Abstract
Hypoxia-inducible factor-1α (HIF-1α) overexpression was shown to be associated with invasion and metastasis of tumors and tumor cell lines. The identification of molecular targets that contribute to HIF-1α-mediated invasion is under intensive investigation. We have analyzed the role of recepteur d'origine nantais (RON), a tyrosine kinase receptor for macrophage-stimulating protein (MSP) that plays a role in breast cancer cell invasion as one of the molecular targets of HIF-1α. Analysis of a panel of breast cancer cell lines indicated a correlation between HIF-1α and RON expression. Treatment of HIF-1α- and RON-positive breast cancer cells with HIF-1α inhibitor, echinomycin, led to the inhibition of HIF-1α activity and RON expression. We have identified HIF-1α binding site on the RON promoter. Chromatin immunoprecipitation analysis and site-directed mutagenesis of the RON promoter confirmed the binding of HIF-1α to RON promoter. HIF-1α inhibitor-, echinomycin-, or short hairpin RNA-mediated selective knockdown of HIF-1α or HIF-1α target RON tyrosine kinase abrogated RON gene expression, and the RON ligand macrophage-stimulating protein mediated invasion of breast cancer cells. Consequently, the data presented herein demonstrated RON as a novel molecular target of HIF-1α and suggest a potential therapeutic role for HIF-1α or RON tyrosine kinase inhibitors in the blockade of RON tyrosine kinase-mediated invasion of carcinoma cells.
The hypoxic response is mainly regulated by the hypoxia-inducibl efactor-1 (HIF-1),2 a basic helix-loop-helix transcription factor composed of two subunits HIF-1α and HIF-1β (1). HIF-1α forms heterodimers with HIF-1β, and this complex binds to hypoxia-responsive element (HRE: 5′-RCGTG-3′) within the promoter regions of target genes. Multiple studies of HIF-1α and breast cancer have shown a significant association between HIF-1α overexpression and poor prognosis coupled to increased patient mortality (2–6). The levels of HIF-1α in human primary breast tumors increased with the progression of the pathologic stage (7). In a large retrospective study of 745 patients with high levels of HIF-1α at diagnosis, early relapse and metastatic disease were predicted (5). HIF-1α expression is closely linked to an aggressive phenotype in breast cancer, and HIF-1α expression enhanced osteolytic bone metastasis of breast cancer (8, 9). After prolonged treatment hormone-sensitive breast tumors frequently become resistant to hormonal therapy, and it was hypothesized that hypoxia may promote estrogen-independent growth. Deletion of HIF-1α in the mammary epithelium resulted in delayed tumor onset and retarded tumor growth as well as decreased pulmonary metastasis (10). These results suggest that HIF-1α is a negative prognostic factor in breast cancer progression. The HIF-1β subunit is constitutively expressed, whereas expression of HIF-1α is regulated by oxygen tension. HIF-1α protein is not detected in cells under normoxic conditions (20–22% O2) and is rapidly induced by hypoxic conditions (1–2% O2). However, in the invasive carcinoma cells, including breast, steady-state HIF-1α expression can be detected even under normoxia. The synthesis of HIF-1α protein has been shown to be regulated in an O2-independent fashion, for example, through activation of the receptor tyrosine kinase pathways (11, 12). The molecular targets of HIF-1α that contribute to breast tumorigenesis are under active investigation.
Macrophage-stimulating protein (MSP) is the only known ligand for recepteur d'origine nantais (RON), a tyrosine kinase receptor. MSP is an 80-kDa heterodimer consisting of a 53-kDa α-chain and a 30-kDa β-chain linked by a disulfide bond. The β-chain of MSP binds to RON (13). RON is initially synthesized as a single chain precursor, 170-kDa pro-RON, which is subsequently cleaved into 40-kDa alpha chain and 150-kDa beta chain. The alpha chain is completely extracellular, whereas the beta chain traverses the cell membrane and contains the intracellular tyrosine kinase (13). The RON receptor also participates in cross-talk with other receptor tyrosine kinases such as MET and epidermal growth factor receptor. Several human tumor tissues show increased RON expression, including tumors of the breast, colon, lung, liver, kidney, ovary, stomach, pancreas, bladder, and prostate (14). Gene expression analyses indicated increase in RON expression is associated with metastatic disease. Transgenic mice that overexpress a wild-type or constitutively active RON receptor in the mammary epithelium induced mammary transformation and associated with a high degree of metastasis with metastatic foci detected in the liver and lungs of >86% of all the transgenic animals (15). These studies demonstrated that RON overexpression can be a causative factor for metastatic breast cancer. RON overexpression in human breast cancer is associated with an aggressive cancer phenotype with decreased disease free survival time in patients and an increase in breast cancer metastasis (16).
We have recently shown that MSP promotes invasion of RON expression positive but not RON-negative breast cancer cells (17). Since the published clinical data suggested a correlation between HIF-1α, RON expression, and metastatic status of tumors, we have investigated whether HIF-1α/RON axis plays a role in breast cancer cell invasion. Screening of a panel of breast cancer cell lines indicated a correlation between steady-state HIF-1α and RON protein expression. A small molecule inhibitor, echinomycin, has been shown to specifically inhibit HIF-1α DNA-binding activity to the HRE site on the vascular endothelial growth factor (VEGF) promoter resulting in the loss of VEGF expression (18). Treatment of HIF-1α/RON expression-positive MDA MB 231 and MDA MB 468 breast cancer cells with echinomycin resulted in the inhibition of HIF-1α activity and RON protein expression. We have identified a novel HRE site on the RON promoter. ChIP analysis and site-directed mutagenesis of the RON promoter confirmed the authenticity of the HIF-1α binding site on the RON gene. HIF-1α inhibitor echinomycin- as well as shRNA-mediated selective knockdown of HIF-1α or HIF-1α target RON tyrosine kinase demonstrated the role of HIF-1α in the regulation of RON tyrosine kinase and the RON ligand MSP-associated invasion of breast cancer cells. These data presented herein identified RON tyrosine kinase as a novel target of HIF-1α and suggest a potential therapeutic role in the inhibition of HIF-1α/RON axis for the blockade of invasion and metastasis of carcinoma cells.
EXPERIMENTAL PROCEDURES
Cell Culture—Breast cancer cells were obtained from the American Type Culture Collection (ATCC). Cells were grown in McCoy's 5A medium supplemented with 10% fetal bovine serum (Atlanta Biologicals), amino acids, antibiotics, pyruvate, and vitamins (Invitrogen). All the cell lines were cultured in a 37 °C humidified atmosphere containing 5% CO2.
Generation of HIF-1α and RON Knockdown Stable Cell Lines—Scramble shRNA and shRNA HIF-1α expression plasmid was provided by Dr. John Copland and Dr. Jenny Wang provided the scramble and shRNA RON expression plasmids. We have transfected control scramble shRNA or shRNA HIF-1α or shRNA RON expression plasmids into HIF-1α/RON-positive MDA MB 231 breast cancer cells and, following selection in the puromycin medium, obtained HIF-1α and RON expression knockdown clones for characterization.
Western Blot Analysis—Equal amounts of cell lysates from breast cancer cells were resolved by 7.5% SDS-PAGE, and Western analysis was performed as described previously (17). Rabbit anti-human RON, actin polyclonal, and HIF-1α monoclonal antibodies were purchased from Santa Cruz Biotechnology. To analyze the effect of echinomycin cells were treated for 16 h with 0.5 and 1.0 μm echinomycin prior to Western analysis. To determine the effect of shRNA HIF-1α or shRNA RON knockdown on RON expression, MDA MB 231 cells stably expressing scrambled shRNA control or shRNA HIF-1α or shRNA RON were analyzed for RON expression by Western blots. To determine if PI3K/AKT pathways are involved in the RON receptor-mediated signaling, cells were starved for 16 h and stimulated with RON ligand, MSP either in the presence or absence of PI3K inhibitor, LY294002 and ERK kinase inhibitor, PD98059. Western analysis using pAKT, total AKT, pERK, and total ERK was performed.
Real-time RT-PCR—Total RNA was isolated from cells using TRIzol reagent (Invitrogen). 100 ng of total RNA was subjected to real-time RT-PCR reaction with SYBR Green real-time PCR mix (Bio-Rad Laboratories) according to the manufacturer's instruction. The Primers for RON were: sense primers, 5′-AGC CCA CGC TCA GTG TCT AT-3′; antisense primers, 5′-GGG CAC TAG GAT CAT CTG TCA-3′. Primers for HIF-1α were: sense primers, 5′-CTCAAAGTCGGACAGCCTCA-3′; antisense primers, 5′-CCCTGCAGTAGGTTTCTGCT-3′. Primers for actin were: sense primers, 5′-ACA CTG TGC CCA TCT ACG AGG-3′; antisense primers, 5′-AGG GGC CGG ACT CGT CAT ACT-3′. The β-actin mRNA was amplified simultaneously for an endogenous control.
In Vitro Kinase Assay—In vitro kinase assay to analyze RON kinase activity was performed as described previously for RON protein (19). Briefly, RON was immunoprecipitated from cell lysates by RON polyclonal antibody. IgG antibody was used as a negative control for the immunoprecipitation. The exogenous substrate, myelin basic protein was added to the kinase reaction mixture at a concentration of 1.0 μg/reaction tube. Reactions were stopped with 5 μl of 4% sample buffer. Phosphorylated myelin basic protein was visualized after SDS-PAGE by autoradiography.
RT-PCR—Total RNA from breast cancer cells was reverse transcribed into cDNA. PCR analysis was then performed to determine RON expression using the cDNA as template. Primers for actin were used as a control to determine the RON expression levels. RT-PCR analysis allows a rough estimate of the differences in RON expression in the cell lines tested. A total of 30 cycles of amplification was performed. For the studies involving echinomycin MDA MB 231, MDA MB 468 cells were treated for 16 h with 0.5 and 1.0 μm echinomycin before RNA isolation and RT-PCR analysis. Primers for RON generate a 246-bp fragment as follows: sense primers, 5′-AGC CCA CGC TCA GTG TCT AT-3′; antisense primers, 5′-GGG CAC TAG GAT CAT CTG TCA-3′. Primers for actin generate a 621-bp fragment as follows: sense primers, 5′-ACA CTG TGC CCA TCT ACG AGG-3′; antisense primers, 5′-AGG GGC CGG ACT CGT CAT ACT-3′.
Invasion Assay—The invasive behavior of breast cancer cells was analyzed in a trans-well Boyden chamber (Costar, Bethesda, MD) using a polycarbonate filter (8-μm pores) and a 0.1% gelatin matrix in the upper chamber. Briefly, 3 × 104 cells/well were plated in Matrigel invasion upper chambers in 0.3 ml of serum-free medium. The lower chambers contained 0.7 ml of 10% fetal bovine serum medium. The cells were treated with 2.5 ng of MSP. After 24-h incubation at 37 °C the cells on the top surface of the chamber were gently removed with cotton swabs. The migrant cells on the undersurface of the membrane were fixed in 70% methanol and stained with crystal violet. Images of migrant cells were captured by a photomicroscope (Nikon Eclipse-TE 2000-U, Japan). The invasion values were determined by eluting crystal violet in 10% acetic acid, and the absorbance was taken using a Fluorostar Optima plate reader at 585 nm. Quantification data represent the mean ± S.D. of three separate invasion assays. To assess the effect of echinomycin, an inhibitor of HIF-1α binding MDA MB 231, MDA MB 468 cells were treated with 0.5 and 1.0 μm echinomycin for 16 h, trypsinized, and plated for Matrigel invasion assay. To determine the effect of shRNA HIF-1α- or shRNA RON-mediated RON expression knockdown on MSP-mediated invasion, MDA MB 231 cells stably expressing scramble shRNA or shRNA HIF-1α or shRNA RON were used for the Matrigel invasion assay.
ChIP—The ChIP assay was performed as described previously (17). RON promoter primers covering the HRE site were used to carry out PCR on DNA isolated from chromatin immunoprecipitation using control IgG or HIF-1α antibodies. The optimal reaction conditions for PCR were determined for each primer pair. Parameters included denaturation at 95 °C for 1 min and annealing at 64 °C for 1 min followed by elongation at 72 °C for 1 min. PCR products were analyzed by 1.0% agarose/ethidium bromide gel electrophoresis. The following primers were used for PCR to generate a 293-bp fragment covering the HIF-1α binding site on the RON promoter. Forward: 5′-CTC CAA GGG CCG GAA GAG TCG GAT GG-3′; reverse: 5′-TTA AGC AGC GGT CCC GAC AGC CCC AA-3′.
Construction of –400-bp Wild-type/Mutant RON Promoter-luciferase Reporter Constructs—The 400-bp RON promoter-luciferase reporter was described previously (17). The HRE site-specific mutation construct of the RON promoter was generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) based on the 400-bp RON promoter-luciferase reporter construct as a template. The mutation was confirmed by DNA sequencing. The oligonucleotide sequences used to generate HRE mutant construct were: wild-type primer, 5′-GCGTTGCTGTCGTGCGTCCGCAGG-3′; mutant primer, 5′-GCGTTGCTGTAAAGCGTCCGCAGG-3′.
Luciferase Assay—Breast cancer cells were seeded into 12-well plates at a density of 15 × 104 cells/well the day before transfection. The wild-type or HRE mutant RON-Luc constructs (1 μg) or control null vector without the RON promoter insert (pGL3) were transiently transfected into cells using Lipofectamine 2000 (Invitrogen). Cells were harvested 24 h following transfection, and luciferase activity was measured following normalization to protein levels (Luciferase Assay System, Promega, Madison, WI). To determine the effect of echinomycin on wild-type RON promoter activity, cells transfected with –400-bp wild-type RON promoter were treated for 16 h with varying concentrations of echinomycin prior to luciferase assay.
RESULTS
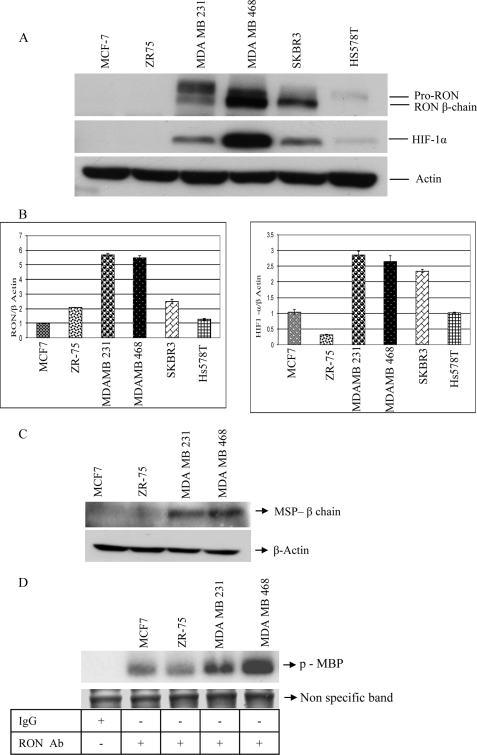
Expression of HIF-1α, RON, MSP, and RON Kinase Activity in Breast Cancer Cells—We have recently shown that the RON ligand, MSP, promotes invasion of RON-positive but not RON-negative breast cancer cells (17). To determine if HIF-1α-mediated RON expression is the contributing factor for the invasive phenotype of breast cancer cells, we have determined HIF-1α and RON expression levels in these cell lines. We have carried out Western analysis on equal amounts of total cell lysates from breast cancer cells using rabbit anti-human RON, actin polyclonal, and HIF-1α monoclonal antibodies (Fig. 1A). RON antibody recognizes 170-kDa pro-RON and intracellular 150-kDa β-chain of mature RON receptor. HIF-1α antibody recognizes 120-kDa protein species. RON expression correlated with HIF-1α protein expression levels in these cells. We have carried out real-time RT-PCR analysis to determine RON and HIF-1α mRNA expression. Total RNA from breast cancer cells was reverse-transcribed into cDNA. We have performed real-time RT-PCR analysis using RON, HIF-1α, and actin primers (Fig. 1B). Differential expression of RON and HIF-1α message was detected in the breast cancer cells. Actin mRNA was amplified simultaneously for an endogenous control and represented as RON/actin mRNA and HIF-1α/actin mRNA. To determine if the invasive breast cancer cells express RON ligand, MSP, we have carried out Western analysis on the total cell lysates. High MSP expression was detected in the invasive MDA MB 231 and MDA MB 468 cells in comparison to MCF-7 and ZR75, which are non-invasive in the presence of RON ligand, MSP (Fig. 1C). We next carried out an in vitro kinase assay to determine the RON kinase activity in the invasive breast cancer cells by immunoprecipitating the total cell lysates with control IgG or RON polyclonal antibody and performed kinase assay using myelin basic protein as a substrate as previously described for RON protein (19). Higher RON kinase activity was detected in the invasive MDA MB 231 and MDA MB 468 cells in comparison to MCF-7 and ZR75 cells. A nonspecific band is shown to represent loading control (Fig. 1D).
FIGURE 1.
A, RON and HIF-1α protein expression: equal amounts of total cell lysates from a panel of breast cancer cells were resolved by 7.5% SDS-PAGE and Western analysis using rabbit anti-human RON, actin polyclonal antibody, and HIF-1α mouse monoclonal antibody was performed. B, expression of RON and HIF-1α message: total RNA from a panel of breast cancer cell lines was subjected to real-time RT-PCR reaction with SYBR Green real-time PCR mix. Primers for RON, HIF-1α, and actin were described under “Experimental Procedures.” C, expression of MSP protein: equal amounts of cell lysates were resolved by 7.5% SDS-PAGE and Western analysis using rabbit anti-human MSP and actin polyclonal antibodies was performed. D, RON kinase activity: immunoprecipitation using control IgG or RON polyclonal antibody was performed on the total cell lysates and in vitro kinase assay using myelin basic protein as a substrate was carried out as described under “Experimental Procedures.” Nonspecific band was shown to represent loading control.
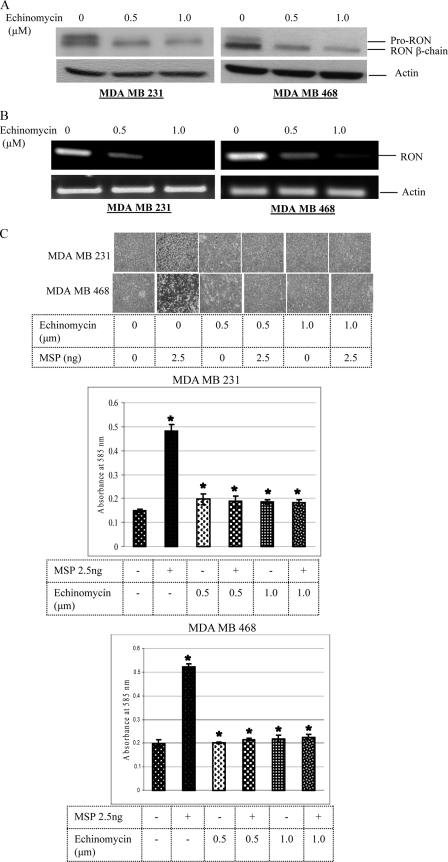
Echinomycin Inhibits RON Expression and MSP-induced Invasion of Breast Cancer Cells—A small molecule inhibitor, echinomycin, has been shown to specifically inhibit HIF-1α DNA-binding activity to the HRE site on the vascular endothelial growth factor (VEGF) promoter resulting in the loss of VEGF expression (18). To determine if echinomycin-mediated loss of HIF-1α-binding activity abrogates RON expression in the HIF-1α/RON expression-positive invasive breast cancer cells, we have treated MDA MB 231 and MDA MB 468 cells with echinomycin for 16 h and carried out Western analysis on the total cell lysates using RON and actin polyclonal antibodies. RON protein expression was decreased in both the cell lines tested (Fig. 2A). We have carried out RT-PCR analysis to determine if the echinomycin-mediated decrease in the RON protein levels reflected RON message levels. Total RNA from control and echinomycin-treated (16 h) breast cancer cells was reverse-transcribed into cDNA, and PCR using RON and actin primers was performed. Echinomycin treatment reduced RON message levels in both the cell lines (Fig. 2B). To determine if echinomycin-mediated blockade in RON gene expression abrogates RON ligand, MSP-induced invasion of breast cancer cells, we carried out an in vitro Matrigel assay. Matrigel invasion assay is a reliable test to evaluate cancer cell invasiveness. This assay elegantly reflects the actual ability of cancer cells to invade and metastasize in vivo in response to activation of the RON receptor. MDA MB 231 and MDA MB 468 cells were treated for 16 h with echinomycin, trypsinized, and plated along with untreated control cells for the Matrigel assay either in the presence or absence of 2.5 ng MSP for 24 h. We have stained the migrant cells with crystal violet and photographed the cells (Fig. 2C). Quantification was done by eluting crystal violet in 10% acetic acid, and the absorbance was taken using a Fluostar Optima plate reader at 585 nm. MSP induced the invasion through Matrigel of MDA MB 231 and MDA MB 468 control cells but not echinomycin-treated cells where RON expression was blocked.
FIGURE 2.
A, effect of echinomycin on RON protein expression: equal amounts of total cell lysates from control or echinomycin-treated cells were resolved by 7.5% SDS-PAGE, and Western analysis using rabbit anti-human RON and actin polyclonal antibodies was performed. B, effect of echinomycin on RON message: total RNA from control or echinomycin-treated cells was reverse-transcribed into cDNA and PCR analysis was performed using primers for RON and actin as described under “Experimental Procedures.” C, echinomycin blocks MSP-mediated invasion: control or echinomycin-treated breast cancer cells were plated in Matrigel invasion chamber (upper). The cells were treated with 2.5 ng MSP for 24 h. The migrant cells on the undersurface of the membrane were fixed in 70% methanol and stained with crystal violet. Images of migrant cells were captured by a photomicroscope (Nikon Eclipse-TE 2000-U, 10×). Quantification data represents the mean ± S.D. of three separate invasion assays.
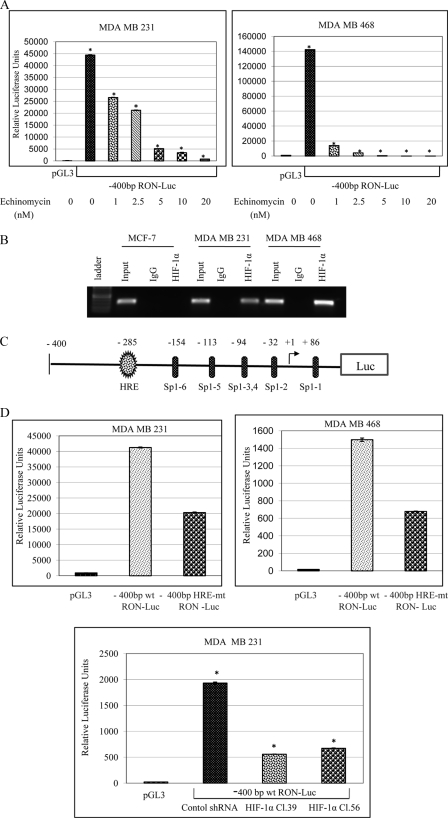
HIF-1α Directly Regulates RON Gene Expression—To determine if the decreased RON expression levels in the echinomycin-treated cells was due to decreased RON transcription, we analyzed RON promoter activities using a –400-bp RON promoter deletion construct (Fig. 3C) in control and echinomycin-treated breast cancer cells. The –1.2-kb full-length and –400-bp RON promoter deletion constructs were described previously (17). We have identified two novel HIF-1α binding sites (HRE) at –285 bp and –1176 bp relative to the transcription start site. Our previous work indicated both –1.2-kb full-length and –400-bp RON promoter deletion constructs exhibited similar activities in the MDA MB 231 and MDA MB 468 breast cancer cells suggesting all the regulatory elements are present in the –400-bp region of the RON promoter. Consequently, we have analyzed the activity of the –400-bp RON promoter in the control and echinomycin-treated breast cancer cells. A dose-dependent decrease in the RON promoter activity was observed in echinomycin-treated MDA MB 231 and MDA MB 468 breast cancer cells (Fig. 3A). To demonstrate that HIF-1α binds to RON promoter in vivo we carried out ChIP analysis with control IgG or HIF-1α antibodies on the chromatin fragments from MCF-7, MDA MB 231, and MDA MB 468 breast cancer cells (Fig. 3B). DNA from the immunoprecipitates was isolated and PCR using primers covering the HIF-1α binding site on the RON promoter was performed on the isolated DNA and non-immunoprecipitated starting chromatin material (input). RON was detected in the input chromatin fragments from all the three cell lines. However, accumulation of RON was detected in the HIF-1α immunoprecipitates from MDA MB 231 and MDA MB 468 cells but not MCF-7 where HIF-1α expression was not observed by Western analysis (Fig. 1A). RON was not detected in the control IgG immunoprecipitates demonstrating the specificity of HIF-1α binding to RON promoter. To confirm if HIF-1α binding to the HRE site on the RON promoter was contributing factor to the RON gene expression we have used site-directed mutagenesis to generate site-specific HRE mutant RON promoter construct. Wild-type and mutant primer sequences to generate the promoter reporter plasmids were described under “Experimental Procedures.” We have analyzed the activities of the –400-bp wild-type and HRE mutant RON promoter constructs in MDA MB 231 and MDA MB 468 breast cancer cells (Fig. 3D). HRE mutant RON promoter plasmid exhibited a 50–60% decrease in the activity in comparison to the wild-type promoter. This promoter construct contains 6 Sp1 binding sites in addition to the HIF-1α binding site (HRE). Our previous work indicated the overlapping Sp1 sites at –94 bp and Sp1 site at –113 bp relative to the transcription start site are required for the basal RON promoter activity, which were retained in the HRE mutant construct. We have further confirmed the contribution of HIF-1α binding to RON promoter activity by analyzing the wild-type RON promoter activity in the scramble shRNA control and shRNA HIF-1α-mediated HIF-1α expression knockdown MDA MB 231 breast cancer cells. The HIF-1α knockdown clones showed a significant reduction in the wild-type RON promoter activity, thus validating the role of HIF-1α in the regulation of RON gene expression. These experiments were repeated several times and the results are very consistent.
FIGURE 3.
A, effect of echinomycin on RON promoter activity: cells were transfected with pGL3 control vector or –400-bp wild-type RON promoter. 24 h following transfection, cells were treated with varying concentrations of echinomycin for 16 h, and luciferase activity was measured following normalization to protein levels. B, ChIP assay on the chromatin fragments from breast cancer cells using control IgG or HIF-1α antibodies was performed as described under “Experimental Procedures.” C, illustration: RON promoter-luciferase reporter construct depicting the HIF-1α binding site (HRE) and Sp1 binding sites. D, effect of HRE binding site mutation on RON promoter activity: cells were transfected with either pGL3 control vector or –400-bp wild-type RON promoter or HRE binding site mutant RON promoter-luciferase reporter constructs and luciferase activity was measured after 48 h following normalization to protein levels. To determine the effect of HIF-1α expression knockdown on RON promoter activity control or –400-bp wild-type RON promoter activity was analyzed in control shRNA or shRNA HIF-1α-expressing clones.
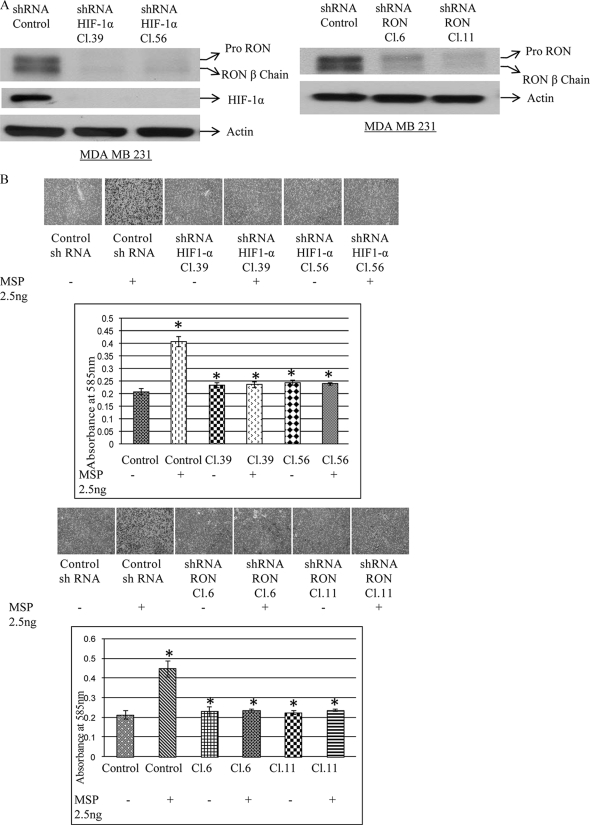
Selective Knockdown of HIF-1α or HIF-1α Target RON Blocks RON Gene Expression and MSP-induced Invasion of Breast Cancer Cells—To validate the findings that echinomycin-mediated loss of HIF-1α binding was contributing to the inhibition of RON gene expression and MSP-mediated invasion of breast carcinoma cells, we selectively knocked down HIF-1α or HIF-1α target RON tyrosine kinase in the MDA MB 231 cells. Puromycin-resistant clones were selected for characterization. Western analysis using RON, actin polyclonal, and HIF-1α monoclonal antibodies was performed on the total cell lysates from scramble shRNA control and HIF-1α, RON knockdown clones (Fig. 4A). RON protein expression was almost completely inhibited in both the HIF-1α knockdown and HIF-1α target RON knockdown MDA MB 231 clones. To determine if the inhibition of RON expression in the HIF-1α and RON knockdown clones abrogates RON ligand, MSP-induced invasive phenotype of breast carcinoma cells, we carried out an in vitro Matrigel assay (Fig. 4B). Although MSP promoted invasion through Matrigel of the scrambled shRNA expressing control MDA MB 231 clones, both shRNA HIF-1α-mediated HIF-1α expression knockdown clones, where RON expression was inhibited, and shRNA RON-mediated HIF-1α target RON knockdown clones did not exhibit the invasive phenotype. These molecular data complement the pharmacological data using echinomycin where loss of HIF-1α binding and RON expression also blocked invasion of breast carcinoma cells (Fig. 2).
FIGURE 4.
A, shRNA HIF-1α or shRNA RON blocks RON expression: equal amounts of cell lysates from control, scrambled shRNA or shRNA HIF-1α, shRNA RON-expressing MDA MB 231 clones were resolved by 7.5% SDS-PAGE, and Western analysis using rabbit anti-human RON, actin polyclonal antibody, and HIF-1α mouse monoclonal antibody was performed. B, shRNA HIF-1α or shRNA RON blocks MSP-mediated invasion: control scrambled shRNA, shRNA HIF-1α or shRNA RON-expressing MDA MB 231 clones were plated in Matrigel invasion chamber (upper). The cells were treated with 2.5 ng of MSP for 24 h. The migrant cells on the undersurface of the membrane were fixed in 70% methanol and stained with crystal violet. Images of migrant cells were captured by a photomicroscope (Nikon Eclipse-TE 2000-U, 10×). Quantification data represent the mean ± S.D. of three separate invasion assays.
PI3K and ERK Mediate RON Tyrosine Kinase-mediated Cell Invasion—To determine if PI3K and ERK play a role in the RON tyrosine kinase pathway-mediated invasion of breast carcinoma cells, we serum-starved MDA MB 231 and MDA MB 468 cells for 16 h and stimulated with RON ligand, MSP either in the presence or absence of PI3K inhibitor, LY294002, and ERK kinase inhibitor, PD98059. We carried out Western analysis on the total cell lysates using pAKT, total AKT, pERK, and total ERK antibodies. RON ligand, MSP treatment stimulated PI3K activity, which was effectively blocked by the PI3K inhibitor, LY294002. However, both MDA MB 231 and MDA MB 468 cells showed basal ERK activity, which was not further elevated by MSP treatment. ERK kinase inhibitor, PD98059, inhibited the basal ERK activity (Fig. 5A). In vitro Matrigel analysis indicated that MSP promoted invasion of MDA MB 231 cells in the absence of PI3K, LY294002, and ERK kinase inhibitor, PD98059 (Fig. 5B). However, both inhibitors blocked the RON ligand, MSP-induced invasion of MDA MB 231 cells.
FIGURE 5.
A, effect of PI3K and ERK inhibitors on RON-mediated signaling: cells were serum-starved for 16 h and stimulated with RON ligand, MSP, either in the absence or presence of the PI3K inhibitor, LY294002, or the ERK inhibitor, PD98059. Western analysis using pAKT, total AKT, pERK, and total ERK antibody was performed on the total cell lysates. B, effect of PI3K and ERK inhibitors on RON-mediated invasion: cells were starved for 16 h and plated for Matrigel invasion assay either in the absence or presence of RON ligand, MSP, and PI3K and ERK inhibitors. 24 h later migrant cells were stained and photographed. Quantification data represent the mean ± S.D. of three separate invasion assays.
DISCUSSION
Tumor microenvironment especially hypoxia contributes to the progression and metastasis of tumors and tumor cell lines. HIF-1α protein is a key player in the regulation of genes involved in tumor hypoxia. HIF-1α protein is stabilized under hypoxia but is rapidly degraded through ubiquitin proteasome pathway in normoxic conditions. However, in the invasive carcinoma cells, including breast cancer cells, HIF-1α expression can be detected even under normoxia due to the activation of growth factor receptor pathways or due to the defects in the protein degradation pathways (1). Multiple studies noted the clinical relevance between HIF-1α overexpression and progression of breast tumors (2–6). Deletion of HIF-1α in the mammary epithelium resulted in delayed tumor onset and retarded tumor growth as well as decreased pulmonary metastasis (10). The identification of molecular targets of HIF-1α that play an important role in tumor cell invasion is under active investigation. The role of receptor tyrosine kinases in tumor progression is well documented. Multiple human tumor types show elevated expression of RON tyrosine kinase receptor. Gene expression analyses suggested that increased RON expression is associated with metastatic disease. Mammary-specific RON receptor overexpression induced metastatic mammary tumors suggesting that RON overexpression can be a causative factor for metastatic breast cancer (15). We have recently shown that RON tyrosine kinase receptor is involved in the RON ligand MSP-induced invasive phenotype of breast carcinoma cells (17). These results demonstrated the role of RON tyrosine kinase in the acquisition of invasive phenotype of breast carcinoma cells.
Analysis of a panel of breast cancer cell lines displayed a correlation between HIF-1α and RON tyrosine kinase expression (Fig. 1A). Although MCF-7 cells did not show HIF-1α and RON under normoxia, hypoxia induced expression of both of these proteins (supplemental Fig. S4). However, MDA MB 231 and MDA MB 468 cells, which show steady-state HIF-1α under normoxia, exhibited a slight increase in HIF-1α under hypoxia with out any affect on RON expression (supplemental Fig. S1). The invasive cells express RON ligand, MSP, and also exhibited higher RON tyrosine kinase activity. A small molecule inhibitor, echinomycin, was shown to inhibit HIF-1α-binding activity to the HRE site on the VEGF promoter resulting in loss of VEGF expression (18). Echinomycin was reported to selectively inhibit in a sequence-specific fashion HIF-1α-binding activity but not the binding of activator protein 1 or nuclear factor-κB (NF-κB) p65 subunit to the corresponding promoters. Further, echinomycin appears to affect only the HIF-1α DNA-binding activity and not HIF-1α transcription and translation. Echinomycin treatment resulted in the decrease in RON protein expression levels as a consequence of the decrease in RON message (Fig. 2, A and B). Significantly, echinomycin-mediated inhibition of RON expression abrogated RON ligand, MSP-induced invasion of breast carcinoma cells (Fig. 2C).
Mutations or amplification of the RON gene were/was not reported. However, abnormal expression of RON was documented in various cancers of epithelial origin (14). Overexpression and activation of RON in epithelial cancers in contrast to normal epithelium indicates that cellular mechanisms that control RON expression may be dysfunctional in the primary tumors and tumor cell lines. The promoter for RON gene has been partially characterized (17, 20–21). The RON promoter lacks a distinct TATA box or CCAAT sequences and is dependent on Sp1 transcription factor for basal promoter activity and gene expression. HIF-1α binds to HRE (5′-RCGTG-3′) on the target genes. We have now identified two novel HIF-1α binding sites (at –285 bp; –1176 bp relative to the transcription start site) on the RON promoter. Our previous work indicated that both –1.2-kb full-length and –400-bp RON promoter deletion constructs exhibited similar activities in the MDA MB 231 and MDA MB 468 cells suggesting that all the regulatory elements are present in the –400-bp region of the promoter (17). Consequently, we have analyzed the activity of the –400-bp RON promoter, which contains HRE at –285 bp in addition to the Sp1 binding sites. HIF-1α inhibitor echinomycin exhibited a dose-dependent decrease in the RON promoter activity (Fig. 3A). In vivo ChIP analysis revealed the binding of HIF-1α to the native RON promoter in the RON expression-positive MDA MB 231- and MDA MB 468-invasive breast cancer cells but not in HIF-1α and RON expression-negative MCF-7 cells (Fig. 3B). This result confirms the authenticity of HIF-1α binding in the regulation of RON gene expression. To better evaluate the contribution of HIF-1α to the activity of RON promoter the putative HIF-1α binding site was mutated. HRE mutant RON promoter showed a significant loss in RON promoter activity. Also, in the reverse experiment wild-type RON promoter exhibited reduced activity in the HIF-1α expression knockdown breast cancer cells (Fig. 3D), thus demonstrating the role of HIF-1α in the regulation of RON promoter activity. The HIF-1α mutant RON plasmid retained the basal promoter activity. This was not surprising, because the construct contains six Sp1 binding sites, and we have shown recently that the overlapping Sp1 site at –94 bp and Sp1 site at –113 bp are critical for basal RON promoter activity (17). Triple mutations at these sites resulted in the complete loss of RON promoter activity.
Our results using pharmacological inhibitor echinomycin clearly demonstrated the role of HIF-1α in the regulation of RON gene expression and RON ligand MSP-mediated invasion of breast carcinoma cells. However, it may be plausible that the sequence specificity of echinomycin may not be restricted to HIF-1α, and there may be other known/unknown transcription factors that have similar core-binding sites in their DNA sequences. In addition, HIF-1α has been shown to regulate genes involved in cell proliferation, survival, angiogenesis, and motility. Consequently, to corroborate the results obtained using HIF-1α inhibitor echinomycin and also to demonstrate the specificity of RON gene regulation by HIF-1α and the contribution of HIF-1α/RON axis in the promotion of invasive phenotype of carcinoma cells we have selectively knocked down HIF-1α and HIF-1α target RON tyrosine kinase. Specific shRNA-mediated knockdown of HIF-1α or HIF-1α target RON resulted in the loss of RON tyrosine kinase expression in invasive MDA MB 231 breast cancer cells (Fig. 4A). These data clearly complement the result demonstrating that treatment of breast cancer cells with HIF-1α inhibitor, echinomycin, reduced RON tyrosine kinase expression (Fig. 2A). Most significantly, the loss of RON tyrosine kinase expression due to the shRNA HIF-1α-mediated inhibition of HIF-1α activity or through shRNA RON-mediated direct knockdown of the HIF-1α target RON resulted in the abrogation of RON ligand MSP-induced invasion of breast carcinoma cells. Hypoxia induced RON ligand, MSP-mediated invasion of MDA MB 231 cells, which was abrogated in the RON knockdown cells (supplemental Fig. S2). The blockade in cell invasion in the HIF-1α inhibitor, echinomycin-treated, or shRNA HIF-1α-mediated selective HIF-1α knockdown MDA MB 231 cells was not due to a change in cell survival, because apoptosis analysis using annexin V-FITC indicated no change at 24 h, the time point used for the invasion assay (supplemental Fig. S3). A previous report also indicated that RON ligand, MSP, promoted metastasis in a mouse model of breast cancer without affecting cell proliferation or cell survival (22). So, the RON tyrosine kinase pathway may promote invasion and metastasis by influencing the ability of tumor cells to migrate and invade extracellular matrix and blood vessels, and not by changing cell proliferation or cell survival. Our data indicated that both PI3K and ERK pathways are required for RON signaling-mediated breast cancer cell invasion. To summarize, the results presented in this report unequivocally identified RON tyrosine kinase as a novel target of HIF-1α and HIF-1α or RON tyrosine kinase inhibitors may have a potential therapeutic role in the blockade of RON tyrosine kinase-induced invasion of carcinoma cells.
Supplementary Material
Acknowledgments
We thank Drs. John Copland and Jenny Wang for kindly providing us the shRNA HIF-1α and shRNA RON plasmids, respectively, and Dr. Pothana Saikumar for the photomicroscope facilities.
This work was supported, in whole or in part, by National Institutes of Health Grant P30 CA54174 from the NCI.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: HIF-1α, hypoxia-inducible factor-1α; MSP, macrophage-stimulating protein; RON, recepteur d'origine nantais; HRE, hypoxia responsive element; shRNA, short hairpin RNA; RT, reverse transcription; ChIP, chromatin immunoprecipitation; VEGF, vascular endothelial growth factor; PI3K, phosphatidylinositol 3-kinase; ERK, extracellular signal-regulated kinase.
References
- 1.Kimbro, K. S., and Simons, J. W. (2006) Endocr. Relat. Cancer 13 739–749 [DOI] [PubMed] [Google Scholar]
- 2.Bos, R., Vander Groep, P., Greijer, A. E., Shvartis, A., Meijer, S., Pinedo, H. M., Semenzi, G. L., Van Diest, P. J., and Van der Wall, E. (2003) Cancer 97 1573–1581 [DOI] [PubMed] [Google Scholar]
- 3.Vleugel, M. M., Greijer, A. E., Shvarts, A., Van der groep, P., Van Berkel, M., Aarbodem, Y., Van Tinteren, H., Harris, A. L., Van Diest, P. J., and Van der Wall, E. (2005) J. Clin. Pathol. (Lond.) 58 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gruber, G., Greiner, R. H., Hlushchuk, R., Aebersold, D. M., Altermatt, H. J., Berclaz, G., and Djonov, V. (2004) Breast Cancer Res. 6 R191–R198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dales, J. P., Garcia, S., Meunier-Carpentier, S., Andrae-Meyer, L., Haddad, O., Lavaut, M. N., Allasia, C., Bonnier, P., and Charpin, C. (2005) Int. J. Cancer 116 734–739 [DOI] [PubMed] [Google Scholar]
- 6.Kronbald, A., Sirstrom, K., Ryden, L., Nordenskjold, B., and Landberg, G. (2006) Int. J. Cancer 118 2609–2616 [DOI] [PubMed] [Google Scholar]
- 7.Bos, R., Zhong, H., Hanrahan, C. F., Mommers, E. C. M., Semenza, G. L., Pinedo, H. M., Abeloff, M. D., Simons, J. W., Van diest, P. J., and Van der Wall, E. (2001) J. Natl. Cancer Inst. 93 309–314 [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto, Y., Ibusuki, M., Okumura, Y., Kawasoe, T., Kai, K., Iyama, K., and Iwase, H. (2008) Breast Cancer Res. Treat. 110 465–475 [DOI] [PubMed] [Google Scholar]
- 9.Hiraga, T., Kizaka-Kondoh, S., Hirota, K., Hiraoka, M., and Yoneda, T. (2007) Cancer Res. 67 4157–4163 [DOI] [PubMed] [Google Scholar]
- 10.Liao, D., Corele, C., Seagroves, T. N., and Johnson, R. (2007) Cancer Res. 67 563–572 [DOI] [PubMed] [Google Scholar]
- 11.Fukuda, R., Hirota, K., Fan, F., Jung, Y. D., Ellis, L. M., and Semenza, G. L. (2002) J. Biol. Chem. 277 38295–38311 [DOI] [PubMed] [Google Scholar]
- 12.Zhong, H., Chiles, K., Feldser, D., Laughner, E., Hanrahan, C., Georgescu, M.-M., Simons, J. W., and Semenza, G. L. (2000) Cancer Res. 60 1541–1545 [PubMed] [Google Scholar]
- 13.Wang, M. H., Lee, W., Luo, Y. L., Weis, M. T., and Yao, H. P. (2007) J. Pathol. 213 402–411 [DOI] [PubMed] [Google Scholar]
- 14.Leonis, M. A., Thobe, M. N., and Waltz, S. (2007) Future Oncol. 3 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zinser, G. M., Leonis, M. A., Toney, K., Pathrose, P., Thobe, M., Kader, S. A., Peace, B. F., Beauman, S. R., Collins, M. H., and Waltz, S. E. (2006) Cancer Res. 66 11967–11974 [DOI] [PubMed] [Google Scholar]
- 16.Lee, W.-Y., Chen, H. H., Chow, N. H., Su, W. C., Lin, P. W., and Guo, H. R. (2005) Clin. Cancer Res. 11 2222–2228 [DOI] [PubMed] [Google Scholar]
- 17.Thangasamy, A., Rogge, J., and Ammanamanchi, S. (2008) J. Biol. Chem. 283 5335–5343 [DOI] [PubMed] [Google Scholar]
- 18.Kong, D., Park., E. J., Stephen, A. G., Calvani, M., Cardellina, J. H., Monks, A., Fisher, R. J., Shoemaker, R. H., and Melillo, G. (2005) Cancer Res. 65 9047–9055 [DOI] [PubMed] [Google Scholar]
- 19.Danilkovitch-Miagkova, A., Miagkov, A., Skeel, A., Nakaigawa, N., Zbar, B., and Leonard, E. J. (2001) Mol. Cell. Biol. 21 5857–5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Gatto, F., Gilbert, E., Ronsin, C., and Breathnach, R. (1995) Biochim. Biophys. Acta 1263 93–95 [DOI] [PubMed] [Google Scholar]
- 21.Narasimhan, M., and Ammanamanchi, S. (2008) Cancer Res. 68 5185–5192 [DOI] [PubMed] [Google Scholar]
- 22.Welm, A. L., Sneddon, J. B., Taylor, C., Nuyten, D. S. A., Van de Vijver, M. J., Hasegawa, B. H., and Bishop, M. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7570–7575 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.