Abstract
Intramembrane-cleaving proteases are required for reverse signaling and membrane protein degradation. A major class of these proteases is represented by the GXGD-type aspartyl proteases. GXGD describes a novel signature sequence that distinguishes these proteases from conventional aspartyl proteases. Members of the family of the GXGD-type aspartyl proteases are the Alzheimer disease-related γ-secretase, the signal peptide peptidases and their homologs, and the bacterial type IV prepilin peptidases. We will describe the major biochemical and functional properties of the signal peptide peptidases and their relatives. We then compare these properties with those of γ-secretase and discuss common mechanisms but also point out a number of substantial differences.
During the last years, a number of intramembrane-cleaving proteases termed I-CLiPs3 have been identified (1). I-CLiPs are generally involved in regulated intramembrane proteolysis (2). Upon shedding of a large part of the ectodomain of membrane proteins, the remaining membrane-retained stub is cleaved by specialized proteases within the hydrophobic lipid membrane. Generally, this cleavage can have two predominant biological functions: first, signaling via the liberated ICD within the substrate-expressing cell (reverse signaling) (2); and second, degradation of membrane-retained stubs, which are not required for any further biological function (3). I-CLiPs of three protease classes, metalloproteases, serine proteases, and aspartyl proteases, have been discovered so far (see accompanying minireview by Wolfe (44)).
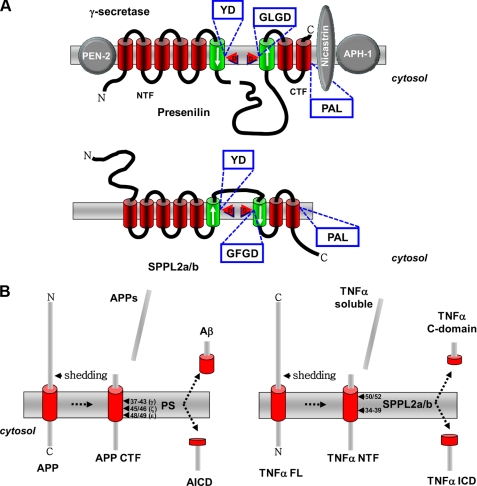
Intramembrane-cleaving aspartyl proteases are represented by the class of the GXGD-type proteases (4). These are unconventional aspartyl proteases that, like the conventional aspartyl proteases, utilize two critical aspartyl residues for peptide bond cleavage. However, in contrast to the conventional proteases, the critical aspartyl residues are located within two TMDs (Fig. 1A). Moreover, these aspartyl residues are embedded in active-site motifs that are completely different from those of conventional aspartyl proteases. The class of GXGD-type aspartyl proteases is currently represented by three different protease families, the most prominent of which is the PS family, providing the catalytically active subunit of γ-secretase (Fig. 1A) (4). PS/γ-secretase is the I-CLiP that liberates amyloid β-peptide, the major component of senile plaques in Alzheimer disease patients (5). In addition, the bacterial type IV prepilin peptidases also belong to the class of the GXGD-type proteases (6). Besides these two protease families, two additional subfamilies of related proteases that also belong to the GXGD-type aspartyl protease family have been identified. These include SPP as well as the SPP homologs, the SPP-like (SPPL) proteases (Fig. 1A) (7, 8).
FIGURE 1.
A, schematic representation of SPPL2a/b, a member of the SPP/SPPL family, and PS, the catalytic core of theγ-secretase. Note the opposite topology of the active sites (indicated by arrows) of the two proteases and their substrates, APP for PS and TNFα for SPPL2a/b. B, proteolytic processing of APP and TNFα. Shedding releases the extracellular part of APP (APPs) and TNFα (TNFα soluble). In the case of APP, a C-terminal fragment (APP CTF), and in case of TNFα, an N-terminal fragment (TNFα NTF) are produced. These membrane-bound fragments are substrate to intramembrane cleavage by PS or SPPL2a/b, respectively, releasing small peptides to the extracellular space (Aβ and TNFα C-domain, respectively) and to the cytosol (APP intracellular domain (AICD) and TNFα ICD), respectively). TNFα FL, full-length TNFα.
We will first describe the biochemical, functional, and structural properties of SPP family members. By comparison of these properties, we will then identify common mechanisms of intramembrane proteolysis by GXGD-type proteases but also point out some fundamental differences.
Intramembrane Proteolysis by SPP and SPPL Family Members
An SPP activity capable of cleaving within the hydrophobic environment of the membrane has been proposed for a long time and was thought to be involved in signal peptide degradation (9), generation of cell-surface histocompatibility antigen HLA–E epitopes (10), and processing of the hepatitis C virus polyprotein (11). In these cases, SPP generates peptide fragments either to ensure their release from the membrane and to allow their subsequent degradation or to form functional small peptides involved in immune surveillance and virus invasion. Human SPP was biochemically identified upon affinity labeling using a photocross-linkable derivative of the SPP inhibitor 1,3-bis[(benzyloxycarbonyl-l-leucyl-l-leucyl)amino]acetone ((Z-LL)2-ketone) (7). This revealed a 42-kDa multi-TMD protein termed SPP. A data bank search identified five additional SPP homologs, SPPL2a/b/c, SPPL3, and SPPL4 (occurring only in yeast), defining the SPPL family (7, 8, 12). Although SPP/SPPL proteases lack obvious sequence homologies to other proteins, they all share a YD motif as well as the GXGD motif in two neighboring TMDs and additionally the PAL sequence, located in their most C-terminal TMD (Fig. 1A) (7, 8, 12, 13). These three conserved domains are required for the proteolytic activity of PS (5) and SPP (7, 14). The YD and GXGD motifs provide the critical aspartate residues required for the protease activity and represent novel active-site signature sequences not present in any of the conventional aspartyl proteases. A direct contribution of the PAL motif to the catalytic center is supported by the finding that a transition state analog inhibitor fails to bind to SPP and PS upon mutagenesis of the PAL sequence (15). It is currently unknown how the PAL domain affects SPP activity; however, one may assume a close proximity of all three conserved domains in the native enzyme forming the catalytic center. Indeed, this was recently demonstrated for PS (16). As for PS (17), mutagenesis of the aspartate residue within the GXGD motif leads to inhibition of SPP/SPPL (7, 14), and in zebrafish, expression of GXGD aspartate mutants phenocopies a morpholino-mediated knockdown of the respective SPP family member (14). Interestingly, the two active-site domains are in opposite membrane orientation in SPPs compared with PS (Fig. 1A) (18, 19). In line with the findings described above, the C-terminal region of SPP, which contains all three highly conserved motifs, was found to be sufficient for proteolytic activity (20).
SPP family members apparently do not require any other cofactors for their proteolytic activity (7) because, in contrast to PS, which needs three additional proteins to exert intramembrane proteolysis (Fig. 1A) (5, 21), overexpression of SPP family members leads to a significant increase in proteolytic turnover of a substrate (18, 22, 23). However, it has been claimed that SPP forms homodimers (24). Homodimer formation was demonstrated by glycerol gradient centrifugation and co-immunoprecipitation of differentially tagged SPP species (24). Strikingly, the homodimer was selectively labeled by an active-site inhibitor, strongly supporting the notion that dimerization is required for biological activity. However, one has to keep in mind that SPP is a very hydrophobic membrane protein; thus, artificial dimer formation may occur. Indeed, in a later study using a different inhibitor, selective labeling of the monomer was observed (25). Thus, more research is required to prove the functional relevance of dimer formation.
Members of the SPP family apparently accept only membrane proteins of type II orientation as substrates (Fig. 1B). Obviously, all signal peptides adopt a type II orientation during cotranslation into the ER and are therefore preferred substrates of SPP (7). Consistent with their substrate preference, SPP is located predominantly within the ER (14, 22). Whether or not SPP may be actively retained within the ER by its putative KKXX retention signal (7) is currently unknown. In Caenorhabditis elegans, the IMPAS gene, which encodes the worm SPP, is required for molting. Interestingly, the IMPAS knockdown phenotype is rescued by the homolog of the human lipoprotein receptor-related protein, which is known to be involved in cholesterol and lipoprotein homeostasis (12). However, a substrate of IMPAS has so far not been identified. Substrates for the ER-located SPPL family members, SPPL3 (14) and SPPL2c (22), are currently also not known, although it is tempting to speculate that they, in analogy to SPP, turn over signal peptides as well. However, one needs to keep in mind that SPPL3 may also occur within later compartments (22). SPPL2a and SPPL2b accumulate in the Golgi apparatus, on the plasma membrane, and within endosomes (14, 22). TNFα (Fig. 1B) (22, 23), the Fas ligand (26), and Bri2 (Itm2b) (27) are the only known substrates for SPPL2a/b. All three substrates undergo ectodomain shedding by a protease of the ADAM (a disintegrin and metalloprotease) family (Fig. 1B) (22, 23, 26, 27). Apparently, shedding of these substrates is not absolutely required for subsequent intramembrane proteolysis, but it may greatly facilitate this cleavage (45). In that regard, it is interesting to note that phorbol ester-induced shedding of TNFα significantly stimulates intramembrane proteolysis of TNFα (22), suggesting that shedding supports subsequent proteolysis. In line with this finding, SPP cleavage of the preprolactin signal sequence is very efficient upon previous cleavage by signal peptidase. However, even a mutant preprolactin substrate that is not processed by signal peptidase still undergoes intramembrane proteolysis to some extent (28).
Because ADAM-mediated shedding occurs predominantly on the plasma membrane, SPPL2a and SPPL2b may favor cleaving their truncated substrates at the cell surface or upon re-internalization within endosomes. The resulting ICD of the SPPL2a/b cleavage may be involved in nuclear signaling. Specifically, the ICD of TNFα has been shown to stimulate expression of the pro-inflammatory cytokine interleukin-12 (22). Similarly, the ICD of the Fas ligand translocates to the nucleus, where it may suppress gene transcription (26). Although specific signaling mechanisms have been identified for SPPL2a and SPPL2b, one may still expect that these proteases also perform a “membrane-proteasome”-like function for the degradation of type II-oriented membrane protein stubs.
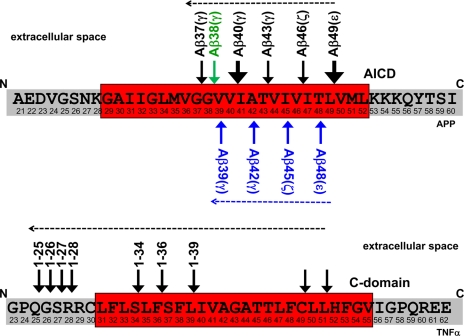
The substrates of SPP/SPPL family members may require unwinding of their α-helical TMD prior to endoproteolysis. For SPP, this is apparently promoted by helix-breaking residues within the hydrophobic core of the signal peptides (28), although such residues can be deleted in the TMD of TNFα without significantly affecting intramembrane proteolysis by SPPL2b.4 Unfortunately, structural information is not available for any of the SPP family members. Therefore, it is difficult to predict how the intramembrane cleavage is mechanistically performed and how the essential water molecules gain access to the hydrophobic environment. However, a single intramembrane cut does not seem to be sufficient for the release of the TNFα ICD (Fig. 2). In that case, multiple cleavages occurring within the membrane have been reported (23). These multiple cuts may be required to facilitate the efficient release of the cleavage products from the membrane. Whether such multiple cleavages occur with other substrates as well is unknown. However, a synthetic substrate for SPP was shown to undergo one major as well as several other minor intramembrane cuts (25). Nevertheless, it is too early to speculate if such multiple intramembrane cleavages are a general phenomenon for all SPP family members, although very similar cleavage patterns were observed for γ-secretase substrates such as APP and Notch. Here, these cleavages are termed ε, ζ, and γ and occur in a sequential manner, from ε to γ (Fig. 2) (see also Ref. 5).
FIGURE 2.
Multiple intramembrane cleavages are mediated byγ-secretase (upper) and SPPL2a/b (lower). The known cleavage sites of γ-secretase in APP (upper) and SPPL2a/b in TNFα (lower) are indicated by arrows. Major cleavage sites in the APP transmembrane domain are indicated by enlarged arrows. Dashed arrows indicate potential direction of the cleavages by the respective intramembrane protease. For APP, two parallel product lines have been suggested, which are initiated by cleavages after amino acid 48 or 49 (5). The initiating cleavage of γ-secretase liberates the APP intracellular domain (AICD) into the cytosol, whereas that of SPPL2a/b liberates the TNFα C-domain into the extracellular space.
Comparison of the Biochemical and Cellular Properties of GXGD Proteases
The above-described biochemical, structural, and functional properties of γ-secretase and members of the SPP/SPPL family clearly point out some striking similarities, but there are also obvious differences. The sequence homologies between the two proteases are very limited and predominantly restricted to three domains (Fig. 1A), which apparently all contribute to the active site. The very limited homology suggests that SPP/SPPL family members and PSs may have evolved by convergence and are probably not directly related. Moreover, γ-secretase clearly requires four different proteins (PS, Aph-1 (anterior pharynx-defective 1), Pen-2 (presenilin enhancer 2), and NCT) (21) for its activity, whereas SPP family members work either on their own or as homodimers (7, 23, 24). Currently, there is no evidence suggesting that any additional cofactors are required for SPP/SPPL function, and studies on functional expression of SPP/SPPL family members in mammalian cells and even in bacteria (20) may rule out essential cofactors.
Another very important difference is the opposite orientation of the active-site domains YD and GXGD (Fig. 1A) (7, 10, 19). It has been strongly argued that this difference is related to the different orientation of the individual substrates: type I for γ-secretase and type II for SPP/SPPL family members. However, although it is indeed tempting to argue in this direction, a formal proof has so far not been obtained. If that hypothesis is true, one has to find an opposite sequential order of the intramembrane cleavages discussed above. An opposite orientation of both substrate and active sites would suggest an ε-like cleavage liberating the secreted C-domain and a γ-like cleavage finally releasing the ICD of an SPP/SPPL substrate (Figs. 1B and 2).
The regulated intramembrane proteolysis pathway implies that initial shedding is required to allow substrate binding (2). The only exception of this rule so far is the family of Rhomboid proteases (see accompanying minireview by Wolfe (44)). γ-Secretase strictly follows this rule (Fig. 1B) because only N-terminally truncated substrates are accepted (29). Moreover, NCT is believed to mediate this size selection and to serve as the initial substrate-docking site (30), although that has recently been challenged (31). So far, no NCT-like factor has been identified for any of the SPP/SPPL family members. Nevertheless, as discussed above, initial shedding events have been reported for all known substrates of SPP/SPPL family members. Do these substrates require shedding to allow subsequent intramembrane proteolysis, or can the full-length protein be cleaved as well? SPPL2b has been shown to bind substantial amounts of its full-length substrates (23, 27), quite in contrast to γ-secretase, which co-isolates predominantly with the truncated substrate only (32). One may therefore speculate that both substrates can bind to SPPL2b, but only the trimmed substrate can be efficiently converted. Such a mechanism indicates that a truncated substrate may have chemical or structural properties by itself to facilitate intramembrane proteolysis. In the absence of an NCT-like activity, unwinding of the TMD of a truncated substrate may be energetically favored as soon as the ectodomain has been removed. However, more research and probably even a resolution of the structure of SPP with its substrate bound may be required to finally address this point.
Major hallmarks of PS are the many FAD-related mutations, which shift the γ-cleavage to amino acid 42 (or, in some rare cases, to amino acid 43) (5, 33). These mutations generally occur at highly conserved positions, and “artificial” mutations can be created by mutagenesis of such amino acids. Are SPP/SPPL family members similarly affected by amino acid exchanges of homologous amino acids? This has so far been addressed only by Sato et al. (25). They found that “FAD-like” mutations inserted in the YD and GXGD motifs of SPP apparently decreased the general activity to some extent. This “loss of function” or better “reduced function” correlates well with observations made for FAD-associated PS1 mutations expressed in C. elegans, where they affected the physiological function of γ-secretase in Notch signaling. In this case, the mutants showed a significant reduction of their ability to rescue the Notch phenotype of worms lacking their endogenous PS homolog (SEL-12) (34, 35). Moreover, it is known that certain very aggressive PS mutations selectively affect the ε-cleavage and therefore inhibit the efficient release of the ICD required for Notch signaling (36, 37). However, so far at least for SPP, no shift in the cleavage precision has been observed (25). Nevertheless, the reduced function of similar mutants in SPP and PS clearly suggests biochemical similarities of both types of GXGD-type proteases. Such similarities, which are probably related to structural homologies, are supported by the finding that certain γ-secretase inhibitors such as L-685,458 and helical peptides mimicking the substrate-docking site block SPP activity as well (15, 24, 25, 38, 39). Interestingly, whereas L-685,458 significantly reduced SPP activity, N-(N-(3,5-difluorophenacetyl)-l-alanyl-S-phenylglycine t-butyl ester (DAPT) failed to do so (38). This may suggest different binding sites of such inhibitors. As discussed in Ref. 5, a subset of NSAIDs and other compounds have been shown to exhibit a modulatory effect on γ-secretase specificity by shifting the pathological cleavage after amino acid 42 of the Aβ domain to position 38, which results in the production of a rather benign shorter Aβ peptide (40). Interestingly, the NSAIDs sulindac sulfide and indomethacin also shifted the SPP cleavage site of an artificial substrate by one amino acid (25). This may demonstrate again similar biochemical/structural properties of γ-secretase and SPP/SPPL. Furthermore, this also suggests that most likely the GXGD-type protease itself, the lipid environment of these proteases, or both are targeted by NSAIDs. Whether NSAIDs affect the structure of the active site, as has been proposed for PS (41), needs to be shown for SPP/SPPL family members. Because of the substantial differences of γ-secretase and SPP/SPPL substrates, selective NSAID binding to substrates seems to be unlikely. Similarly, the findings by Sato et al. (25) may also rule out Pen-2, NCT, and Aph-1 as potential binding sites because these proteins do not interact with SPP/SPPL.
Functionally, both γ-secretase and SPP/SPPL family members are I-CLiPs involved in reverse signaling (42). Such a function has now been best studied for the γ-secretase substrate Notch (43) and the SPPL2a/b substrate TNFα (22). In both cases, ICDs are generated from a membrane-bound precursor and are then employed in gene regulation. Whereas nuclear signaling has been clearly shown for the Notch ICD, such a signaling pathway has been only indirectly demonstrated for the TNFα ICD (22). As for γ-secretase, for which many substrates have been observed, we also expect in the near future the description of a much larger repertoire of SPP/SPPL substrates. Whether these are all involved in a physiologically relevant signaling pathway is unlikely. As for γ-secretase, we also expect that SPP family members may degrade membrane stubs (in type II orientation) by exhibiting a membrane-proteasome-like function, a biologically very important function required to exclude the clustering of cellular membranes with protein stubs. Finally, essential functions of SPP family members may be affected by γ-secretase inhibitors, a fact that needs to be kept in mind for the development of safe Alzheimer disease therapeutics.
Supplementary Material
This work was supported in part by the Deutsche Forschungsgemeinschaft (a Gottfried Wilhelm Leibniz award (to C. H.), Collaborative Research Center (SFB596) “Molecular Mechanisms of Neurodegeneration” (to H. S. and C. H.), and Grant HA1737-11 (to C. H. and R. F.)), the Bundesministerium für Bildung und Forschung (“Degenerative Dementias: Target Identification, Validation and Translation into Treatment Strategies” (to C. H. and H. S.)), the Alzheimer Research Award of the Hans and Ilse Breuer Foundation (to H. S.), and Boehringer Ingelheim Pharma GmbH & Co. KG. The first article in this minireview series on proteases was published in the November 7, 2008 issue; the second and third articles are published in this issue. Additional articles will be published in future issues. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: I-CLiP, intramembrane-cleaving protease; ICD, intracellular domain; TMD, transmembrane domain; PS, presenilin; SPP, signal peptide peptidase; ER, endoplasmic reticulum; TNFα, tumor necrosis factor α; APP, β-amyloid precursor protein; NCT, nicastrin; FAD, familial Alzheimer disease; NSAID, nonsteroidal anti-inflammatory drug; Aβ, amyloid β-protein.
R. Fluhrer and C. Haass, unpublished data.
References
- 1.Wolfe, M. S., and Kopan, R. (2004) Science 305 1119–1123 [DOI] [PubMed] [Google Scholar]
- 2.Brown, M. S., Ye, J., Rawson, R. B., and Goldstein, J. L. (2000) Cell 100 391–398 [DOI] [PubMed] [Google Scholar]
- 3.Kopan, R., and Ilagan, M. X. (2004) Nat. Rev. Mol. Cell Biol. 5 499–504 [DOI] [PubMed] [Google Scholar]
- 4.Haass, C., and Steiner, H. (2002) Trends Cell Biol. 12 556–562 [DOI] [PubMed] [Google Scholar]
- 5.Steiner, H., Fluhrer, R., and Haass, C. (2008) J. Biol. Chem. 283 29627–29631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LaPointe, C. F., and Taylor, R. K. (2000) J. Biol. Chem. 275 1502–1510 [DOI] [PubMed] [Google Scholar]
- 7.Weihofen, A., Binns, K., Lemberg, M. K., Ashman, K., and Martoglio, B. (2002) Science 296 2215–2218 [DOI] [PubMed] [Google Scholar]
- 8.Ponting, C. P., Hutton, M., Nyborg, A., Baker, M., Jansen, K., and Golde, T. E. (2002) Hum. Mol. Genet. 11 1037–1044 [DOI] [PubMed] [Google Scholar]
- 9.Weihofen, A., Lemberg, M. K., Ploegh, H. L., Bogyo, M., and Martoglio, B. (2000) J. Biol. Chem. 275 30951–30956 [DOI] [PubMed] [Google Scholar]
- 10.Lemberg, M. K., Bland, F. A., Weihofen, A., Braud, V. M., and Martoglio, B. (2001) J. Immunol. 167 6441–6446 [DOI] [PubMed] [Google Scholar]
- 11.McLauchlan, J., Lemberg, M. K., Hope, G., and Martoglio, B. (2002) EMBO J. 21 3980–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grigorenko, A. P., Moliaka, Y. K., Soto, M. C., Mello, C. C., and Rogaev, E. I. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 14955–14960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martoglio, B., and Golde, T. E. (2003) Hum. Mol. Genet. 12 R201–R206 [DOI] [PubMed] [Google Scholar]
- 14.Krawitz, P., Haffner, C., Fluhrer, R., Steiner, H., Schmid, B., and Haass, C. (2005) J. Biol. Chem. 280 39515–39523 [DOI] [PubMed] [Google Scholar]
- 15.Wang, J., Beher, D., Nyborg, A. C., Shearman, M. S., Golde, T. E., and Goate, A. (2006) J. Neurochem. 96 218–227 [DOI] [PubMed] [Google Scholar]
- 16.Tolia, A., Horré, K., and De Strooper, B. (2008) J. Biol. Chem. 283 19793–19803 [DOI] [PubMed] [Google Scholar]
- 17.Wolfe, M. S., Xia, W., Ostaszewski, B. L., Diehl, T. S., Kimberly, W. T., and Selkoe, D. J. (1999) Nature 398 513–517 [DOI] [PubMed] [Google Scholar]
- 18.Nyborg, A. C., Jansen, K., Ladd, T. B., Fauq, A., and Golde, T. E. (2004) J. Biol. Chem. 279 43148–43156 [DOI] [PubMed] [Google Scholar]
- 19.Friedmann, E., Lemberg, M. K., Weihofen, A., Dev, K. K., Dengler, U., Rovelli, G., and Martoglio, B. (2004) J. Biol. Chem. 279 50790–50798 [DOI] [PubMed] [Google Scholar]
- 20.Narayanan, S., Sato, T., and Wolfe, M. S. (2007) J. Biol. Chem. 282 20172–20179 [DOI] [PubMed] [Google Scholar]
- 21.Edbauer, D., Winkler, E., Regula, J. T., Pesold, B., Steiner, H., and Haass, C. (2003) Nat. Cell Biol. 5 486–488 [DOI] [PubMed] [Google Scholar]
- 22.Friedmann, E., Hauben, E., Maylandt, K., Schleeger, S., Vreugde, S., Lichtenthaler, S. F., Kuhn, P. H., Stauffer, D., Rovelli, G., and Martoglio, B. (2006) Nat. Cell Biol. 8 843–848 [DOI] [PubMed] [Google Scholar]
- 23.Fluhrer, R., Grammer, G., Israel, L., Condron, M. M., Haffner, C., Friedmann, E., Bohland, C., Imhof, A., Martoglio, B., Teplow, D. B., and Haass, C. (2006) Nat. Cell Biol. 8 894–896 [DOI] [PubMed] [Google Scholar]
- 24.Nyborg, A. C., Kornilova, A. Y., Jansen, K., Ladd, T. B., Wolfe, M. S., and Golde, T. E. (2004) J. Biol. Chem. 279 15153–15160 [DOI] [PubMed] [Google Scholar]
- 25.Sato, T., Nyborg, A. C., Iwata, N., Diehl, T. S., Saido, T. C., Golde, T. E., and Wolfe, M. S. (2006) Biochemistry 45 8649–8656 [DOI] [PubMed] [Google Scholar]
- 26.Kirkin, V., Cahuzac, N., Guardiola-Serrano, F., Huault, S., Luckerath, K., Friedmann, E., Novac, N., Wels, W. S., Martoglio, B., Hueber, A. O., and Zornig, M. (2007) Cell Death Differ. 14 1678–1687 [DOI] [PubMed] [Google Scholar]
- 27.Martin, L., Fluhrer, R., Reiss, K., Kremmer, E., Saftig, P., and Haass, C. (2008) J. Biol. Chem. 283 1644–1652 [DOI] [PubMed] [Google Scholar]
- 28.Lemberg, M. K., and Martoglio, B. (2002) Mol. Cell 10 735–744 [DOI] [PubMed] [Google Scholar]
- 29.Struhl, G., and Adachi, A. (2000) Mol. Cell 6 625–636 [DOI] [PubMed] [Google Scholar]
- 30.Shah, S., Lee, S. F., Tabuchi, K., Hao, Y. H., Yu, C., LaPlant, Q., Ball, H., Dann, C. E., 3rd, Sudhof, T., and Yu, G. (2005) Cell 122 435–447 [DOI] [PubMed] [Google Scholar]
- 31.Chávez-Gutiérrez, L., Tolia, A., Maes, E., Li, T., Wong, P. C., and De Strooper, B. (2008) J. Biol. Chem. 283 20096–20105 [DOI] [PubMed] [Google Scholar]
- 32.Esler, W. P., Kimberly, W. T., Ostaszewski, B. L., Ye, W., Diehl, T. S., Selkoe, D. J., and Wolfe, M. S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2720–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haass, C., and Selkoe, D. J. (2007) Nat. Rev. Mol. Cell Biol. 8 101–112 [DOI] [PubMed] [Google Scholar]
- 34.Levitan, D., Doyle, T. G., Brousseau, D., Lee, M. K., Thinakaran, G., Slunt, H. H., Sisodia, S. S., and Greenwald, I. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 14940–14944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baumeister, R., Leimer, U., Zweckbronner, I., Jakubek, C., Grunberg, J., and Haass, C. (1997) Genes Funct. 1 149–159 [DOI] [PubMed] [Google Scholar]
- 36.Moehlmann, T., Winkler, E., Xia, X., Edbauer, D., Murrell, J., Capell, A., Kaether, C., Zheng, H., Ghetti, B., Haass, C., and Steiner, H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 8025–8030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bentahir, M., Nyabi, O., Verhamme, J., Tolia, A., Horre, K., Wiltfang, J., Esselmann, H., and De Strooper, B. (2006) J. Neurochem. 96 732–742 [DOI] [PubMed] [Google Scholar]
- 38.Weihofen, A., Lemberg, M. K., Friedmann, E., Rueeger, H., Schmitz, A., Paganetti, P., Rovelli, G., and Martoglio, B. (2003) J. Biol. Chem. 278 16528–16533 [DOI] [PubMed] [Google Scholar]
- 39.Iben, L. G., Olson, R. E., Balanda, L. A., Jayachandra, S., Robertson, B. J., Hay, V., Corradi, J., Prasad, C. V., Zaczek, R., Albright, C. F., and Toyn, J. H. (2007) J. Biol. Chem. 282 36829–36836 [DOI] [PubMed] [Google Scholar]
- 40.Weggen, S., Eriksen, J. L., Das, P., Sagi, S. A., Wang, R., Pietrzik, C. U., Findlay, K. A., Smith, T. E., Murphy, M. P., Bulter, T., Kang, D. E., Marquez-Sterling, N., Golde, T. E., and Koo, E. H. (2001) Nature 414 212–216 [DOI] [PubMed] [Google Scholar]
- 41.Lleo, A., Berezovska, O., Herl, L., Raju, S., Deng, A., Bacskai, B. J., Frosch, M. P., Irizarry, M., and Hyman, B. T. (2004) Nat. Med. 10 1065–1066 [DOI] [PubMed] [Google Scholar]
- 42.Weihofen, A., and Martoglio, B. (2003) Trends Cell Biol. 13 71–78 [DOI] [PubMed] [Google Scholar]
- 43.Selkoe, D., and Kopan, R. (2003) Annu. Rev. Neurosci. 26 565–597 [DOI] [PubMed] [Google Scholar]
- 44.Wolfe, M. S. (2009) J. Biol. Chem. 284 13969–13973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin, L., Fluhrer, R., and Haass, C. (2009) J. Biol. Chem. 284 5662–5670 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.