Abstract
Cytokinesis in bacteria depends upon the contractile Z ring, which is composed of dynamic polymers of the tubulin homolog FtsZ as well as other membrane-associated proteins such as FtsA, a homolog of actin that is required for membrane attachment of the Z ring and its subsequent constriction. Here we show that a previously characterized hypermorphic mutant FtsA (FtsA*) partially disassembled FtsZ polymers in vitro. This effect was strictly dependent on ATP or ADP binding to FtsA* and occurred at substoichiometric levels relative to FtsZ, similar to cellular levels. Nucleotide-bound FtsA* did not affect FtsZ GTPase activity or the critical concentration for FtsZ assembly but was able to disassemble preformed FtsZ polymers, suggesting that FtsA* acts on FtsZ polymers. Microscopic examination of the inhibited FtsZ polymers revealed a transition from long, straight polymers and polymer bundles to mainly short, curved protofilaments. These results indicate that a bacterial actin, when activated by adenine nucleotides, can modify the length distribution of bacterial tubulin polymers, analogous to the effects of actin-depolymerizing factor/cofilin on F-actin.
Bacterial cell division requires a large number of proteins that colocalize to form a putative protein machine at the cell membrane (1). This machine, sometimes called the divisome, recruits enzymes to synthesize the septum cell wall and to initiate and coordinate the invagination of the cytoplasmic membrane (and in Gram-negative bacteria, the outer membrane). The most widely conserved and key protein for this process is FtsZ, a homolog of tubulin that forms a ring structure called the Z ring, which marks the site of septum formation (2, 3). Like tubulin, FtsZ assembles into filaments with GTP but does not form microtubules (4). The precise assembly state and conformation of these FtsZ filaments at the division ring is not clear, although recent electron tomography work suggests that the FtsZ ring consists of multiple short filaments tethered to the membrane at discrete junctures (5), which may represent points along the filaments bridged by membrane anchor proteins.
In Escherichia coli, two of these anchor proteins are known. One of these, ZipA, is not well conserved but is an essential protein in E. coli. ZipA binds to the C-terminal tail of FtsZ (6–8), and purified ZipA promotes bundling of FtsZ filaments in vitro (9, 10). The other, FtsA, is also essential in E. coli and is more widely conserved among bacterial species. FtsA is a member of the HSP70/actin superfamily (11, 12), and like ZipA, it interacts with the C-terminal tail of FtsZ (7, 13–15). FtsA can self-associate (16, 17) and bind ATP (12, 18), but reports of ATPase activity vary, with Bacillus subtilis FtsA having high activity (19) and Streptococcus pneumoniae FtsA exhibiting no detectable activity (20). There are no reports of any other in vitro activities of FtsA, including effects on FtsZ assembly.
Understanding how FtsA affects FtsZ assembly is important because FtsA has a number of key activities in the cell. It is required for recruitment of a number of divisome proteins (21, 22) and helps to tether the Z ring to the membrane via a C-terminal membrane-targeting sequence (23). FtsA, like ZipA and other divisome proteins, is necessary to activate the contraction of the Z ring (24, 25). In E. coli, the FtsA:FtsZ ratio is crucial for proper cell division, with either too high or too low a ratio inhibiting septum formation (26, 27). This ratio is roughly 1:5, with ∼700 molecules of FtsA and 3200 molecules of FtsZ per cell (28), which works out to concentrations of 1–2 and 5–10 μm, respectively.
Another interesting property of FtsA is that single residue alterations in the protein can result in significant enhancement of divisome activity. For example, the R286W mutation of FtsA, also called FtsA*, can substitute for the native FtsA and divide the cell. However, this mutant FtsA causes E. coli cells to divide at less than 80% of their normal length (29) and allows efficient division of E. coli cells in the absence of ZipA (30), indicating that it has gain-of-function activity. FtsA* and other hypermorphic mutations such as E124A and I143L can also increase division activity in cells lacking other essential divisome components (31–33). The R286W and E124A mutants of FtsA also bypass the FtsA:FtsZ ratio rule, allowing cell division to occur at higher ratios than with WT2 FtsA. This may be because the altered FtsA proteins self-associate more readily than WT FtsA, which may cause different changes in FtsZ assembly state as compared with WT FtsA (17, 34).
In this study, we use an in vitro system with purified FtsZ and a purified tagged version of FtsA* to elucidate the role of FtsA in activating constriction of the Z ring in vivo. We show that FtsA*, at physiological concentrations in the presence of ATP or ADP, has significant effects on the assembly of FtsZ filaments.
EXPERIMENTAL PROCEDURES
Medium and Chemicals—PIPES, isopropyl-β-d-thiogalactopyranoside, GTP, ATP, and ADP were obtained from Sigma Co. All other chemicals used were of analytical grade. Bacterial cells were grown in LB (0.5% yeast extract, 0.5% NaCl, 1% Tryptone).
Protein Purification—FtsZ and FtsZ(F268C) were purified from WM971 and WM3261, respectively, as described previously (35). To purify FtsA or FtsA*, the E. coli ftsA or ftsA* genes were expressed from the T7 RNA polymerase promoter in pET28a (pWM1260 or pWM1690, respectively) in strain C43(DE3) (30, 36). The resulting proteins contain a His6-T7 tag (HT) at their N termini and are referred to as HT-FtsA proteins. An overnight culture was inoculated 1:100 in 3 liters of LB at 37 °C, grown to an A600 of 0.2–0.3 and induced with 1 mm isopropyl-β-d-thiogalactopyranoside. The cells were further grown for 4 h, collected at 10,000 × g, and resuspended in 50 ml of lysis buffer (50 mm Tris, pH 8.0, 0.3 m NaCl, 10 mm MgCl2, and 1 mm EDTA) containing 0.4 mg/ml lysozyme, 1 mm phenylmethylsulfonyl fluoride, and 0.1% β-mercaptoethanol and incubated for 1 h on ice. Cells were broken by sonication with a Branson Sonifier 250 (output setting of 20, 30-s pulse, 10 cycles) and centrifuged at 20,000 × g. The pellet was resuspended in 20 ml of 25 mm PIPES (pH 7.4) containing 0.3 m NaCl, 0.1 mm CHAPS, and 1% Nonidet P-40, incubated for 1 h, and then centrifuged in a Beckman TLA 100 centrifuge with a TLA 100.3 rotor at 80,000 rpm for 30 min. The clear supernatant fraction was collected and loaded on a Talon cobalt column. The column was washed with 25 mm PIPES, pH 7.4, containing 0.3 m NaCl and 25 mm imidazole and eluted with 150 mm imidazole. The protein preparation was desalted using a P6 Biogel column. The concentration of HT-FtsA* was measured by Coomassie Blue staining after SDS-PAGE with bovine serum albumin standards, and the protein was stored at –80 °C.
Sedimentation Assay—Purified FtsZ (12 μm) was polymerized in FtsZ assembly buffer (25 mm PIPES, pH 7.4, with 10 mm MgCl2 and 100 mm potassium glutamate) plus 2 mm GTP and HT-FtsA* (0–5μm), in the absence or presence of 2 mm ATP, ADP, or AMP-PNP. The polymerization reaction took place at 37 °C for 20 min. FtsZ polymers were then sedimented with a Beckman TLA 100.3 ultracentrifuge rotor at 80,000 rpm for 30 min, and the pellet was washed and resuspended in warm buffer and subjected to SDS-PAGE. FtsZ band intensity was measured using ImageJ software (Wayne Rasband, National Institutes of Health).
Measurement of Phosphate Release—Phosphate release was measured with the PiPer phosphatase assay kit (Invitrogen) using the manufacturer's protocol. Briefly, in a 96-well plate, FtsZ was polymerized in 25 mm PIPES buffer (pH 7.4) containing 10 mm MgCl2, 100 mm potassium glutamate, 2 mm ATP, and 2 mm GTP with or without FtsA* for 30 min at 37 °C. After 30 min, an equal volume of PiPer reagent was added and incubated at 37 °C for another 1 h. Fluorescence emission was measured at 590 nm while exciting at 530 nm in a Tecan Infinite M200 fluorescent microplate reader. Phosphate standards were run in the same set to calculate the moles of phosphate released.
Microscopy—For electron microscopy, FtsZ (12 μm) was polymerized in 25 mm PIPES (pH 7.0), 10 mm MgCl2, 100 mm potassium glutamate, 2 mm GTP, 2 mm ATP in the absence or presence of HT-FtsA* for 20 min at 37 °C. Samples were transferred to a glow-discharged Formvar carbon-coated copper grid and negatively stained using 0.25% methylamine tungstate. Digital images of the specimen were captured with a 100-kV JEOL JEM1200 electron microscope equipped with a Gatan 1K × 1K CCD camera.
For fluorescence microscopy, purified FtsZ(R268C) was labeled with fluorescein maleimide and assembled in 25 mm PIPES buffer (pH 7.0) containing 10 mm MgCl2, 1 mm GTP, and 100 mm potassium glutamate, with or without 2.5 μm HT-FtsA* + ATP. After 15 min of assembly at 37 °C, 3 μl of the sample was placed on a microscope slide, covered with a glass coverslip, and examined with a ×100 oil objective on an Olympus BX-60 microscope with a green fluorescent protein/fluorescein isothiocyanate filter set. Higher concentrations of HT-FtsA* led to visible aggregates.
ATP Binding Assays—To measure ATP binding, HT-FtsA or HT-FtsA* (3 μm) was incubated with 50 μm mantATP in FtsZ assembly buffer at room temperature for 30 min. The sample was excited at 356 nm, and fluorescence emission spectra were monitored from 410 to 490 nm on a Photon Technology International fluorometer. The effects of free ATP on the spectra of HT-FtsA or HT-FtsA* bound to mantATP were initially measured by adding 1 mm free ATP to the sample. The HT-FtsA*-mantATP complex was subsequently titrated against free ATP, and the change in the fluorescence intensity at 448 nm was used to calculate dissociation constants using a double reciprocal curve as described previously (35).
To calculate the dissociation constant of the HT-FtsA*-ATP/ADP complex, we measured the intrinsic tryptophan fluorescence of HT-FtsA*. Binding was measured by titrating ATP or ADP (0–1.5 mm) against HT-FtsA* (1 μm). The sample was excited at 290 nm to minimize the inner filter effects, and the fluorescence spectra were monitored between 310 and 370 nm on a Photon Technology International fluorometer. The change in fluorescence intensity at 340 nm was used to calculate dissociation constants with a double reciprocal curve as described previously (35). Inner filter correction was done for all spectra prior to calculation.
RESULTS
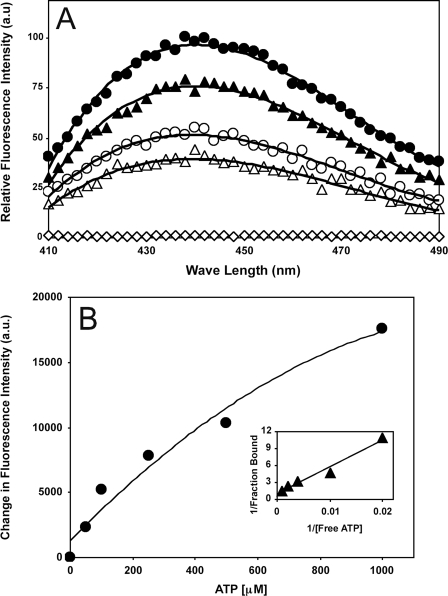
Purified HT-FtsA* Binds ATP—To understand how FtsA might affect FtsZ assembly, we purified soluble WT FtsA with an N-terminal His6-T7 tag (HT-FtsA) from pWM1260 in C43(DE3). Purified HT-FtsA bound ATP as measured by the increase in fluorescence of a fluorescent ATP derivative (mantATP) upon the addition of HT-FtsA and the strong quenching of this fluorescence by excess unlabeled ATP (Fig. 1A). However, this HT-FtsA preparation displayed very low ATPase activity and had no detectable effect on FtsZ assembly or GTPase activity under a number of conditions (data not shown).
FIGURE 1.
Purified HT-FtsA and HT-FtsA* bind ATP. A, fluorescence intensity plots of HT-FtsA-mantATP without (filled triangles) and with (open triangles) 1 mm ATP or HT-FtsA*-mantATP without (filled circles) and with (open circles) 1 mm ATP. Only HT-FtsA* with 1 mm ATP (open diamonds) shows no fluorescence. a.u., arbitrary units. B, change in the HT-FtsA*-mantATP fluorescence intensity at 440 nm with increasing concentrations of ATP. The inset shows a double reciprocal plot of the binding data.
These problems prompted us to try HT-FtsA* instead as in vivo evidence suggests that it is an activated form of FtsA that has a distinct conformation (30). Like HT-FtsA, HT-FtsA* was able to complement an ftsA mutant (30), indicating that it was functional. HT-FtsA* was overproduced and purified from pWM1690 in C43(DE3), using the same procedure as for HT-FtsA. As with HT-FtsA (data not shown), the Coomassie Blue-stained HT-FtsA* band was >95% pure (supplemental Fig. S1). The one prominent band below HT-FtsA* was confirmed by mass spectrometry to be a breakdown product of HT-FtsA* (data not shown). Like HT-FtsA, HT-FtsA* bound ATP efficiently (Fig. 1, A and B). Most importantly, unlike HT-FtsA, HT-FtsA* regulated FtsZ assembly (see below). Therefore, we used HT-FtsA* for the remainder of this study.
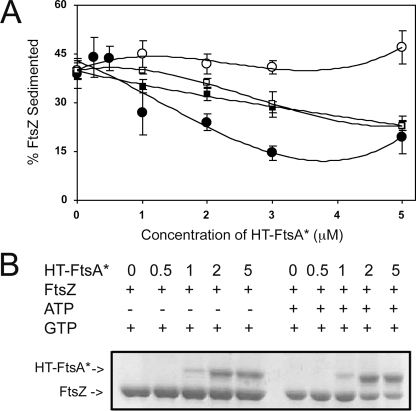
HT-FtsA* Decreases the Polymer Mass of FtsZ in the Presence of ATP—The effects of HT-FtsA* on FtsZ polymerization were examined by sedimentation, which is a widely used and sensitive assay for the extent of FtsZ assembly. FtsZ (12 μm) was polymerized in a physiological buffer at pH 7.4, and polymer mass was measured by the fraction of FtsZ in the pellet after ultracentrifugation. In the absence of HT-FtsA*, ∼40% of the total FtsZ reproducibly appeared in the pellet fraction (Fig. 2B and data not shown), giving a baseline level of assembly under these conditions.
FIGURE 2.
ATP activates HT-FtsA* inhibition of FtsZ assembly. A, FtsZ sedimentation efficiency with increasing concentration of HT-FtsA*, without added nucleotide (open circles) or with 2 mm ATP (filled circles), 2 mm AMP-PNP (open squares), and 2 mm ADP (filled squares). Error bars indicate S.E. B, Coomassie Blue-stained SDS-PAGE of a representative sedimentation experiment, showing levels of HT-FtsA* or FtsZ in the pellets; ATP and GTP were at 2 mm, FtsZ was at 12 μm, and HT-FtsA* was at the concentrations in μm shown.
When HT-FtsA* was added to the reaction mixture at concentrations up to 5 μm, the FtsZ in the pellet fraction did not change significantly from ∼40%, indicating that the HT-FtsA* preparation had little or no negative effect on FtsZ assembly (Fig. 2, A, open circles, and B). However, the addition of ATP had dramatic inhibitory effects; when 5 μm HT-FtsA* was added to FtsZ in the presence of ATP, only 19% of the total FtsZ was found in the pellet as compared with 39% in the absence of ATP (Fig. 2, A, closed circles, and B). ATP alone had no effect on FtsZ sedimentation.
Concentrations of HT-FtsA* below 5 μm also inhibited FtsZ assembly. For example, 1 μm HT-FtsA* + ATP resulted in ∼25% of FtsZ in the pellet fraction, and HT-FtsA* levels between 2 and 3 μm further reduced FtsZ pelleting (Fig. 2A, open circles). In contrast, concentrations of HT-FtsA* at 0.5 μm and below had no significant effect on FtsZ assembly with this assay (Fig. 2, A and B), suggesting that a critical concentration of HT-FtsA* between 0.5 and 1 μm may be necessary to mediate a detectable effect. Therefore, we can conclude that HT-FtsA* concentrations above ∼1 μm inhibit FtsZ sedimentation but only when ATP is present. Importantly, this ∼1 μm threshold level of HT-FtsA* is similar to measured levels of native FtsA in E. coli cells (28), assuming that the cytoplasmic volume of E. coli is ∼2 fl.
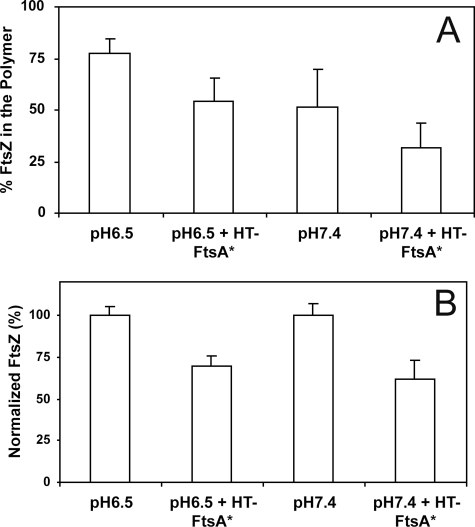
To test the robustness of the inhibition of FtsZ assembly by HT-FtsA*, we measured the effect of HT-FtsA* on FtsZ polymers formed at lower pH, a less physiological but more permissive condition for in vitro assembly (37, 38). We polymerized FtsZ (12 μm) at pH 6.5 and pH 7.4 with GTP and ATP, with or without 5 μm HT-FtsA*. As expected, somewhat higher levels of FtsZ were pelleted at pH 6.5 than at pH 7.4 in the absence of HT-FtsA* (Fig. 3A). The addition of HT-FtsA* reduced FtsZ polymerization at both pH conditions, but similar normalized percentages of FtsZ were pelleted in both cases (Fig. 3B). These results indicate that HT-FtsA*-mediated depolymerization of FtsZ occurs not only under conditions of moderate FtsZ assembly at physiological pH but also under conditions of more complete FtsZ assembly at lower pH.
FIGURE 3.
Role of pH in HT-FtsA*-mediated inhibition of FtsZ sedimentation. A, effect of HT-FtsA* on FtsZ sedimentation efficiency at different pH values. Error bars indicate S.E. B, FtsZ sedimentation efficiencies from panel A, normalized to 100%. Error bars indicate S.E.
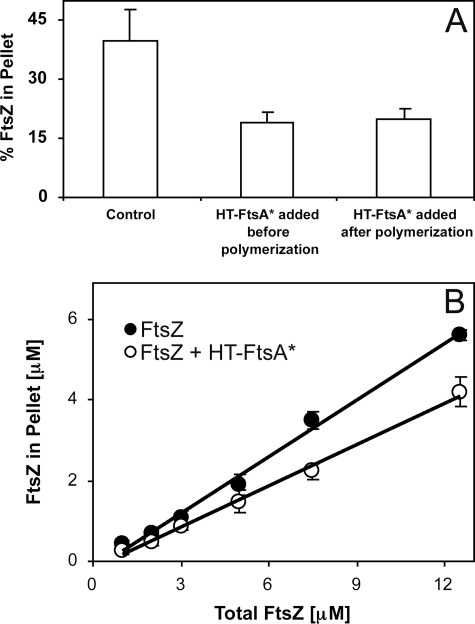
HT-FtsA* Can Disassemble Preformed FtsZ Polymers and Does Not Sequester FtsZ Monomers—The above results show that HT-FtsA* can reduce FtsZ polymer mass when it is present in the reaction mixture prior to FtsZ assembly in GTP. To determine whether it can also disassemble preformed FtsZ polymers, FtsZ was first polymerized for 10 min at 37 °C followed by the addition of HT-FtsA*. The sample was incubated at 37 °C for another 20 min, and FtsZ polymers were sedimented. As shown in Fig. 4A, HT-FtsA* was found to decrease the sedimentable polymer mass of preformed FtsZ polymers to a level similar to that when added prior to FtsZ assembly (∼20%). This suggests that HT-FtsA* is able to depolymerize preformed FtsZ polymers quite efficiently.
FIGURE 4.
Mechanisms of FtsZ disassembly by ATP-activated HT-FtsA*. A, FtsZ sedimentation efficiency without HT-FtsA* (Control), with 5 μm HT-FtsA* and ATP added prior to FtsZ assembly (middle column), or after assembly (right column). Error bars indicate S.E. B, FtsZ sedimentation efficiency at different FtsZ concentrations, with (open circles) or without (filled circles) added 5 μm HT-FtsA* + ATP. The x-intercept represents the apparent cc for FtsZ assembly. Error bars indicate S.E.
FtsZ exhibits a critical concentration (cc) for assembly, which is increased significantly by inhibitors such as SulA that probably sequester FtsZ monomers. To ask whether HT-FtsA* might similarly sequester FtsZ, we calculated the cc for FtsZ assembly over a range of FtsZ concentrations in the presence of HT-FtsA* and ATP. Without HT-FtsA*, the cc for FtsZ polymerization was 0.43 ± 0.17 μm (Fig. 4B, filled circles), which is similar to other published values (39, 40). In the presence of 5 μm HT-FtsA* and ATP, which efficiently inhibits FtsZ assembly as shown above, the cc was calculated to be 0.52 ± 0.25 μm (Fig. 4B, open circles). The insensitivity of the cc to HT-FtsA* suggests that HT-FtsA* does not inhibit FtsZ polymerization by sequestering FtsZ monomers.
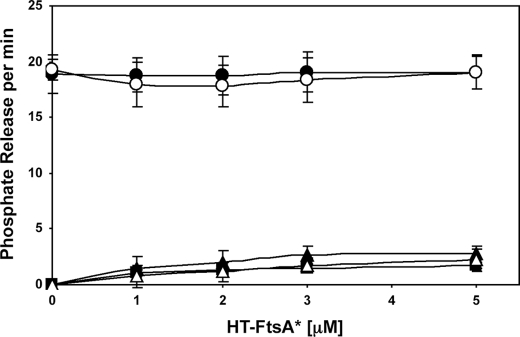
HT-FtsA* Does Not Significantly Affect the GTPase Activity of FtsZ—Because of its inhibition of FtsZ assembly, we asked whether HT-FtsA* might affect FtsZ GTPase activity. Using a sensitive fluorescent phosphate release assay with a fixed concentration of 6 μm FtsZ plus GTP and ATP, we did not detect any changes in nucleotide hydrolysis with increasing concentrations of HT-FtsA* (Fig. 5, upper plots). The same was true with GTP in the absence of ATP. In both cases, the phosphate release rate held steady at ∼3 mol of phosphate released per minute per mole of FtsZ, which is similar to previously reported levels of GTPase activity for FtsZ.
FIGURE 5.
HT-FtsA* does not affect FtsZ GTPase activity. The upper plots show GTPase activity of 6 μm FtsZ at different concentrations of HT-FtsA*, either with GTP alone (open circles) or with GTP+ATP (filled circles), measured as phosphate release per min with the PiPer assay. The lower plots show phosphate release per min measured at different concentrations of HT-FtsA*, with ATP (filled triangles), GTP (filled squares), or FtsZ + ATP (open triangles) after subtraction of the no-nucleotide control values. Error bars indicate S.E.
Because ATP hydrolysis of E. coli FtsA has not yet been characterized, we also tested HT-FtsA* for ATPase activity. Using the same phosphate release assay, we detected very weak activity that increased linearly with increasing HT-FtsA* (Fig. 5, lower plots). This low activity was similar in the presence of ATP, GTP, or FtsZ + ATP, indicating that FtsZ does not stimulate ATP hydrolysis by HT-FtsA*. The maximal activity of ∼0.3 mol of phosphate released per minute per mol of HT-FtsA* was calculated at 5 μm HT-FtsA*.
ADP or AMP-PNP Also Activates FtsZ Disassembly by HT-FtsA* but Less Efficiently—An ATP molecule is present in the crystal structure of Thermotoga maritima FtsA, and our preparation of HT-FtsA* has weak ATPase activity. We therefore asked whether ATP hydrolysis was important for the HT-FtsA*-mediated FtsZ disassembly activity by testing the roles of ADP or non-hydrolyzable ATP.
We assayed FtsZ sedimentation in the presence of ADP and different concentrations of HT-FtsA*. Although ADP alone or ADP + 1 μm HT-FtsA* resulted in the background sedimentable FtsZ polymer mass of ∼40%, increasing concentrations of HT-FtsA* induced a gradual decrease in sedimentable FtsZ polymer mass to 24%, as compared with the 19% of FtsZ polymer mass for 5 μm HT-FtsA* and ATP (Fig. 2A, compare filled squares with filled circles). The relationship between FtsZ disassembly and HT-FtsA* concentration fits a linear curve with ADP instead of the more hyperbolic curve with ATP.
We then tested the ability of non-hydrolyzable ATP (AMP-PNP) to promote FtsZ disassembly by HT-FtsA*. The data for disassembly with increasing HT-FtsA* concentration was similar to the ADP data, although there was no measurable effect on FtsZ assembly at 1 μm HT-FtsA* (Fig. 2A, open squares). This suggests that adenine nucleotide binding is sufficient to allow HT-FtsA* to disassemble FtsZ, but ATP hydrolysis may promote FtsZ disassembly at lower HT-FtsA* concentrations.
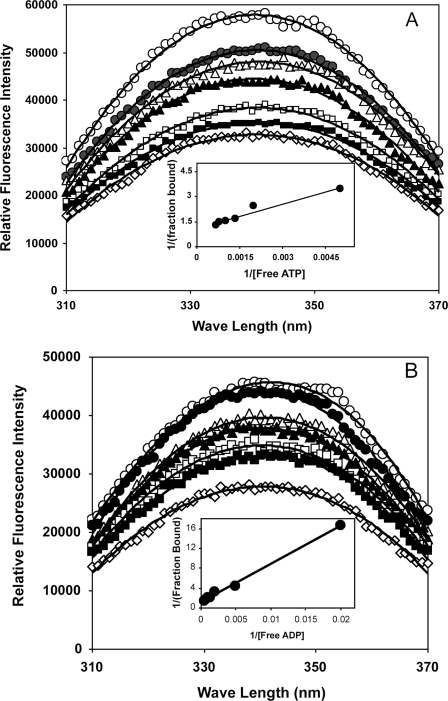
The above results indicate that the effects of HT-FtsA* on FtsZ assembly were attenuated with ADP as compared with ATP. One explanation could be lower binding affinity of ADP to HT-FtsA* as compared with ATP. To address this, we calculated the binding affinities of ATP and ADP to HT-FtsA* by monitoring tryptophan fluorescence of HT-FtsA* with different concentrations of nucleotide. HT-FtsA* contains three tryptophan residues, with two in the membrane-targeting sequence at the extreme C terminus plus the tryptophan at residue 286 that marks the FtsA* mutation. The fluorescence data for ATP (Fig. 6A) or ADP (Fig. 6B) were used to generate the corresponding double reciprocal plots (Fig. 6, A and B, insets). The calculated Kd was 319 ± 67 μm for ATP, similar to the Kd calculated from the mantATP experiments in Fig. 1 (549 ± 80 μm). The Kd for ADP was 928 ± 155 μm, indicating that HT-FtsA* binds ATP with 2–3 times higher affinity than ADP. The difference in binding affinities may explain why HT-FtsA* has less effect on FtsZ assembly in the presence of ADP as compared with ATP.
FIGURE 6.
Binding of ADP and ATP to HT-FtsA*. A, HT-FtsA* (1 μm) was titrated against different concentrations of ATP by measuring changes in tryptophan fluorescence emission between 310 and 370 nm with excitation of 290 nm followed by correction for inner filter effects. Each plot shows HT-FtsA* with 0 (open circles), 50 (filled circles), 100 (open triangles), 500 (filled triangles), 750 (open squares), 1250 (filled squares), or 1500 μm (open diamonds) added ATP or ADP, with a corresponding double reciprocal plot for calculating binding constants in the inset. B, the same, except that ADP was used instead of ATP.
Despite its ability to bind ATP, WT FtsA (HT-FtsA) did not exhibit detectable changes in tryptophan fluorescence upon the addition of ATP or ADP (data not shown). This suggests that the two tryptophans in the FtsA membrane-targeting sequence (23) do not undergo measurable conformational shifts upon nucleotide addition, and it is the Trp-286 residue unique to FtsA* that is most influenced.
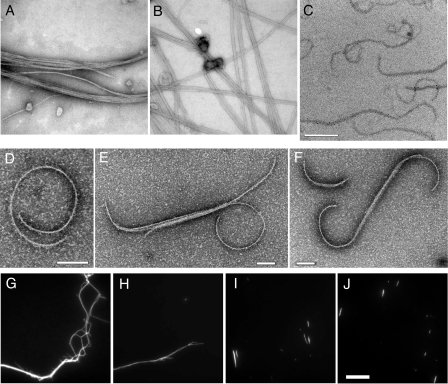
Effects of HT-FtsA* on the Morphology of FtsZ Polymers—We next explored the mechanism of FtsZ assembly inhibition by HT-FtsA* by direct observation of FtsZ polymer morphologies. FtsZ (12 μm) was polymerized with GTP as described above, and the polymers were examined by transmission electron microscopy. In the absence of HT-FtsA*, FtsZ formed long straight polymers, sometimes in bundles, which were well distributed over most of the carbon surface on the copper grid (Fig. 7A). The addition of ATP without HT-FtsA* or HT-FtsA* without ATP had no detectable effect on the morphology of the FtsZ polymers (Fig. 7B and data not shown).
FIGURE 7.
HT-FtsA* and ATP exert significant effects on FtsZ polymer morphology. FtsZ filaments were examined by electron microscopy (A–F) or fluorescence microscopy for fluorescein-labeled FtsZ268C (G–J). Shown are FtsZ with GTP (A, G, and H); FtsZ + GTP + HT-FtsA* without ATP (B); and FtsZ + GTP + HT-FtsA* + ATP (C–F and I–J). All panels represent experiments with FtsZ, HT-FtsA*, and ATP described under “Experimental Procedures.” Scale bar in panel C = 500 nm for panels A–C; scale bars in panels D–F = 100 nm; scale bar in panel J = 10 μm for panels G–J.
When HT-FtsA* was added along with ATP, however, fewer areas in the grid contained visible polymers, and most of the polymers were significantly shorter. The few long polymers were not straight like the control polymers but instead frequently were curved (Fig. 7, C–F). The curves were at a fairly consistent angle, resulting in circles with an average diameter of 311 ± 52 nm. As the measured width of the stained polymers was 6 ± 1 nm, these likely represent single protofilaments. Assuming that each subunit is about 4 nm in size, the average cyclized protofilament would comprise ∼200 subunits, each offset from the next by ∼2 degrees (as compared with essentially 0 degrees for straight polymers).
To confirm the above results and to rule out fixation artifacts, we fluorescently labeled a cysteine-containing FtsZ mutant, F268C (a gift from Dr. Harold Erickson, Duke University) with fluorescein maleimide in the absence and presence of HT-FtsA* and ATP and observed the polymers with a fluorescence microscope. FtsZ formed mostly long fluorescent filaments in the absence of HT-FtsA* (Fig. 7, G–H), whereas in the presence of HT-FtsA* and ATP, the majority of the FtsZ structures formed were short (Fig. 7, I–J), although there was a small percentage of longer polymers (data not shown). We could not detect the curved or circular polymers, probably because they are too small to visualize with light microscopy. Nevertheless, the significantly shorter polymers seen only in the presence of HT-FtsA* and ATP support the electron microscopic data and the idea that HT-FtsA* mediates a major alteration in FtsZ assembly that is inducible by adenine nucleotides.
DISCUSSION
The actin homolog FtsA is essential for cell division in E. coli, and mutants of FtsA can have significant negative or positive effects on the division process. It is likely that the tubulin homolog FtsZ is a target of these effects because FtsA interacts with the C-terminal tail of FtsZ (7). A large portion of the FtsA molecule is probably in intimate contact with FtsZ as mutations that inhibit the ability of FtsA to localize to the Z ring cluster around a putative binding face (41). We therefore surmised that any activities of FtsA on the Z ring should directly regulate the assembly state of FtsZ. In this study, we have shown that a hypermorphic, fully functional derivative of FtsA, HT-FtsA*, induces major changes in FtsZ assembly state in vitro.
The dynamic assembly and disassembly of FtsZ is driven by GTP binding and hydrolysis (42). Such dynamics are reflected in vivo by rapid FtsZ subunit turnover at the Z ring (43, 44) and by direct observation of rapidly moving fluorescent FtsZ in cells between polymers in the ring and monomers confined to helical tracks on the membrane (45–47). In vitro, the dynamics of FtsZ assembly have been well documented, including direct observation of polymer formation in time-lapse studies (48, 49). When GTP hydrolysis is drastically decreased by mutation, FtsZ filaments generally become more stable and less functional, often mislocalizing as stable spirals in the cell instead of dynamic Z rings (50–52).
Our initial studies of the effects of HT-FtsA* on FtsZ assembly showed that HT-FtsA* and ATP reduced FtsZ polymer mass. This effect was directly observed as an increase in the proportion of shorter, curved FtsZ protofilaments as compared with the generally straight, often bundled protofilaments in the untreated samples. Interestingly, the curved FtsZ filaments cyclized to form ∼300 nm diameter circles, which were severalfold larger than FtsZ minirings formed on a lipid monolayer (53) or cyclized FtsZ filaments on mica (49) but smaller than the rings formed by FtsZ expressed in fission yeast (54). These effects could be potentially caused by HT-FtsA*-mediated severing of FtsZ protofilaments or destabilization of protofilament bundles, which are supported by the ability of HT-FtsA* + ATP to reduce the polymer mass of preassembled FtsZ.
The effects of HT-FtsA* on FtsZ assembly were substoichiometric, with effects being observed at greater than a 1:10 FtsA: FtsZ ratio in the presence of ATP. It is important to emphasize that the gain-of-function effect of HT-FtsA* in vivo is also substoichiometric as levels of FtsA (or chromosomally produced FtsA*) should be about 1 μm, about 5–10-fold lower than the concentration of FtsZ. Below this 1:10 ratio, HT-FtsA* had no detectable effect on FtsZ assembly in vitro, suggesting that HT-FtsA* needs to be at a critical concentration to exert its effects. A 1:3 ratio of FtsA:FtsZ had maximal activity, with further increases in HT-FtsA* having little effect, possibly because of protein aggregation.
The other well characterized FtsZ inhibitors of E. coli, SulA and MinC, exhibit key differences and similarities with HT-FtsA*. For example, SulA increases the cc for FtsZ assembly, and a 1:1 stoichiometry is required for maximum inhibition of FtsZ assembly in vitro by a MalE fusion to SulA (39). MinC, a key FtsZ inhibitor in E. coli and other bacteria, also requires a 1:1 stoichiometry for maximum inhibition of FtsZ assembly, but like HT-FtsA*, exhibits a dose-dependent partial inhibition at stoichiometries much lower than 1:1 and does not inhibit GTP hydrolysis by FtsZ (55, 56). For both HT-FtsA* and MinC, FtsZ assembly is not completely abolished under any condition. Intriguingly, a small domain in T. maritima FtsA shares modest structural similarity with the N-terminal domain of MinC (11, 12), underscoring the potential similarity of mechanisms between FtsA and MinC. EzrA and ClpX, two FtsZ assembly inhibitors of B. subtilis, also differ from HT-FtsA* in their requirement for a near 1:1 stoichiometry with FtsZ for maximum inhibition of FtsZ assembly (57–59).
By analogy, substoichiometric levels of the eukaryotic actin-depolymerizing factor/cofilin actin-binding proteins distort and sever actin filaments allosterically by binding to the side of the filaments (60, 61). As a result of severing by actin-depolymerizing factor/cofilin and other factors, overall actin filament length is decreased, resulting in more rapid recycling of actin into new actin cables. As was postulated for MinC (55), we speculate that HT-FtsA* and WT FtsA may have a similar role in FtsZ dynamics and recycling.
The HT-FtsA*-mediated effects on FtsZ assembly require ATP or ADP. Apart from FtsZ itself, this is the first demonstration of a nucleotide-dependent effect of a divisome protein on FtsZ assembly. The modest effectiveness of ADP and the ability of the non-hydrolyzable analog AMP-PNP to mimic the ADP state suggest that nucleotide binding itself is sufficient to induce a conformational change in the protein. The lack of significant ATP hydrolysis by HT-FtsA* is consistent with this idea, as is the complete lack of function in vivo of a mutant FtsA unable to bind ATP (18).3 Because the sensitive fluorescence assay for phosphate release detected weak hydrolysis of either GTP or ATP with increasing HT-FtsA*, it suggests either that minor contaminating proteins in the HT-FtsA* preparation with high nucleotide hydrolysis activities contributed to the effect or that HT-FtsA* itself has weak ATPase and GTPase activities or a combination of both. Because ADP and AMP-PNP do not stimulate FtsZ disassembly activity as efficiently as ATP at lower concentrations of HT-FtsA*, we favor the idea that HT-FtsA* may have very weak ATP hydrolysis activity and that this activity promotes more efficient FtsZ disassembly than nucleotide binding alone. One possibility is that each HT-FtsA* molecule hydrolyzes ATP only once, without exchange.
Purified WT FtsA (HT-FtsA) had no detectable effect on FtsZ assembly despite its ability to bind ATP in vitro and to affect Z ring stability in vivo. This suggests that WT FtsA may depend on other factors, either proteins or lipids, for its activation. Other factors present at the Z ring may also be required to induce FtsA ATP hydrolysis and/or nucleotide exchange, which would potentially regulate FtsA activity during the formation and constriction of the ring. Further studies of cell division proteins in a purified system clearly will be necessary to address these and other possibilities.
Supplementary Material
Acknowledgments
We thank Harold Erickson (Duke University) for providing the FtsZ R268C mutant, William Dubinsky (University of Texas Medical School at Houston) for the mass spectrometry analysis, and members of the Margolin laboratory for useful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-GM61074 (to W. M.) and R37-GM20478 (to W. D.). This work was also supported by a grant from the John S. Dunn research foundation (to W. D.).
The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure.
Footnotes
The abbreviations used are: WT, wild-type; HT, histidine tag; AMP-PNP, 5′-adenylylimido-diphosphate; mantATP, 2′(3′)-O-(N-methylanthraniloyl-ATP; PIPES, piperazine-N,N-bis[2-ethanesulfonicacid]; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; cc, critical concentration.
Y. Wang and W. Margolin, unpublished results.
References
- 1.Weiss, D. S. (2004) Mol. Microbiol. 54 588–597 [DOI] [PubMed] [Google Scholar]
- 2.Bi, E., and Lutkenhaus, J. (1991) Nature 354 161–164 [DOI] [PubMed] [Google Scholar]
- 3.Margolin, W. (2005) Nat. Rev. Mol. Cell Biol. 6 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michie, K. A., and Lowe, J. (2006) Annu. Rev. Biochem. 75 467–492 [DOI] [PubMed] [Google Scholar]
- 5.Li, Z., Trimble, M. J., Brun, Y. V., and Jensen, G. J. (2007) EMBO J. 26 4694–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hale, C. A., and de Boer, P. A. (1997) Cell 88 175–185 [DOI] [PubMed] [Google Scholar]
- 7.Ma, X., and Margolin, W. (1999) J. Bacteriol. 181 7531–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosyak, L., Zhang, Y., Glasfeld, E., Haney, S., Stahl, M., Seehra, J., and Somers, W. S. (2000) EMBO J. 19 3179–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raychaudhuri, D. (1999) EMBO J. 18 2372–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hale, C. A., Rhee, A. C., and de Boer, P. A. (2000) J. Bacteriol. 182 5153–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bork, P., Sander, C., and Valencia, A. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 7290–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Den Ent, F., and Lowe, J. (2000) EMBO J. 19 5300–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, X., Huang, J., Mukherjee, A., Cao, C., and Lutkenhaus, J. (1997) J. Bacteriol. 179 5551–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Din, N., Quardokus, E. M., Sackett, M. J., and Brun, Y. V. (1998) Mol. Microbiol. 27 1051–1063 [DOI] [PubMed] [Google Scholar]
- 15.Yan, K., Pearce, K. H., and Payne, D. J. (2000) Biochem. Biophys. Res. Commun. 270 387–392 [DOI] [PubMed] [Google Scholar]
- 16.Yim, L., Vandenbussche, G., Mingorance, J., Rueda, S., Casanova, M., Ruysschaert, J. M., and Vicente, M. (2000) J. Bacteriol. 182 6366–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiomi, D., and Margolin, W. (2007) Mol. Microbiol. 66 1396–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez, M., Valencia, A., Ferrandiz, M.-J., Sandler, C., and Vicente, M. (1994) EMBO J. 13 4919–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feucht, A., Lucet, I., Yudkin, M. D., and Errington, J. (2001) Mol. Microbiol. 40 115–125 [DOI] [PubMed] [Google Scholar]
- 20.Lara, B., Rico, A. I., Petruzzelli, S., Santona, A., Dumas, J., Biton, J., Vicente, M., Mingorance, J., and Massidda, O. (2005) Mol. Microbiol. 55 699–711 [DOI] [PubMed] [Google Scholar]
- 21.Corbin, B. D., Geissler, B., Sadasivam, M., and Margolin, W. (2004) J. Bacteriol. 186 7736–7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rico, A. I., Garcia-Ovalle, M., Mingorance, J., and Vicente, M. (2004) Mol. Microbiol. 53 1359–1371 [DOI] [PubMed] [Google Scholar]
- 23.Pichoff, S., and Lutkenhaus, J. (2005) Mol. Microbiol. 55 1722–1734 [DOI] [PubMed] [Google Scholar]
- 24.Addinall, S. G., Bi, E., and Lutkenhaus, J. (1996) J. Bacteriol. 178 3877–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen, S. O., Thompson, L. S., and Harry, E. J. (2005) J. Bacteriol. 187 6536–6544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dewar, S. J., Begg, K. J., and Donachie, W. D. (1992) J. Bacteriol. 174 6314–6316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai, K., and Lutkenhaus, J. (1992) J. Bacteriol. 174 6145–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rueda, S., Vicente, M., and Mingorance, J. (2003) J. Bacteriol. 185 3344–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geissler, B., Shiomi, D., and Margolin, W. (2007) Microbiology (Read.) 153 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geissler, B., Elraheb, D., and Margolin, W. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4197–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goehring, N. W., Petrovska, I., Boyd, D., and Beckwith, J. (2007) J. Bacteriol. 189 633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geissler, B., and Margolin, W. (2005) Mol. Microbiol. 58 596–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernard, C. S., Sadasivam, M., Shiomi, D., and Margolin, W. (2007) Mol. Microbiol 64 1289–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiomi, D., and Margolin, W. (2008) Mol. Microbiol. 67 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beuria, T. K., Santra, M. K., and Panda, D. (2005) Biochemistry 44 16584–16593 [DOI] [PubMed] [Google Scholar]
- 36.Miroux, B., and Walker, J. E. (1996) J. Mol. Biol. 260 289–298 [DOI] [PubMed] [Google Scholar]
- 37.Mukherjee, A., and Lutkenhaus, J. (1999) J. Bacteriol. 181 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheffers, D. J. (2008) FEBS Lett. 582 2601–2608 [DOI] [PubMed] [Google Scholar]
- 39.Dajkovic, A., Mukherjee, A., and Lutkenhaus, J. (2008) J. Bacteriol. 190 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen, Y., and Erickson, H. P. (2005) J. Biol. Chem. 280 22549–22554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pichoff, S., and Lutkenhaus, J. (2007) Mol. Microbiol. 64 1129–1138 [DOI] [PubMed] [Google Scholar]
- 42.Mukherjee, A., and Lutkenhaus, J. (1998) EMBO J. 17 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stricker, J., Maddox, P., Salmon, E. D., and Erickson, H. P. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson, D. E., Gueiros-Filho, F. J., and Erickson, H. P. (2004) J. Bacteriol. 186 5775–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun, Q., and Margolin, W. (1998) J. Bacteriol. 180 2050–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thanedar, S., and Margolin, W. (2004) Curr. Biol. 14 1167–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu, L., and Yu, J. (2008) Biophys. J. 95 2009–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, X.-C., and Margolin, W. (1997) EMBO J. 16 5455–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mingorance, J., Tadros, M., Vicente, M., Gonzalez, J. M., Rivas, G., and Velez, M. (2005) J. Biol. Chem. 280 20909–20914 [DOI] [PubMed] [Google Scholar]
- 50.Addinall, S. G., and Lutkenhaus, J. (1996) Mol. Microbiol. 22 231–237 [DOI] [PubMed] [Google Scholar]
- 51.Ma, X., Ehrhardt, D. W., and Margolin, W. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 12998–13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Redick, S. D., Stricker, J., Briscoe, G., and Erickson, H. P. (2005) J. Bacteriol. 187 2727–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Erickson, H. P., Taylor, D. W., Taylor, K. A., and Bramhill, D. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivasan, R., Mishra, M., Wu, L., Yin, Z., and Balasubramanian, M. K. (2008) Genes Dev. 22 1741–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu, Z., Mukherjee, A., Pichoff, S., and Lutkenhaus, J. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dajkovic, A., Lan, G., Sun, S. X., Wirtz, D., and Lutkenhaus, J. (2008) Curr. Biol. 18 235–244 [DOI] [PubMed] [Google Scholar]
- 57.Haeusser, D. P., Schwartz, R. L., Smith, A. M., Oates, M. E., and Levin, P. A. (2004) Mol. Microbiol. 52 801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weart, R. B., Nakano, S., Lane, B. E., Zuber, P., and Levin, P. A. (2005) Mol. Microbiol. 57 238–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chung, K. M., Hsu, H. H., Yeh, H. Y., and Chang, B. Y. (2007) J. Biol. Chem. 282 14891–14897 [DOI] [PubMed] [Google Scholar]
- 60.Andrianantoandro, E., and Pollard, T. D. (2006) Mol. Cell 24 13–23 [DOI] [PubMed] [Google Scholar]
- 61.Carlier, M. F., Laurent, V., Santolini, J., Melki, R., Didry, D., Xia, G. X., Hong, Y., Chua, N. H., and Pantaloni, D. (1997) J. Cell Biol. 136 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.