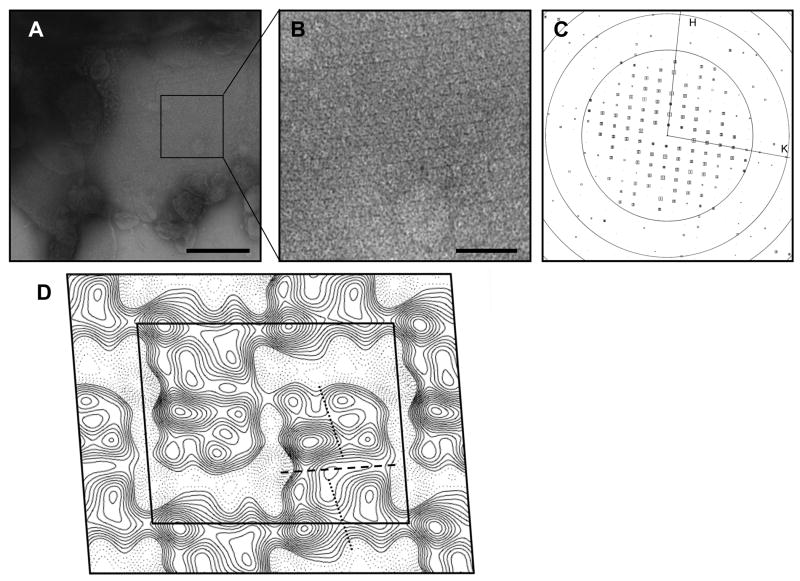
2D crystallization trials of ApUT were carried out in various buffer conditions and different lipid environments. The channel incorporated into membranes with all tested lipids, but 2D crystals only formed in DMPC and DOPC membranes at a lipid-to-protein ratio of 0.5 at a protein concentration of 1 mg/ml. ApUT formed crystalline sheets that typically grew out of clustered proteoliposomes and tended to form multilayered stacks. The salt concentration (50 – 500 mM NaCl) and pH (4-9) of the dialysis buffer were varied. The salt concentration did not affect 2D crystallization, but crystals only formed in a pH range of 6-7. Addition of divalent cations (Mg2+ or Ca2+) did not improve the crystal order or size. The crystals exhibited a tetragonal lattice (Fig. 3A - C) with unit cell dimensions of a = 177.1 Å, b = 140.0 Å and γ = 85.8°. The program ALLSPACE 16 indicated p121 symmetry, which features a screw axis along one of the lattice directions. Figure 3D shows a 24 Å projection map calculated from the best-ordered 2D crystal with p121 symmetry and a temperature factor (-150 Å2) applied. Each unit cell contains two rhomboidal densities of 12.0 nm × 6.7 nm. Each rhomboidal density in Figure 3D appears to consist of a top and a bottom half (indicated by the dashed line in Fig. 3D) of approximately the same size, separated by a region of lower density. These two halves should represent the two ApUT subunits in the dimer. Each monomer, in turn, appears to consist of a left and a right half (indicated by dotted lines in Fig. 3D).
Fig. 3. 2D crystals of ApUT.
(A) Negatively stained 2D crystal of ApUT. The scale bar is 200 nm. (B) Close-up view of the area marked in (A) clearly showing the tetragonal lattice. The scale bar is 50 nm. (C) Calculated Fourier transform of the crystal shown in (B) after unbending of the crystal lattice. The squares indicate the signal-to-noise ratio of the individual reflections; the larger the square and the lower the number, the better is the signal-to-noise ratio of the reflection. Concentric rings indicate the zero transitions of the contrast transfer function. (D) Projection map of a negatively stained ApUT crystal at 24 Å resolution. p121 symmetry and a temperature factor of -150 Å2 were applied. The black outline indicates a unit cell with dimensions of a = 177.1 Å, b = 140.0 Å and γ = 85.8°. The dashed line indicates the putative boundary between the two monomers in the crystallized ApUT dimer, and the two dotted lines indicate the two halves of the monomers.
Reconstituted ApUT exhibits specific urea transport
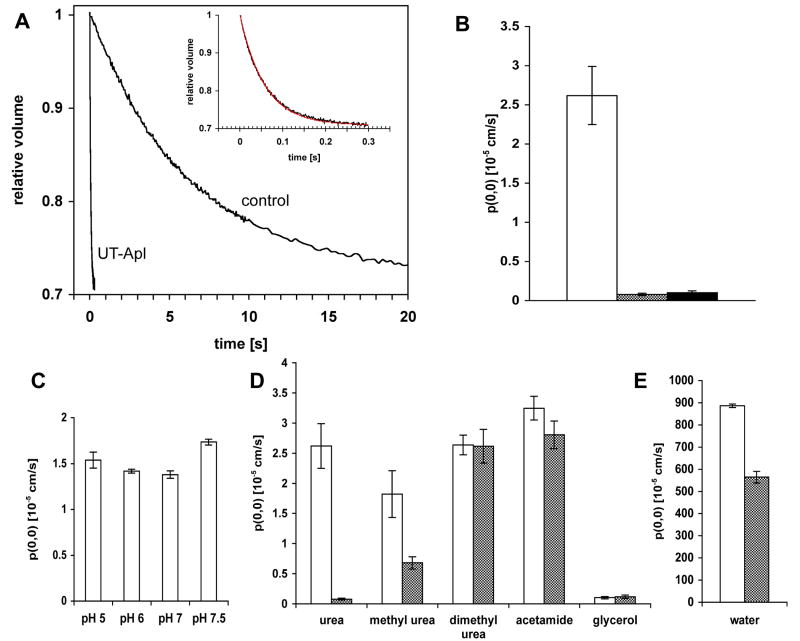
To determine whether the recombinant ApUT protein forms a functional urea channel, the purified protein was reconstituted into preformed E. coli polar lipid vesicles, and urea permeability was measured by stopped-flow fluorometry. Proteoliposomes and control vesicles, preloaded with 5,6-carboxyfluoresceine (CF) and urea, were rapidly mixed with urea-free isoosmotic buffer. Urea efflux and concomitant shrinkage of the vesicles was measured by CF self-quenching 17. The urea permeability coefficient of ApUT proteoliposomes was ~33 times higher (2.62 ± 0.37 × 10-5 cm/s) than that of the vesicles lacking the urea transporter (0.08 ± 0.015 × 10-5 cm/s) (Fig. 4A, B). ApUT mediated urea permeation was completely abolished when the reversible urea transport inhibitor phloretin was added to the bath solution (Fig. 4B). Urea permeability of ApUT was measured in a pH range from 5 to 7.5 and was found to be unaffected by the pH (Fig. 4C).
Fig. 4. Solute and water permeability of ApUT.
(A) Time trace (average of 8 traces) and nonlinear regression curve of urea efflux from proteoliposomes and control liposomes measured by fluorescence quenching. Liposomes were equilibrated in buffer (850 mosmol/kg) containing 500 mM urea. Liposomes were rapidly mixed with an equal volume of a solution with identical osmolality containing 250 mM urea. Inset shows a magnified tracing of urea efflux from the ApUT proteoliposomes. (B) The average urea permeability of vesicles containing ApUT is ~33-fold higher (Purea = 2.62 × 10-5 cm/s, white) than that of control vesicles (Purea = 0.08 × 10-5 cm/s, grey). Urea transport is abolished (Purea = 0.1 × 10-5 cm/s, black) in the presence of 2 mM phloretin. Each measurement was repeated at least three times. Error bars indicate the standard deviation. (C) Urea permeability under different pH conditions, showing no effect of the pH on urea transport by ApUT. The volume change of the liposomes was measured by light scattering. (D) Coefficients of solute permeabilities and (E) osmotic water permeability of proteoliposomes containing ApUT (white bars) and control vesicles (grey bars). Each measurement was repeated at least three times. Error bars indicate the standard deviation.
To test the specificity of the transporter for urea, we measured permeabilities of several urea analogs, methyl urea: 1,2-dimethyl urea, and acetamide, as well as glycerol (Fig. 4D). 1,2-dimethyl urea, acetamide and glycerol permeation was not facilitated by ApUT. Methyl urea permeability, however, was ~2.7 times higher in comparison to control vesicles. This result suggests that methyl urea may permeate the ApUT pore, although with lower efficiency than urea.
Water permeability of ApUT was measured by monitoring the volume change of proteoliposomes and control vesicles subjected to a doubled external osmotic pressure. The osmotic water permeability coefficient of proteoliposomes (0.90 ± 0.0076 × 10-2 cm/s) was ~1.6 times higher than that of control vesicles (0.56 ± 0.027 × 10-2 cm/s), indicating that ApUT is permeable to water (Fig. 4E). The water permeability was also inhibited by phloretin (data not shown).
Urea concentration dependence of ApUT mediated urea permeation
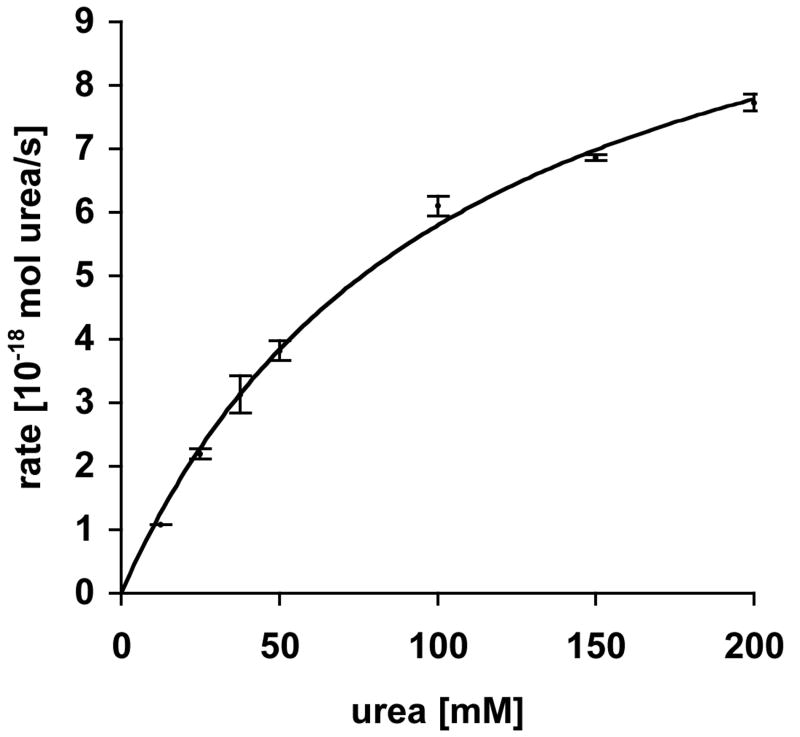
To determine the half-saturation concentration for ApUT-mediated urea permeation (K1/2), we measured urea permeability of liposomes at different urea concentrations (Fig. 5A). The efflux of urea increased with increasing urea concentration. At higher substrate concentrations the rate of urea efflux reached saturation and showed typical Michaelis-Menten kinetics. A nonlinear regression analysis resulted in a K1/2 value of 104.6 ± 9.96 mM and a Vmax of 11.85 ± 0.55 × 10-18 mol urea/(s * vesicle).
Fig. 5. Kinetics of urea permeation through ApUT.
Concentration dependence of urea efflux. The nonlinear regression analysis gave a half-saturation constant K1/2 of 104.6 ± 9.96 mM and a Vmax of 11.85 ± 0.55 × 10-18 mol urea/(s*vesicle). Each measurement was repeated at least three times. Error bars indicate the standard deviation.
Discussion
Experimental determination of the molecular mass of a membrane protein oligomer is not as straightforward as it is for a soluble protein, because membrane proteins are either embedded in a lipid bilayer or otherwise surrounded by a detergent micelle. Blue-native gel electrophoresis and cross-linking are two methods that have previously been used to determine the oligomeric state of detergent-solubilized membrane proteins 15; 18; 19; 20; 21. Here, we used both methods to show that the prokaryotic urea transporter ApUT is a dimer in detergent solution. While cross-linking stabilized some, but not all, of the ApUT dimers (Fig. 2A), presumably due to incomplete cross-linking, blue-native gel electrophoresis in the absence of SDS showed that virtually all the protein exists as a dimer in DDM (Fig. 2B, lane 1). Gel filtration chromatography of DDM-solubilized ApUT also showed a single elution peak, presumably representing the dimer (Fig. 1B). A projection map at 24 Å resolution revealed that a unit cell consists of two densities indicating that the crystalline array is formed by ApUT dimers. The dimensions of the projection of the ApUT dimer corresponds well to those of projections of other dimeric membrane proteins with similar numbers of transmembrane helices, such as the Na+/H+ transporter from E. coli 22 and the bacterial ClC chloride channel 23. Since a 2D crystal, which contains the membrane protein in a lipid bilayer, is a good representation of the in vivo situation, the dimer may indeed be the physiological unit of ApUT.
Even if ApUT exists as a dimer in the membrane, this observation does not imply that oligomerization is required for urea transport. Although the monomers are the functional units, dimeric association has been reported, for example, for the bacterial ClC chloride channel 23 and the bacterial protein translocation complex SecYEG 24. Similarly, aquaporins are only stable as tetramers, although each subunit forms an independent water pore 25. In contrast, oligomerization is essential for K+ channels to form a central ion conduction channel 26. The question thus arises whether dimerization is required for ApUT to form a urea conducting channel or whether each subunit forms an independent channel. Because of the limited resolution, our projection map does not provide an answer to this question. However, hydropathy analysis suggests that the sequences of the N- and C-terminal halves of UTs are related, with each half consisting of five membrane-spanning domains and the two halves being connected by a hydrophilic extracellular loop 27. It has been suggested that this topology arose from a gene duplication of a five transmembrane domain segment early in the evolutionary history of these genes 13. The largest urea transporter, UT-A1, has 20 transmembrane segments and may thus have evolved by an additional gene duplication 28. This situation is reminiscent of some ion channels. Ca2+ and Na+ channels are monomers containing four internal repeats, and are thought to have evolved from K+ channels, which form homo-tetrameric channels, through two gene duplications 29. It is therefore tempting to speculate that analogous to ion channels, UTs may contain a central urea channel that is either formed by the four repeats in UT-A1 or, in the case of all other UTs, by two monomers coming together and each contributing half of the channel. Mutational and further structural studies will be needed, however, to resolve the question of whether UTs must oligomerize to form a channel or whether each subunit in the oligomer forms an independent urea pathway.
The urea efflux observed by stopped-flow measurements demonstrates that recombinant ApUT forms functional channels upon reconstitution into liposomes. Urea transport was abolished by phloretin, proving that the transporter is specific for urea with a small permeability for water. It was found that urease activity is needed to establish A. pleuropneumoniae infection in the respiratory tract of pigs 30, suggesting that it may use a similar survival mechanism as that of H. pylori. The urea channel of H. pylori, UreI, is pH-gated and opens at low pH (below ~6.5). Urea permeates through the channel and is converted into NH3 and CO2 by urease inside the bacteria. Ammonia diffuses to the periplasm and therefore buffers this space. However, unlike UreI from H. pylori, which belongs to the urea/amide channel family and is thus distinct from the UTs, our data show that urea transport mediated by ApUT does not change in the pH range of pH 5 – 7.5. This characteristic has also been found for urea transport mediated by other prokaryotic and eukaryotic UTs 31; 32 and thus seems to be conserved in the UT family. Nevertheless, the UT from Y. pseudotuberculosis, Yut, can substitute the function of UreI, as the urea uptake ability is restored in a UreI-deficient H. pylori strain transcomplemented with yut 14.
Glycerol and urea analogs such as 1,2-dimethyl urea and acetamide did not permeate ApUT. Methyl urea permeability, however, was ~2.7 times higher compared to control vesicles, and ApUT is also permeable to water. Results from other studies performed on eukaryotic UTs differ. Transport studies on purified plasma membranes from Xenopus laevis oocytes expressing UT-A2 and UT-A3 revealed that these transporters neither permeate any urea analogs, including methyl urea, nor water 33. UT-B expressed in erythrocytes, however, showed high permeabilities for acetamide and methyl urea 34 as well as water 35. ApUT, which shares 26% and 21% identical and 44% and 45% similar residues with UT-A2 and UT-B, respectively, seems to be more specific than UT-B but less specific than UT-As. The permeability of urea analogs through ApUT decreases with the molecular size of the compound (urea < methyl urea < 1.2-dimethyl urea). Since acetamide, with a size comparable to that of urea, is not transported by ApUT, the presence of two amide groups seems to be another selection criterion of the transporter. It is surprising that UT-As are impermeable to water, because the high polarity and water solubility of urea suggests that a polar, hydrophilic conduit would best facilitate transmembrane urea transport,through which one would expect some water permeability. Phloretin also inhibited water transport by ApUT (data not shown), suggesting that both urea and water permeate through the same channel.
The ApUT-mediated efflux of urea increased with increasing urea concentration. At higher urea concentrations, the efflux rate reached saturation and showed typical Michaelis-Menten kinetics. We measured a half-activation constant K1/2 of ~110 mM, which is in the range of K1/2 values reported for UT-B (K1/2= 396 mM 32 and 218 mM 36). For UT-A and Yut, however, a saturable urea flux was not observed, even at urea concentrations as high as 1 M 6; 14; 33. A K1/2 could therefore not be determined for UT-A. This discrepancy between members of the UT family may be related to the systems used to measure the urea efflux rates. UT-B kinetics were measured in erythrocytes, whereas UT-A-mediated efflux was measured in frog oocytes. Since we used purified protein reconstituted into liposomes to characterize urea transport by ApUT, our measurements should not be affected by other membrane proteins present in membranes purified from erythrocytes or in Xenopus oocytes.
In summary, our results show that ApUT from A. pleuropneumoniae is a dimer in detergent solution and in the membrane and that it forms a urea specific channel with some permeability for water. The urea flux can be saturated, is inhibited by phloretin, and is independent of pH. Our projection map of ApUT provides the first structural information on UTs. A high-resolution structure will be required, however, to fully understand the mechanism of UT-mediated urea transport.
Material and Methods
Cloning and cell culture
The gene encoding the urea transporter was amplified by PCR using genomic DNA from A. pleuropneunomiae (ATCC, catalog no. 27088D) as a template. The gene was cloned into the pET-15b plasmid (Novagen) with a 6xHis-tag fused to the N terminus. The recombinant plasmid was sequenced, and the inserted gene was found to be identical to the published sequence 31. ApUT was expressed in E. coli strain C-43. 12 l of 2x YT medium were inoculated at 37°C with an overnight culture, followed by induction with 0.5 mM isopropyl-ß,D-1-thiogalactopyranoside (IPTG) at an OD600 of 0.8. Cells were grown at 25°C and harvested 2-3 h after induction.
Protein purification
Cells were resuspended in homogenization buffer (10 mM Tris-HCl (pH 7.5), 150 mM NaCl and 1 mM PMSF) and broken by one passage through a microfluidizer operated at 1.5 kbar. Unbroken cells were removed by centrifugation at 6,500g for 15 min at 4°C, and the membrane fraction was collected by centrifugation at 150,000g for 90 min at 4°C. Membranes were resuspended in homogenization buffer and stored at −80°C until further use. Membranes were solubilized in buffer A (1% (w/v) dodecyl-ß,D-maltoside (DDM) (Anatrace) in 50 mM Tris-HCl (pH 7.5), 300 mM NaCl). After stirring for 1 h at 4 C, the solution was clarified by centrifugation at 150,000g for 25 min at 4°C and incubated with Ni2+-NTA resin (Qiagen) (1 ml per 100 mg of membranes) for 1 h at 4°C. The resin was poured into a column and washed with 10 column volumes of buffer B (0.1% DDM in 20 mM Tris-HCl (pH 7.5), 500 mM NaCl, 30 mM imidazole). The protein was eluted in buffer C (0.05% DDM in 20 mM Tris-HCl (pH 7.5), 300 mM NaCl, 300 mM imidazole). Peak fractions were combined and further purified by size-exclusion chromatography using a Superose-12 column (GE Healthcare) equilibrated with buffer C. Typically, up to 0.7 mg of pure ApUT could be purified from one liter of E. coli cell culture.
Cross-linking and blue-native gel electrophoresis
Prior to cross-linking experiments, the protein was dialyzed against 0.05% DDM in 25 mM NaPi (pH 7.4), 300 mM NaCl. Glutardialdehyde was added to final concentrations ranging from 1.5 to 10 mM. After a 30-min incubation at room temperature, the reactions were quenched by addition of 200 mM Tris-HCl (pH 7.4). The cross-linked samples were analyzed on a Coomassie-stained polyacrylamide-SDS gel.
For blue native gel electrophoresis 37 linear 4-16% Bis-Tris NativePAGE gels (Invitrogen) were used. Protein samples were supplemented with a 10-fold concentrated loading dye (5% Coomassie Brilliant Blue, 100 mM Bis-Tris (pH 7.0), 500 mM 6-amino-n-caproic acid).
Two-dimensional (2D) crystallization
Purified ApUT was diluted to 1 mg/ml with 0.05% DDM in 20 mM Tris-HCl (pH 7.5), 300 mM NaCl. 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC), 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC), 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), and E. coli polar lipids (all from Avanti) were each dissolved at 10 mg/ml in 1.5%–5% (w/v) decyl-ß,D-maltoside (DM) and mixed with protein at lipid-to-protein ratios of 0.1 – 1.5 (w/w). After an overnight incubation on ice, the mixtures were dialyzed at 30°C against 50 mM MES (pH 6), 200 mM NaCl, 3 mM NaN3 for 7 days with buffer exchanges every other day.
Electron microscopy and image processing
2D crystals were negatively stained with uranyl formate as described 38. Electron micrographs were recorded with an FEI Tecnai T12 electron microscope at an acceleration voltage of 120 kV. Images were taken on imaging plates at a magnification of 67,000x and a defocus of −1.5 μm using low-dose conditions. Imaging plates were read out with a DITABIS micron imaging plate scanner using a step size of 15 μm, a gain setting of 20,000, and a laser power setting of 30%. 2 × 2 pixels were averaged, yielding a pixel size of 4.5 Å on the specimen level. Selected images were processed using the program 2dx 39, which is based on the MRC image processing programs 40.
Reconstitution of purified ApUT into liposomes
Liposomes were prepared as described previously 19. 50 mg of E. coli polar lipids (Avanti) were resuspended in 1 ml of dialysis buffer (50 mM Tris-HCl (pH 7.5), 0.9% (w/v) NaCl). A 20-ml aliquot of purified protein solution (1.4 mg/ml) was mixed with 112 ml of liposomes (50 mg/ml) at a lipid-to-protein ratio of 200 (w/w), 10 ml of 20% (w/v) sodium cholate and 17 ml of 5 M NaCl, in a final volume of 205 ml. The mixture was incubated on ice for 10 minutes, followed by a passage through a 1-ml Sephadex G-50 (fine) column equilibrated with dialysis buffer.
Solute and water permeability measurements
Liposomes were incubated overnight in dialysis buffer containing 15 mM 5,6-carboxyfluoresceine (CF). External CF was removed by passing the liposomes through a PD-10 desalting sephadex column (GE). Osmotic water permeability (Pf) was measured at 25°C as described earlier 41; 42; 43. Briefly, liposomes were abruptly exposed to a doubling of external osmolarity in a stopped-flow fluorometer (SF.17 MV, Applied Photophysics, Leatherhead, United Kingdom), which has a measurement dead time of less than 2 ms. The rate of water efflux from the liposomes was measured as a decrease of CF fluorescence due to self-quenching of the fluorophore. Data from 8-10 measurements were averaged and fit to a single exponential curve. A family of single exponential curves was generated by simulation of the water permeability equation, in which only the Pf value was varied using the MathCad software (MathSoft Inc., Cambridge, MA) as described earlier 44.
The permeability for urea and urea analogs was measured by pre-equilibrating the vesicles at a solute concentration of 300 mM in buffer (650 mosmol/kg) for 30 min and then rapidly diluting the vesicle suspension two-fold in the stopped-flow device to create an isoosmotic solute gradient 45. An appropriate amount of NaCl was added to maintain the isoosmotic conditions upon dilution. Vesicle shrinkage due to solute efflux was measured by recording CF quenching over time. Since CF is sensitive to pH, for measurements of pH dependence of solute efflux, light scattering at 600 nm was used to measure vesicle shrinkage.
Osmolalities of all solutions were confirmed and, if necessary, adjusted by measuring freezing point depression on a Precision Instruments Osmette A osmometer. Vesicle sizes were determined by using quasi-elastic light scattering with a DynaPro particle sizer. Permeability coefficients were calculated using the Mathcad software as described earlier 45; 46. Urea transport inhibitor studies were performed by incubating the vesicles for 30 min in buffer containing 2 mM phloretin. Kinetic studies of ApUT were performed by incubating the proteoliposomes in urea solutions ranging in concentration from 25 mM to 500 mM. The urea flux per average vesicle was calculated based on Eq. (1):
| (1) |
where Jurea is the rate of uptake in mol/s, SA is the surface area of a vesicle in cm2, and [urea] is the urea concentration in mol/cm3. Purea was determined experimentally. The surface area of a single average vesicle was determined from the diameter measured by quasi-elastic light scattering as described above.
Acknowledgments
This work was supported by NIH grant RO1 GM082927 (to T.W.) and DK43955 (to M.L.Z. and J.C.M). The molecular EM facility at Harvard Medical School was established with a generous donation from the Giovanni Armenise Harvard Center for Structural Biology and is maintained with funds from NIH Grant PO1 GM62580 (to Stephen C. Harrison). T.W. is an investigator of the Howard Hughes Medical Institute. S.R. was a fellow of the German Academy of Sciences Leopoldina (BMBF-LPD 9901/8-163).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mills J, Wyborn NR, Greenwood JA, Williams SG, Jones CW. An outer-membrane porin inducible by short-chain amides and urea in the methylotrophic bacterium Methylophilus methylotrophus. Microbiology. 1997;143(Pt 7):2373–9. doi: 10.1099/00221287-143-7-2373. [DOI] [PubMed] [Google Scholar]
- 2.ElBerry HM, Majumdar ML, Cunningham TS, Sumrada RA, Cooper TG. Regulation of the urea active transporter gene (DUR3) in Saccharomyces cerevisiae. J Bacteriol. 1993;175:4688–98. doi: 10.1128/jb.175.15.4688-4698.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacAulay N, Gether U, Klaeke DA, Zeuthen T. Passive water and urea permeability of a human Na(+)-glutamate cotransporter expressed in Xenopus oocytes. J Physiol. 2002;542:817–28. doi: 10.1113/jphysiol.2002.020586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valladares A, Montesinos ML, Herrero A, Flores E. An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol Microbiol. 2002;43:703–15. doi: 10.1046/j.1365-2958.2002.02778.x. [DOI] [PubMed] [Google Scholar]
- 5.Weeks DL, Eskandari S, Scott DR, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–5. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- 6.You G, Smith CP, Kanai Y, Lee WS, Stelzner M, Hediger MA. Cloning and characterization of the vasopressin-regulated urea transporter. Nature. 1993;365:844–7. doi: 10.1038/365844a0. [DOI] [PubMed] [Google Scholar]
- 7.Borgnia M, Nielsen S, Engel A, Agre P. Cellular and molecular biology of the aquaporin water channels. Annu Rev Biochem. 1999;68:425–58. doi: 10.1146/annurev.biochem.68.1.425. [DOI] [PubMed] [Google Scholar]
- 8.Rojek A, Praetorius J, Frokiaer J, Nielsen S, Fenton RA. A current view of the mammalian aquaglyceroporins. Annu Rev Physiol. 2008;70:301–27. doi: 10.1146/annurev.physiol.70.113006.100452. [DOI] [PubMed] [Google Scholar]
- 9.Saier MH., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Athmann C, Zeng N, Kang T, Marcus EA, Scott DR, Rektorschek M, Buhmann A, Melchers K, Sachs G. Local pH elevation mediated by the intrabacterial urease of Helicobacter pylori cocultured with gastric cells. J Clin Invest. 2000;106:339–47. doi: 10.1172/JCI9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith CP, Fenton RA. Genomic organization of the mammalian SLC14a2 urea transporter genes. J Membr Biol. 2006;212:109–17. doi: 10.1007/s00232-006-0870-z. [DOI] [PubMed] [Google Scholar]
- 12.Dobson JG, Jr, Shea LG, Fenton RA. Adenosine A2A and beta-adrenergic calcium transient and contractile responses in rat ventricular myocytes. Am J Physiol Heart Circ Physiol. 2008;295:H2364–72. doi: 10.1152/ajpheart.00927.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minocha R, Studley K, Saier MH., Jr The urea transporter (UT) family: bioinformatic analyses leading to structural, functional, and evolutionary predictions. Receptors Channels. 2003;9:345–52. doi: 10.3109/714041015. [DOI] [PubMed] [Google Scholar]
- 14.Sebbane F, Bury-Mone S, Cailliau K, Browaeys-Poly E, De Reuse H, Simonet M. The Yersinia pseudotuberculosis Yut protein, a new type of urea transporter homologous to eukaryotic channels and functionally interchangeable in vitro with the Helicobacter pylori UreI protein. Mol Microbiol. 2002;45:1165–74. doi: 10.1046/j.1365-2958.2002.03096.x. [DOI] [PubMed] [Google Scholar]
- 15.Heuberger EH, Veenhoff LM, Duurkens RH, Friesen RH, Poolman B. Oligomeric state of membrane transport proteins analyzed with blue native electrophoresis and analytical ultracentrifugation. J Mol Biol. 2002;317:591–600. doi: 10.1006/jmbi.2002.5416. [DOI] [PubMed] [Google Scholar]
- 16.Valpuesta JM, Carrascosa JL, Henderson R. Analysis of electron microscope images and electron diffraction patterns of thin crystals of phi 29 connectors in ice. J Mol Biol. 1994;240:281–7. doi: 10.1006/jmbi.1994.1445. [DOI] [PubMed] [Google Scholar]
- 17.Zeidel ML, Ambudkar SV, Smith BL, Agre P. Reconstitution of functional water channels in liposomes containing purified red cell CHIP28 protein. Biochemistry. 1992;31:7436–40. doi: 10.1021/bi00148a002. [DOI] [PubMed] [Google Scholar]
- 18.Raunser S, Appel M, Ganea C, Geldmacher-Kaufer U, Fendler K, Kühlbrandt W. Structure and Function of Prokaryotic Glutamate Transporters from Escherichia coli and Pyrococcus horikoshii. Biochemistry. 2006;45:12769–12805. doi: 10.1021/bi061008+. [DOI] [PubMed] [Google Scholar]
- 19.Raunser S, Haase W, Bostina M, Parcej DN, Kuhlbrandt W. High-yield expression, reconstitution and structure of the recombinant, fully functional glutamate transporter GLT-1 from Rattus norvegicus. J Mol Biol. 2005;351:598–613. doi: 10.1016/j.jmb.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 20.Vinothkumar KR, Raunser S, Jung H, Kuhlbrandt W. Oligomeric structure of the carnitine transporter CaiT from Escherichia coli. J Biol Chem. 2006;281:4795–801. doi: 10.1074/jbc.M508993200. [DOI] [PubMed] [Google Scholar]
- 21.Bessonneau P, Besson V, Collinson I, Duong F. The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. Embo J. 2002;21:995–1003. doi: 10.1093/emboj/21.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams KA, Geldmacher-Kaufer U, Padan E, Schuldiner S, Kühlbrandt W. Projection structure of NhaA, a secondary transporter from Escherichia coli, at 4.0 A resolution. EMBO Journal. 1999;18:3558–63. doi: 10.1093/emboj/18.13.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R. X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature. 2002;415:287–94. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 24.Breyton C, Haase W, Rapoport TA, Kuhlbrandt W, Collinson I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature. 2002;418:662–5. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- 25.Gonen T, Walz T. The structure of aquaporins. Q Rev Biophys. 2006;39:361–96. doi: 10.1017/S0033583506004458. [DOI] [PubMed] [Google Scholar]
- 26.MacKinnon R. Potassium channels. FEBS Lett. 2003;555:62–5. doi: 10.1016/s0014-5793(03)01104-9. [DOI] [PubMed] [Google Scholar]
- 27.Shayakul C, Hediger MA. The SLC14 gene family of urea transporters. Pflugers Arch. 2004;447:603–9. doi: 10.1007/s00424-003-1124-x. [DOI] [PubMed] [Google Scholar]
- 28.Fenton RA, Stewart GS, Carpenter B, Howorth A, Potter EA, Cooper GJ, Smith CP. Characterization of mouse urea transporters UT-A1 and UT-A2. Am J Physiol Renal Physiol. 2002;283:F817–25. doi: 10.1152/ajprenal.00263.2001. [DOI] [PubMed] [Google Scholar]
- 29.Miller C. An overview of the potassium channel family. Genome Biol. 2000;1:REVIEWS0004. doi: 10.1186/gb-2000-1-4-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bosse JT, MacInnes JI. Urease activity may contribute to the ability of Actinobacillus pleuropneumoniae to establish infection. Can J Vet Res. 2000;64:145–50. [PMC free article] [PubMed] [Google Scholar]
- 31.Bosse JT, Gilmour HD, MacInnes JI. Novel genes affecting urease acivity in Actinobacillus pleuropneumoniae. J Bacteriol. 2001;183:1242–7. doi: 10.1128/JB.183.4.1242-1247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brahm J. Urea permeability of human red cells. J Gen Physiol. 1983;82:1–23. doi: 10.1085/jgp.82.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maciver B, Smith CP, Hill WG, Zeidel ML. Functional characterization of mouse urea transporters UT-A2 and UT-A3 expressed in purified Xenopus laevis oocyte plasma membranes. Am J Physiol Renal Physiol. 2008;294:F956–64. doi: 10.1152/ajprenal.00229.2007. [DOI] [PubMed] [Google Scholar]
- 34.Zhao D, Sonawane ND, Levin MH, Yang B. Comparative transport efficiencies of urea analogues through urea transporter UT-B. Biochim Biophys Acta. 2007;1768:1815–21. doi: 10.1016/j.bbamem.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Yang B, Verkman AS. Analysis of double knockout mice lacking aquaporin-1 and urea transporter UT-B. Evidence for UT-B-facilitated water transport in erythrocytes. J Biol Chem. 2002;277:36782–6. doi: 10.1074/jbc.M206948200. [DOI] [PubMed] [Google Scholar]
- 36.Mayrand RR, Levitt DG. Urea and ethylene glycol-facilitated transport systems in the human red cell membrane. Saturation, competition, and asymmetry. J Gen Physiol. 1983;81:221–37. doi: 10.1085/jgp.81.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–31. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 38.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gipson B, Zeng X, Zhang ZY, Stahlberg H. 2dx--user-friendly image processing for 2D crystals. J Struct Biol. 2007;157:64–72. doi: 10.1016/j.jsb.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 40.Crowther RA, Henderson R, Smith JM. MRC image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 41.Lande MB, Donovan JM, Zeidel ML. The relationship between membrane fluidity and permeabilities to water, solutes, ammonia, and protons. J Gen Physiol. 1995;106:67–84. doi: 10.1085/jgp.106.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Negrete HO, Rivers RL, Goughs AH, Colombini M, Zeidel ML. Individual leaflets of a membrane bilayer can independently regulate permeability. J Biol Chem. 1996;271:11627–30. doi: 10.1074/jbc.271.20.11627. [DOI] [PubMed] [Google Scholar]
- 43.Rivers RL, Dean RM, Chandy G, Hall JE, Roberts DM, Zeidel ML. Functional analysis of nodulin 26, an aquaporin in soybean root nodule symbiosomes. J Biol Chem. 1997;272:16256–61. doi: 10.1074/jbc.272.26.16256. [DOI] [PubMed] [Google Scholar]
- 44.Grossman EB, Harris HW, Jr, Star RA, Zeidel ML. Water and nonelectrolyte permeabilities of apical membranes of toad urinary bladder granular cells. Am J Physiol. 1992;262:C1109–18. doi: 10.1152/ajpcell.1992.262.5.C1109. [DOI] [PubMed] [Google Scholar]
- 45.Mathai JC, Sprott GD, Zeidel ML. Molecular mechanisms of water and solute transport across archaebacterial lipid membranes. J Biol Chem. 2001;276:27266–71. doi: 10.1074/jbc.M103265200. [DOI] [PubMed] [Google Scholar]
- 46.Mathai JC, Mori S, Smith BL, Preston GM, Mohandas N, Collins M, van Zijl PC, Zeidel ML, Agre P. Functional analysis of aquaporin-1 deficient red cells. The Colton-null phenotype. In J Biol Chem. 1996;271:1309–13. doi: 10.1074/jbc.271.3.1309. [DOI] [PubMed] [Google Scholar]