Abstract
Chemical cross-linking is a potentially useful technique for probing the architecture of multiprotein complexes. However, analyses using typical bifunctional cross-linkers often suffer from poor yields, and large-scale modification of nucleophilic side chains can result in artifactual results attributable to structural destabilization. We report here the de novo design and development of a type of protein cross-linking reaction that uses a photogenerated oxidant to mediate rapid and efficient cross-linking of associated proteins. The process involves brief photolysis of tris-bipyridylruthenium(II) dication with visible light in the presence of the electron acceptor ammonium persulfate and the proteins of interest. Very high yields of cross-linked products can be obtained with irradiation times of <1 second. This chemistry obviates many of the problems associated with standard cross-linking reagents.
Large multiprotein complexes mediate most important biological processes. A central question in the study of these “protein machines” (ref. 1, p. 291) is to elucidate the network of protein–protein interactions and, ideally, to ascertain how this architecture might change during the course of the catalytic cycle in question. Chemical cross-linking (2) (3) (4) is a potentially useful technique for this purpose. However, cross-linkers are most often applied to the analysis of interactions between a few proteins (5–7); successful analysis of contacts in large complexes is more difficult (8–10). This is largely because typical bifunctional cross-linkers, comprised of two electrophiles connected by a linker arm, have many drawbacks with regard to applications to complex systems. They are constitutively reactive and cannot be “triggered” at a desired time. Poor yields are often obtained even with long incubation times. Even more seriously, large-scale modification of nucleophilic side chains, such as the acylation of lysines, on the surface of the proteins during the extended incubation times required for typical reagents raises the concern of artifactual results attributable to structural destabilization. The purpose of this study was to design a photo-activatable reagent that would cross-link closely associated proteins very rapidly and in high yield. If cross-linking were very rapid, a photo-activated reagent might be useful for probing the dynamics of protein–protein interactions in a particular complex as it proceeds through a catalytic cycle. This kind of application is beyond the reach of current cross-linking technology. In particular, a visible light-triggered reaction would be useful for probing protein–protein interactions in living cells or in crude extracts because cells contain few visible chromophores but many molecules that absorb UV light.
MATERIALS AND METHODS
Proteins.
UvsY protein (11), the 180-aa C-terminal domain of yeast TATA box-binding protein (TBP) (12), Gal80 protein (13), the glutathione S-transferase (GST)–Gal4 activation domain fusion protein (14), and the radiolabeled polypeptide containing the Gal4 activation domain (15) were purified as described in the literature. Antibody raised against yeast TBP was a kind gift from Stephen A. Johnston (University of Texas Southwestern).
General Cross-Linking Protocol.
Cross-linking reactions were carried out in a total volume of 20 μl in a buffer comprised of 15 mM sodium phosphate (pH 7.5), 150 mM NaCl, and 0.125 mM Ru(bpy)3Cl2 (Aldrich). Easily oxidized buffer components such as β-mercaptoethanol and DTT should be avoided. Protein concentrations varied depending on the experiment between 0.01 and 20 μM. The solution was placed in a 1.7-ml Eppendorf tube positioned parallel to the beam of light at a distance of 50 cm from a 150-W xenon arc lamp (Oriel, Stamford, CT). Ammonium persulfate (APS) was added to 2.5 mM just before irradiation. Light was filtered first through 10 cm of distilled water and then through a 380- to 2,500-nm cut-on filter (Oriel 49470). Exposure time was controlled by shining the light through timed shutters of a single lens reflex camera with the lens and back cover removed from the camera body. In most experiments, the sample was irradiated for 0.5 seconds. Immediately after irradiation, samples were quenched with 7 μl of 4× gel loading buffer (0.2 M Tris/8% SDS/2.88 M β-mercaptoethanol/40% glycerol/0.4% xylene cyanol/0.4% bromophenol blue), were heated to 95°C for 5 min, and then were separated by electrophoresis through a 10% tricine SDS polyacrylamide gel. The reducing conditions used preclude the observation of products linked by disulfide bonds. Proteins were visualized by staining with Coomassie brilliant blue or by Western blot analysis.
RESULTS AND DISCUSSION
Design of a Visible Light-Initiated Protein Cross-Linking Reaction.
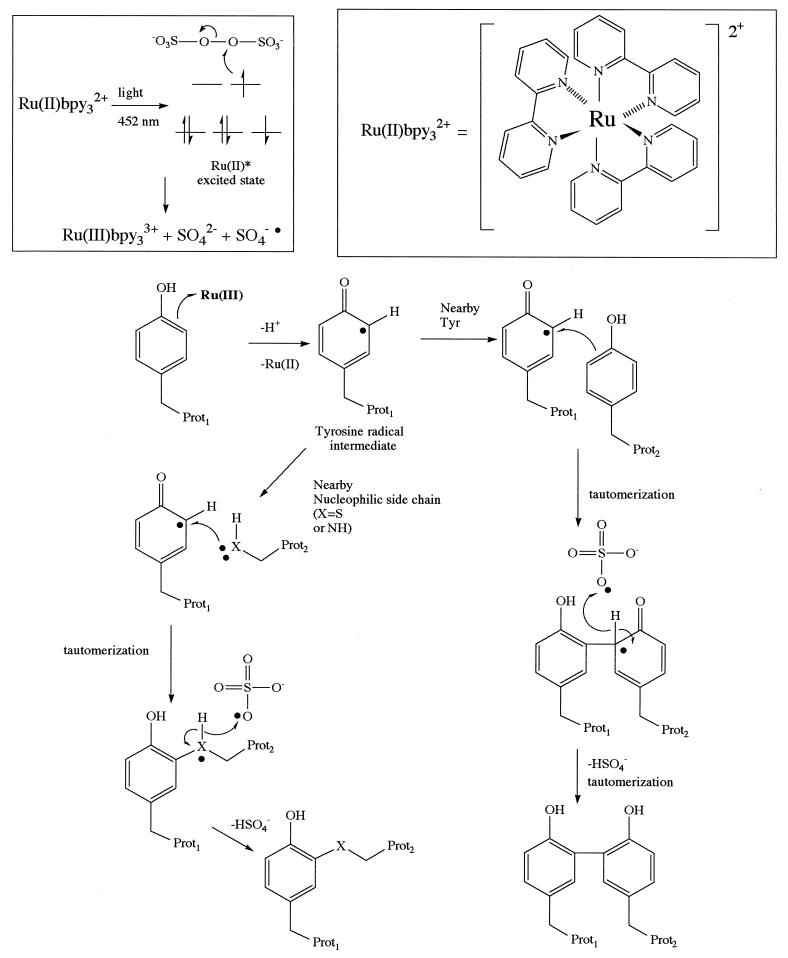
The photolysis of the ruthenium(II) tris-bipyridyl dication (Ru(II)bpy32+) in the presence of ammonium persulfate was explored as a method to generate reactive intermediates that might bring about efficient cross-linking of associated proteins. The logic behind this choice is as follows. Ru(II)bpy32+ is an efficient visible light-harvesting molecule, with a λmax of 452 nm in water and a molar extinction coefficient of ≈14,700 M−1. Photolysis of this metal complex is known to produce an excited state able to donate an electron to persulfate, resulting in cleavage of the O-O bond (16). The products are Ru(III), a potent one-electron oxidant (17–19), the sulfate radical, which should be a good hydrogen atom abstraction agent, and sulfate anion. As shown in Fig. 1, reasonable mechanisms can be proposed by which these species can bring about protein cross-linking. Ru(III)-mediated formation of a tyrosyl radical is proposed as an initiating step. Coupling of this radical with one of several possible nucleophiles and subsequent removal of a hydrogen atom by the sulfate radical could consummate the reaction. This scheme is precedented by previous work on the oxidative cross-linking of proteins mediated by chemically activated metal complexes from our laboratory and others (5, 15, 20–27) as well as intramolecular reactions that occur naturally in certain proteins (see ref. 28 for a review). Oxidative coupling is an appealing reaction on which to base a photo-initiated reaction because it results in the direct coupling of nearby residues without an intervening linker arm (29).
Figure 1.
Mechanism-based design of a photo-initiated protein cross-linking reaction. The top left part of the figure represents schematically the electron flow when Ru(II)bpy32+ (structure shown at the top right of the figure) is photolyzed in the presence of a persulfate, generating Ru(III) and sulfate radical. Ru(III) is a potent one-electron oxidant and would be expected to oxidize residues such as tyrosine. The resultant radical could proceed to form cross-linked products between two associated proteins by at least two mechanisms, as shown in the figure. If another tyrosine residue is nearby, then arene coupling would be expected. Alternatively, a nearby nucleophilic lysine or cysteine group could attack the radical to eventually provide a heteroatom–arene linkage. In each case, a hydrogen atom must be lost to form stable products, and the sulfate radical produced during Ru(III) formation could play a key role in this step. This figure represents only mechanistic hypotheses on which the reaction was designed. The true mechanism of Ru(II)bpy32+/persulfate-mediated cross-linking remains to be determined experimentally.
Ru(II)bpy32+/Persulfate-Mediated Protein Cross-Linking Is Rapid and Efficient.
UvsY protein, a native hexamer involved in phage T4 recombination (11, 30), was photolyzed in the presence of Ru(II)bpy32+ and APS by using a 150-W Xe arc lamp and a filter that cut off light below 380 nm. As seen in Fig. 2, lane 6, photolysis resulted in the production of covalently coupled UvsY multimers. Remarkably, only a 0.5-second irradiation of visible light was required to achieve the ≈60% yield observed. No reaction was observed in the dark or when the metal complex was omitted. An ≈20-fold lower yield was observed in the absence of APS, demonstrating the requirement of Ru(III) and/or the sulfate radical for efficient cross-linking. Photoexcited Ru(bpy)32+ is known to be an efficient generator of singlet oxygen (31, 32), which is expected to be the dominant reaction in the absence of APS. Not surprisingly, this pathway also yields cross-linked products (see Fig. 2, lane 7), but of a different nature (as evidenced by their distinct mobilities) and only after much longer irradiation.
Figure 2.
Treatment of the hexameric UvsY protein with Ru(II)bpy32+, APS, and light (>380 nm) results in extremely rapid and efficient covalent cross-linking. Shown is a Coomassie-stained denaturing gel. The bands observed in lanes 6 and 7 have mobilities consistent with multimers of the UvsY protein. The metal complex, APS, and photolysis are required for the formation of detectable products for short irradiation times (0.5 seconds for lanes 1–6). However, readily detectable levels of cross-linking can be obtained in the absence of APS with longer irradiation times (lane 7, 60 seconds).
We have coined the term photo-induced cross-linking of unmodified proteins (PICUP) for this process to distinguish it from very different azide- and benzophenone-based light-initiated reactions sometimes used to probe biomolecular interactions (33, 34). These species, when photolyzed with UV light, generate intermediates that are able to insert into C-H bonds of proteins. They are sometimes used to probe protein–protein interactions by linking them covalently to the protein of interest, often through an engineered cysteine side chain. When the modified protein is docked with the factor(s) with which it interacts, photolysis can lead to protein cross-linking if the azide or benzophenone moiety is near the protein–protein interface. Azide- and benzophenone-based reagents cannot be used “in trans” like the Ru(bpy)32+-based system described here because they are photoactivated insertion reagents, rather than true cross-linkers. Finally, the n to π* transitions, which must be excited to activate these reagents, have very low extinction coefficients compared with Ru(bpy)32+, requiring much longer irradiation times and λmaxs in the UV, not the visible, region.
Photoinitiated Cross-Linking of Transcription Factors.
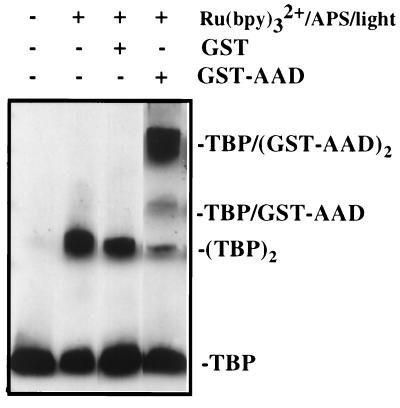
A particular interest of this laboratory (35), and many others, is to elucidate the interactions between transcriptional regulatory factors and the RNA polymerase II transcriptional machinery that is critical for controlling the expression of most eukaryotic genes. To ask whether PICUP would be useful for probing interactions between transcription factors, binding of the 34-residue core acidic activation domain (AAD) of the yeast Gal4 protein (36, 37), a regulator of galactose-metabolizing genes (38), to the conserved C-terminal 180-residue domain of TBP was examined. TBP is a basal transcription factor that recognizes the TATA element located in many promoters and has been proposed to be an important target of many activators (39), including Gal4 protein (14, 40). Free TBP is a native homodimer that must dissociate for the protein to bind DNA (41). The complex is comprised of a 2:1 molar ratio of AAD to TBP (S.-H. Yang and T.K., unpublished work). TBP was mixed with two equivalents of either GST or a GST-Gal4 AAD fusion protein (14). Ru(II)bipy32+ and APS were added, and the sample was illuminated for 0.5 seconds. The results were analyzed by denaturing gel electrophoresis and Western blotting by using an antibody specific for TBP. Lane 3 of Fig. 3 shows that, in the presence of GST, TBP homodimer was the only TBP-containing product, as expected. The results of this experiment were essentially identical to that in which GST was omitted (Fig. 3, lane 2), demonstrating that PICUP does not couple GST and TBP, two proteins that do not associate stably. Photolysis of the GST-AAD fusion and TBP resulted in the production of three products. The TBP homodimer was present, but at a reduced level compared with that formed in the absence of the AAD. The majority of product was represented by two bands, with the expected apparent molecular masses of GST-AAD-TBP and (GST-AAD)2-TBP, respectively, consistent with a 2:1 (GST-AAD⋅TBP) complex. The overall yield of products was ≈65%.
Figure 3.
Photo-initiated cross-linking of the Gal4 activation domain with TBP. The samples exposed to light were irradiated for 0.5 seconds with light of >380 nm by using a 150-W xenon arc lamp. Shown is a Western blot using antibody raised against TBP.
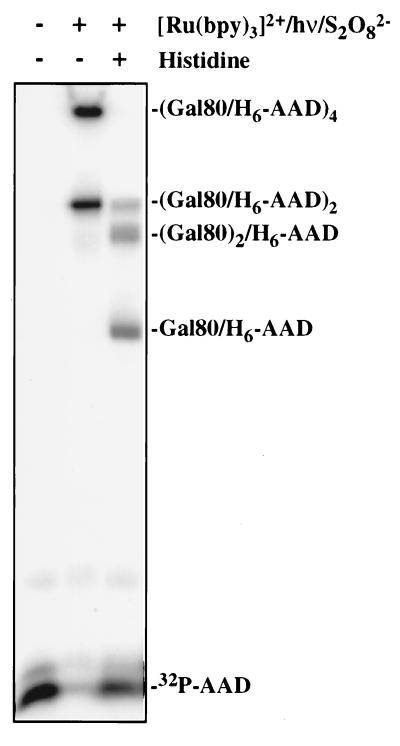
Another factor known to bind the Gal4 AAD is Gal80 protein (Gal80p), a specific repressor of Gal4p (36, 42). Like Gal4p, Gal80p is a native homodimer (37), and these dimers associate weakly to form homotetramers (K. Melcher, T.K., and S. A. Johnston, unpublished work). Gal80p and the Gal4 AAD form a tight complex with a 2:2 stoichiometry. As shown in Fig. 4, when a radiolabeled 54-residue polypeptide containing the Gal4 AAD (15) was mixed with a slight excess of purified Gal80p and photolyzed in the presence of Ru(bpy)32+ and APS for half a second, almost all of the AAD monomer was converted to cross-linked products. Fig. 4 is a phosphorimage and shows only AAD-containing bands. The lower band in Fig. 4, lane 2 has an apparent molecular mass consistent with an AAD2-Gal802 complex. The upper band corresponds to an (AAD-Gal80p)4 species. The formation of this product was not surprising because this experiment used micromolar protein concentrations and PICUP was able to trap the weak associations between Gal80p dimers that occur at these levels. As shown in Fig. 4, the degree of cross-linking can be “toned down” if desired by adding electron-donating amino acids such as histidine to the solution (Fig. 4, lane 3). In this case, three product bands were observed, with the apparent molecular masses expected of the AD-Gal80, AD-Gal802, and AD2-Gal802 species. Approximately 60% of the monomeric AD remained. This is the spectrum of products expected from the native 2:2 complex. Presumably, the added histidine either competes with the proteins for Ru(III) or sulfate radical or quenches tyrosyl radicals on the surface of the proteins before diffusional collision with another protein can lead to a cross-link. Histidine does not interfere with the AD-Gal80p association (data not shown). Because PICUP is so efficient, the addition of histidine is a useful method to sort out tight contacts from weak or transient associations, because cross-links resulting from the latter are quenched completely whereas those resulting from the former are less affected. This “histidine tuning” strategy also should be useful in controlling the extent of multimer formation in experiments using large multiprotein complexes. Exogenous tyrosine also inhibited cross-linking (data not shown), consistent with this residue being a target of Ru(III), but this inhibition was so efficient, it blocked all cross-linking and so is not useful as a tuning agent.
Figure 4.
Cross-linking of a 32P-labeled H6-tagged Gal4 activation domain (1 μM) to Gal80 protein (0.5 μM). Shown is a phosphorimager scan. In the absence of histidine, cross-linked (Gal4 AAD⋅Gal80)2 and (Gal4 AAD⋅Gal80)4 are produced in almost quantitative yield. Addition of histidine (7.5 mM) modulates the reaction, producing three products corresponding to the AAD-Gal80p heterodimer, the AAD cross-linked to the Gal80p dimer, and the (AAD-Gal80p)2 species.
Efficient Cross-Linking Using a Flashlight as the Light Source.
Because PICUP should be of utility to many biochemistry and molecular biology laboratories that do not have access to an intense light source such as the 150-W lamp we used, it was of interest to determine whether more readily available light sources would support the reaction. This was done by using GST, which is known to exist as a homodimer (43) that further associates into larger aggregates. Fig. 5 shows that, even when using a standard flashlight, good yields of cross-linking can be achieved under conditions identical to those used with the 150-W lamp, except that the light must be closer to the tube (5 cm vs. 50 cm for the high-intensity lamp) and the irradiation time must be increased to 5–30 seconds. This is still very rapid compared with traditional cross-linking techniques.
Figure 5.
PICUP does not require an intense light source. Cross-linking of GST under standard conditions was carried out by using a 150-W xenon lamp or a common flashlight as the light source. Excellent yields can be obtained in the latter case by simply increasing the photolysis time and reducing the lamp–sample distance from 50 to 5 cm.
PICUP should be a useful tool for the analysis of protein–protein contacts in multiprotein complexes. The reaction uses commercially available reagents and is fast and extremely efficient. By using GST as a model substrate, detectable yields (5–10%) of homodimers can be achieved with only a 30-millisecond irradiation using the Xe lamp (data not shown). It may be possible to reduce this time even further by using a laser as the light source. This suggests that the reaction could be very useful for kinetic studies of the dynamics of protein–protein associations. Therefore PICUP is an analytically useful system in which protein cross-linking can be triggered with visible light (see ref. 44 for a slow, low-level photocoupling of proteins mediated by protoporphyrin). The use of long-wavelength light is attractive because few biomolecules absorb outside of the UV region, an important point in carrying out cross-linking in crude extracts or eventually in living cells.
Acknowledgments
We thank Dr. Kathlynn Brown for helpful discussions. This work was funded by a grant from the National Institutes of Health (GM-58175).
ABBREVIATIONS
- TBP
TATA box-binding protein
- GST
glutathione S-transferase
- APS
ammonium persulfate
- Ru(II)bpy32+
ruthenium(II) tris-bipyridyl dication
- PICUP
photo-induced cross-linking of unmodified proteins
- AAD
acidic activation domain
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Alberts B. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 2.Mattson G, Conklin E, Desai S, Nielander G, Savage M D, Morgensen S. Mol Biol Rep. 1993;17:167–183. doi: 10.1007/BF00986726. [DOI] [PubMed] [Google Scholar]
- 3.Ji T H, Ji I. Pharmacol Ther. 1989;43:321–332. doi: 10.1016/0163-7258(89)90013-2. [DOI] [PubMed] [Google Scholar]
- 4.Gaffney B J. Biochim Biophys Acta. 1985;822:289–317. doi: 10.1016/0304-4157(85)90012-7. [DOI] [PubMed] [Google Scholar]
- 5.Bertrand R, Derancourt J, Kassab R. Biochemistry. 1997;36:9703–9714. doi: 10.1021/bi970615h. [DOI] [PubMed] [Google Scholar]
- 6.Jiang H, Giedroc D, Kodadek T. J Biol Chem. 1993;268:7904–7911. [PubMed] [Google Scholar]
- 7.Jiang H, Kodadek T. J Biol Chem. 1993;268:7904–7911. [PubMed] [Google Scholar]
- 8.Friedrichson T, Kurzchalia T V. Nature (London) 1998;394:802–805. doi: 10.1038/29570. [DOI] [PubMed] [Google Scholar]
- 9.Schere P E, Krieg U C. Methods Cell Biol. 1991;34:419–426. doi: 10.1016/s0091-679x(08)61696-9. [DOI] [PubMed] [Google Scholar]
- 10.Norcum M T, Warrington J A. Protein Sci. 1998;7:79–87. doi: 10.1002/pro.5560070108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kodadek T, Gan D C, Stemke-Hale K. J Biol Chem. 1989;264:16451–16457. [PubMed] [Google Scholar]
- 12.Hoopes B C, LeBlanc J F, Hawley D. J Biol Chem. 1992;267:11539–11547. [PubMed] [Google Scholar]
- 13.Platt A, Reece R J. EMBO J. 1998;17:4086–4091. doi: 10.1093/emboj/17.14.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melcher K, Johnston S. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fancy D, Kodadek T. Tetrahedron. 1997;53:11953–11960. [Google Scholar]
- 16.Nickel U, Chen Y-H, Schneider S, Silva M I, Burrows H D, Forosinho S J. J Phys Chem. 1994;98:2883–2888. [Google Scholar]
- 17.Gray H B, Winkler J R. Annu Rev Biochem. 1996;65:537–561. doi: 10.1146/annurev.bi.65.070196.002541. [DOI] [PubMed] [Google Scholar]
- 18.Berglund J, Pascher T, Winkler J R, Gray H B. J Am Chem Soc. 1997;119:2464–2469. [Google Scholar]
- 19.Yocom K M, Shelton J B, Shelton J R, Schroeder W A, Worosila G, Isied S S, Bordignon E, Gray H B. Proc Natl Acad Sci USA. 1982;79:7052–7055. doi: 10.1073/pnas.79.22.7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stadtman E. Free Radical Biol Med. 1990;9:315–325. doi: 10.1016/0891-5849(90)90006-5. [DOI] [PubMed] [Google Scholar]
- 21.Gill G, Richter-Rusli A A, Ghosh M, Burrows C J, Rokita S E. Chem Res Toxicol. 1997;10:302–309. doi: 10.1021/tx960170i. [DOI] [PubMed] [Google Scholar]
- 22.Heinecke J W, Li W, Francis G A, Goldstein J A. J Clin Invest. 1993;91:2866–2872. doi: 10.1172/JCI116531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malencik D A, Anderson S R. Biochemistry. 1996;35:4375–4386. doi: 10.1021/bi9526037. [DOI] [PubMed] [Google Scholar]
- 24.Brown K C, Yang S-H, Kodadek T. Biochemistry. 1995;34:4733–4739. doi: 10.1021/bi00014a030. [DOI] [PubMed] [Google Scholar]
- 25.Fancy D, Melcher K, Johnston S A, Kodadek T. Chem Biol. 1996;3:551–559. doi: 10.1016/s1074-5521(96)90146-5. [DOI] [PubMed] [Google Scholar]
- 26.Tew D, Ortiz de Montellano P R. J Biol Chem. 1988;263:17880–17886. [PubMed] [Google Scholar]
- 27.Wilks A, Ortiz de Montellano P R. J Biol Chem. 1992;267:8827–8833. [PubMed] [Google Scholar]
- 28.Stubbe J, Riggs-Gelasco P. Trends Biochem Sci. 1998;23:438–443. doi: 10.1016/s0968-0004(98)01296-1. [DOI] [PubMed] [Google Scholar]
- 29.Brown K C, Yu Z, Burlingame A L, Craik C S. Biochemistry. 1998;37:4397–4406. doi: 10.1021/bi9728046. [DOI] [PubMed] [Google Scholar]
- 30.Beernick H T, Morrical S W. Biochemistry. 1998;37:5673–5681. doi: 10.1021/bi9800956. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Rodgers M A J. J Phys Chem. 1995;99:12797–12803. [Google Scholar]
- 32.Tanielian C, Wolff C, Esch M. J Phys Chem. 1996;100:6555–6560. [Google Scholar]
- 33.Chen Y, Ebright Y W, Ebright R H. Science. 1994;265:90–92. doi: 10.1126/science.8016656. [DOI] [PubMed] [Google Scholar]
- 34.Dorman G, Prestwich G D. Biochemistry. 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 35.Denison C, Kodadek T. Chem Biol. 1998;5:R129–R145. doi: 10.1016/s1074-5521(98)90167-3. [DOI] [PubMed] [Google Scholar]
- 36.Johnston S A, Salmeron J M, Jr, Dincher S S. Cell. 1987;50:143–146. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- 37.Van Hoy M, Leuther K K, Kodadek T, Johnston S A. Cell. 1993;72:587–594. doi: 10.1016/0092-8674(93)90077-4. [DOI] [PubMed] [Google Scholar]
- 38.Johnston M. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Struhl K. Annu Rev Genet. 1995;29:651–674. doi: 10.1146/annurev.ge.29.120195.003251. [DOI] [PubMed] [Google Scholar]
- 40.Wu Y, Reece R J, Ptashne M. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- 41.Coleman R A, Taggart A K P, Benjamin L R, Pugh B F. J Biol Chem. 1995;270:13842–13849. doi: 10.1074/jbc.270.23.13842. [DOI] [PubMed] [Google Scholar]
- 42.Ma J, Ptashne M. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 43.Lim K, Ho J X, Keeling K, Gilliland G L, Ruker F, Carter D C. Protein Sci. 1994;3:2233–2244. doi: 10.1002/pro.5560031209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verweij H, Dubbelman T M A R, Van Steveninck J. Biochem Biophys Acta. 1981;647:87–94. doi: 10.1016/0005-2736(81)90297-2. [DOI] [PubMed] [Google Scholar]