In the past decade or so, membrane-embedded proteases that carry out hydrolysis on the transmembrane region of their substrates have been discovered. These I-CLiPs2 (1) somehow create an environment for water and the hydrophilic residues needed for catalysis and bend or unwind their helical substrates to make the amide bonds susceptible to hydrolysis. Despite the distinction of being membrane-embedded and cleaving TMDs, the residues essential for catalysis by these I-CLiPs are virtually the same as those found in aqueous proteases, clear examples of convergent evolution toward a common mechanism. Described herein are the different types of I-CLiPs and an update on their structural and mechanistic features and biological roles.
S2P Metalloproteases
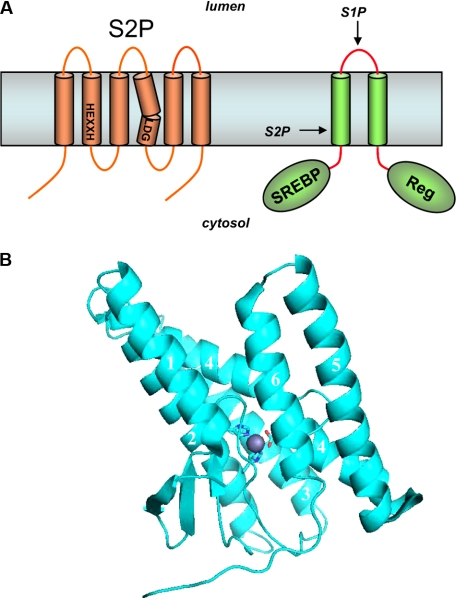
SREBPs are transcription factors that promote expression of genes involved in the synthesis of cholesterol and fatty acids (reviewed in Ref. 2). SREBPs are synthesized as a two-TMD precursor protein (Fig. 1A) that undergoes proteolytic release. The luminal loop between the two TMDs is first cleaved by the membrane-tethered S1P when cholesterol levels are low. Release of the transcription factor requires subsequent cleavage by S2P, which performs a hydrolysis three residues within the TMD. Complementation cloning identified S2P as a multipass membrane protein containing a conserved HEXXH sequence characteristic of zinc metalloproteases. Sequential processing by S1P and S2P likewise occurs for the transcription factor ATF6 during the ER stress response.
FIGURE 1.
Intramembrane metalloproteases. A, S2P contains conserved metalloprotease HEXXH and LDG motifs. SREBP is first cleaved by S1P in the luminal loop. The regulatory domain (Reg) helps to ensure that S1P proteolysis occurs when cholesterol levels are low. Subsequent intramembrane proteolysis releases this transcription factor for expression of genes essential to cholesterol and fatty acid synthesis. B, crystal structure of an archaeal S2P in the closed conformation. Zinc (gray sphere) is proximal to two histidines in TMD2, one aspartate in the kinked TMD4 (side chain sticks), and one glutamate in TMD2 (not shown). In the open conformation, TMD1 and TMD6 are considerably farther apart, suggesting the site of lateral gating by which substrate TMD accesses the internal active site.
The two histidines and the glutamate are required for S2P activity, consistent with known metalloprotease biochemistry in which the two histidines coordinate with zinc, the zinc activates the scissile amide bond, and the glutamate activates the catalytic water. A conserved aspartate located quite distant from HEXXH in the linear sequence is likewise critical for S2P activity and coordinates with the zinc atom (see below). Regarding the substrate, SREBP contains a conserved and helix-destabilizing asparagine-proline sequence within its TMD1 that is critical for proteolytic processing of the nearby leucine–cysteine bond by S2P.
S2P-like proteases are also found in bacteria (3) and archaea (4). These prokaryotic proteins play an essential role in the proteolysis of membrane-bound transcription factors needed for sporulation, controlling gene expression in the mother cell after engulfment of the forespore. Cleavage of pro-σk and release of the membrane-tethered transcription factor requires SpoIVFB in Bacillus subtilis. Another bacterial S2P family member, YaeL (also called RseP) in Escherichia coli, plays a role in coordinating cell growth and cell division through intramembrane proteolysis of RseA, a factor critical for responding to extracytoplasmic stress (5). Interestingly, the membrane orientations of substrates SREBP and σk are opposite each other, correlating with those of their respective enzymes, S2P and SpoIVFB, which are similarly thought to have opposite orientations (3). This implies that the catalytic region must align with peptide substrate with proper relative directionality.
Although SpoIVFB and YaeL are both bacterial S2P-like enzymes that cleave transmembrane proteins, the regulation of intramembrane proteolysis via these two I-CLiPs is quite different. For RseA cleavage by YaeL/RseP, the regulation is similar to that for SREBP cleavage by S2P: intramembrane proteolysis requires a prior cleavage event outside the membrane by another protease called DegS (6). In contrast, SpoIVFB apparently does not require prior proteolysis, and regulation occurs more directly at the level of SpoIVFB. Two membrane proteins, BofA and SpoIVFA, serve to inhibit SpoIVFB activity, and this inhibition is released by proteolysis of SpoIVFA by other proteases (7, 8).
The E. coli YaeL/RseP protease has been purified with preservation of proteolytic activity (9). Most recently, a high resolution crystal structure of an S2P family member has been reported (10), confirming the presence of zinc and its proximity to the key transmembrane histidine, glutamate, and aspartate residues (Fig. 1B). The protease crystallized in two conformations, one in which the active site appears more accessible through lateral gating (“open”) and one in which it is less accessible (“closed”). TMD2–4 are highly conserved, contain the catalytic residues, and do not vary much between the two conformations. TMD2–4 are thus thought to represent the core domain, whereas TMD1, TMD5 and TMD6, which are more conformationally flexible, are thought to be important for substrate gating from the lipid bilayer into the internal, water-containing active site. In the open conformation, TMD1 and TMD6 are spaced farther apart, suggesting that substrate enters the active site by traversing between these two TMDs.
γ-Secretase Aspartyl Protease Complexes
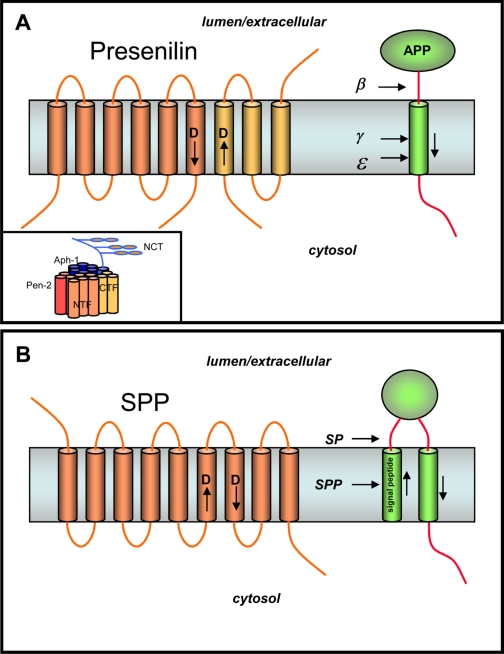
Proteolytic production of the Aβ peptide is a critical step in the pathogenesis of Alzheimer disease (reviewed in Ref. 11; see also the Thematic Minireview Series on the Molecular Basis of Alzheimer Disease published in the October 31, 2008, February 20, 2009, and March 6, 2009 issues of the Journal of Biological Chemistry). The N terminus of Aβ is produced from APP by β-secretase, which leads to membrane shedding of the large luminal/extracellular APP domain (Fig. 2A). The remaining protein stub is then cleaved at least twice by the membrane-embedded γ-secretase complex, in the middle of the TMD at the γ-site to release Aβ and near the inner leaflet at the ε-site to release the APP intracellular domain.
FIGURE 2.
Intramembrane aspartyl proteases. A, PS, the γ-secretase complex, and the proteolysis of APP. PS is cut into two pieces, an NTF (dark portion) and a CTF (light portion), that remain associated. Each fragment donates one aspartate essential for γ-secretase activity. APP is first cleaved in the extracellular domain by β-secretase, and the remnant is cleaved twice within the membrane by γ-secretase to produce the Aβ peptide of Alzheimer disease (secreted) and the intracellular domain (freed into the cytosol). Inset, PS interacts with three other membrane proteins, nicastrin (NCT), Aph-1, and Pen-2, to form active γ-secretase. B, SPP. Signal peptides are removed from membrane proteins via signal peptidase (SP), and these peptides are released from the membrane by SPP-mediated intramembrane proteolysis. Like PS, SPP contains two aspartates essential for protease activity, but the conserved aspartate-containing motifs are in the opposite orientation compared with their PS counterparts.
The nine-TMD PS is the catalytic component of an unusual aspartyl protease complex (reviewed in Ref. 12). PS is required for γ-secretase activity and is cut into two pieces, an NTF and a CTF, the formation of which is gated by limiting cellular factors. The NTF and CTF remain associated in a high molecular weight complex and are metabolically stable. Both PS fragments contain one conserved transmembrane aspartate that is essential for γ-secretase activity, and transition state analog inhibitors of γ-secretase bind directly to the NTF and CTF, suggesting that the γ-secretase active site is at the interface between these two PS fragments.
In addition to PS, γ-secretase is composed of three other integral membrane proteins: nicastrin, Aph-1, and Pen-2. Coexpression, RNA interference, and the identification of assembly intermediates suggest the order in which these four subunits come together, and partial dissociation of the protease complex with detergent offers a model for how these subunits interact (Fig. 2A, inset). Nicastrin and Aph-1 together can stabilize full-length PS, and the final addition of Pen-2 triggers PS endoproteolysis and γ-secretase activity. Pen-2 is also required to stabilize the PS subunits. A recent study demonstrated that one of each of these four components per complex is sufficient for γ-secretase proteolytic activity (13).
Although the specific biochemical functions of these PS cofactors are mostly unknown, nicastrin is thought to play a role in substrate recognition (14). The ectodomain of nicastrin resembles a catalytically inactive aminopeptidase and putatively recognizes the N terminus of γ-secretase substrates. Mutation of the aminopeptidase domain was reported to prevent this interaction, suggesting that nicastrin is a gatekeeper for the γ-secretase complex: type I membrane proteins that have not shed their ectodomains cannot interact properly with nicastrin and do not gain access to the active site. However, a new study contradicts this view with evidence that mutation of the aminopeptidase domain can interfere with the maturation of the γ-secretase complex, not the activity of the mature complex (15).
In addition to APP, PS/γ-secretase cuts a growing list of other type I integral membrane protein stubs (16). The protease displays poor substrate specificity and apparently serves a major degradative function, clearing protein stubs from the membrane. However, γ-secretase proteolysis is known to be critical for several important signaling events. 1) Ligand-activated proteolysis of the Notch receptor is essential for signaling, which is crucial to many cell differentiation events in all metazoans (17); 2) proteolysis of N-cadherin leads to degradation of the transcriptional activator CBP (cAMP-responsive element-binding protein-binding protein) (18); and 3) neuregulin-1-triggered cleavage of ErbB4 inhibits astrocyte differentiation by interacting with repressors of astrocyte gene expression (19).
Since their discovery, I-CLiPs have been envisioned to have an internal active site, sequestered from the hydrophobic lipid tails but with a pore or cavity that could allow entry of water (1). Substrate access to the active site would therefore require initial docking on the outer surface of the protease and lateral gating. Initial evidence for such a mechanism came from isolation of the γ-secretase complex with an immobilized transition state analog inhibitor in which an endogenous APP substrate copurified (20), suggesting the existence of a separate substrate-binding site distinct from the active site. Designed helical peptides based on the TMD of APP apparently interact with this docking site, specifically at the PS NTF/CTF interface (21), suggesting that upon binding to the outer surface of PS at the NTF/CTF interface, the substrate can pass, either in whole or in part, between these two PS subunits to access the internal active site.
Purification of the γ-secretase complex (22) has allowed the first glimpse into its structure. Electron microscopy with negative staining and single particle analysis reveals that the complex has a globular structure that at low resolution (∼15 Å) appears rather amorphous (23). Nevertheless, two important features can be gleaned: a large interior region of low electron density and of ∼20-Å diameter that is presumably where the active site resides and the presence of two small openings that may be the site of entry for water. Other structural features have been revealed by cysteine mutagenesis with cross-linking of chemical probes (24, 25). The generation of a cysteine-less PS that retains the ability to assemble with other complex members, undergo endoproteolysis to the NTF and CTF, and process APP allowed incorporation of single cysteine resides at various sites near the key aspartates. Disulfide formation with thiol-containing reagents then provided information about the relative accessibility of these sites from the aqueous milieu, allowing the construction of a model in which water can funnel down to where the aspartates reside. Using this same approach, two recent studies suggest that TMD9 serves as a gatekeeper for lateral entry of the substrate TMD (26, 46). More detailed structural information will likely require a crystal structure of PS or a PS homolog. Purified γ-secretase has also recently been reconstituted into lipid vesicles with detergent removal, allowing characterization of lipid requirements for activity within membranes (27), and this advance may also ultimately prove useful for structure elucidation.
SPP Aspartyl Proteases
The discovery of SPP as a PS-like intramembrane aspartyl protease solidified the concept of PS as the catalytic component for γ-secretase (see accompanying minireview by Fluhrer et al. (45) for more details about SPP and its homologs and for references). SPP clears remnant signal peptides from the membrane after their production by signal peptidase (Fig. 2B). However, this process also plays a role in immune surveillance and hepatitis C virus maturation. SPP was identified by affinity labeling with a peptidomimetic inhibitor and was found to possess two conserved aspartate-containing TMD sequences that resemble those found in PS (Fig. 2B). As with S2P compared with its bacterial relatives, the orientation of the aspartate-containing TMDs of SPP is apparently opposite that of PS, again in correlation with the orientation of SPP substrates, which is opposite that of γ-secretase substrates. Just prior to the identification of SPP, an entire family of so-called PS homologs had been discovered through a bioinformatics approach; however, it is still not clear if all of these proteins have catalytic activity. Two homologs, SPP-like proteases SPPL2a and SPPL2b, have been found to cleave tumor necrosis factor-α, the Fas ligand, and the dementia-associated Bri2 protein, although the biological roles of these proteolytic events are unclear.
Expression of human SPP in yeast reconstituted the protease activity, suggesting that, unlike γ-secretase, the protein has activity on its own and does not require other mammalian protein cofactors. This has recently been confirmed by the expression of various SPP orthologs in E. coli and purification of active enzyme to homogeneity. Moreover, unlike PS, SPP is not processed into two pieces. Thus, SPP may be a more tractable enzyme for understanding this type of aspartyl I-CLiP and may shed light on γ-secretase structure and function. Indeed, the catalytic sites of the two proteases appear remarkably similar: their activities are inhibited by some of the same active site-directed peptidomimetics and helical peptides, and activity can be modulated by other compounds that similarly affect γ-secretase. In terms of substrate recognition, however, SPP may display an important difference from γ-secretase: a putative requirement for helix-breaking residues that are thought to facilitate the ability of the enzyme to access the site of hydrolysis.
Rhomboid Serine Proteases
Investigation of a conserved growth factor signaling pathway in Drosophila also led to intramembrane proteolysis. Proteolysis of EGF receptor ligands is required for interaction with the cognate receptor. In vertebrates, this is accomplished by membrane-tethered metalloproteases. Genetic analysis in Drosophila identified, however, two essential factors, called Star and Rhomboid-1, for proteolysis of the EGF ortholog Spitz. Star ushers Spitz from the ER to the Golgi, where it encounters Rhomboid-1 (28). Rhomboid-mediated proteolysis in the Golgi is then followed by secretion for intercellular communication.
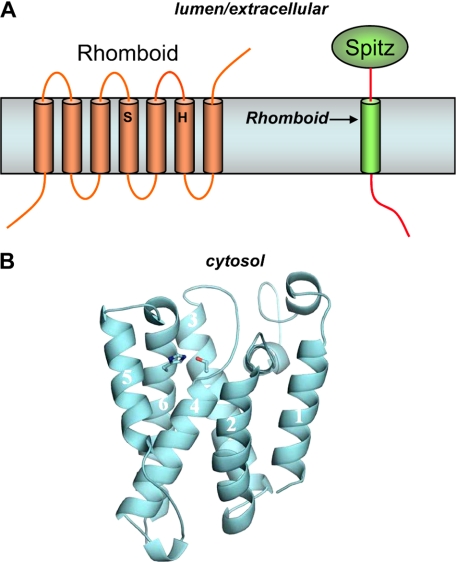
A requirement for a transmembrane serine, histidine, and asparagine suggested a catalytic triad typically found in serine proteases (29), although subsequent studies support a Ser-His dyad (Fig. 3A) (30). The cleavage site of Spitz was estimated to be at an equivalent depth in the TMD to these Rhomboid residues. Moreover, Spitz cleavage was sensitive only to serine protease inhibitors, and catalytic amounts of Rhomboid-1 still allowed Spitz proteolysis. Thus, Rhomboid-1 appears to be a novel intramembrane serine protease.
FIGURE 3.
Intramembrane serine proteases. A, Rhomboid proteins contain a conserved serine and histidine, which compose the catalytic dyad of a serine protease. Rhomboid-1 cleaves within the transmembrane region of the Drosophila EGF-like growth factor Spitz. B, structure of E. coli Rhomboid GlpG. The serine in TMD4 and the histidine in TMD6 are coordinated in a manner consistent with known serine proteases and at a depth within the membrane consistent with the site of proteolysis of Rhomboid substrates.
Although Rhomboid-1 does not display much sequence specificity within the Spitz TMD, a glycine-alanine motif is apparently critical for substrate specificity (31). This finding suggests that, as with S2P and SPP, Rhomboid seems to require helix-destabilizing residues within the TMD of its substrates. Unlike most other I-CLiPs, however, substrate cleavage by Rhomboid does not require prior cleavage by another protease. Rhomboid regulation apparently occurs mainly by translocation of the substrate from the ER to the Golgi (mediated by Star) and spatial control of Rhomboid transcription.
Like S2P, Rhomboid genes have been conserved throughout evolution. The natural substrates for prokaryotic and archaeal Rhomboid proteins are unknown, with one exception: Providencia stuartii Rhomboid protease AarA cleaves a protein called TatA as part of a quorum-sensing signal (32). As for eukaryotic Rhomboid proteins, two mitochondrial membrane proteins were identified as substrates for yeast Rhomboid Rbd1p (33–35). Rbd1p-mediated release of one of these substrates (dynamin-like GTPase Mgm1p) is essential for remodeling of the mitochondrial membrane, and the human ortholog of Rbd1p, PARL, could restore substrate proteolysis and proper growth rates and mitochondrial morphology in a yeast Rbd1p mutant (34), suggesting conservation of this role. Indeed, a later study identified the mitochondrial protein OPA1 as a likely substrate for PARL, the cleavage of this substrate being critical to crista remodeling and cytochrome c release during apoptosis (36). In Toxoplasma, TgROM5, one of five non-mitochondrial Rhomboid proteins in these parasites, cleaves a cell-surface adhesion protein as a key step in cell invasion (37), and similar findings in the related Plasmodium falciparum, the malaria parasite, have recently been reported (38), suggesting that Rhomboid proteins are potential targets for treating infections by these deadly pathogens.
Rhomboid provided the first crystal structures of an I-CLiP, with four reports on the E. coli Rhomboid GlpG (39–42) and one on the Hemophilus influenza Rhomboid HiGlpG (43). These structures show remarkable similarities and important differences that provide insight into how this class of membrane-embedded proteases carries out hydrolysis in the lipid bilayer. The structures all reveal that the serine and histidine implicated as the catalytic dyad are indeed coordinated with each other and lie at a depth within the membrane consistent with where Rhomboid proteins cleave their transmembrane substrates (Fig. 3B). A cavity is open to the periplasmic space, with the catalytic dyad at the bottom of this opening, and this cavity contains multiple water molecules.
How substrate enters this cavity is not entirely clear, but the position of TMD5 varies in the different structures, and movement of this domain can provide a space through which substrate may reach the catalytic dyad. Indeed, one of the reported structures contains a bound lipid in this space (41), with the phosphate group residing near the Ser-His dyad and a key Asn residue that may contribute to the oxyanion hole that stabilizes intermediates and transition states during serine protease catalysis. Mutational analysis revealed that altering residues predicted to disrupt the role of TMD5 as a gate led to increased proteolytic activity (44), providing further validation for the site of lateral entry of the substrate. These exciting structural findings in turn validate the prior molecular and biochemical studies on Rhomboid proteins and suggest that such approaches have been providing true mechanistic insight into the workings of other I-CLiPs. These structures offer details that should inspire other specific hypotheses about how Rhomboid proteins handle substrates to hydrolyze TMDs.
Perspective
The membrane-embedded I-CLiPs appear to recapitulate the mechanisms of soluble proteases, and the first crystal structures of Rhomboid and S2P support this notion, at least for the serine and metallo-I-CLiPs. The I-CLiPs discovered so far all play critical roles in biology and are closely regulated, but the means of control vary. They are all involved in cell signaling but in a variety of ways. Membrane topology apparently dictates the types of substrates that can be cleaved. Most I-CLiPs appear to require helix-breaking residues near the cleavage sites of their substrates, although γ-secretase may be a notable exception.
Critical remaining issues include the identity of substrates for the I-CLiP family members whose roles are unknown. For instance, although entire families of PS homologs and Rhomboid proteins have been discovered, natural substrates are known only for a handful of these proteins. Another key issue is the specific mechanisms of these proteases (e.g. elucidating conformational changes that take place in both enzyme and substrate during proteolysis, identifying enzyme residues that directly interact with substrate). Structural biology is clearly the emerging frontier in the study of I-CLiPs, with Rhomboid and S2P providing the first fruits of such endeavors. Detailed structural understanding should provide a clearer appreciation of how these remarkable enzymes work, and this should include cocrystal structures with inhibitors that offer further insight into mechanisms and that pave the way for structure-based design in cases in which the target has high therapeutic relevance.
This work was supported, in whole or in part, by National Institutes of Health Grants GM79555, AG17574, and NS41355 (to M. S. W.). The first article in this minireview series on proteases was published in the November 7, 2008 issue; the second and third articles are published in this issue. Additional articles will be published in future issues. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: I-CLiP, intramembrane-cleaving protease; TMD, transmembrane domain; S2P, site-2 protease; SREBP, sterol regulatory element-binding protein; S1P, site-1 protease; ER, endoplasmic reticulum; Aβ, amyloid β-protein; APP, amyloid β-protein precursor; PS, presenilin; NTF, N-terminal fragment; CTF, C-terminal fragment; SPP, signal peptide peptidase; EGF, epidermal growth factor.
References
- 1.Wolfe, M. S., De Los Angeles, J., Miller, D. D., Xia, W., and Selkoe, D. J. (1999) Biochemistry 38 11223–11230 [DOI] [PubMed] [Google Scholar]
- 2.Rawson, R. B. (2003) Nat. Rev. Mol. Cell Biol. 4 631–640 [DOI] [PubMed] [Google Scholar]
- 3.Rudner, D. Z., Fawcett, P., and Losick, R. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 14765–14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinch, L. N., Ginalski, K., and Grishin, N. V. (2006) Protein Sci. 15 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanehara, K., Akiyama, Y., and Ito, K. (2001) Gene (Amst.) 281 71–79 [DOI] [PubMed] [Google Scholar]
- 6.Alba, B. M., Leeds, J. A., Onufryk, C., Lu, C. Z., and Gross, C. A. (2002) Genes Dev. 16 2156–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou, R., and Kroos, L. (2005) Mol. Microbiol. 58 835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campo, N., and Rudner, D. Z. (2006) Mol. Cell 23 25–35 [DOI] [PubMed] [Google Scholar]
- 9.Akiyama, Y., Kanehara, K., and Ito, K. (2004) EMBO J. 23 4434–4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng, L., Yan, H., Wu, Z., Yan, N., Wang, Z., Jeffrey, P. D., and Shi, Y. (2007) Science 318 1608–1612 [DOI] [PubMed] [Google Scholar]
- 11.Goedert, M., and Spillantini, M. G. (2006) Science 314 777–781 [DOI] [PubMed] [Google Scholar]
- 12.Wolfe, M. S. (2006) Biochemistry 45 7931–7939 [DOI] [PubMed] [Google Scholar]
- 13.Sato, T., Diehl, T. S., Narayanan, S., Funamoto, S., Ihara, Y., De Strooper, B., Steiner, H., Haass, C., and Wolfe, M. S. (2007) J. Biol. Chem. 282 33985–33993 [DOI] [PubMed] [Google Scholar]
- 14.Shah, S., Lee, S. F., Tabuchi, K., Hao, Y. H., Yu, C., LaPlant, Q., Ball, H., Dann, C. E., 3rd, Sudhof, T., and Yu, G. (2005) Cell 122 435–447 [DOI] [PubMed] [Google Scholar]
- 15.Chávez-Gutiérrez, L., Tolia, A., Maes, E., Li, T., Wong, P. C., and De Strooper, B. (2008) J. Biol. Chem. 283 20096–20105 [DOI] [PubMed] [Google Scholar]
- 16.Beel, A. J., and Sanders, C. R. (2008) CMLS Cell. Mol. Life Sci. 65 1311–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selkoe, D., and Kopan, R. (2003) Annu. Rev. Neurosci. 26 565–597 [DOI] [PubMed] [Google Scholar]
- 18.Marambaud, P., Wen, P. H., Dutt, A., Shioi, J., Takashima, A., Siman, R., and Robakis, N. K. (2003) Cell 114 635–645 [DOI] [PubMed] [Google Scholar]
- 19.Sardi, S. P., Murtie, J., Koirala, S., Patten, B. A., and Corfas, G. (2006) Cell 127 185–197 [DOI] [PubMed] [Google Scholar]
- 20.Esler, W. P., Kimberly, W. T., Ostaszewski, B. L., Ye, W., Diehl, T. S., Selkoe, D. J., and Wolfe, M. S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2720–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornilova, A. Y., Bihel, F., Das, C., and Wolfe, M. S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 3230–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraering, P. C., Ye, W., Strub, J. M., Dolios, G., LaVoie, M. J., Ostaszewski, B. L., Van Dorsselaer, A., Wang, R., Selkoe, D. J., and Wolfe, M. S. (2004) Biochemistry 43 9774–9789 [DOI] [PubMed] [Google Scholar]
- 23.Lazarov, V. K., Fraering, P. C., Ye, W., Wolfe, M. S., Selkoe, D. J., and Li, H. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 6889–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tolia, A., Chávez-Gutiérrez, L., and De Strooper, B. (2006) J. Biol. Chem. 281 27633–27642 [DOI] [PubMed] [Google Scholar]
- 25.Sato, C., Morohashi, Y., Tomita, T., and Iwatsubo, T. (2006) J. Neurosci. 26 12081–12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tolia, A., Horré, K., and De Strooper, B. (2008) J. Biol. Chem. 283 19793–19803 [DOI] [PubMed] [Google Scholar]
- 27.Osenkowski, P., Ye, W., Wang, R., Wolfe, M. S., and Selkoe, D. J. (2008) J. Biol. Chem. 283 22529–22540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, J. R., Urban, S., Garvey, C. F., and Freeman, M. (2001) Cell 107 161–171 [DOI] [PubMed] [Google Scholar]
- 29.Urban, S., Lee, J. R., and Freeman, M. (2001) Cell 107 173–182 [DOI] [PubMed] [Google Scholar]
- 30.Lemberg, M. K., Menendez, J., Misik, A., Garcia, M., Koth, C. M., and Freeman, M. (2005) EMBO J. 24 464–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Urban, S., and Freeman, M. (2003) Mol. Cell 11 1425–1434 [DOI] [PubMed] [Google Scholar]
- 32.Stevenson, L. G., Strisovsky, K., Clemmer, K. M., Bhatt, S., Freeman, M., and Rather, P. N. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esser, K., Tursun, B., Ingenhoven, M., Michaelis, G., and Pratje, E. (2002) J. Mol. Biol. 323 835–843 [DOI] [PubMed] [Google Scholar]
- 34.McQuibban, G. A., Saurya, S., and Freeman, M. (2003) Nature 423 537–541 [DOI] [PubMed] [Google Scholar]
- 35.Herlan, M., Vogel, F., Bornhovd, C., Neupert, W., and Reichert, A. S. (2003) J. Biol. Chem. 278 27781–27788 [DOI] [PubMed] [Google Scholar]
- 36.Frezza, C., Cipolat, S., Martins de Brito, O., Micaroni, M., Beznoussenko, G. V., Rudka, T., Bartoli, D., Polishuck, R. S., Danial, N. N., De Strooper, B., and Scorrano, L. (2006) Cell 126 177–189 [DOI] [PubMed] [Google Scholar]
- 37.Brossier, F., Jewett, T. J., Sibley, L. D., and Urban, S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 4146–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker, R. P., Wijetilaka, R., and Urban, S. (2006) PLoS Pathog. 2 e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Y., Zhang, Y., and Ha, Y. (2006) Nature 444 179–180 [DOI] [PubMed] [Google Scholar]
- 40.Wu, Z., Yan, N., Feng, L., Oberstein, A., Yan, H., Baker, R. P., Gu, L., Jeffrey, P. D., Urban, S., and Shi, Y. (2006) Nat. Struct. Mol. Biol. 13 1084–1091 [DOI] [PubMed] [Google Scholar]
- 41.Ben-Shem, A., Fass, D., and Bibi, E. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 462–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, Y., and Ha, Y. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2098–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemieux, M. J., Fischer, S. J., Cherney, M. M., Bateman, K. S., and James, M. N. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 750–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker, R. P., Young, K., Feng, L., Shi, Y., and Urban, S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 8257–8262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fluhrer, R., Steiner, H., and Haass, C. (2009) J. Biol. Chem. 284 13975–13979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato, C., Takaqi, S., Tomita, T., and Iwatsubo, T. (2008) J. Neurosci. 28 6264–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]