Abstract
Recent studies have identified RNA transcripts arising from mammalian telomeres with the transcript of the C-rich strand (r(5′-UUAGGG-3′)n) being much more abundant than transcripts of the G-rich strand. Here we used transmission electron microscopy, CD, and nuclease digestion to investigate the structure of ∼640-nucleotide (nt) RNA transcripts of the C-rich and G-rich strands of mammalian telomeric DNA. The CD spectrum of the C-rich RNA in low salt (10 mm KCl) or high salt (100 mm KCl) was typical of mixed sequence RNA, whereas the CD spectrum for the G-rich RNA differed with changes characteristic of parallel G-quadruplexes at the higher salt concentration. Electron microscopy visualization of the C-rich RNA revealed relatively extended unstructured molecules 59.7 ± 17.8 nm in length and with a width consistent with single-stranded RNA following metal coating. In contrast, the G-rich RNA was observed as round particles and short, thick rods with the rods being most prevalent in high salt conditions and absent in low salt. The rods were 22.7 ± 4.8 nm in length and 7.6 nm in width. Digestion of the G-rich RNA with T1 RNA nuclease revealed a ladder of bands whose sizes were integral multiples of 24 nt plus a 4-nt overhang. These observations suggest a model in which G-rich telomeric RNA folds into chains of particles each consisting of four (UUAGGG) repeats stabilized by parallel G-quartets and joined by UUA linkers. These chains further condense to form short rods and round particles.
Telomeres are the terminal elements of eukaryotic chromosomes. They maintain the integrity of linear chromosomes by blocking misrecognition of the DNA ends as double strand breaks. They also provide a mechanism to overcome the progressive erosion of DNA sequences at chromosome ends due to the end replication problem, and they serve to control transcription of genes near the telomere. These complex functions reflect the interplay between a large set of both telomere-specific and nonspecific factors and the unique structural organization of the telomere that sets it apart from the remainder of the genome. Indeed telomeres have long been known to be heterochromatic and to show telomere position effects in which transcription of genes located near the telomere is repressed (reviewed in Ref. 1).
Telomeric DNA of most higher members of the animal kingdom consists of ∼5–20 kb of the repeat (5′-TTAGGG-3′)n, whereas in plants, the repeat is (5′-TTTAGGG-3′)n with plant telomeres frequently exceeding 50 kb (2, 3). The normal cellular core histones comprise the major protein species at plant and animal telomeres. In addition, there are six telomere-specific proteins termed the shelterin proteins that have been identified largely in mammalian cells, and they consist of TRF1 and TRF2, which bind directly to duplex telomeric repeats, Pot1, which binds to the single-stranded 3′ overhang on the G-rich strand, and additional factors including hRap1, Tin2, and TPP1, which scaffold onto TRF1 and TRF2 (reviewed in Ref. 4).
The organization of telomeres into chromatin and higher-order structures is believed to be central to their function. We showed that the 3′ single strand overhang at telomere ends will strand-invade back into the preceding duplex telomeric repeats to form a lasso-like structure termed a t-loop (5). T-loops provide a simple mechanism to hide telomere ends from the double strand break repair machinery, and these structures have been observed over a large range of eukaryotes (Refs. 5 and 6 and references therein). What factors contribute to the heterochromatinization of higher eukaryotic telomeres, the telomere position effects, and higher-order telomere structure remains unclear but may involve long range structural organization, histone modifications, the telomere-specific proteins, and possibly, structural RNA molecules.
Until recently, telomeres were thought to be transcriptionally silent. However, recent work from Azzalin et al. (7) and Schoeftner and Blasco (8) has revealed the presence of telomeric transcripts in mammalian cells with the transcript of the C-rich strand, r(UUAGGG)n being much more abundant than the transcript of the G-rich strand, r(CCCUAA)n (7, 8). Transcription of the G-rich RNA is thought to be initiated from subtelomeric loci by RNA polymerase II and can range in size up to 9 kb. Interestingly, G-rich RNA molecules localize to telomeres, where they may play a role in inhibiting telomerase activity (8). The G-rich transcripts also functionally interact with the SMG class of proteins, and this is required for telomere maintenance (7). Thus, the G-rich telomeric RNA could represent a new structural component of the telomere resembling X-chromosome inactivation in females via Xist RNA (9).
The G-rich telomeric RNA contains repeats of three guanines, a motif that has been shown to form G-quadruplexes in single-stranded DNA (10–13). G-quadruplexes consist of multiple quartets of guanine bases hydrogen-bonded to one another and associated with either a monovalent or a divalent cation. If G-quadruplexes were to form in the G-rich telomeric RNA, this would result in a very different structure as contrasted to the C-rich telomeric RNA, which would not be predicted to form similar structures. G-quadruplexes may shield the G-rich telomeric transcripts from nucleases, as well as providing a conformation different from normal transcripts. Single-stranded DNA-containing tracts of guanines can fold into a variety of quadruplex structures with the strands folding into parallel or antiparallel arrangements, depending on sequence and ionic environment. G-quadruplexes can form in an intramolecular or intermolecular fashion (reviewed in Refs. 10 and 14).
In a study of human and Oxytricha 12–72-nt2-long single-stranded G-rich telomeric DNA molecules, Yu et al. (10) concluded that although the Oxytricha telomeric repeat with four guanines can exhibit both intermolecular and intramolecular interactions, the human DNA with three guanines per repeat showed only intramolecular interactions. They proposed that the human DNA was arranged into a “beads on a string” structure with each “bead” consisting of three G-quartet bundles stacked in an antiparallel arrangement, joined by flexible TTA linkers.
Although studies of short G-rich telomeric DNA oligonucleotides provide a guide for understanding the structure of short G-rich telomeric RNA transcripts, less is known about G-quadruplex formation in RNA at lengths consistent with those found in vivo. Furthermore, RNA can be stabilized by base pairs that are not allowed in DNA, and runs of guanines in RNA may be sterically inhibited from forming antiparallel quadruplexes (15, 16). Finally, the telomeric RNA transcripts in human cells range from ∼100 nt to nearly 9000 nt, and such long RNA molecules could form complex structures not possible with short RNA molecules (7, 8). For these reasons, we have examined the structure of G-rich and C-rich telomeric RNA transcripts of the size found in vivo.
In this study, we transcribed a block of 5′-TTAGGG-3′ duplex repeats in a plasmid to generate G-rich and C-rich telomeric RNA ∼640 nt long. CD analysis and transmission electron microscopic (EM) examination revealed striking differences between the G-rich and C-rich telomeric RNA. EM analysis showed that the G-rich molecules were highly compacted into round particles and short thick rods, and the CD spectral signature of the same molecules was consistent with the presence of G-quartets. On the other hand, similar CD spectra and compacted structures were not observed in the C-rich RNA molecules. Additionally, T1 nuclease fragments the G-rich RNA, which is resolved on gels into a ladder of bands separated by a 24-nt repeat. These results suggest a model for the intramolecular folding of the RNA. Recently, Xu et al. (17) published a study of synthetic r(UUAGGG) RNA oligonucleotides 12 and 22 nt long and reported G-quadruplex formation based on CD and NMR data. As discussed below, their suggestion of intermolecular associations is not supported by our findings.
EXPERIMENTAL PROCEDURES
RNA Synthesis—A pGEM-based plasmid, pRST5, containing 576 bp of telomeric repeats was linearized with NotI and transcribed with T7 RNA polymerase to generate the G-rich transcript or linearized with HindIII and transcribed with T3 RNA polymerase to generate the C-rich transcript (18). The transcription reactions were performed using a MEGAscript® kit (Ambion/AB, Austin, TX) according to the manufacturer's procedures. The RNA was purified using two phenol:chloroform extractions followed by an isopropyl alcohol precipitation in ammonium acetate. The samples were then treated with proteinase K (Roche Diagnostics GmbH, Mannheim, Germany) at 50 mg/ml in the presence of 0.5% SDS for 3 h at 55 °C. This was followed by two phenol:chloroform extractions and isopropyl alcohol precipitation in ammonium acetate. The integrity of the RNA was determined using 8 m urea/4% polyacrylamide gel electrophoresis followed by staining with ethidium bromide.
Gel Electrophoretic Analysis of the RNA—To determine the integrity of the RNA after transcription, the RNA was melted for 5 min at 95 °C in a 95% formaldehyde loading buffer and then run on an 8 m urea/4% polyacrylamide gel at 125 V for 30 min. The gel was stained with ethidium bromide and visualized with a UV transilluminator in a Gel Doc XR system (Bio-Rad).
RNase T1 Digestion—The RNA molecules were prepared for digestion by melting at 95 °C for 5 min followed by slow cooling to 4 °C at a rate of 1 °C/min; the G-rich RNA was incubated in 0, 25, 50, 100, and 150 mm KCl to promote quadruplex formation before subsequent RNase digestion. Both the C-rich and the G-rich RNA molecules were incubated with RNase T1 (Ambion/AB) according to the manufacturer's procedures for RNA structure analysis and then electrophoresed on denaturing 8 m urea/4% polyacrylamide gels followed by staining with ethidium bromide. The gels were visualized as described above, and the lengths of RNA bands in the gel were determined using a 0.1–2-kb RNA ladder (Invitrogen) and Quantity One software (Bio-Rad).
CD Spectral Analysis—Circular dichroism measurements were performed on C-rich and G-rich RNA at the University of North Carolina (UNC) Macromolecular Interactions Facility using a Pistar-180 CD spectropolarimeter fitted with a Peltier temperature control unit (Applied Photophysics Ltd., Leatherhead, UK). Each RNA sample was diluted to a final concentration of 40 μg/ml in 10 mm potassium phosphate buffer (pH 7.5) with or without 100 mm KCl. The RNA samples were heated to 95 °C for 5 min and then slowly cooled to 4 °C at a rate of 1.0 °C/min. CD spectra were recorded at 23 °C from 200 to 320 nm with a 0.5-nm step size using a 1-mm path length cuvette. A baseline CD measurement of 10 mm potassium phosphate buffer was subtracted from each spectrum. Baseline-subtracted spectra were smoothed using the Pistar Pro-Data software.
Electron Microscopy—Preparation of the RNA followed the procedure of Griffith and Christiansen with modifications (19). The RNA molecules were folded by heating to 95 °C in 10 mm Tris, pH 7.7, 0.1 mm EDTA, and 100 mm KCl buffer for 5 min and then cooled to 4 °C at 1 °C per min in a Mastercycler gradient thermocycler (Eppendorf AG, Hamburg, Germany). The RNA was diluted to 2 μg/ml in the same buffer and adsorbed to thin glow-charged carbon foils supported by 400 mesh copper grids in the presence of 2.5 mm spermidine for 1 min. This was followed by dehydration through 0, 25, 50, 75, 100% ethyl alcohol/water washes, air drying, and finally rotary shadowcasting with tungsten at 1 × 10–6 torr. Samples were imaged in an FEI Tecnai 12 EM instrument at 20 or 40 kV, and images were recorded using a Gatan Ultrascan 4000 CCD camera (Gatan Inc., Pleasanton CA). Lengths of the RNA molecules were determined from the digital images using Gatan digital micrograph software.
RESULTS
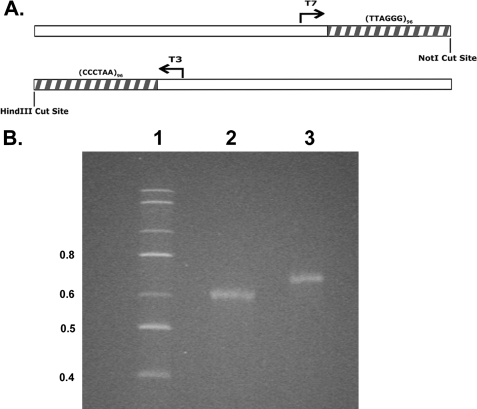
Synthesis of Long C-rich and G-rich RNA Molecules—To synthesize C-rich and G-rich telomeric RNA molecules of a size found in vivo, we utilized a plasmid generated in this laboratory containing a cassette of 96 telomeric repeats (18). This telomeric repeat sequence is flanked by T3 and T7 RNA polymerase promoters oriented into the repeat block in opposite directions, allowing transcription of the G-rich (T7 RNA polymerase) or C-rich (T3 RNA polymerase) strands (Fig. 1A). By cleaving the plasmid immediately adjacent to the telomeric repeat sequence, RNA molecules consisting of (5′-CCCUAA-3′)96 or (5′-UUAGGG-3′)96 sequence can be synthesized. Following in vitro transcription and purification of the RNA, denaturing polyacrylamide gel electrophoresis (see “Experimental Procedures”) revealed full-length transcripts with few degradation products (Fig. 1B). Under these conditions, the C-rich RNA, which is 20 nt shorter, migrated slightly faster than the G-rich RNA.
FIGURE 1.
Plasmid map and synthesis of telomeric RNA transcripts. A, a map of the linearized plasmid containing 96 TTAGGG repeats and the cleavage sites used to generate G-rich (T7 RNA polymerase) and C-rich (T3 RNA polymerase) transcripts. B, electrophoresis of the C-rich RNA (lane 2) and G-rich RNA (lane 3) on an 8 m urea/4% polyacrylamide gel (see “Experimental Procedures”). Lane 1 shows makers of 0.4 kbases and higher.
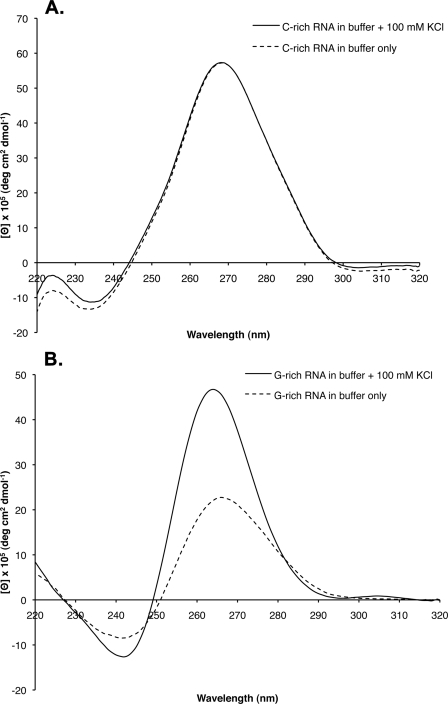
CD Spectral Analysis Reveals G-quartet Formation in the G-rich Telomeric RNA—G-quadruplex formation in RNA has been shown to generate a characteristic CD spectral change (16, 20–22). To measure the CD spectra, the C-rich and G-rich RNA molecules were melted at 95 °C for 5 min and allowed to slowly cool to 4 °C. In 100 mm KCl, the spectrogram of the G-rich RNA revealed a positive peak at 265 nm and a negative peak at 240 nm, characteristic of parallel G-quadruplex formation in RNA (16, 20–22) (Fig. 2A). Furthermore, the peaks were enhanced as the concentration of KCl was increased from 10 to 100 mm, suggesting that the increased ionic strength is responsible for inducing quadruplex formation in the RNA. The CD spectra of the C-rich RNA (Fig. 2B) showed no change after the addition of 100 mm KCl.
FIGURE 2.
CD spectral analysis of the G-rich and C-rich telomeric RNA molecules. The CD spectra of the G-rich (A) and C-rich (B) RNA transcripts were determined (see “Experimental Procedures” and “Results”) in low salt buffer or buffer containing 100 mm KCl. deg, degree.
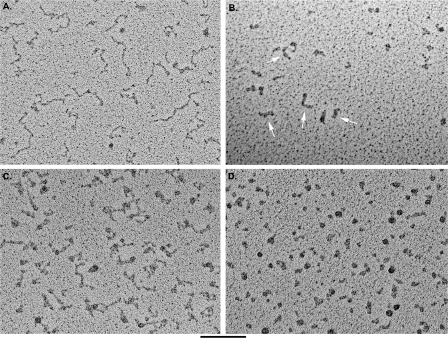
Visualization of the Telomeric RNA Molecules Reveals Highly Compacted G-rich but Not C-rich Telomeric RNA Molecules—C-rich and G-rich telomeric RNAs were prepared for EM using methods we applied previously to visualize other RNA molecules (Refs. 23 and 24; see also “Experimental Procedures”). Both the G-rich and the C-rich telomeric RNA were melted at 95 °C for 5 min and then slowly cooled to 4 °C in TE buffer (10 mm Tris, 0.1 mm EDTA, pH 7.5) or in TE plus 100 mm KCl or TE plus 2 mm MgCl2. The samples were further diluted in thea same buffer used during folding and prepared for EM visualization (see “Experimental Procedures”). Examination of the C-rich RNA in all salt conditions revealed fields of nucleic acid molecules (Fig. 3A) with an extended, kinked appearance, typical of single-stranded RNA or DNA lacking extensive secondary structure. Measurement of 50 typical molecules from the C-rich RNA sample folded in TE buffer with 100 mm KCl yielded an average length of 59.7 ± 17.8 nm. The measured width of the molecules was 3.9 ± 0.9 nm, but this included the kinks and was thus larger than the RNA strand alone. Although the length of the RNA is severalfold shorter than what would be expected for extended linear duplex RNA of 640 bp (∼0.3–0.4 nm/nt), it was not possible to estimate the foreshortening due to the kinking. Nonetheless, the RNA was significantly more extended than typical mRNA molecules we have examined in the past. The length and appearance of the C-rich RNA was not significantly different when the RNA was prepared for EM in buffers containing lower or higher KCl or magnesium.
FIGURE 3.
Visualization of C-rich and G-rich telomeric transcripts. C-rich (A) and G-rich (B) RNA molecules in 100 mm KCl were prepared for EM by mounting on thin carbon supports, dehydrating, and rotary shadowcasting with tungsten. The C-rich RNA appears as an extended thread with kinks and compact regions. The G-rich RNA appears as a mixture of balls and thick rods (arrows). The thickness of the rods is significantly greater than that of the C-rich RNA or duplex DNA. C-rich (C) and G-rich (D) RNA molecules were mounted in 10 mm KCl as in A and B. The C-rich RNA again appears extended with kinks, whereas the G-rich RNA appears as mostly balls as opposed to a mixture of rods and balls. The bar is equivalent to 100 nm.
Examination of the G-rich RNA folded in a buffer containing 100 mm KCl revealed fields of RNA molecules with a very different appearance than the C-rich RNA. The G-rich RNA molecules appeared as either round particles or short thick rods (Fig. 3B). Quantification of 160 G-rich RNA molecules folded in 100 mm KCl revealed that 53% were folded into rod-like structures, 32% were folded into round particles, and 11% were linear with knots or small beads along the molecule length. Only 3% of the molecules were extended linear structures typical of the C-rich molecules. When the RNA was prepared in 100 mm NaCl, 30% were rods, 49% were round particles, 17% were linear molecules with knots, and 4% were kinked linear molecules typical of the C-rich RNA. The most distinct species were the rod-like molecules. Measurement of 50 rods provided a mean length of 22.7 ± 4.8 nm, roughly one-third of the length of the C-rich RNA molecules folded in the same conditions. Width of the shadowcasted rod-like molecules measured 7.6 ± 1.2 nm, which is twice the thickness of shadowcasted duplex DNA (∼3–4 nm).
When the C-rich and G-rich RNA molecules were folded in low salt (TE with 10 mm KCl), the C-rich RNA appeared extended and kinked as observed in C-rich molecules folded in 100 mm KCl (Fig. 3C). In contrast, the G-rich RNA folded in low salt appeared as fields of round or ring-like particles with few if any rods or extended RNA fibers (Fig. 3D). The round particles were the same size as those observed in molecules folded in the higher salt concentration (100 mm KCl).
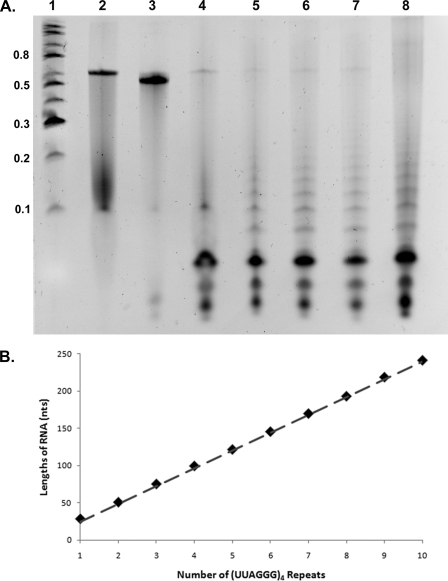
T1 Nuclease Cleaves the G-rich RNA into a Ladder of Fragments Consistent with Parallel G-quadruplex Formation—T1 nuclease cleaves RNA after a single-stranded guanine residue. Thus, it would not be expected to cleave the C-rich RNA but may or may not cleave the G-rich RNA depending on whether the guanines are sequestered into G-quadruplexes. Both the C-rich and the G-rich RNA molecules were incubated with T1 nuclease for 15 min at 23 °C. The products were ethyl alcohol-precipitated and then electrophoresed on an 8 m urea/4% acrylamide gel, and the RNA was stained with ethidium bromide. As shown in Fig. 4A (lane 3), the C-rich RNA was not cleaved by T1. Digestion of the G-rich RNA, however, produced a striking ladder of bands (Fig. 4A, lanes 5–8). The ladders were more pronounced when the G-rich RNA was folded in 150 mm KCl as contrasted to 25 mm KCl (compare lanes 5 and 8). Comparison with markers showed that the G-rich RNA was digested into products, the smallest of which was 28–29 nt followed by bands migrating close to 52, 75, 100, 122, 146, 170, 193, 219, and 242 nt. These values indicate that the RNA had been cleaved into multiples of 24 nt with the smallest fragment representing four (UUAGGG) repeats folding into a quadruplex inaccessible to RNase T1 plus one UUAG extension beyond the quadruplex in the 3′ direction. As illustrated in Fig. 4B, the sizes of the ladder of bands fit very well with the predicted sizes for multiples of 24 nt plus a 4-nt overhang. In contrast, as expected, the C-rich RNA was not cleaved by T1 nuclease, and a single RNA band is observed after T1 nuclease digestion.
FIGURE 4.
T1 RNase digestion of G-rich and C-rich telomeric transcripts. A, the C-rich (lane 3) and G-rich (lanes 4–8) RNA molecules were treated with T1 RNase in the presence of buffer only (lane 4), 25 mm KCl (lane 5), 50 mm KCl (lane 6), 100 mm KCl (lane 7), and 150 mm KCl (lane 8). Lane 2 contains untreated G-rich RNA. The C-rich RNA lacks cleavage sites and is not digested. The G-rich RNA is cleaved into a ladder of bands, which is more pronounced under conditions favoring G-quadruplex formation. B, the sizes of the cleavage products in lanes 5–8 were determined based on the markers (lane 1, 0.1–0.8 kb). These lengths (solid diamonds) are plotted as size in nt (y axis) versus the number of (UUAGGG)4 repeats each band would contain. The dotted line shows the predicted size of cleavage products, each containing multiples of (UUAGGG)4 repeats plus a single UUAG overhang.
DISCUSSION
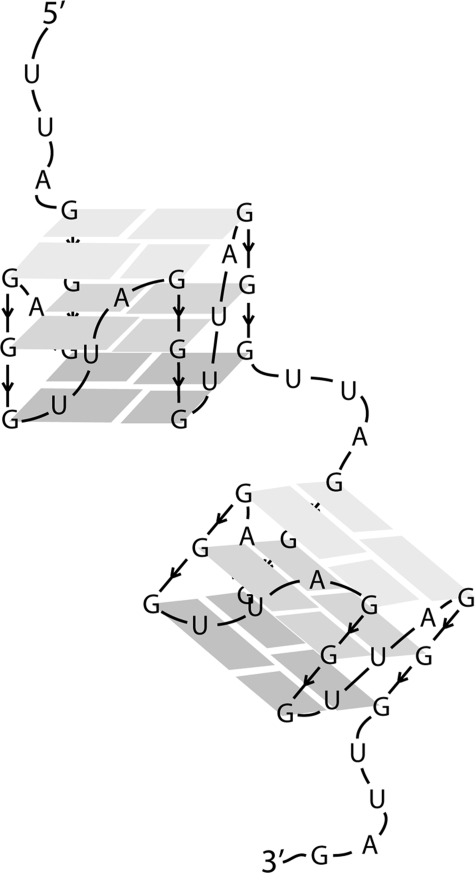
In this study, we examined the structure of telomeric RNA transcripts of a length typically found in mammalian cells. We generated both the C-rich and the G-rich transcripts using bacterial RNA polymerases and a plasmid containing a 576-bp tract of pure telomeric repeats. Although the C-rich RNA was relatively featureless, as seen by EM, and showed a CD spectrum typical of mixed sequence RNA, the G-rich RNA showed spectral signatures of parallel G-quadruplex formation in 100 mm salt, and as seen by EM, was condensed into compact rods and round balls. Further, although the C-rich RNA was resistant to digestion by T1 nuclease, the G-rich RNA was cleaved into a series of fragments of increasing size, beginning with ∼28 nt followed by multiples of 24 nt. Thus, we conclude that the G-rich RNA is organized into a chain of G-quadruplex beads, where each bead is stabilized by G-quartets. The G-quadruplexes are joined by UUA linkers, and the EM data suggest that this chain of beads then further folds on itself into highly compact structures.
The RNA particles detected by T1 nuclease digestion would be less than 10 kDa in mass. This is below the limit of resolution of the EM methods used here, even if the G-rich RNA were extended into a linear string of beads. It was not obvious from the EM images how the round particles are related to the short thick rods. The observation that the rods are absent under conditions that do not promote G-quartet formation (annealing in 10 mm KCl) might argue that the round particles lack G-quadruplexes and may be compacted via only base pairings between UUA segments. On the contrary, the RNA in the round particles could be arranged into G-quadruplex beads but may represent a different, higher-order folding pathway less favored in the higher salt. We could not determine whether the round particles were spherical or more disk-shaped. However, the observation that some of the round balls appeared as donuts with holes in the center and the fact that the projected area of the round particles and short thick rods were roughly similar might suggest a relation as simple as a curling of the rods into rings or disks. Were the G-rich RNA extended to 0.3–0.4 nm/nt, then the chain would measure ∼200 nm in length. Very rough modeling suggests that condensation into a string of G-quartet beads, as illustrated in Fig. 5, would add a ∼3-fold compaction, reducing the length to 60 nm. The short thick rods seen by EM measure 22 nm, arguing that the G-rich RNA has undergone a 3-fold further compaction.
FIGURE 5.
Model of the folding of the G-rich RNA into a string of RNA beads. The G-rich RNA transcript is shown folded into a string of RNA beads, with each bead consisting of a parallel G-quadruplex formed by three G-quartets joined by a UUA linker. Interactions between the UUA linkers may lead to higher-order folding, as seen by EM.
In the recent study of Xu et al. (17), an elegant combination of CD, NMR, and mass spectrometry was used to examine the structure of a 12-nt RNA oligonucleotide (r(UAGGGUUAGGGU) in the presence of sodium ions. They interpreted their results as indicating that two such oligonucleotides interact to generate an intermolecular, parallel G-quadruplex structure. No studies were carried out on the analogous C-rich RNA. The G-rich oligonucleotide was resistant to T1 nuclease in sodium and potassium ions. Major differences between their study and the results reported here have to do with the size of the telomeric RNA and concentrations used. The high concentration of oligonucleotides they used would favor intermolecular interactions, whereas their short size would exclude intramolecular interactions. In our study, the large size of the RNA molecules used allows ample opportunity for complex intramolecular folding and G-quadruplex formation.
The study of Yu et al. (17) used longer telomeric G-rich DNA oligonucleotides ranging up to 72 nt and employed CD and gel electrophoretic analysis (10). Single-stranded DNA has been shown to form both parallel and antiparallel G-quadruplex structures (10–13). For the human telomeric repeat, their results excluded intermolecular interactions and were consistent with a model in which the DNA was condensed into a linear chain of particles to generate a “string of beads” conformation. Each bead, they suggested, consists of a block of four telomeric repeats organized in an antiparallel arrangement, and these beads are joined by TTA linkers.
From the evidence presented in this report, we propose a beads on a string model for long telomeric RNA folding where individual (UUAGGG)4 units form quadruplex beads, connected by UUA linkers. Evidence from CD suggests that in the presence of potassium ions, parallel G-quartets will form in the G-rich RNA, in agreement with previous studies on telomeric RNA. After folding the G-rich RNA in 100 mm KCl, RNase T1 digestion produced a ladder of bands from 28–242 nt with an average distance between bands of 23.8 nt, representing RNase T1 degrading the RNA one bead at a time. In addition, the presence of the smallest band at ∼28 nucleotides represents the maximum length of one bead, likely a (UUAGGG)4UUAG sequence (RNase T1 cuts after single-stranded guanine residues). Evidence from EM suggests that the RNA further folds onto itself to form a more complex structure, likely involving interactions between the UUA linkers. In the crystal structure of a quadruplex-forming telomeric DNA (PDB 1K8P), the analogous TTA linkers were found to extend laterally from the G-quadruplex, thereby becoming available for base pairing or other intermolecular interactions (12). Thus, it is conceivable that a similar folding motif is formed in telomeric RNA, where base pairing through U·A or U·U base pairs leads to further compaction in the long RNA molecules.
In vivo the G-rich RNA is localized to the telomere (7, 8). This likely reflects specific binding of the G-rich RNA to shelterin or other proteins, which then mediate telomeric association. A recent study by Pedroso et al. (25) concluded that TRF2 can modulate G-quadruplex formation in telomeric DNA sequences. High concentrations of K+ inhibited a d(TTAGGG)n oligonucleotide uptake into the telomeric sequence of pRST5, suggesting that G-quadruplexes may inhibit the invasion of the single-stranded 3′ overhang of telomeres into duplex telomeres, thereby preventing t-loop formation. However, this inhibition was overcome by TRF2. Whether the telomeric RNA is involved in a similar interaction between DNA and TRF2 is unknown. In the future, it will be important to examine the structure of RNA-protein complexes formed with the G-rich telomeric RNA.
Acknowledgments
We thank SezginÖzgür, Sarah Compton, Gökhan Tolun, and Smaranda Willcox for aid and advice in carrying out these experiments and Ashutosh Tripathy in the UNC Macinfac core facility for aid in the CD spectral analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants GM31819 and ES013773 (to J. D. G.). This work was also supported by an award from the Glenn Foundation.
Footnotes
The abbreviations used are: nt, nucleotide; EM, electron microscopic; TE, Tris EDTA.
References
- 1.Nosek, J., and Tomaska, L. (2008) Origin and Evolution of Telomeres, Landes Bioscience, Austin TX
- 2.Fajkus, J., Kovarik, A., Kralovics, R., and Bezdek, M. (1995) Mol. Gen. Genet. 247 633–638 [DOI] [PubMed] [Google Scholar]
- 3.Cesare, A. J., Quinney, N., Willcox, S., Subramanian, D., and Griffith, J. D. (2003) Plant J. 36 271–279 [DOI] [PubMed] [Google Scholar]
- 4.de Lange, T. (2005) Genes Dev. 19 2100–2110 [DOI] [PubMed] [Google Scholar]
- 5.Griffith, J., Comeau, L., Rosenfield, S., Stansel, R., Bianchi, A., Moss, H., and de Lange, T. (1999) Cell 97 503–514 [DOI] [PubMed] [Google Scholar]
- 6.Raices, M., Verdun, R. E., Compton, S. A., Haggblom, C. I., Griffith, J. D., Dillin, A., and Karlseder, J. (2008) Cell 132 745–757 [DOI] [PubMed] [Google Scholar]
- 7.Azzalin, C., Reichenbach, P., Khoriauli, L., Giulotto, E., and Lingner, J. (2007) Science 318 798–801 [DOI] [PubMed] [Google Scholar]
- 8.Schoeftner, S., and Blasco, M. A. (2008) Nat. Cell Biol. 10 228–236 [DOI] [PubMed] [Google Scholar]
- 9.Ng, K., Pullirsch, D., Leeb, M., and Wutz, A. (2007) EMBO Rep. 8 34–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu, H. Q., Miyoshi, D., and Sugimoto, N. (2006) J. Am. Chem. Soc. 48 15461–15468 [DOI] [PubMed] [Google Scholar]
- 11.Wang, Y., and Patel, D. J. (1993) Structure (Lond.) 1 263–282 [DOI] [PubMed] [Google Scholar]
- 12.Parkinson, G. N., Lee, M. P., and Neidle, S. (2002) Nature 417 876–880 [DOI] [PubMed] [Google Scholar]
- 13.Phan, A. T., Kuryavyi, V., Luu, K. N., and Patel, D. J. (2007) Nucleic Acids Res. 35 6517–6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keniry, M. (2001) Biopolymers 56 123–146 [DOI] [PubMed] [Google Scholar]
- 15.Westhof, E., and Fritsch, V. (2000) Structure (Lond.) 8 R55–R65 [DOI] [PubMed] [Google Scholar]
- 16.Tang, C. F., and Shafer, R. H. (2006) J. Am. Chem. Soc. 128 5966–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu, Y., Kaminaga, K., and Komiyama, M. (2008) J. Am. Chem. Soc. 130 11179–11184 [DOI] [PubMed] [Google Scholar]
- 18.Stansel, R., de Lange, T., and Griffith, J. D. (2001) EMBO J. 20 5532–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffith, J., and Christiansen, G. (1978) Annu. Rev. Biophys. Bioeng. 7 19–35 [DOI] [PubMed] [Google Scholar]
- 20.Qi, J., and Shafter, R. H. (2007) Biochemistry 46 7599–7606 [DOI] [PubMed] [Google Scholar]
- 21.Kumari, S., Bugaut, A., Huppert, J. L., and Balasubramanian, S. (2007) Nat. Chem. Biol. 3 218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wieland, M., and Hartig, J. S. (2007) Chem. Biol. 7 757–763 [DOI] [PubMed] [Google Scholar]
- 23.Wang, Y.-H., Murphy, F., Cech, T., and Griffith, J. (1994) J. Mol. Biol. 236 64–71 [DOI] [PubMed] [Google Scholar]
- 24.Nakamura, T., Wang, Y.-H., Zaug, A., Griffith, J., and Cech, T. (1995) EMBO J. 14 4849–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedroso, I. N., Hayward, W., and Fletcher, T. M. (2008) Nucleic Acids Res. 37 1541–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]