Abstract
Wnt11 signals through both canonical (β-catenin) and non-canonical pathways and is up-regulated during osteoblast differentiation and fracture healing. In these studies, we evaluated the role of Wnt11 during osteoblastogenesis. Wnt11 overexpression in MC3T3E1 pre-osteoblasts increases β-catenin accumulation and promotes bone morphogenetic protein (BMP)-induced expression of alkaline phosphatase and mineralization. Wnt11 dramatically increases expression of the osteoblast-associated genes Dmp1 (dentin matrix protein 1), Phex (phosphate-regulating endopeptidase homolog), and Bsp (bone sialoprotein). Wnt11 also increases expression of Rspo2 (R-spondin 2), a secreted factor known to enhance Wnt signaling. Overexpression of Rspo2 is sufficient for increasing Dmp1, Phex, and Bsp expression and promotes bone morphogenetic protein-induced mineralization. Knockdown of Rspo2 abrogates Wnt11-mediated osteoblast maturation. Antagonism of T-cell factor (Tcf)/β-catenin signaling with dominant negative Tcf blocks Wnt11-mediated expression of Dmp1, Phex, and Rspo2 and decreases mineralization. However, dominant negative Tcf fails to block the osteogenic effects of Rspo2 overexpression. These studies show that Wnt11 signals through β-catenin, activating Rspo2 expression, which is then required for Wnt11-mediated osteoblast maturation.
Wnt signaling is a key regulator of osteoblast differentiation and maturation. In mesenchymal stem cell lines, canonical Wnt signaling by Wnt10b enhances osteoblast differentiation (1). Canonical Wnt signaling through β-catenin has also been shown to enhance the chondroinductive and osteoinductive properties of BMP22 (2, 3). During BMP2-induced osteoblast differentiation of mesenchymal stem cell lines, cross-talk between BMP and Wnt pathways converges through the interaction of Smad4 with β-catenin (2).
Canonical Wnt signaling is also critical for skeletal development and homeostasis. During limb development, expression of Wnt3a in the apical ectodermal ridge of limb buds maintains cells in a highly proliferative and undifferentiated state (4, 5). Disruption of canonical Wnt signaling in Lrp5/Lrp6 compound knock-out mice results in limb- and digit-patterning defects (6). Wnt signaling is also involved in the maintenance of post-natal bone mass. Gain of function in the Wnt co-receptor Lrp5 leads to increased bone mass, whereas loss of Lrp5 function is associated with decreased bone mass and osteoporosis pseudoglioma syndrome (7, 8). Mice with increased Wnt10b expression have increased trabecular bone, whereas Wnt10b-deficient mice have reduced trabecular bone (9). Similarly, mice nullizygous for the Wnt antagonist sFrp1 have increased trabecular bone accrual throughout adulthood (10).
Although canonical Wnt signaling regulates osteoblastogenesis and bone formation, the profile of endogenous Wnts that play a role in osteoblast differentiation and maturation is not well described. During development, Wnt11 is expressed in the perichondrium and in the axial skeleton and sternum (11). Wnt11 expression is increased during glucocorticoid-induced osteogenesis (12), indicating a potential role for Wnt11 in osteoblast differentiation. Interestingly, Wnt11 activates both β-catenin-dependent as well as β-catenin-independent signaling pathways (13). Targeted disruption of Wnt11 results in late embryonic/early post-natal death because of cardiac dysfunction (14). Although these mice have no reported skeletal developmental abnormalities, early lethality obfuscates a detailed examination of post-natal skeletal modeling and remodeling.
In murine development, Wnt11 expression overlaps with the expression of R-spondin 2 (Rspo2) in the apical ectodermal ridge (11, 15). R-spondins are a novel family of proteins that share structural features, including two conserved cysteinerich furin-like domains and a thrombospondin type I repeat (16). The four R-spondin family members can activate canonical Wnt signaling (15, 17–19). Rspo3 interacts with Frizzled 8 and Lrp6 and enhances Wnt ligand signaling. Rspo1 enhances Wnt signaling by interacting with Lrp6 and inhibiting Dkk-mediated receptor internalization (20). Rspo1 was also shown to potentiate Wnt3a-mediated osteoblast differentiation (21). Rspo2 knock-out mice, which die at birth, have limb patterning defects associated with altered β-catenin signaling (22–24). However, the role of Rspo2 in osteoblast differentiation and maturation remains unclear.
Herein we report that Wnt11 overexpression in MC3T3E1 pre-osteoblasts activates β-catenin and augments BMP-induced osteoblast maturation and mineralization. Wnt11 increases the expression of Rspo2. Overexpression of Rspo2 in MC3T3E1 is sufficient for augmenting BMP-induced osteoblast maturation and mineralization. Although antagonism of Tcf/β-catenin signaling blocks the osteogenic effects of Wnt11, Rspo2 rescues this block, and knockdown of Rspo2 shows that it is required for Wnt11-mediated osteoblast maturation and mineralization. These studies identify both Wnt11 and Rspo2 as novel mediators of osteoblast maturation and mineralization.
EXPERIMENTAL PROCEDURES
Induction of Osteoblast Maturation—Murine MC3T3 E1 (clone 14) cells were plated at 2 × 104 cells/well of a 24-well plate (or scaled up to larger wells based on this surface area ratio) and cultured for 2 days in α-minimum Eagle's medium supplemented with 10% fetal bovine serum (Hyclone, characterized), penicillin/streptomycin, and l-glutamine. The media were subsequently replaced with fresh media supplemented with β-glycerol phosphate (4 mm) and ascorbic acid 2-phosphate (25 μg/ml). BMP2 (dimer concentration) was added where indicated for only the first 3 days of induction. Fresh tissue culture medium, supplemented with β-glycerol phosphate and ascorbic acid 2-phosphate, was added every 2–3 days after the initial period of BMP induction. The mineralization experiments were harvested on days 12–16. For the microarray and Q-RT-PCR-based gene expression analysis, cells were harvested at days 3, 6, and 9.
Retroviral Vectors—RNA was harvested from MC3T3E1 (for Wnt11 cDNA cloning) or Wnt11 overexpressing MC3T3 E1 pre-osteoblasts (for Rspo2 cDNA cloning) and reverse-transcribed to cDNA using Superscript III reverse transcriptase (Invitrogen). The cDNAs encoding Wnt11 and mRspo2 were amplified, sequenced for mutations, and subcloned into a murine myeloproliferative sarcoma virus-based retroviral vector (pRet) (25) containing an IRES-EGFP1 cassette. The cDNA encoding dominant negative Tcf4 was kindly provided by the laboratory of Ormond Macdougald (26). The microRNAi-based knockdown cassettes were designed using the Invitrogen miRNAi designer tool. Two separate miRNAi vectors against Rspo2 (Rspo2 959 and Rspo2 1390, corresponding to the location in the coding sequence) were designed and cloned into the pCDNA6.2 GW/miR vector. After sequencing, the knockdown cassette was subcloned into the pRet retroviral vector. Retroviral supernatant was produced as described below.
Generation of Stable Retroviral Producer Lines—Stable retroviral producer lines were established using the trans-infection method as follows: phoenix A (amphotropic) cells were plated at a density of 4 × 105 cells/well of a 6-well plate and cultured overnight. On the following day, the culture media were aspirated and exchanged with fresh media (Dulbecco's modified Eagle's medium, high glucose, 10% fetal bovine serum, penicillin/streptomycin, l-glutamine). Chloroquine dihydrochloride (Sigma, S764663) was added to phoenix cells 5 min prior to transfection. Plasmid DNA (1–2 μg/ml) was transfected into the phoenix cells using the Promega Profection kit (E1200). The media were changed 8–10 h later and replaced with fresh medium. On the following day (day 1), the media were changed, and the cells were placed at 32 °C, 5% CO2 overnight to harvest viral supernatant on day 2 post-transfection. Fresh media were added every 24 h until 4 days post-transfection.
On day 1 of transfection, the ecotropic retroviral packaging line, GP+E86, was placed in a 6-well dish (4 × 104 cells/well) and cultured overnight. On day 2, retroviral supernatant was harvested from phoenix cells, filtered (0.45 μm), and supplemented with the retroviral attachment promoter protamine sulfate (5 μg/ml). Approximately 2 ml of retroviral supernatant was added to the GP+E86 cells on day 2 and again on days 3 and 4. The entire transfection protocol was performed twice, for a total of six harvests of retroviral supernatant. Stable producer lines were selected in puromycin when applicable (1 μg/ml). GP+E86 derived from this protocol were considered stable retroviral producer cell lines with an approximate viral titer of 1 × 107 colony-forming units/ml.
Retroviral Transduction of MC3T3 E1 Cells—Cells were plated in a 6-well plate (4–5 × 104 cells/well) and cultured overnight at 37 °C, 5% CO2. On the following day, retroviral supernatant was harvested from GP+E 86 ecotropic retroviral producers, filtered (0.45 μm), and supplemented with protamine sulfate (5 μg/ml). Alternatively, filtered retroviral supernatant was frozen, thawed, and supplemented with protamine sulfate. Tissue culture medium was subsequently removed from each well and replaced with 2 ml of retroviral supernatant. Cells were cultured with retroviral supernatant overnight (37 °C, 5% CO2). This procedure was repeated the following day. Retrovirally transduced cells were selected for 5–7 days in puromycin (1 μg/ml) when applicable. Using this protocol, ∼100% of MC3T3 cells were transduced as determined by enhanced GFP fluorescence (data not shown). EGFP1 transduced MC3T3 cells were used as control cells for all overexpression studies, whereas an miRNAi specific for LacZ was used as a control for all miRNAi-based knockdown studies.
Alkaline Phosphatase Colorimetric Assay—MC3T3 cells (5 × 103) were plated in flat-bottom 96-well plates in 200 μl of α-minimum Eagle's medium (Invitrogen). After culturing for 12 days (37 °C, 5% CO2), the media were aspirated, and alkaline phosphatase activity was measured using a p-nitrophenyl phosphate-based assay kit (Sigma). Briefly, 50 μl of alkaline reaction buffer was added to each well followed by 50 μl of 2× p-nitrophenyl phosphate substrate solution. The plates were subsequently incubated for 15–20 min at 37 °C, 5% CO2. p-Nitrophenyl phosphate production was measured using a Molecular Devices Spectramax microplate reader (405 nm) and compared with a standard curve. Alkaline phosphatase activity was subsequently normalized to cell number using the rapid cell proliferation kit (EMD Biosciences). At least three independent experiments were performed with multiple biological replicates. Statistical significance was determined using analysis of variance follow by the Tukey-Kramer multiple comparisons test.
Calcium Phosphate Cytochemistry—For Alizarin Red S staining, cells were harvested between days 12 and 16 (unless otherwise indicated). The cells were washed in PBS and fixed in ethanol (50%) for 5 min. The ethanol solution was removed and an Alizarin Red S solution (1%) was added for 5 min. Each well was rinsed three times with PBS, once (rapidly) with deionized water to reduce background staining, and once again with 1× PBS.
Quantitative RT-PCR—MC3T3 cells were cultured for 2 days in 6-well plates (8 × 104 cells/well). The cells were subsequently induced with 160 ng/ml (4 nm dimer concentration) recombinant human BMP2, β-glycerol phosphate (4 mm), and ascorbic acid 2 phosphate (25 μg/ml). Total RNA was isolated at various time points using the Qiagen RNeasy kit with on-column DNase I digestion. All primers were designed using the Primer3 program (Whitehead Institute, Cambridge, MA). The sequences are available upon request from the corresponding author. cDNAs were synthesized using Superscript III reverse transcriptase (Invitrogen). The cDNAs were subsequently diluted (∼1:2), and 0.5 μl was added to each PCR using the ABI Power SYBR 2× SYBR green master mix. All PCR samples were prepared in duplicate wells of a 96-well plate. Quantitative PCR was performed on an ABI 7500 fast thermal cycler. To ensure primer specificity, melt curves were performed after 40 cycles of PCR. Agarose gel electrophoresis was performed after melt curve analysis to further validate each primer set. Fold- differences in gene expression were calculated using the ΔΔCt method (27, 28), comparing untreated controls to BMP-treated samples and normalizing to β-actin. Statistical tests were performed as mentioned previously.
Microarray Analysis—RNA was isolated from cells as described previously. Samples were digested with DNase I to remove any residual DNA contamination. After UV spectrophotometric quantification, a minimum of 12 μg of total RNA was submitted to the University of Michigan Cancer Center Microarray Core Facility at a concentration of no less than 500 ng/μl. All subsequent sample processing was performed by the Microarray Core Facility. Quality control on samples was performed by the Microarray Core Facility using an Agilent Bioanalyzer. 5 μg of RNA was used for each cDNA synthesis reaction. Expression values and fold-change of treatment relative to controls were provided by the Microarray core Facility. Clustering analysis was performed after processing the raw data (.cel files). Genes with an expression level ≥400 and an interquartile range ≥0.4 were selected. The list was limited to the top 300 differentially expressed genes.
Western Blotting—Tissue culture medium was aspirated, and cells were rinsed once in PBS. Subcellular protein fractionation was performed in three steps. First, cells were lysed in hypotonic buffer (25 mm HEPES, pH 7.4, 5 mm MgCl2, 5 mm EDTA, 5 mm dithiothreitol) for 10 min, scraped, and Dounce-homogenized (20 strokes, B pestle). Following 5 min of centrifugation at 4000 × g, the supernatant was removed and collected as the cytosolic fraction. The supernatant was then centrifuged for 3 min at 15,000 × g to remove any carryover from the pellet. The pellet was subsequently rinsed in PBS with protease and phosphatase inhibitors (as above) and incubated for 10 min (room temperature) in a second buffer containing 10 mm HEPES, pH 7.4, 10 mm KCl, 0.1 mm EDTA, 1 mm dithiothreitol, 0.2% IGE-PAL, protease and phosphatase inhibitors, sodium fluoride, and sodium orthovanadate, as above. After incubation, this fraction was pelleted at 15,000 × g for 3 min. The cytosol was removed and collected as the membrane fraction. Finally, the pellet was rinsed once in PBS with protease and phosphatase inhibitors and resuspended in 20 mm HEPES, pH 7.4, 0.4 m sodium chloride, 1 mm EDTA, 10% glycerol, 1 mm dithiothreitol, and protease and phosphatase inhibitors. The pellet was subsequently shaken for 4 h (4 °C, 1000 rpm). The extract was centrifuged (15,000 × g, 5 min) to generate the nuclear fraction. Protein was quantified using the BCA assay (Pierce). Equal amounts of total protein were separated using SDS-PAGE and electrotransferred to a nitrocellulose membrane. The blots were probed using anti-β-catenin (Upstate, 06734), anti-NFATc4 (Santa Cruz Biotechnology, sc-13036), anti-NFATc1 (Santa Cruz Biotechnology, sc-7294), anti-phospho-protein kinase Cα (Upstate, 06822), anti-phospho-Smad1, -5, and -8 (Chemicon, AB3848), anti-β-tubulin (Sigma, T7816), or C-terminal anti-lamin A (Sigma, L1293). The blots were subsequently stripped with restore stripping buffer (Pierce 21059) and re-probed 1–2 times. Autoradiography was performed, and films were scanned. Photoshop was used to generate composite images for treatment groups in the same experiment but run in nonadjacent lanes (i.e. control versus Wnt11 versus Rspo2). Brightness and contrast levels were uniformly adjusted across all groups. All Western blots were performed on lysates from at least three independent experiments.
RESULTS
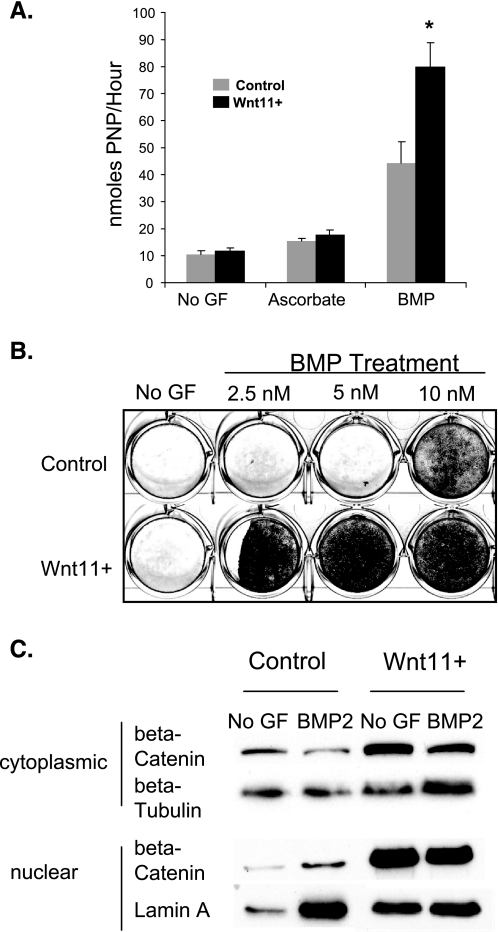
Wnt11 Enhances BMP2-induced Osteoblast Maturation and Mineralization—Wnt11 was expressed by MC3T3 E1 pre-osteoblasts under basal growth conditions and was increased with BMP treatment (supplemental Fig. 1A). Overexpression of Wnt11 (Wnt11+) increased levels 1000-fold. We examined alkaline phosphatase expression in MC3T3 E1 pre-osteoblasts overexpressing Wnt11 (Wnt11+) with and without BMP2 treatment. BMP-treated control cells cultured in ascorbate/BGP showed significantly increased alkaline phosphatase activity relative to ascorbate/BGP-treated cells and untreated controls (Fig. 1A). BMP-treated Wnt11+ cells showed the greatest increase in alkaline phosphatase activity, ∼2-fold higher than the BMP-treated control cells (Fig. 1A). There was no significant difference in Wnt11+ cell number relative to controls (data not shown).
FIGURE 1.
Wnt11 enhances BMP-induced osteoblast differentiation. A, MC3T3 E1 cells were cultured in osteogenic conditions with or without BMP. Alkaline phosphatase activity was evaluated 6 days after the start of BMP induction. Alkaline phosphatase activity was normalized to cell number using the rapid cell proliferation kit. Analysis of variance followed by the Tukey-Kramer multiple comparisons test were used to determine statistical significance between groups. n >3, * = p < 0.01, BMP-treated versus all other groups. B, MC3T3 cells were cultured with ascorbate and BGP with the indicated concentrations of recombinant human BMP2. The cells were harvested at day 12–16 for Alizarin Red S staining. Greater than three independent experiments were conducted. C, cells were harvested after 3 days of osteoinduction, with and without BMP, and lysed by Dounce homogenization. Subcellular fractions were collected, and Western blots were performed. Equivalent amounts of protein were loaded per lane, depending on the experiment (5–10 μg). The blots for the cytosolic and nuclear fractions were probed with anti-β-catenin, stripped, and re-probed with β-tubulin and lamin A, respectively. Western blots were performed on three independent experiments with duplicate wells in each experiment. The figure shows protein lysates harvested from representative wells. No GF, no growth factor.
To examine the effect of Wnt11 expression on osteoblast maturation, we performed a mineralization assay. Similar to the alkaline phosphatase results, Wnt11 overexpression enhanced the effects of BMP2 (Fig. 1B). At lower concentrations, BMP2 treatment of control cells failed to induce mineralization, whereas Wnt11 cells showed robust mineralization in the presence of BMP2.
Wnt11 Increases β-Catenin Levels in MC3T3 E1.14 Cells—Given that canonical Wnt signaling plays a prominent role in osteoblast differentiation, and that Wnt11 has been shown to signal in a canonical manner, we evaluated the levels of cytosolic and nuclear β-catenin. Levels of nuclear and cytosolic β-catenin were elevated in Wnt11+ cells relative to controls (Fig. 1C), indicative of increased canonical Wnt signaling. BMP treatment had no effect on β-catenin levels.
Wnt11 Regulates Osteoblast Gene Expression—To evaluate changes in MC3T3E1 gene expression with Wnt11, we performed a microarray analysis of gene expression using the mouse 430A 2.0 chip (Affymetrix). Gene expression in control cells treated with ascorbate and BGP, with or without 4 nm BMP2, was compared with Wnt11+ cells under identical conditions, 3, 6, and 9 days after the initiation of osteoblast induction. A set of genes shows changes in expression in BMP2-treated control cells consistent with previously published studies (supplemental Table S1). Nine days post-treatment, there are increases in Id1, Sp7 (osterix), Bsp (Ibsp), osteocalcin (Bglap1), and Phex, indicative of osteoblast maturation. Interestingly, expression of Tcf7, a Wnt-associated transcription factor, is also increased in BMP-treated cells.
A relatively discrete number of genes show changes with Wnt11 overexpression (without BMP treatment). Dmp1 (dentin matrix acidic phosphoprotein 1) is among the extracellular matrix genes showing increased expression in Wnt11 cells relative to control cells (supplemental Table S2). Dmp1 regulates extracellular matrix mineralization, calcium-phosphate homeostasis, and also serves as transcriptional regulator of dentin sialo-phosphoprotein (29, 30). Similarly, otoraplin/cartilage-derived retinoic acid-sensitive protein (Otor/CD-RAP), a proteoglycan involved in chondrogenesis (31), is increased by Wnt11 cells at all time points examined. Interestingly, in control cells, BMP treatment results in repression of Otor expression (supplemental Table S1). Expression of Phex, a gene mutated in hypophosphatemic rickets (32), is also increased in Wnt11+ cells relative to controls, independent of BMP treatment (supplemental Table S2). Rspo2 expression is increased in Wnt11 cells relative to controls (supplemental Table S2). Genes that show further enhancement with Wnt11 overexpression in the presence of BMP treatment are shown in supplemental Table S3. Dmp1 and Phex are both increased, suggesting either an additive or synergistic effect of BMP and Wnt11.
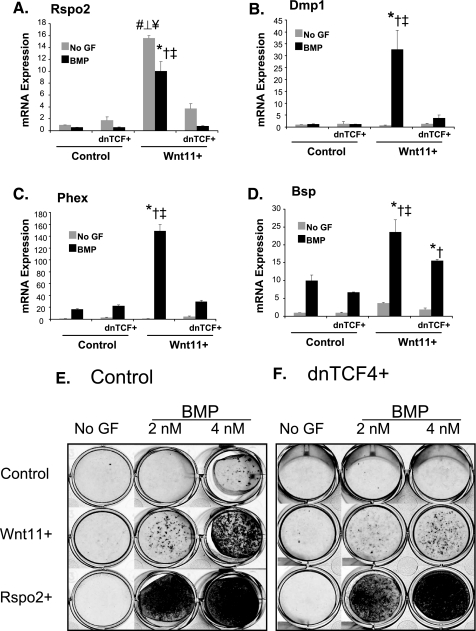
Increased expression of some of these genes was validated using qPCR (Fig. 2 for Rspo2, Phex, Dmp, and Bsp; others not shown). BMP and Wnt11 synergistically increase expression of Phex, Bsp, and Dmp1, a result that is consistent with the microarray data. By contrast, Rspo2 expression is greatest in Wnt11-overexpressing cells that are not treated with BMP, whereas BMP appears to decrease Rspo2 expression (Fig. 2 and supplemental Fig. 1B).
FIGURE 2.
Dominant negative Tcf inhibits Wnt11-induced gene expression and osteoblast maturation. MC3T3 control (GFP) and MC3T3 Wnt11+ cells were transduced with a retrovirus encoding dnTcf. The cells were cultured for 2 days and placed in osteo-inductive conditions with and without BMP2. The cells were harvested for RNA at day 9 of induction. cDNA was synthesized for each sample, and gene expression was analyzed by Q-RT-PCR. A, Rspo2; B, Dmp1; C, Phex; D, Bsp. Values represent fold-change in gene expression relative to non-BMP-treated GFP controls, as described previously. Statistical analysis was performed using analysis of variance with subsequent Tukey-Kramer multiple comparison testing. n >3, p < 0.01. # ⊥ ¥ indicates statistical significantly increased expression in non-BMP (no GF)-treated Wnt11 cells relative to the other non-BMP treated groups (#, control; ⊥, control dnTcf; ¥, Wnt11 dnTcf). *†‡ indicates statistically significant differences between BMP-treated Wnt11+ cells and all other BMP-treated groups (*, control; †, control dnTcf; ‡, Wnt11+ dnTcf). E, MC3T3 control (GFP), Wnt11+, and Rspo2+ cells were placed in osteogenic media with or without BMP (at the indicated concentration). The cells were harvested after the onset of mineralization (days 12–16) and stained with Alizarin Red S. F, control/dnTcf, Wnt11+/dnTcf, and Rspo2+/dnTcf were cultured in parallel with the MC3T3 cells above. Results repeated three independent times. No GF, no growth factor.
β-Catenin Signaling Is Required for Wnt11-mediated Osteoblast Maturation and Mineralization—To determine whether β-catenin signaling was required for the pro-osteoblastic effects of Wnt11, we introduced a dominant negative form of Tcf (dnTcf) and examined osteoblast differentiation and gene expression. Overexpression of dnTcf (dnTcf+ cells) has no statistically significant effect on gene expression in either non-BMP or BMP-treated control cells (Fig. 2). However, BMP treated Wnt11+/dnTcf+ cells show significantly lower levels of Rspo2, Bsp, Dmp1, and Phex (Fig. 2) relative to BMP-treated Wnt11+ cells. Furthermore, dnTcf+/Wnt11+ cells showed reduced mineralization (Fig. 2, E and F). Thus, β-catenin/Tcf signaling mediates at least some of the pro-osteoblast effects of Wnt11.
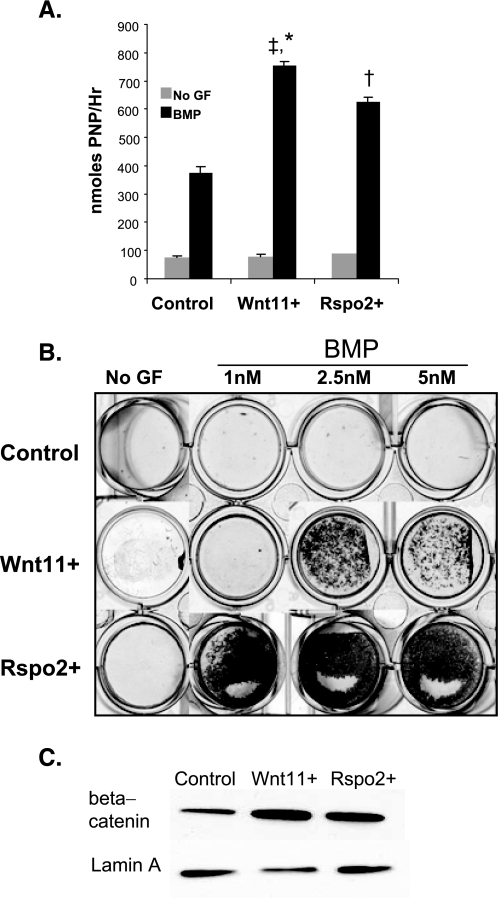
Rspo2 Enhances Osteoblast Maturation and Mineralization and Stabilizes β-Catenin—Because R-spondin expression is both induced by Wnt signaling (33) and an activator of β-catenin (17, 33), we examined if some of the osteogenic effects of Wnt11 were mediated by Rspo2. We cloned the cDNA encoding murine Rspo2 and retrovirally overexpressed it in MC3T3 E1.14 cells (Rspo2+ cells). Overexpression increased Rspo2 levels 50-fold over control conditions, which was increased ∼5-fold over Rspo2 expression in Wnt11+ cells (supplemental Fig. 1B). Osteoblast maturation and mineralization were subsequently evaluated in Wnt11+, Rspo2+, and control cells. BMP-treated Rspo2- and Wnt11-overexpressing cells show statistically significant increased levels of alkaline phosphatase activity relative to control cells. However, neither Wnt11 nor Rspo2 overexpression is sufficient for induction of alkaline phosphatase (Fig. 3A). Rspo2 overexpression potently enhances mineralization at all concentrations of BMP (Fig. 3B). Furthermore, Rspo2 is sufficient for β-catenin activation with consequent nuclear accumulation (Fig. 3C).
FIGURE 3.
Rspo2 increases BMP-induced osteoblast differentiation and increases β-catenin stabilization. A, MC3T3 control (GFP), Wnt11+, and Rspo2+ pre-osteoblasts were cultured in osteogenic media with or without BMP. The plates were harvested after the onset of mineralization at day 6 of induction and evaluated for alkaline phosphatase induction. Alkaline phosphatase activity was normalized to total cell number. *‡ indicates statistical significance relative to BMP-treated controls (*) and Rspo2+ (‡) cells. † indicates statistical significance relative to BMP-treated control cells n >3. B, MC3T3 control (GFP), Wnt11+, and Rspo2+ cells were placed in osteogenic conditions with or without varying doses of BMP. The plates were harvested after the onset of mineralization (days 12–16) and stained with Alizarin Red S. Representative wells shown; repeated more than three times. C, MC3T3 control (GFP), Wnt11+, and Rspo2+ were cultured in osteogenic media for 3 days. The cells were harvested for protein and membrane; cytosolic and nuclear fractions were collected. Western blots were performed on the nuclear fraction, and the membranes were probed with anti-β-catenin and anti-lamin A as a loading control. Western blots were performed on three independent experiments with duplicate wells in each experiment. The figure shows protein lysates harvested from representative wells.
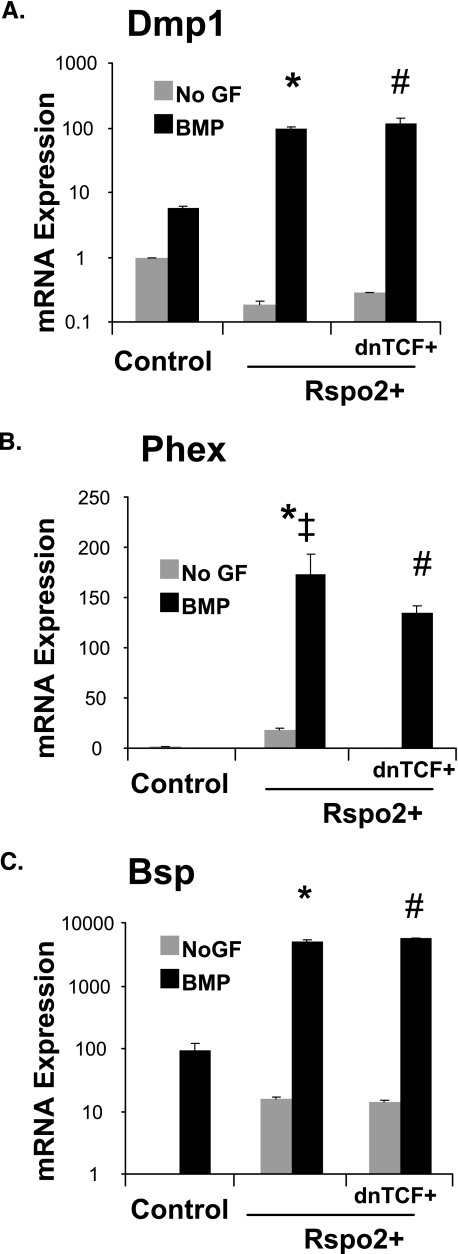
Rspo2 Enhances Osteoblast Gene Expression in a Partially Tcf-independent Manner—To determine how Rspo2 enhanced osteoblast maturation and mineralization, we examined osteoblast-associated gene expression in Rspo2+ cells (Fig. 4). BMP-treated Rspo2+ cells show significantly higher levels of osteoblast-associated genes Bsp, Dmp1, and Phex expression relative to BMP2-treated control cells. Phex expression is partially decreased in Rspo2+/dnTcf4+ cells relative to Rspo2+ cells but remains much higher than BMP-treated control cells. Interestingly, expression levels of Dmp1 and Bsp are unchanged in BMP treated Rspo2+/dnTcf4+ cells relative to Rspo2+ cells. Importantly, dnTcf4 does not block Rspo2-mediated mineralization (Fig. 2F).
FIGURE 4.
Rspo2 enhances osteoblast-associated gene expression independent of β-catenin. MC3T3 control (GFP), control/dnTcf+, Rspo2+, and Rspo2+/dnTcf+ were cultured for 9 days in osteogenic media, with or without BMP. The cells were harvested, and Q-RT-PCR was performed as described previously. Statistical analysis was performed as mentioned previously. A, Dmp1; B, Phex; C, Bsp. Values represent fold-change in gene expression relative to non-BMP (No GF)-treated controls, as described previously. *‡ indicates statistically significant increased gene expression in BMP-treated Rspo2+ cells relative to BMP-treated control (*) and Rspo2+/dnTcf+ (‡) groups. # indicates statistically significantly different levels of gene expression in Rspo2+/dnTcf+ relative to BMP-treated control. n >3, p < 0.01. No GF, no growth factor.
Rspo2 Expression Is Required for Osteoblast Maturation and Mineralization—To determine whether Rspo2 expression was required for Wnt11-enhanced osteoblast maturation and mineralization, we used retrovirus-based microRNAi to knock down expression of Rspo2. Two retrovirus-based miRNAi vectors directed against mRspo2 were designed and tested (Rspo2 959 and Rspo2 1390). Both viruses specifically knocked down expression of Rspo2 and not that of other Rspo family members (data not shown). Our initial experiments indicated that Rspo2 959 was more effective (data not shown) at knocking down expression of Rspo2. All subsequent experiments were therefore performed with Rspo2 959 (Rspo2 miR+).
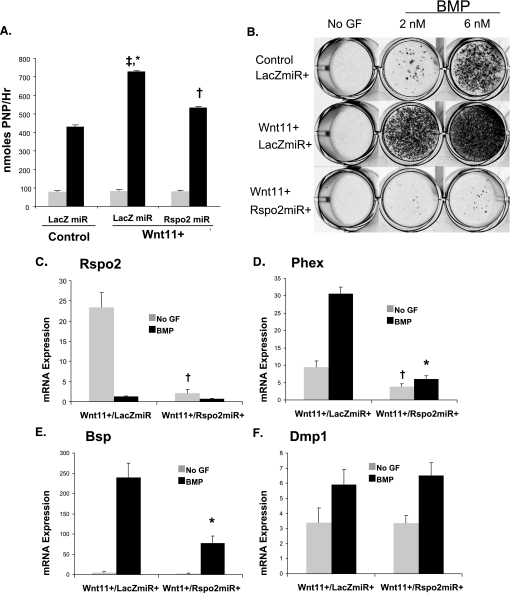
Control cells were infected with a LacZ miRNAi retrovirus (miRNAi control, LacZmiR+), whereas Wnt11 cells were infected with either a LacZ control miRNAi retrovirus or the Rspo2 miRNAi retrovirus. Alkaline phosphatase induction and mineralization were evaluated as markers of osteoblast maturation and function. Knockdown of Rspo2 expression in Wnt11 cells decreases Wnt11-enhanced alkaline phosphatase induction to near control levels (Fig. 5A). Importantly, knockdown of Rspo2 expression resulted in a nearly complete blockade of mineralization; levels of mineralization in Wnt11+/Rspo2miR+ cells were lower than both control/LacZmiR+ and Wnt11+/LacZmiR+ cells (Fig. 5B).
FIGURE 5.
Rspo2 is required for Wnt11-regulated osteoblast maturation and associated gene expression MC3T3 cells. Control and Wnt11+ cells were infected with an miRNAi LacZ control retrovirus. Wnt11+ cells were infected in parallel with an Rspo2-specific miRNAi retrovirus (control/LacZmiR+, Wnt11+/LacZmiR+, and Wnt11+/Rspo2miR+). A, cells were placed in osteogenic media, with or without BMP, for 6 days and harvested for an alkaline phosphatase assay as described previously. *‡ indicates statistical significance relative to BMP-treated control cells (*) and Rspo2 (‡) cells. † indicates statistical significance relative to BMP-treated control cells. B, control/LacZmiR+, Wnt11+/LacZmiR+, and Wnt11+/Rspo2miR+ cells were placed in osteogenic conditions with the indicated concentration of BMP. The cells were harvested after the onset of mineralization and stained with Alizarin Red S. Representative wells are shown and repeated more than three times. C–F, cells were harvested for RNA after 9 days of osteo-induction. Gene expression was subsequently analyzed by Q-RT-PCR. C, Rspo2; D, Dmp1; E, Phex; F, Bsp. Expression values represent fold-change in gene expression relative to non-BMP-treated controls, normalized to β-actin. Statistical analysis was performed as mentioned previously. †,* indicate that Rspo2miR significantly decreased gene expression in non-BMP-treated (†) or BMP-treated groups (*) n >3, p < 0.01. No GF, no growth factor.
We also evaluated levels of osteoblast-associated gene expression in Wnt11+/Rspo2 miR+ cells. By quantitative PCR, Rspo2 expression in knockdown cells was reduced by 99% relative to the Wnt11+/LacZmiR+ control cells (Fig. 5C). BMP-treated Wnt11+/Rspo2miR+ cells demonstrate significantly reduced expression of BSP and Phex (Fig. 5, D and E). However, Dmp1 levels were unaffected by Rspo2 knockdown (Fig. 5F).
DISCUSSION
We have determined that Wnt11 enhances BMP-induced osteoblast maturation and mineralization in a Tcf/β-catenin-dependent manner and that this osteogenic effect is mediated, in part, through increased expression of Rspo2. Wnt11 increases Rspo2 expression, and Rspo2 overexpression potently enhanced osteoblast mineralization and maturation.
dnTcf blocks Rspo2 expression by Wnt11 suggesting that the increase in Rspo2 by Wnt11 requires Tcf/β-catenin signaling. In silico promoter analysis (supplemental Fig. S2) provides evidence to support this conclusion, as three Tcf consensus sites are located within a 1.5-kb region upstream of the start codon in the Rspo2 gene. Decreased expression of Rspo2 as a result of Tcf antagonism in Wnt11+ is a likely explanation for decreased levels of osteoblast differentiation in the Wnt11+/dnTcf+ cells.
A requirement for Rspo2 in Wnt11-mediated osteoblast maturation and mineralization is supported by Rspo2 knockdown data. BMP-treated Wnt11+/Rspo2miR+ cells demonstrate a near complete absence of mineral in response to BMP induction. These cells also show decreased alkaline phosphatase activity, as well as decreased expression of Bsp and Phex, genes associated with osteoblast maturation and mineralization. These studies clearly identify Rspo2 as a novel and potent osteogenic factor.
Similarly, a recent study has shown that Rspo1 potentiates osteoblast differentiation secondary to Wnt3a (21). However, our study suggests that the effects of Rspo2 are Tcf-independent. Although dnTcf blocks both Rspo2 expression and mineralization in Wnt11+ cells, it has no effect on enhanced mineralization by Rspo2. Furthermore, in Rspo2+/dnTcf+ cells, Bsp and Phex remain elevated but are decreased in the Wnt11+/dnTcf+ cells.
These data have allowed us to construct a model for the relationship between Wnt11, Rspo2, and BMP signaling during osteoblast maturation and mineralization (supplemental Fig. S3). Whereas β-catenin signaling is required for inducing Rspo2 expression, Rspo2 has effects independent of β-catenin-Tcf.
It is interesting to note that BMP treatment down-regulates expression of Rspo2 in MC3T3 pre-osteoblasts and also down-regulates expression of Rspo2 in Wnt11+ cells (although levels of expression remain elevated relative to BMP-treated controls). Given the potent effects of Rspo2 on osteoblast maturation, down-regulation of Rspo2 expression by BMPs may be a mechanism to limit the extent of mineralization by fully differentiated osteoblasts. Similarly, BMP has been shown to induce expression of BMP pathway inhibitors, including the soluble antagonists Noggin and Chordin, as well as inhibitory Smad. Thus, BMP may reduce Rspo2 as part of a negative feedback loop to “fine-tune” the extent of mineralization.
We postulate that decreases in Bsp and Phex expression may be responsible for decreased mineral observed with Wnt11+/Rspo2mir+ cells and Wnt11+/dnTcf+ cells. Bsp has a high affinity for hydroxyapatite and is involved in biomineralization (34). Conditional Phex deletion in osteoblasts results in severe osteomalacia, indicating an important role for Phex in bone mineralization and calcium-phosphate homeostasis (35). Antagonism of Phex and Bsp expression therefore may disrupt the process of biomineralization.
Wnt11 and Rspo2, acting downstream of Wnt11, modulate the expression of additional extracellular matrix proteins, particularly the bone/dentin-associated extracellular matrix protein Dmp1. Dmp1 is known to regulate osteoblast gene expression and may also influence hydroxyapatite crystal nucleation (29, 36). β-Catenin and Smad family members, along with AP1 family members, form a transcriptional complex that cooperatively regulates Dmp1 expression. Such complexes have been previously shown to regulate expression of target genes (37). Although dnTcf decreased Wnt11-mediated Dmp1 expression, it had no effect on Rspo2-induced Dmp1 expression. Furthermore, Rspo2 knockdown had no effect on Wnt11-mediated Dmp1 expression. This result suggests that Dmp1 expression by Wnt11 is regulated through β-catenin but is independent of Rspo2 (supplemental Fig. S3). Although Dmp1 remains elevated in Wnt11+/Rspo2miR+ cells, it is not sufficient for the pro-osteogenic effects of Wnt11.
Our results in MC3T3 E1 cells, a committed pre-osteoblast line, show that BMP stimulation is required for the pro-osteogenic effects of Wnt11 and Rspo2. This suggests that Wnt11 and Rspo2 do not have a direct effect on osteoblast differentiation, but act to modulate osteoblast maturation and function downstream from BMP signaling. It is possible that the Wnt11 and Rspo2 may have a different function in uncommitted osteoprogenitors, relative to more committed cells, such as the MC3T3 cells. Two lines of evidence support this hypothesis. First, during development, Wnt11 and Rspo2 are expressed in the apical ectodermal ridge, a zone of highly proliferative progenitor cells in the developing limbs. Second, during fracture healing, Wnt signaling promotes osteoprogenitor proliferation and inhibits differentiation (38). Studies examining the function of Wnt11 and Rspo2 in mesenchymal stem cells will allow a better understanding of these factors in regulating proliferation and differentiation of uncommitted osteoprogenitors, as opposed to regulating osteoblast maturation in terminally differentiated cells.
These studies identify Wnt11 and Rspo2 as novel osteogenic factors. Furthermore, Rspo2 is expressed downstream of Wnt11 and is both necessary and sufficient to mediate the osteogenic effects of Wnt11. Although the viral expression approach used in this study allows for sustained expression and release of Wnt11 and Rspo2, it does not allow for fine control of growth factor kinetics and timing of release. Future studies utilizing purified Wnt11 and Rspo2 will allow for better determinations of how growth factor kinetics and timing influence osteoprogenitor proliferation and differentiation and osteoblast maturation. If the downstream mediators of Rspo2 signaling are identified, this may serve as another pathway to target for development of bone anabolic agents.
Supplementary Material
Acknowledgments
We thank Weijun Luo for analyzing the microarray data and Susan Volk, Ormond MacDougald, and Hailu Shitaye for helpful discussions and critical comments concerning the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DE017471 and R01AR028922.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1–S3.
Footnotes
The abbreviations used are: BMP, bone morphogenetic protein; Rspo2, R-spondin 2; Bsp, bone sialoprotein; Phex, phosphate regulating gene with homologies to endopeptidases on the X-chromosome; GFP, green fluorescent protein; PBS, phosphate-buffered saline; miRNAi, microRNA interference; dn, dominant negative; Tcf, T-cell factor; BGP, β-glycerol phosphate.
References
- 1.Kennell, J. A., and Macdougald, O. A. (2005) J. Biol. Chem. 280 24004–24010 [DOI] [PubMed] [Google Scholar]
- 2.Fischer, L., Boland, G., and Tuan, R. S. (2002) J. Biol. Chem. 277 30870–30878 [DOI] [PubMed] [Google Scholar]
- 3.Mbalaviele, G., Sheikh, S., Stains, J. P., Salazar, V. S., Cheng, S. L., Chen, D., and Civitelli, R. (2005) J. Cell Biochem. 94 403–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kengaku, M., Capdevila, J., Rodriguez-Esteban, C., De La, P. J., Johnson, R. L., Belmonte, J. C., and Tabin, C. J. (1998) Science 280 1274–1277 [DOI] [PubMed] [Google Scholar]
- 5.Kawakami, Y., Capdevila, J., Buscher, D., Itoh, T., Rodriguez Esteban, C., and Izpisua Belmonte, J. C. (2001) Cell 104 891–900 [DOI] [PubMed] [Google Scholar]
- 6.Holmen, S. L., Giambernardi, T. A., Zylstra, C. R., Buckner-Berghuis, B. D., Resau, J. H., Hess, J. F., Glatt, V., Bouxsein, M. L., Ai, M., Warman, M. L., and Williams, B. O. (2004) J. Bone Miner. Res. 19 2033–2040 [DOI] [PubMed] [Google Scholar]
- 7.Gong, Y., Slee, R. B., Fukai, N., Rawadi, G., Roman-Roman, S., Reginato, A. M., Wang, H., Cundy, T., Glorieux, F. H., Lev, D., Zacharin, M., Oexle, K., Marcelino, J., Suwairi, W., Heeger, S., Sabatakos, G., Apte, S., Adkins, W. N., Allgrove, J., Arslan-Kirchner, M., Batch, J. A., Beighton, P., Black, G. C., Boles, R. G., Boon, L. M., Borrone, C., Brunner, H. G., Carle, G. F., Dallapiccola, B., De Paepe, A., Floege, B., Halfhide, M. L., Hall, B., Hennekam, R. C., Hirose, T., Jans, A., Juppner, H., Kim, C. A., Keppler-Noreuil, K., Kohlschuetter, A., LaCombe, D., Lambert, M., Lemyre, E., Letteboer, T., Peltonen, L., Ramesar, R. S., Romanengo, M., Somer, H., Steichen-Gersdorf, E., Steinmann, B., Sullivan, B., Superti-Furga, A., Swoboda, W., van den Boogaard, M. J., Van Hul, W., Vikkula, M., Votruba, M., Zabel, B., Garcia, T., Baron, R., Olsen, B. R., and Warman, M. L. (2001) Cell 107 513–523 [DOI] [PubMed] [Google Scholar]
- 8.Boyden, L. M., Mao, J., Belsky, J., Mitzner, L., Farhi, A., Mitnick, M. A., Wu, D., Insogna, K., and Lifton, R. P. (2002) N. Engl. J. Med. 346 1513–1521 [DOI] [PubMed] [Google Scholar]
- 9.Bennett, C. N., Longo, K. A., Wright, W. S., Suva, L. J., Lane, T. F., Hankenson, K. D., and MacDougald, O. A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 3324–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodine, P. V., Zhao, W., Kharode, Y. P., Bex, F. J., Lambert, A. J., Goad, M. B., Gaur, T., Stein, G. S., Lian, J. B., and Komm, B. S. (2004) Mol. Endocrinol. 18 1222–1237 [DOI] [PubMed] [Google Scholar]
- 11.Lako, M., Strachan, T., Bullen, P., Wilson, D. I., Robson, S. C., and Lindsay, S. (1998) Gene (Amst.) 219 101–110 [DOI] [PubMed] [Google Scholar]
- 12.Boland, G. M., Perkins, G., Hall, D. J., and Tuan, R. S. (2004) J. Cell. Biochem. 93 1210–1230 [DOI] [PubMed] [Google Scholar]
- 13.Tao, Q., Yokota, C., Puck, H., Kofron, M., Birsoy, B., Yan, D., Asashima, M., Wylie, C. C., Lin, X., and Heasman, J. (2005) Cell 120 857–871 [DOI] [PubMed] [Google Scholar]
- 14.Majumdar, A., Vainio, S., Kispert, A., McMahon, J., and McMahon, A. P. (2003) Development (Camb.) 130 3175–3185 [DOI] [PubMed] [Google Scholar]
- 15.Kazanskaya, O., Glinka, A., del Barco Barrantes, I., Stannek, P., Niehrs, C., and Wu, W. (2004) Dev. Cell 7 525–534 [DOI] [PubMed] [Google Scholar]
- 16.Kim, K. A., Zhao, J., Andarmani, S., Kakitani, M., Oshima, T., Binnerts, M. E., Abo, A., Tomizuka, K., and Funk, W. D. (2006) Cell Cycle 5 23–26 [DOI] [PubMed] [Google Scholar]
- 17.Nam, J. S., Turcotte, T. J., Smith, P. F., Choi, S., and Yoon, J. K. (2006) J. Biol. Chem. 281 13247–13257 [DOI] [PubMed] [Google Scholar]
- 18.Bouwman, P., Gollner, H., Elsasser, H. P., Eckhoff, G., Karis, A., Grosveld, F., Philipsen, S., and Suske, G. (2000) EMBO J. 19 655–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, K. A., Wagle, M., Tran, K., Zhan, X., Dixon, M. A., Liu, S., Gros, D., Korver, W., Yonkovich, S., Tomasevic, N., Binnerts, M., and Abo, A. (2008) Mol. Biol. Cell 19 2588–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binnerts, M. E., Kim, K. A., Bright, J. M., Patel, S. M., Tran, K., Zhou, M., Leung, J. M., Liu, Y., Lomas, W. E., III, Dixon, M., Hazell, S. A., Wagle, M., Nie, W. S., Tomasevic, N., Williams, J., Zhan, X., Levy, M. D., Funk, W. D., and Abo, A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 14700–14705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wenyan, L., Kyung-Ah, K., Jianzhong, L., Arie, A., Xu, F., Xu, C., and Yonghe, L. (2008) FEBS Lett. 582 643–65018242177 [Google Scholar]
- 22.Bell, S. M., Schreiner, C. M., Wert, S. E., Mucenski, M. L., Scott, W. J., and Whitsett, J. A. (2008) Development (Camb.) 135 1049–1058 [DOI] [PubMed] [Google Scholar]
- 23.Aoki, M., Kiyonari, H., Nakamura, H., and Okamoto, H. (2008) Dev. Growth Differ. 50 85–95 [DOI] [PubMed] [Google Scholar]
- 24.Nam, J. S., Park, E., Turcotte, T. J., Palencia, S., Zhan, X., Lee, J., Yun, K., Funk, W. D., and Yoon, J. K. (2007) Dev. Biol. 311 124–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, T. T., Xiong, Q., Enslen, H., Davis, R. J., and Chow, C. W. (2002) Mol. Cell. Biol. 22 3892–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolligs, F. T., Hu, G., Dang, C. V., and Fearon, E. R. (1999) Mol. Cell. Biol. 19 5696–5706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaffl, M. W. (2001) Nucleic Acids Res. 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tichopad, A., Dilger, M., Schwarz, G., and Pfaffl, M. W. (2003) Nucleic Acids Res. 31 e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narayanan, K., Gajjeraman, S., Ramachandran, A., Hao, J., and George, A. (2006) J. Biol. Chem. 281 19064–19071 [DOI] [PubMed] [Google Scholar]
- 30.Ling, Y., Rios, H. F., Myers, E. R., Lu, Y., Feng, J. Q., and Boskey, A. L. (2005) J. Bone Miner. Res. 20 2169–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tscheudschilsuren, G., Bosserhoff, A. K., Schlegel, J., Vollmer, D., Anton, A., Alt, V., Schnettler, R., Brandt, J., and Proetzel, G. (2006) Exp. Cell Res. 312 63–72 [DOI] [PubMed] [Google Scholar]
- 32.HYP Consortium (1995) Nat. Genet. 11 130–136 [DOI] [PubMed] [Google Scholar]
- 33.Wei, Q., Yokota, C., Semenov, M. V., Doble, B., Woodgett, J., and He, X. (2007) J. Biol. Chem. 282 15903–15911 [DOI] [PubMed] [Google Scholar]
- 34.Stubbs, J. T., III, Mintz, K. P., Eanes, E. D., Torchia, D. A., and Fisher, L. W. (1997) J. Bone Miner. Res. 12 1210–1222 [DOI] [PubMed] [Google Scholar]
- 35.Yuan, B., Takaiwa, M., Clemens, T. L., Feng, J. Q., Kumar, R., Rowe, P. S., Xie, Y., and Drezner, M. K. (2008) J. Clin. Investig. 118 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tartaix, P. H., Doulaverakis, M., George, A., Fisher, L. W., Butler, W. T., Qin, C., Salih, E., Tan, M., Fujimoto, Y., Spevak, L., and Boskey, A. L. (2004) J. Biol. Chem. 279 18115–18120 [DOI] [PubMed] [Google Scholar]
- 37.Chakladar, A., Dubeykovskiy, A., Wojtukiewicz, L. J., Pratap, J., Lei, S., and Wang, T. C. (2005) Biochem. Biophys. Res. Commun. 336 190–196 [DOI] [PubMed] [Google Scholar]
- 38.Kim, J. B., Leucht, P., Lam, K., Luppen, C., Ten Berge, D., Nusse, R., and Helms, J. A. (2007) J. Bone Miner. Res. 22 1913–1923 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.