Abstract
NKCC1 is highly expressed in dorsal root ganglion neurons, where it is involved in gating sensory information. In a recent study, it was shown that peripheral nerve injury results in increased NKCC1 activity, not due to an increase in cotransporter expression, but to increased phosphorylation of the cotransporter (Pieraut, S., Matha, V., Sar, C., Hubert, T., Méchaly, I., Hilaire, C., Mersel, M., Delpire, E., Valmier, J., and Scamps, F. (2007) J. Neurosci. 27, 6751–6759). Our laboratory has also identified two Ste20-like kinases that bind and phosphorylate NKCC1: Ste20-related proline-alanine-rich kinase (SPAK) and oxidative-stress response 1 (OSR1). In this study, we show that both kinases are expressed at similar expression levels in spinal cord and dorsal root ganglion neurons, and that both kinases participate equally in the regulation of NKCC1. Using a novel fluorescence method to assay NKCC1 activity in single cells, we show a 50% reduction in NKCC1 activity in DRG neurons isolated from SPAK knockout mice, indicating that another kinase, e.g. OSR1, is present to phosphorylate and activate the cotransporter. Using a nociceptive dorsal root ganglion sensory neuronal cell line, which expresses the same cation-chloride cotransporters and kinases as native DRG neurons, and gene silencing via short hairpin RNA, we demonstrate a direct relationship between kinase expression and cotransporter activity. We show that inactivation of either kinase significantly affects NKCC1 activity, whereas inactivation of both kinases results in an additive effect. In summary, our study demonstrates redundancy of kinases in the regulation of NKCC1 in dorsal root ganglion neurons.
The regulation of intracellular Cl– in neurons is a critical determinant of inhibitory synaptic neurotransmission. Sensory or peripheral neurons express, in abundance, an inwardly poised Na+- and K+-dependent Cl– transport mechanism, NKCC1,2 whose activity drives the uphill accumulation of Cl– ions (2, 3). High intracellular Cl– concentration in dorsal root ganglion (DRG) neurons permits depolarizing γ-aminobutyric acid responses, which mediate presynaptic inhibition and filtration of sensory noise (4). Consequently, the knockout of NKCC1 exhibits a redistribution of internal Cl– in DRG neurons and a pain perception phenotype (3, 5). In addition, peripheral inflammation or nerve injury results in increased NKCC1 function in primary afferents (6, 7). Using a phosphopeptide-specific NKCC1 antibody, it was recently shown that NKCC1 phosphorylation instead of expression level increased in DRG neurons upon nerve injury (1). This observation points to the importance of NKCC1 regulation in the neuropathic pain pathway.
In recent work, our laboratory identified two Ste20-like kinases that directly bind to the cytosolic N-terminal tail of NKCC1 (8). The binding is a pre-requisite for NKCC1 phosphorylation and activation (9–13). The kinases, named SPAK (Ste20-related proline-alanine-rich kinase) and OSR1 (oxidative-stress response 1), share high homology in both their catalytic and regulatory domains. Expression of the two proteins has been examined in tissues using both Northern and Western blot analysis. These studies established that both kinases are widely expressed and that their expression pattern often overlaps (14–18). Co-expression of the kinases has been confirmed using Western blot analysis of well established cultured cell lines (17).
Heterologous expression of SPAK and OSR1 in Xenopus laevis oocytes demonstrates that both kinases are able to bind and activate NKCC1 (10). Whether or not they equally participate to the regulation of NKCC1 in vivo under normal physiological conditions has not yet been determined. We chose to address the role of SPAK and OSR1 by using an established nociceptive dorsal root ganglion sensory neuronal cell line (19), as well as isolated mouse DRG neurons. We demonstrate that undifferentiated 50B11 cells express the same cotransporters and regulatory kinases as native DRG cells. To manipulate the kinases, we decreased their expression via shRNA knockdown and used a SPAK knockout mouse for the isolated DRG neurons. To measure NKCC1 activity, we used unidirectional 86Rb-uptake with the established cell line and developed a novel fluorescence assay to assess cotransporter activity in single isolated DRG neurons. Our data demonstrate that both kinases are expressed to similar levels in both the cell line and isolated mouse DRG neurons, and that the kinases participate in concert to regulate NKCC1 function.
EXPERIMENTAL PROCEDURES
Cell Culture—In this study, we used a novel immortalized rat dorsal root ganglion sensory neuronal cell line, the 50B11 cells. The cells were grown in DMEM/F-12 medium (Invitrogen) supplemented with 10% bovine serum, 100 units/ml penicillin-100 μg/ml streptomycin, 0.2% glucose, 0.5 mm l-glutamine, 1× B-27 supplement (Invitrogen), and 5 μg/ml blasticidin (Invitrogen).
RT-PCR—RNA was isolated from 50B11 cells or dorsal root ganglions from mice or rats using the RNeasy kit from Qiagen (Valencia, CA). Total RNA (2 μg) was then reverse-transcribed into cDNA using reverse transcriptase (SuperScript™ II, Invitrogen). PCR was performed using standard protocols and primers selected according to consensus sequences between rat and mouse. The sequences of these primers are listed in Table 1.
TABLE 1.
Sequence of PCR primers for cation-chloride cotransporters and kinases
Sizes of the PCR products in base pairs (bp) are indicated in the last column.
| Gene | Sense (5′ to 3′) | Antisense (5′ to 3′) | Size |
|---|---|---|---|
| NKCC | TGGTGTCAGGATTTGCACCC | GCCCACCAGTTAATGACAAAC | 418 |
| KCC1 | CGGACAGGGTAACCACAGAG | AGCCGCAGGAAGAGGATGACCC | 309 |
| KCC2 | ACGGACTGCGAGGACGGCGATGG | TGCAGGCACGGCAGGTACACACC | 311 |
| KCC3 | AGCTCAAGGCAGGAAAGGGAC | ATCTTCACTCTGACGCCAGCC | 263 |
| KCC4 | GGAGATGGAAACCCCAGGGA | TCCGATGAAGGTGCCCATGC | 246 |
| SPAK | AAGTCATGGAACAGGTGAGAGGC | TTCAGTCTTATGAAGGTGACCGC | 410 |
| OSR1 | TTTCTCAGCTGAGGTCTCCCCG | TGCCATCATCAGGAATATCCGAGCC | 489 |
| WNK1 | AGCAGCGGCAGTTGGTTCGAGAAG | GCTCACTCTGGATTCTGTGAATAC | 305 |
| WNK2 | GAGAGGATCTGGCCTGCACTGCAG | TGCTCTGGGAGTCCGAGTACACGG | 262 |
| WNK3 | AAAAAAAGCCTGCTGGATGCTTGG | CTAGTTGTCTGGTTTGAAGAATAG | 301 |
| WNK4 | GCCCCAGCTTGTTGGCCGTTTCC | TGCTGTAGGAGTAGTTCATCCAC | 317 |
Western Blot—Control and transfected 50B11 cells were incubated at 37 °C/5% CO2 until confluent, rinsed with phosphate-buffered saline, and lysed with a buffer containing 10 mm NaCl, 0.05 mm Tris-HCl, 2 mm EDTA, 1% Triton X-100, and protease inhibitors (Roche Applied Science). Total proteins (200–250 μg/lane) were resolved by 9% SDS-PAGE and electroblotted onto polyvinylidene difluoride membranes. Membranes were blocked for 2 h at room temperature with 5% nonfat dry milk in TBST (150 mm NaCl, 10 mm Tris-HCl, 0.5% Tween 20 (polyoxyethylenesorbitan monolaurate)), and then incubated overnight at 4 °C with 1:1500 dilution anti-OSR1 (17) or 1:100 dilution anti-SPAK (8) antibodies (rabbit polyclonal) in TBST/5% nonfat dry milk. Membranes were then washed extensively in TBST, and protein bands were visualized by enhanced chemiluminescence (ECL plus, Amersham Biosciences).
Gene Knockdown by shRNA—The specific RNAi sequences against OSR1/SPAK were designed by BLOCK-iT™ RNAi Designer (Invitrogen) (Table 2). The RNAi constructs were then synthesized as short hairpins that adopted the human micro-RNA miR-30 structure (Zhou et al. 27) with AgeI and EcoRI restriction sites on each end, respectively. The shRNA encoding sequence was then inserted into the mammalian expression vector pLKO2 (modified from pLKO1 vector (Open Biosystems) by introducing an AgeI restriction site via QuikChange Mutagenesis (Stratagene, La Jolla, CA)). To create knockdown cell lines, 50B11 cells were trypsinized and plated at 30% density the day prior to transfection. The shRNA-containing plasmids were then transfected into the cells with FuGENE 6 (Roche Applied Science) at a 3:1 ratio (DNA:transfection reagent). Transfected cells were incubated at 37 °C/5% CO2 for 48 h prior to addition of puromycin 1–10 μg/ml. Under puromycin selection, the expressed siRNA molecules permanently knock down their targets. The knockdown is passed down through generations and cell passages without affecting cell viability.
TABLE 2.
RNA sequence of shRNA molecules used to knock down the kinases
A sequence targeting EGFP (not expressed in cells) was chosen as an irrelevant sequence to control for unspecific effects of shRNA expression (see Refs. 28, 29). Mir30 is a regulatory micro-RNA, SPAK, and OSR1 specific sequences were inserted in place of “NNN...” in the human mir30 structure.
Rescuing NKCC1 Activity in Knockdown Clones—The cDNAs encoding mouse SPAK (mSPAK) and mouse OSR1 (mOSR1) were cloned into the mammalian expression vector pcDNA3 in-frame with an N-terminal HA epitope. The constructs were then transfected (as described above) into the OSR1-KD and SPAK-KD cells, respectively. After neomycin (100 μg/ml) and puromycin (1–10 μg/ml) double selection, individual clones were isolated, and mSPAK and mOSR1 overexpression in cells was assessed by Western blot analysis using a mouse polyclonal anti-HA antibody.
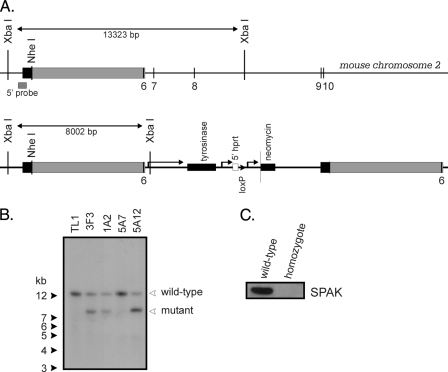
SPAK Knockout Mouse—The Mutagenic Insertion and Chromosome Engineering Resource (MICER) clone MHPN27a7, which contains a fragment of the mouse SPAK gene, was obtained from the Wellcome Trust Sanger Institute (Hinxton, Cambridge, UK), linearized with NheI and electroporated in TL1 (20) mouse embryonic stem cells. The transfected cells were maintained on fibroblast feeder cells in DMEM supplemented with 15% fetal bovine serum, 50 mg/ml gentamicin, 1000 units/ml leukemia inhibitory factor, 90 mm β-mercaptoethanol, and 0.2 mg/ml G418. Upon recombination, exon 6 (110 bp) was duplicated, and the tyrosinase, neomycin resistance, and 5′ hprt (hypoxanthine phosphoribosyltransferase gene) genes were inserted between the two exons (see Fig. 1A). We picked ∼500 independent neomycin-resistant colonies and grew them in 96-well plates on feeder layers, expanded them, and analyzed them for the presence of the mutant gene by performing Southern blot analysis using genomic DNA digested with XbaI and hybridized with a 32P-labeled probe consisting of 418 bp downstream of the left arm of recombination (see Fig. 1, B and C). A positive clone (3F3) was injected into C57B6J blastocysts, and six chimeric males (>60% brown fur) were obtained. Several chimeric males produced brown fur offspring demonstrating germ line transmission. Animals carrying the mutant allele were identified by PCR genotyping of tail DNA. After one additional mating with C57BL6 mice, heterozygous mice were crossed to obtain homozygous animals. As no PCR strategy could differentiate heterozygous from homozygous animals, we mated each offspring with C57BL6 animals and assessed the number of mutant and wild-type animals. Animals generating 100% heterozygous offspring were mated together, and a homozygote line was created. Verification of homozygosity was done by Western blot analysis (Fig. 1C). Wild-type mice were generated by breeding the wild-type offspring of the SPAK heterozygous crosses.
FIGURE 1.
Disruption of the SPAK gene. A, structure of the SPAK gene at exon 6, position of the 5′ probe and structure of the DNA fragment containing the mouse tyrosinase gene, a neomycin resistance gene cassette, and a fragment of the 5′ hprt gene. The position of a unique loxP site is indicated. Note that recombination results in the duplication of a region of the gene (MICER, depicted by a gray box). B, Southern blot analysis of embryonic stem (ES) cell genomic DNA digested with XbaI. The 13.3-kb band represents the control gene, and the 8-kb band originates from the mutant gene. The mutant gene is detected in clones 3F3, 1A2, and 5A12 by the 5′ probe. C, Western blot analysis of 60-mg brain protein isolated from a wild-type and a homozygous mouse. The 60-kDa molecular size band represents SPAK kinase.
Mouse DRG Isolation—Wild-type and homozygote SPAK knockout mice were sacrificed by decapitation under anesthesia. Dorsal root ganglia were dissected from the lower thoracic to mid-lumbar regions of the vertebral column and placed in oxygenated serum-free DMEM/F-12 medium. The connective tissue sheath around the ganglia was manually removed, and the DRGs were minced two to three times with iridectomy scissors. Minced DRGs were placed in a flask containing 5 ml of oxygenated serum-free DMEM/F-12 medium containing 1 mg/ml collagenase D (Roche Applied Science), 0.6 mg/ml deoxyribonuclease I (Sigma, St. Louis, MO), and 0.6 mg/ml trypsin (Sigma). They were incubated in a 37 °C water bath for 30–40 min; gently hand shaking the flask every 10 min. At the end of the incubation period, individual neurons were isolated from the ganglia by vigorous shaking. Soybean trypsin inhibitor (1 mg/ml, Sigma) was then added, and the dissociated cells were centrifuged at 70 × g for 6 min. The cell pellet was resuspended with 1 ml of DMEM/F-12 medium containing 10% serum. Cell suspensions (120 μl) were plated onto 35-mm glass bottom culture dishes (MatTek Corp., Ashland, MA) coated with poly-d-lysine and laminin, and incubated at 37 °C for 30 min prior to the addition of 2.5 ml of complete DMEM/F-12 medium (Invitrogen). Cells were used the following day for imaging experiments.
86Rb Uptake—Control and transfected 50B11 cells were plated on 6-well dishes and incubated at 37 °C/5% CO2 until confluent. For the uptake, cells were first washed twice with 1 ml of isosmotic saline (140 mm NaCl, 5 mm KCl, 2 mm CaCl2, 0.8 mm MgSO4, 5 mm glucose, 5 mm HEPES buffered to pH 7.4, 300 ± 5 mosm). Cells were then preincubated for 15 min in 1 ml of the same isosmotic saline plus 1 mm ouabain (Sigma) ± 20 μm bumetanide (Sigma). The preincubation solution was then aspirated and replaced with an identical solution containing 1 μCi of 86Rb, 1 mm ouabain ± 20 μm bumetanide. Four 5-μl aliquots of flux solution were sampled at the beginning of each 86Rb uptake condition and used as standards. After a 20-min uptake, the radioactive solution was aspirated, and the cells were washed three times with 1 ml of ice-cold solution, lysed for 1 h with 500 μl of 0.25 n NaOH, and neutralized with 250 μl of glacial acetic acid. 86Rb tracer activity was measured by using 150 μl of lysate for β-scintillation counting. NKCC1 flux was expressed in millimoles of K+/μg of protein/min.
Tl-uptake in Single Cells—Dorsal root ganglions of control and knockout mice were isolated, dissociated into single neurons as described, and incubated at 37 °C/5% CO2 for 24 h before use. For the uptake, thallium was used as the tracer of K+ (FluxOR™ Thallium Detection Kits, Invitrogen). Neurons were first loaded with the thallium-sensitive FluxOR™ dye (1×) in a hypotonic (275 ± 5 mosm) low chloride solution (125 mm NaMeSO3, 2 mm KCl, 2 mm CaCl2, 0.8 mm MgSO4, 5 mm glucose, 20 mm HEPES) plus 1× PowerLoad™ concentrate plus 2.7 mm probenecid (supplied by the kit) for 90 min. The loading solution was then aspirated, and neurons were washed three times with the same solution to remove excessive dye. Neurons were then preincubated for 10 min using 1 ml of the same saline containing 1 mm ouabain and 2.7 mm probenecid, in the presence or absence of 20 μm bumetanide (Sigma). For the detection of fluorescence, the preincubation solution was removed, and the neurons were treated with a hypertonic (340 ± 5 mosm) stimulus solution (2.8 mm TlSO4, 140 mm NaCl, 2 mm KCl, 2 mm CaCl2, 0.8 mm MgSO4, 5 mm Glucose, 20 mm HEPES, 27 mm sucrose, 1 mm ouabain, 2.7 mm probenecid) in the presence or absence of 20 μm bumetanide; and imaged using a 40×/1:30 plan-Neofluar oil objective in an LSM 510 META inverted confocal microscope at 488 nm excitation wavelength.
RESULTS
RT-PCR and Expression of Kinases and Transporters—The Na-K-2Cl cotransporter is abundant in sensory neurons where it participates to the gating of sensory information (2, 3, 21). Its activity is modulated by phosphorylation/dephosphorylation of key threonine residues located in its cytoplasmic N-terminal tail. Heterologous expression studies have demonstrated that NKCC1 phosphorylation is mediated by Ste20p-related serine/threonine kinases SPAK and OSR1 (10, 11, 13, 22). To manipulate expression of the kinases in a native cell system, we chose a rat nociceptive cell line, 50B11, developed by Chen and colleagues (19). Using RT-PCR, four cation-chloride cotransporters (NCC1, KCC1, KCC3, and KCC4) as well as six protein kinases (SPAK, OSR1, WNK1, WNK2, WNK3, and WNK4) were identified in 50B11 cells, a pattern similar to that seen in native mouse and rat DRG neurons (Fig. 2). Note that KCC2, which is exclusively expressed in central neurons, was not amplified in 50B11 cells, rat, and mouse DRG neurons. Due to the lack of sequence information on the rat WNK3, we used a pair of mouse primers, which failed to amplify a fragment from 50B11 cells, rat DRG neurons (Fig. 2), as well as rat brain (data not shown). However, the mouse primers were able to amplify a WNK3 fragment from cDNA isolated from mouse DRG neurons (Fig. 2).
FIGURE 2.
Expression patterns of cotransporters and regulatory kinases. A, expression of NKCC1, KCC1, KCC3, KCC4, OSR1, SPAK, WNK1, WNK2, WNK4, in 50B11 cells, and in rat and mouse DRG neurons were demonstrated by RT-PCR. Absence of WNK3 in 50B11 cells and in rat DRG neurons, compared with mouse DRG neurons is due to the lack of rat-specific WNK3 sequence information. B, central nervous system-specific KCC2 was absent in mouse and rat DRG neurons, as well as in the 50B11 cells, but was present in our positive control brain sample.
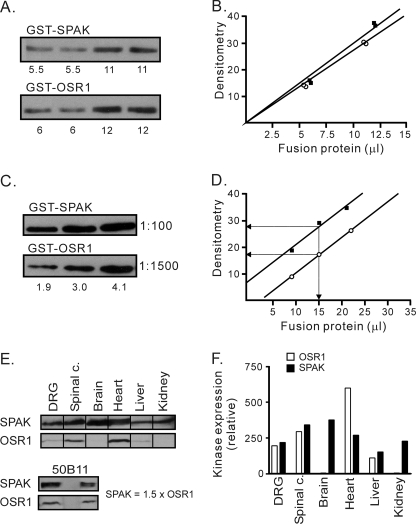
Relative Abundance of SPAK and OSR1 in Tissues—Previous work has shown that expression of OSR1 and SPAK overlaps in tissues (14–17). Because we wanted to assess the relative participation of each kinase in the regulation of NKCC1, we developed a semi-quantitative Western blot method to determine the relative abundance of SPAK and OSR1 kinases. Because purified antibodies have their own affinities toward their targets and have their own titers, it is difficult to compare signals elicited from two distinct antibodies. Therefore, we used GST fusion proteins of full-length SPAK and OSR1 to calibrate each antibody. Purification of GST fusion protein yields not only the full-length protein, but also large amounts of smaller protein fragments. To ensure that equal amounts of GST fusion proteins were utilized for the calibration, we did not rely on Bradford protein assay, but on Western blot analysis of the two fusion proteins using a rabbit polyclonal anti-GST antibody. As seen in Fig. 3 (A and B), Western blot and densitometry analysis showed that GST-SPAK and GST-OSR1 signals were very close to each other (10.2 μl of GST-SPAK = 10 μl of GST-OSR1). We next loaded equal amounts of the two fusion proteins onto SDS-PAGE gel at three different concentrations, transferred to polyvinylidene difluoride membranes, and probed with rabbit polyclonal anti-SPAK (1:100) and anti-OSR1 (1:1500), respectively. To avoid any participation of antibodies directed against GST epitope, the antibodies were preincubated with excess recombinant GST. Fig. 3 (C and D) shows the relative reactivity of the two antibodies, where anti-SPAK at a dilution of 1:100 yielded a signal 1.7 times stronger than anti-OSR1 at a 1:1500 dilution.
FIGURE 3.
Calibration of SPAK and OSR1 antibodies and quantitation of SPAK and OSR1 protein abundance in tissues. A, Western blot shows GST-SPAK and GST-OSR1 at two different amounts, probed with anti-GST antibody (1:1000). B, densitometry analysis shows equivalent signals for both full-length proteins (10.2 μl of GST-SPAK elicits the same signal intensity than 10 μl of GST-OSR1). C, quantitation of anti-SPAK and anti-OSR1 antibodies. The same amounts of GST-SPAK and GST-OSR1 were loaded and run in a gel at three different concentrations, transferred, and immunoblotted with anti-SPAK (1:100) and anti-OSR1 (1:1500) antibodies, respectively. D, densitometry analysis shows that anti-SPAK at 1:100 dilution yields a signal 1.7 times stronger than anti-OSR1 at 1:1500 dilution. E and F, Western blot analysis of SPAK and OSR1 expression in DRG, spinal cord, brain cortex, heart, kidney, liver, and 50B11 cells. Equal amount of total protein (200 μg) was loaded per lane. Primary antibodies were used at dilutions of 1:100 for SPAK and 1:1500 for OSR1 to quantitate the level of expression. Densitometry analysis demonstrates that OSR1 and SPAK have similar expression levels in DRG, spinal cord, and liver. Note that, in brain and kidney, there is very little OSR1 expression, whereas in heart, OSR1 expression was twice that of SPAK. SPAK expression in 50B11 cells was shown to be 1.5 times higher than OSR1 expression.
Comparison of SPAK and OSR1 Protein Abundance in Tissues and 50B11 Cells—To compare the protein expression level of SPAK and OSR1 in native tissues, we collected protein samples from mouse dorsal root ganglia, spinal cord, brain cortex, heart, liver, and kidney. Western blot analysis was performed with 200 μg of total proteins per lane (Fig. 3E). After densitometry analysis and correction based on the specific reactivity of each antibody, we observed similar SPAK and OSR1 expression levels in DRG, spinal cord, and liver. In contrast, OSR1 was barely detectable in brain cortex and kidney, whereas SPAK was nicely expressed in both tissues; OSR1 expression in heart was almost twice the level of SPAK expression. Similar semi-quantitative Western blot analysis was performed with protein extracts isolated from 50B11 cells (Fig. 3E, lower panel), cells used in our kinase knockdown experiments (Fig. 4). In these cells, SPAK expression was slightly higher than OSR1 expression (1.5-fold).
FIGURE 4.
Knockdown of OSR-1 and SPAK in 50B11 cells. A, Western blot analysis of OSR1-KD and wild-type 50B11 cells. An equal amount of total protein (250 μg) was loaded per lane. Anti-SPAK and anti-OSR1 were applied individually. Densitometry analysis revealed that OSR-1 expression in OSR1-KD cells was reduced by 65% (n = 3), whereas SPAK expression remained unaltered (not shown). B, K+ fluxes measured through unidirectional 86Rb uptakes demonstrate that NKCC1 activity (bumetanide-sensitive portion of the flux) was significantly reduced (40%) in OSR1-KD cells. C, Western blot analysis of two SPAK-KD clones and wild-type 50B11 cells. An equal amount of total protein (250 μg) was loaded per lane. Anti-SPAK and anti-OSR1 were applied individually. Densitometry analysis revealed that SPAK expression in both SPAK-KD clones was reduced by 60% (n = 3), whereas OSR1 expression remained unchanged (not shown). D, K+ fluxes demonstrate that NKCC1 activity was reduced by 45% in SPAK-KD cells. Bars represent means ± S.E. (n = 12).
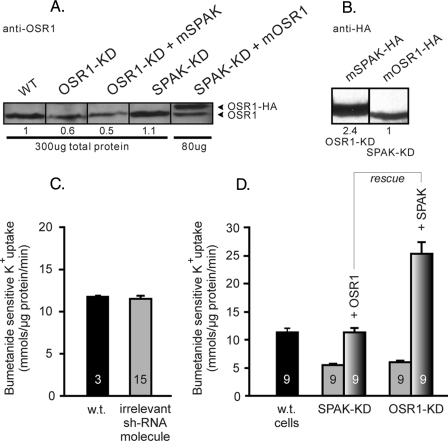
OSR1 and SPAK Knockdown Experiments—Given the fact that SPAK and OSR1 have similar protein expression level in 50B11 cells, we investigated the role of each kinase in regulating NKCC1 function through gene silencing, using the shRNA knockdown technique. First, 50B11 cells were transfected with a vector containing a sequence coding for an OSR1 shRNA molecule. After selection, expression of OSR1 and SPAK as well as NKCC1 function were assessed in these cells relative to untransfected 50B11 cells. As shown in Fig. 4A, expression of OSR1 was reduced by 70–75%, whereas SPAK expression was unchanged. The absence of any effect on SPAK expression argues against any unspecific effect of the OSR1-targeted shRNA. Under these conditions, we observed a ∼50% reduction in NKCC1 function (Fig. 4B). Second, we targeted SPAK in 50B11 cells by transfecting a vector containing a sequence coding for a SPAK shRNA molecule. Fig. 4 (C and D) shows the data obtained with two independent SPAK knockdown clones isolated after antibiotic selection. In both clones, SPAK expression was reduced by some 55% (Fig. 4C), yielding a 50% decrease in NKCC1 function (Fig. 4C). To address the specificity of the knockdown effect, we performed several control experiments. First, to confirm that the observed sh-OSR1 and sh-SPAK data were not due to unspecific effects related to the viral vector, we transfected 50B11 cells with an irrelevant shRNA-containing vector. As seen in Fig. 5C, this vector did not produce any inhibition of NKCC1. Second, to verify the effect of each kinase on NKCC1 function, we overexpressed OSR1 in 50B11 cells where SPAK was knocked down, and inversely, overexpressed SPAK in OSR1 knockdown 50B11 cells. The Western blot data of Fig. 5A show that overexpression of mSPAK in OSR1 knockdown cells does not affect the decreased level of OSR1 and that mOSR1 is overexpressed in SPAK-KD cells. This overexpression is confirmed with the anti-HA antibody (Fig. 5B). Similar results with anti-SPAK antibody were shown for the reverse conditions (data not shown). The data presented in Fig. 5D confirm that each knockdown could be rescued by the other kinase. Note that transfection of mSPAK resulted in very high expression of the kinase (Fig. 5B), leading to an increased activity of the cotransporter that was significantly above control (Fig. 5D).
FIGURE 5.
shRNA control plus rescue experiments via mSPAK and mOSR1 overexpression. A, Western blot analysis of OSR1 expression in wild-type cells, OSR1-KD cells, OSR1-KD cells rescued by SPAK overexpression, SPAK-KD cells, and SPAK-KD cells rescued with OSR1-HA. An equal amount of total protein (300 μg) except for overexpressing cells (80 μg) was loaded per lane. B, Western blot analysis with anti-HA antibody showing expression of SPAK-HA in OSR1-KD cells and OSR-HA in SPAK-KD cells. Note the higher expression level of mSPAK. An equal amount of protein (150 μg) was loaded per lane. C, control with irrelevant shRNA vector transfection shows no effect on NKCC1 activity. D, mOSR1 was overexpressed in sh-SPAK knockdown 50B11 cells, and mSPAK was overexpressed in sh-OSR1 knockdown cells. Note the increased activity upon SPAK overexpression, correlating with the Western data (panel A). Bars in panels C and D represent means ± S.E. All fluxes were measured in triplicate. Five independent clones expressing the irrelevant shRNA molecule were analyzed. N values are indicated inside the bars.
Finally, we co-transfected 50B11 cells with the two vectors and obtained several clones with various degrees of SPAK and OSR1 knockdown. Fig. 6 summarizes the data obtained with several stable clones generated (d-KD1 to d-KD5). As seen in Fig. 6, clone d-KD4 expressed <20% overall kinase expression, resulting in a 70–75% decrease in bumetanide-sensitive K+ uptake. The figure also shows a clear relationship between SPAK and OSR1 expression levels and the level of NKCC1 activity.
FIGURE 6.
Effect of SPAK plus OSR1 knockdown on NKCC1 function. A, Western blot analysis showing decreased levels of kinase expression in the different double knockdown (d-KD) clones. B, quantitation based on signals from panel A, corrected for the antibody calibration. C, NKCC1 activity (bumetanide-sensitive K+ flux) was plotted as a function of kinase expression. Data are from wild-type B5011 cells (100% SPAK plus OSR1 expression) and from different 50B11 clones co-transfected with vectors carrying shRNA targeting both SPAK and OSR1. Data were best fitted with a non-linear regression using GraphPad Prism, version 3. Note the close relationship between kinase abundance and NKCC1 activity.
NKCC1 Function in Native DRG Neurons—As shown in panels E and F of Fig. 3, native DRG neurons also express equivalent amounts of SPAK and OSR1 kinases. To assess the role of SPAK in modulating NKCC1 function in these native cells, we developed a single cellbased fluorescence method to assess cotransport activity. The method is based on the ability of cation-chloride cotransporters to carry the monovalent cation Tl+ (thallium) on their K+ site (23) and on the existence of Tl+-sensitive dyes (24). DRG neurons were isolated from wild-type mice and plated in glass-bottom dishes. After attachment to the substrate, the cells were loaded with the thallium-sensitive dye, washed, and imaged by confocal microscopy. Fig. 7A shows that addition of Tl+ to the bathing solution resulted in a marked increase in fluorescence, part of which was significantly reduced by the presence of 20 μm bumetanide in the bathing solution. The curves shown in Fig. 7B are averaged over a large number of cells. Similar experiments were performed with DRG neurons isolated from SPAK knockout mice. The figure also shows the presence of an ouabain plus bumetanide-insensitive component, which is likely due to Tl+ uptake through K+ channels and other transporters expressed in these cells. Consistent with the 50B11 cells data, absence of SPAK reduced by ∼50% the activity of the Na-K-2Cl cotransporter in DRG neurons (Fig. 8).
FIGURE 7.
NKCC1-mediated Tl+ (thallium) uptake in single isolated DRG neurons. DRG neurons were loaded with the thallium-sensitive dye FluxOR. FluxOR fluorescence (approximate fluorescence excitation and emission: 488/525 nm) increases upon addition of 2.8 mm Tl2SO4 to the external medium. A, photographs of DRG neurons showing increase in fluorescence after a 6 min addition of thallium. The two photographs on the left are differential interference contrast (DIC) pictures, whereas the four photographs on the right are fluorescence pictures. B, fluorescence intensity as a function of time. The top trace represents the fluorescence signal average of 26 DRG neurons under regular conditions. The bottom trace represents the fluorescence signal average of 21 DRG neurons incubated in the presence of 20 μm bumetanide. The difference between the two curves represents the bumetanide-sensitive component of the Tl+ uptake, mediated by NKCC1.
FIGURE 8.
NKCC1 activity in DRG neurons isolated from wild-type and SPAK knockout mice. NKCC1 function was assayed in single DRG cells through bumetanide-sensitive Tl+ uptake. A, individual curves; B, quantitation was performed by averaging the slopes of fluorescent increases calculated using the last baseline data point before addition of TI+ and the first six data points after the addition of Tl+ to the medium. Bars represent mean ± S.E. with six data points for wild-type and three data points for SPAK knockout DRG neurons, respectively.
DISCUSSION
SPAK and OSR1 proteins are both found in tissues such as brain, heart, skeletal muscle, lung, kidney, pancreas, testis, intestine, liver, ovary, thymus, and spleen (14–17). Although we had reported previously higher SPAK than OSR1 expression levels in the nervous system (17), a direct comparison between their expression levels was difficult to make, because each purified antibody has its own titer and affinity for its target. In this study, we developed a semi-quantitative Western blot method to compare relative SPAK and OSR1 expression levels. After precise calibration of each antibody with GST fusion proteins, we were able to determine that tissues such as brain cortex and kidney express high amounts of SPAK, but very little OSR1, whereas heart tissue expresses more OSR1 than SPAK. Interestingly, tissues such as dorsal root ganglia and spinal cord express similar levels of SPAK and OSR1. However, caution in interpreting these data needs to be taken, because tissues are typically not composed of single cell types, but made of a variety of cells with specific properties. This is the reason we sampled superficial brain cortex, to avoid contaminating brain tissue with choroid plexi, which have very high amounts of SPAK. This is also one of the reasons we examined kinase expression levels in a cell line, the 50B11 cells. As demonstrated by our RT-PCR data, these cells have expression pattern of cotransporters and kinases that is similar to that of native dorsal root ganglion neurons. As shown in this study, 50B11 cells express similar levels of SPAK and OSR1 kinases. Our semi-quantitative Western blot method might also be relevant when using antibodies for immunodetection on tissue sections. Although antibodies do not necessarily have the same properties in Western blot and immunofluorescence assays, the method described here provides some quantitative measure of antibody titer and strength.
Based on heterologous expression studies, it is well documented that SPAK and OSR1 phosphorylate and activate NKCC1 (10–12, 25). However, no study has yet examined how SPAK and OSR1, when co-expressed, affect NKCC1 activity in native tissues. The most direct way to address this issue is to generate SPAK and OSR1 knockout mice. Our laboratory has generated both knockout mouse models, and although OSR1 knockout mice are embryonic lethal (data not shown), SPAK knockout mice are viable and present no obvious adverse behavioral phenotype, with the exception of reduced fertility. To study the effect of SPAK deletion on NKCC1 activity, we chose to examine isolated DRG neurons, which have high NKCC1 activity throughout life and gate sensory information. Because DRG neurons are isolated in limited numbers, we needed a method to assess the activity of the cotransporter in single cells. We adapted a fluorescent methodology that we developed for high throughput screening of cation-chloride cotransporters using confocal microscopy. This method makes use of Tl+ as a surrogate of K+ and of a Tl+-sensitive fluorescent dye (FluxOR™) to visualize Tl+ (K+) uptake through NKCC1 in single cells. The method allows for acquisition of signals of several cells in as single microscopic field and follow-up analysis of signal intensity over time. Our data show that we can demonstrate a significant bumetanide-sensitive portion in the Tl+ uptake, indicative of NKCC1 function. When measurements were performed using both wild-type and SPAK knockout DRG neurons, we observed a significant reduction in NKCC1 activity in the knockout neurons, indicating a significant role for SPAK in regulating NKCC1. Interestingly, the function of the cotransporter was not completely abrogated, indicating that another kinase could activate NKCC1 in these SPAK-deficient cells.
In the absence of a viable OSR1 knockout animal, we took another approach and used gene silencing in cultured cells to demonstrate the combined roles of SPAK and OSR1 in regulating NKC1 function. As mentioned above, 50B11 cells have been isolated as a nociceptive dorsal root ganglion sensory cell line. Undifferentiated, these cells might not have all properties of native DRG neurons. However, because the cells express all cotransporters and kinases of native DRG cells and express similar levels of SPAK and OSR1, they constitute an adequate model to address the role of the kinases in NKCC1 regulation.
To silence SPAK and OSR1 expression, we expressed shRNA molecules delivered via transfected vectors. As many studies have indicated, shRNA silencing of native genes is generally not as efficient as knockout animals, because complete silencing is rarely obtained (26, 27). In our experiments, we chose a short hairpin structure, which adopts the structure of human micro-RNA mir30, and which was reported to produce highly efficient RNAi effect (27). With this structure, we successfully reduced SPAK expression level to <50% in 50B11 cells. Consistent with our SPAK knockout experiment, NKCC1 activity was significantly affected by a 50% reduction in SPAK expression (see Fig. 4). Furthermore, as anticipated based on the SPAK knockout experiments, we also demonstrated that OSR1 participated in the regulation of NKCC1, because its knockdown in 50B11 cells resulted also in significant attenuation of NKCC1 activity (see Fig. 4). Thus, reducing either kinase only leads to a partial reduction of NKCC1 function, whereas reducing both kinases resulted in additive effect. During the process of generating double knockdown of SPAK and OSR1, we obtained cell lines with different degrees of SPAK and OSR1 knockdown. Considering that SPAK and OSR1 total to 100% Ste20 kinase expression in wild-type 50B11 cells, and given the fact that SPAK expression was determined to be 1.5 times the level of OSR1 expression (see Fig. 3E), we were able to provide a direct relationship between SPAK plus OSR1 expression and NKCC1 activity, as measured through bumetanide-sensitive K+ flux (see Fig. 8). Taken together, our SPAK knockout data and silencing data in 50B11 cells demonstrate that both SPAK and OSR1 participate in the regulation of NKCC1 in dorsal root ganglion neurons. Our data also argue that expression of the Ste20 kinases is limiting, because function of the cotransporter can be modified by manipulating the expression level of the two kinases. These data do not, however, preclude regulation of these kinases by post-translational modifications. In fact, work performed in our laboratory and others have shown that SPAK and OSR1 are themselves the target of phosphorylation by upstream kinases, such as the WNK kinases.
Aside from demonstrating the redundancy in kinase function, our present study introduced three novel approaches for the study of kinase-cotransporter function: the creation and use of a SPAK-deficient mouse, the development of a semi-quantitative Western blot method, and the development of a method to assess NKCC1 function in single cells. These methods will allow us and others to further address the role of kinases in regulating cotransporter function.
Acknowledgments
We thank Dr. Kenneth Gagnon for the discussions we had during the course of this study and for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM074771 and NS53658 (to E. D.).
Footnotes
The abbreviations used are: NKCC1, Na+-K+-2Cl– cotransporter type 1; SPAK, Ste20-related proline-alanine-rich kinase; OSR1, oxidative-stress response 1; DRG, dorsal root ganglion; DMEM, Dulbecco's modified Eagle's medium; RT, reverse transcription; shRNA, short hairpin RNA; RNAi, RNA interference; m, mouse; HA, hemagglutinin; GST, glutathione S-transferase.
References
- 1.Pieraut, S., Matha, V., Sar, C., Hubert, T., Méchaly, I., Hilaire, C., Mersel, M., Delpire, E., Valmier, J., and Scamps, F. (2007) J. Neurosci. 27 6751–6759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Leefmans, F. J., Gamiño, S. M., Giraldez, F., and Nogueron, I. (1988) J. Physiol. (Lond.) 406 225–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung, K.-W., Kirby, M., McDonald, M. P., Lovinger, D. M., and Delpire, E. (2000) J. Neurosci. 20 7531–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willis, W. D. (1999) Exp. Brain Res. 124 395–421 [DOI] [PubMed] [Google Scholar]
- 5.Laird, J. M., Garcia-Nicas, E., Delpire, E. J., and Cervero, F. (2004) Neurosci. Lett. 361 200–203 [DOI] [PubMed] [Google Scholar]
- 6.Granados-Soto, V., Arguelles, C. F., and Alvarez-Leefmans, F. J. (2005) Pain 114 231–238 [DOI] [PubMed] [Google Scholar]
- 7.Price, T. J., Cervero, F., and de Koninck, Y. (2005) Curr. Top. Med. Chem. 5 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piechotta, K., Lu, J., and Delpire, E. (2002) J. Biol. Chem. 277 50812–50819 [DOI] [PubMed] [Google Scholar]
- 9.Dowd, B. F., and Forbush, B. (2003) J. Biol. Chem. 278 27347–27353 [DOI] [PubMed] [Google Scholar]
- 10.Gagnon, K. B., England, R., and Delpire, E. (2006) Am. J. Physiol. 290 C134–C142 [DOI] [PubMed] [Google Scholar]
- 11.Vitari, A. C., Thastrup, J., Rafiqi, F. H., Deak, M., Morrice, N. A., Karlsson, H. K., and Alessi, D. R. (2006) Biochem. J. 397 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gagnon, K. B., England, R., and Delpire, E. (2006) Mol. Cell Biol. 26 689–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagnon, K. B., England, R., and Delpire, E. (2007) Cell Physiol. Biochem. 20 131–142 [DOI] [PubMed] [Google Scholar]
- 14.Ushiro, H., Tsutsumi, T., Suzuki, K., Kayahara, T., and Nakano, K. (1998) Arch. Biochem. Biophys. 355 233–240 [DOI] [PubMed] [Google Scholar]
- 15.Tamari, M., Daigo, Y., and Nakamura, Y. (1999) J. Hum. Genet. 44 116–120 [DOI] [PubMed] [Google Scholar]
- 16.Johnston, A. M., Naselli, G., Gonez, L. J., Martin, R. M., Harrison, L. C., and Deaizpurua, H. J. (2000) Oncogene 19 4290–4297 [DOI] [PubMed] [Google Scholar]
- 17.Piechotta, K., Garbarini, N. J., England, R., and Delpire, E. (2003) J. Biol. Chem. 278 52848–52856 [DOI] [PubMed] [Google Scholar]
- 18.Yan, Y., Nguyen, H., Dalmasso, G., Sitaraman, S. V., and Merlin, D. (2007) Biochim. Biophys. Acta 1769 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, W., Mi, R., Haughey, N., Oz, M., and Höke, A. (2007) J. Peripher. Nerv. Syst. 12 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labosky, P. A., Barlow, D. P., and Hogan, B. L. (1994) Development 120 3197–3204 [DOI] [PubMed] [Google Scholar]
- 21.Rocha-González, H. I., Mao, S., and Alvarez-Leefmans, F. J. (2008) J. Neurophysiol. 100 169–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moriguchi, T., Urushiyama, S., Hisamoto, N., Iemura, S. I., Uchida, S., Natsume, T., Matsumoto, K., and Shibuya, H. (2006) J. Biol. Chem. 280 42685–42693 [DOI] [PubMed] [Google Scholar]
- 23.Delpire, E., Days, E., Mi, D., Lewis, M., Kim, K., Lidnsley, C., and Weaver, C. D. (March 11, 2009) Proc. Natl. Acad. Sci. U. S. A. 106 5383–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weaver, C. D., Harden, D., Dworetzky, S. I., Robertson, B., and Knox, R. J. (2004) J. Biomol. Screen. 9 671–677 [DOI] [PubMed] [Google Scholar]
- 25.Chen, W., Yazicioglu, M., and Cobb, M. H. (2004) J. Biol. Chem. 279 11129–11136 [DOI] [PubMed] [Google Scholar]
- 26.Zeng, Y., and Cullen, B. R. (2003) RNA 9 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou, H., Xia, X. G., and Xu, Z. (2005) Nucleic Acids Res. 33 e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai, P., Xiong, W. C., and Mei, L. (2006) J. Biol. Chem. 281 927–933 [DOI] [PubMed] [Google Scholar]
- 29.Akai, S., Hosomi, H., Minami, K., Tsuneyama, K., Katoh, M., Nakajima, M., and Yokoi, T. (2007) J. Biol. Chem. 282 23996–24003 [DOI] [PubMed] [Google Scholar]