Abstract
Our previous studies (Howarth, J. W., Meller, J., Solaro, R. J., Trewhella, J., and Rosevear, P. R. (2007) J. Mol. Biol. 373, 706–722) of the unique N-terminal region of human cardiac troponin I (hcTnI), predicted a possible intramolecular interaction near the basic inhibitory peptide. To explore this possibility, we generated single cysteine mutants (hcTnI-S5C and hcTnI-I19C), which were labeled with the hetero-bifunctional cross-linker benzophenone-4-maleimide. The labeled hcTnI was reconstituted to whole troponin and exposed to UV light to form cross-linked proteins. Reversed-phase high-performance liquid chromatography and SDS-PAGE indicated intra- and intermolecular cross-linking with hcTnC and hcTnT. Moreover, using tandem mass spectrometry and Edman sequencing, specific intramolecular sites of interaction were determined at position Met-154 (I19C mutant) and Met-155 (S5C mutant) of hcTnI and intermolecular interactions at positions Met-47 and Met-80 of hcTnC in all conditions. Even though specific intermolecular cross-linked sites did not differ, the relative abundance of cross-linking was altered. We also measured the Ca2+-dependent ATPase rate of reconstituted thin filament-myosin-S1 preparation regulated by either cross-linked or non-labeled troponin. Ca2+ regulation of the ATPase rate was lost when the Cys-5 hcTnI mutant was cross-linked in the absence of Ca2+, but only partially inhibited with Cys-19 cross-linking in either the presence or absence of Ca2+. This result indicates different functional effects of cross-linking to Met-154 and Met-155, which are located on different sides of the hcTnI switch peptide. Our data provide novel evidence identifying interactions of the hcTnI-N terminus with specific intra- and intermolecular sites.
The human cardiac variant of troponin I (hcTnI)2 has structural and functional specializations that are related to its critical role in control of cardiac dynamics. These specializations include variations in amino acids that are significant factors in the response of the heart to: adrenergic stimulation (1), sarcomere length (2, 3), and pH (4, 5). An especially significant region of specialization is a unique N-terminal extension of 30–32 amino acids, which contains serial serines at positions 23/24 that are substrates for kinases that control cardiac dynamics (6–8). Despite its significance in control of cardiac function, molecular mechanisms of how the N-terminal human cardiac troponin I (N-hcTnI) region controls sarcomeric and cardiac function remain poorly understood. There is evidence that upon phosphorylation the interaction between the N-hcTnI and the N-lobe of N-hcTnC is weakened (9, 10). The structure of the N-hcTnI was missing in the crystal structure of cardiac troponin (11). However, we recently reported (12) the structure of the N-terminal peptide using NMR. Docking of this structure into the core troponin structure indicated the potential for a previously unappreciated intramolecular interactions of the N terminus with the regions at or near the highly basic inhibitory peptide region of cardiac troponin (12, 13). This interaction appeared plausible not only on the basis of the structure of hcTnI, but also on the basis of the preponderance of basic amino acids in the inhibitory peptide and the presence of acidic residues in the N terminus.
In experiments reported here, we tested the hypothesis that the unique N-terminal region of hcTnI engages in both intra- and intermolecular interactions. We introduced Cys residues into the N-hcTnI at positions 5 and 19 and labeled the Cys residue with the hetero-bifunctional cross-linker, BP-MAL, which upon UV irradiation cross-links to residues within ∼10 Å of the modified Cys (14). We analyzed the cross-linked peptides by Edman sequencing and mass spectrometry to determine specific sites of interaction. The intramolecular sites of interaction were Met-154 and Met-155 in the hcTnI switch peptide for labeled positions 19 and 5, respectively. The intermolecular cross-linking sites on N-hcTnC were 47 and 80 for hcTnI labeled at either position 5 or 19. Measurement of Ca2+-dependent ATPase rate in reconstituted preparations indicated that allosteric effects of the different specific intramolecular cross-links (position Met-154 versus Met-155) to different hydrophobic positions on the switch peptide may affect hcTnC interaction with the switch peptide.
EXPERIMENTAL PROCEDURES
Protein Purification and Mutagenesis—hcTnT, hcTnI, and hcTnC were expressed and purified as previously described with some modifications (15). The hcTnT cDNA was cloned into the pSBETa vector (16). Two cysteines in hcTnC were changed to serines by site-directed mutagenesis (Invitrogen). The hcTnI was cloned into pET17b (Novagen), and two mutants were generated, C80S/C97S/S5C and C80S/C97S/I19C, using a site-directed mutagenesis kit (Invitrogen). The hcTnI was expressed in Rosetta 2 cells (Novagen), and hcTnT and hcTnC were both expressed using BL21(DE3) cells (Novagen).
Troponin Complex Formation and Photocross-linking—HcTnI was dissolved from a freeze-dried powder in 6 m urea, 25 mm Tris-HCl, pH 7.6, 0.3 m NaCl, 5 mm MgCl. BP-MAL was added to the hcTnI in a 2:1 molar ratio. The labeling reaction was incubated at room temperature for 2 h protected from light, then quenched with 2 mm dithiothreitol (17) and dialyzed against 6 m urea, 25 mm Tris-HCl, pH 7.6, 5 mm MgCl, and 1 m NaCl to remove unincorporated BP-MAL. The hcTnT and hcTnC were resolubilized in 6 m urea, 25 mm Tris-HCl, pH 7.6, 5 mm MgCl, and 1 m NaCl. The subunits were then mixed, and the rest of the troponin complex formation and purification was performed as described (15). The troponin complex was cross-linked in the presence of Ca2+ (0.2 mm) or absence of Ca2+ (2 mm EGTA) by irradiating with 365 nm UV light on ice protected by borosilicate glass for 0–30 min (18).
Reconstituted Thin Filament and ATPase Assay—Tropomyosin was prepared from bovine left ventricle and actin from rabbit skeletal tissue as previously described (19). The myosin subfragment-1 (S1) was prepared from rabbit skeletal tissue as described (20). To reconstitute the thin filament for actin-activated-S1 ATPase assays we mixed: 0.2 μm S1, 5 μm actin, 2 μm tropomyosin, 3.29 μm troponin in 28 mm MOPS, pH 7.0, 140 mm NaCl, 7 mm MgCl. The final reaction conditions were: 21 mm MOPS, pH 7.0, 105 mm NaCl, and 5.25 mm MgCl at 25 °C. Components were mixed in the order given: 85 μl of protein, 20 μl of Ca2+ buffer (0.1 mm Ca or 1 mm EGTA final), 15 μl of 2-amino-6-mercapto-7-methylpurine ribonucleoside and purine-nucleoside phosphorylase (substrate and enzyme ratio 1:17, EnzChek Phosphate assay kit, Molecular Probes), and 20 μl of 7 mm ATP (1 mm ATP final). The inorganic phosphate was measured directly in the cuvette in real-time every 10 s for 5 min at 360 nm (21). To compare ATPase assays, two-tailed t-tests were performed with the level of significance set at p value of <0.05 expressed as the means ± S.E.
Protein and Peptide Separations—Cross-linked and non-cross-linked proteins were separated in 15% total acrylamide, 0.5% cross-linked bis-acrylamide, pH 8.8, and the stacking gel was 2.95% total acrylamide, 15% cross-linked with N,N-diallyltartardiamide, pH 6.8, as previously described (22–24). The troponin units were separated using a Grace Vydac C4 reversed-phase HPLC column 150 mm × 4.6 mm at a flow rate of 1 ml/min using a Dionex U-3000 analytical HPLC. The mobile phase solution A (5% acetonitrile with 0.1% trifluoroacetic acid) was used to equilibrate the column, and protein was eluted using solution B (95% acetonitrile, 0.1% trifluoroacetic acid). The linear gradient was stepped with solution B: 0% B, 0–5 min; 0–30% B, 5–20 min; 30–50% B, 20–80 min; 50–90% B, 80–85 min; 90% B, 85–90 min; 90–0% B, 90–91 min; and 0% B, 91–101 min.
Protein fractions separated in the C4 column as above were digested with proteinases to generate small peptides. Endoproteinase Arg-C from Roche Applied Science was used to digest the intramolecular TnI cross-links. The final Arg-C digestion conditions were incubated overnight at 37 °C in 1 m urea, 56 mm Tris-HCl, pH 7.6, 5.6 mm CaCl, 200 mm NaCl, and 0.5 μgof enzyme. To digest the intermolecular cross-links between TnI and TnC, we used immobilized l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin from Pierce. Then the tryptic digests were sequentially treated with Glu-C to better digest the hcTnC. The final Glu-C digest conditions were 1 m urea, 200 mm NaCl, 40 mm NH4HCO3, pH 7.8, and 0.2 μgof enzyme. The reaction mixtures were incubated overnight at 37 °C.
After the proteins were digested into peptides, they were separated using a Dionex U-3000 capillary LC system with a C18 PepMap 150- × 0.3-mm column and a flow rate of 4 μl/min. The mobile phase solution A (5% acetonitrile with 0.1% trifluoroacetic acid) was used to equilibrate the column, and protein was eluted using solution B (95% acetonitrile, 0.1% trifluoroacetic acid). The linear gradient was stepped with solution B: 0% B, 0–10 min; 0–60% B, 10–130 min; 60–90% B, 130–135 min; 90% B, 135–140 min; 90–0% B, 140–142 min; and 0% B, 140–160 min.
Cross-linked Peptide Analysis—Peptide fractions were analyzed by mass spectrometry using an ABI 4700 MALDI TOF/TOF, LTQ hybrid ion-trap/FTICR (Thermo Scientific Inc.), and a Voyager De Pro MALDI-TOF (Applied Biosystems), or by Edman sequencing using a Procise 494 cLC instrument at the W.M. Keck Foundation, Yale University. The ABI 4700 MALDI TOF/TOF was used to generate tandem mass spectrometer data using an α-cyano-4-hydroxycinnamic acid matrix as previously described (25). The samples used for direct infusion via static spray into the LTQ-FTICR were dissolved in 50% acetonitrile and 0.1% formic acid. The sample was loaded into a coated nanospray emitter with a 4-μm inner diameter (New Objective Inc.) and a 1.5-kV spray voltage. The peptide peaks were manually detected and selected for MS2 fragmentation using collision-induced dissociation (26). The use of the Voyager DE Pro was similar to the ABI 4700, except 150 shots were performed. All mass spectrometry experiments were repeated in full at least four times.
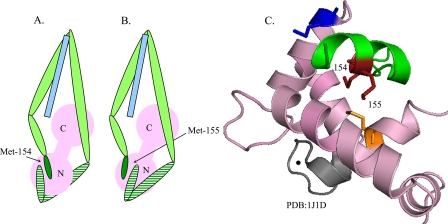
The analysis of the mass spectrometry data were largely done manually. However, Edman sequencing and use of software “Links” were helpful in first determining the cross-linked peptides (27). After determining cross-linked peptides, manual tandem mass spectrometry analysis was manageable. With cross-linking experiments it is not possible to use the normal search algorithms. Using the peptide known to be cross-linked, manual de novo sequencing was done to determine the b/y ions that corresponded to the cross-linked peptide complex and thus determine site-specific cross-links. The suspected cross-linked peptide complex was input into an MS/MS fragment ion calculator from the Institute for Systems Biology available on the web allowing matching of experimental to theoretical b/y ions, and with trial and error we determined the specific cross-linking site. PyMOL software was used to create Fig. 6C using available crystal structure (PDB: 1J1D).3
FIGURE 6.
A model of troponin complex showing change in structural organization associated with intramolecular cross-linking. Pink, troponin C subunit; green, troponin I subunit; light blue, troponin T subunit; green strips, N-terminal troponin I; and darker green, switch peptide of troponin I. A, schematic of the I19C mutant cross-linking to Met-154 of N-hcTnI and N-hcTnC. B, schematic of the S5C mutant cross-linking to Met-155 of N-hcTnI and N-hcTnC. C, N-terminal lobe of TnC along with the switch peptide part of TnI. Gray, site II Ca2+ binding loop; black bead, Ca2+ bound to site II; dark blue, intermolecular cross-linked site at aa 47 of troponin C; red, intramolecular cross-linked sites at aa 154 and 155 of troponin I; orange, intermolecular cross-linked site at aa 80 of troponin C.
RESULTS
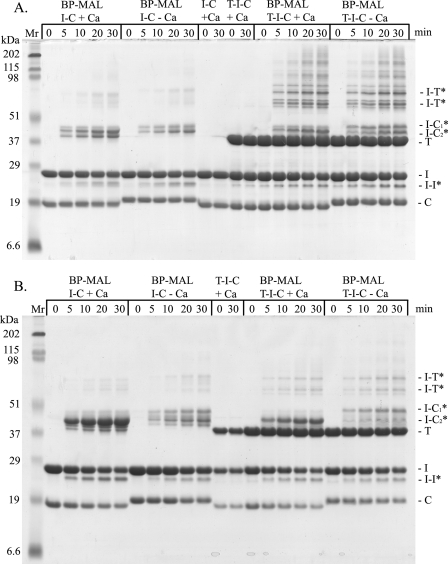
We used SDS-PAGE analysis to determine if the N-hcTnI cross-links to itself or other units of troponin. A time-based response of BP-MAL to exposure of 365 nm light showed cross-linking at positions 5 and 19 of the N-hcTnI with itself and other troponin units (Fig. 1). As shown in Fig. 1A the S5C hcTnI mutant cross-linked to itself (I-I), hcTnC (I-C), and hcTnT (I-T) in the lanes designated “T-I-C ± Ca” from 0- to 30-min exposure to light. In addition, the binary complex of hcTnI and hcTnC (I-C ± Ca) showed similar inter- and intramolecular cross-linking with hcTnI and hcTnC as seen in the left part of Fig. 1A. We included non-labeled hcTnI incorporated into a troponin complex as a negative control for breakdown. The N-hcTnI labeled with BP-MAL at position 19 cross-linked to all troponin subunits, but there were less cross-linked bands compared with position 5. This may be due to the increased reach of position 5 being at a more terminal end compared with position 19.
FIGURE 1.
Representative 15% SDS-PAGE of cross-linked troponin subunits. BP-MAL, benzenophenone-4-malemide cross-linker; min, minutes exposed to 365 nm UV light; T, troponin T; I, troponin I; C, troponin C; I-T, troponin I cross-linked to troponin T; I-C1*, slower migrating troponin I cross-linked to troponin C; I-C2*, faster migrating troponin I cross-linked to troponin C; I-I* intramolecular cross-link of troponin I; I-T*, troponin I cross-linked to troponin T; I-C, troponin I and troponin C complex; T-I-C, whole troponin complex; + Ca, cross-linked in the presence of Ca2+; -Ca, cross-linked in the absence of Ca2+. A, the S5C mutant troponin I was used on all lanes of the gel. B, the I19C mutant troponin I was used in all lanes of the gel. Note: the protein loads were equalized on each gel, but gels were not equalized to each other.
Fig. 2A shows results in which we determined Ca2+-dependent ATPase rate of reconstituted thin filament-myosin-S1 preparations regulated by a control troponin complex containing hcTnI (S5C or I19C) non-labeled or labeled with BP-MAL and cross-linked in the absence or presence of Ca2+ prior to reconstitution into thin filaments. Ca2+ regulation of ATPase rate was similar for controls and for preparations regulated by troponin (hcTnI-S5C) cross-linked in the presence of Ca2+ (Fig. 2A). However, Ca2+ regulation was lost in preparations reconstituted with troponin (hcTnI-S5C) cross-linked in the absence of Ca2+ (Fig. 2A). As shown in Fig. 2B there was regulation of Ca2+ activation of ATPase rate, albeit blunted, in preparations regulated by troponin (hcTnI-I19C) compared with non-labeled troponin (no BP-MAL) independent of whether cross-linked in presence or absence of Ca2+.
FIGURE 2.
ATPase assay using reconstituted thin filament-myosin-S1. ATPase activity normalized relative to non-labeled non-cross-linked troponin in presence of 0.7 mm Ca2+ n = 4 ± S.E. The assay was performed in higher salt (21 mm MOPS, pH 7.0, 105 mm NaCl, 5.25 mm MgCl, and 1 mm ATP) to keep human troponin in solution. X-link Ca2+, cross-linked either with or without Ca2+; NA, not applicable; Assay Ca2+, assay condition with or without Ca2+. A, S5C mutant, assayed with human troponin and S5C troponin I mutation. *, p value = 0.0002; #, p value = 0.005. B, I19C mutant, assayed with human troponin and I18C troponin I mutation. *, p value < 0.0001; #, p value < 0.0001; **, p value = 0.0044.
We also established specific cross-linked sites. Our first step was to separate the cross-linked proteins and subsequent digested peptides by HPLC. To separate the whole troponin cross-linked (dark line) and non-cross-linked subunits (light line), we used a reversed-phase C4 column evaluated at 280 nm as shown in Fig. 3A. The cross-linked proteins and peptides are represented by the horizontal bars through the peaks (Fig. 3). The cross-linked proteins were manually fractionated and digested for determination of sites of intramolecular cross-linking using endoproteinase Arg-C (Fig. 3, B and D) and for intermolecular cross-linking sequentially with immobilized trypsin and endoproteinase Glu-C (Fig. 3, C and E). The intramolecular cross-links illustrated in Fig. 3 (B and D) are represented as dark lines, and the light lines are non-cross-linked and digested TnI used as negative controls to help identify cross-linked peptides. The manually collected peptide fractions were analyzed with MALDI-TOF mass spectrometry to confirm the cross-linked fractions.
FIGURE 3.
HPLC of peptides and whole proteins. The bar indicates either cross-linked proteins or peptides depending on the run. Signal evaluated at 280 nm for A and 260 nm for B–E. Fractions were collected manually, and each fraction from B–E was analyzed by MALDI-TOF to determine which fractions were cross-linked. A, representative separation of cross-linked and non-cross-linked whole troponin subunits. The dark line represents whole troponin complex cross-linked for 30 min and separated by C4 (4.6 × 150 mm) reversed-phase HPLC, and the light line is non-cross-linked negative control. I, troponin I; T, troponin T; C, troponin C; I+I, intramolecular cross-link of troponin I; I+T, intermolecular cross-link of troponin I and T; I+C, intermolecular cross-link of troponin I and C. B, the dark line intramolecular cross-link of the troponin I mutant S5C digested with Arg-C and separated by C18 (0.3 × 150 mm) reversed-phase capillary HPLC, and the light line is non-cross-linked negative control. C, the dark line intermolecular cross-link of the troponin I mutant S5C to troponin C digested sequentially with trypsin and Glu-C and separated by C18 (0.3 × 150 mm) reversed-phase capillary HPLC, and the light line is the enzymes-only control. D, the dark line intramolecular cross-link of the troponin I mutant I19C digested with Arg-C and separated by C18 (0.3 × 150 mm) reversed-phase capillary HPLC, and the light line is non-cross-linked negative control. E, dark line intermolecular cross-links of the troponin I mutant I19C to troponin C digested sequentially with trypsin and Glu-C and separated by C18 (0.3 × 150 mm) reversed-phase capillary HPLC, and the light line is the enzymes-only control.
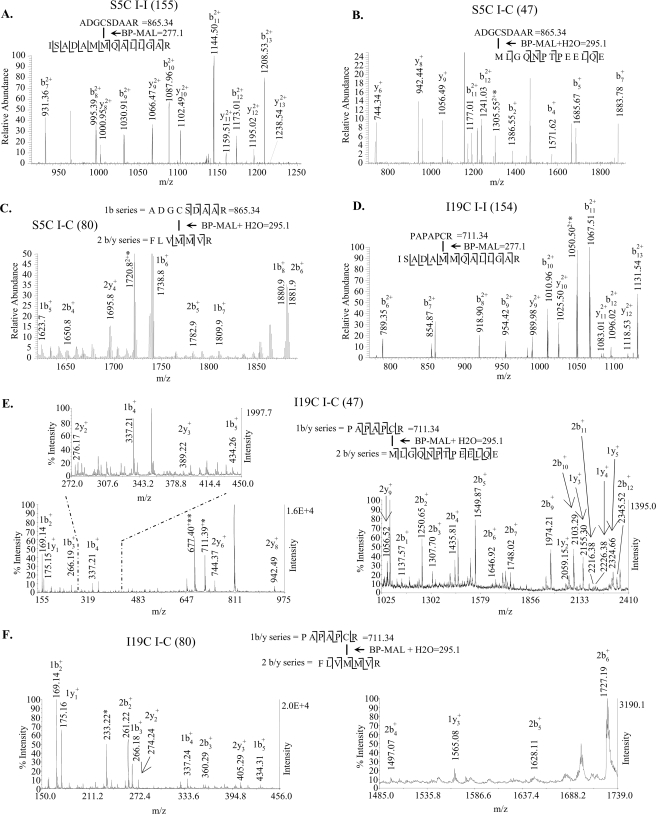
To determine cross-linked peptides, we used MALDI-TOF mass spectrometry (Fig. 4). The cross-linked peptide complex has three components depicted the N-hcTnI on top of the complex, the cross-linker (BP-MAL) in the middle, and the peptide cross-linked on the bottom of the complex. As shown in Fig. 4 (A–F) we were able to fragment the components as well as the intact complex allowing for more confidence in our peptide assignment. Once the cross-linked fractions were determined by MALDI-TOF, fractions were analyzed by either Edman sequencing or tandem mass spectrometry. The intramolecular cross-linked fraction illustrated in Fig. 3B was Edman sequenced, and both cross-linked and non-cross-linked peptides were present; however, the cross-linked peptides were in higher abundance (Table 1). The most abundant intermolecular cross-linked fraction (Fig. 3E) was Edman sequenced, and both cross-linked and non-cross-linked peptides were present (Table 2). The Edman sequencing results supported the cross-linked peptide designations from the MALDI-TOF experiments.
FIGURE 4.
MALDI-TOF spectra of cross-linked peptide complex purified as in Fig. 3 (B–E). BP-MAL, cross-linker mass 277.1 with hydrolyzed maleimide ring 18 = 295.1 (30). Complexes are broken up into major fragments as indicated in the spectrum. The unidentified peaks are non-cross-linked peptides. A, intramolecular cross-link of troponin I at position 5 to troponin I amino acids 149–162. B, intermolecular cross-link of troponin I at position 5 to troponin C amino acids 47–59. C, intermolecular cross-link of troponin I at position 5 to troponin C amino acids 77–83. D, intramolecular cross-link of troponin I at position 19 to troponin I amino acids 149–162. E, intermolecular cross-link of troponin I at position 19 to troponin C amino acids 47–59. F, intermolecular cross-link of troponin I at position 19 to troponin C amino acids 77–83.
TABLE 1.
Edman sequence of peptides from C18 Arg-C digested fraction from Fig. 3B (intramolecular cross-link S5C)
Sequencer yields are in picomoles, and X-link represents cross-link.
| Cycle | X-link TnI (2-6) | X-link TnI (149-163) | Non-X-link TnI (52-66) |
|---|---|---|---|
| 1 | A 35.2 | I 44.4 | T 9.5 |
| 2 | D 20.3 | S 19.5 | L 9.5 |
| 3 | G 7.1 | A 19.8 | L 8.1 |
| 4 | NDa | D 16.9 | L 7.4 |
| 5 | S 4.4 | A 12.6 | Q 5.3 |
| 6 | M 1.6 | I 4.5 | |
| 7 | M 3.2 | A 8.6 | |
| 8 | Q 4.8 | ND | |
| 9 | A 7.8 | Q 4.5 | |
| 10 | L 3.0 | E 2.1 | |
| 11 | L 5.2 | L 5.2 | |
| 12 | G 2.1 | E 1.5 | |
| 13 | A 2.3 | R 0.7 | |
| 14 | R 0.6 | E 1.0 | |
| 15 | A 1.7 |
ND, not determined quantitatively due to low yields.
TABLE 2.
Edman sequence of peptides from C18 trypsin/Glu-C digested fraction from Fig. 3E most abundant fraction (I+C cross-link I19C)
Sequencer yields are in picomoles, and X-link represents cross-link.
| Cycle | X-link TnI (14-20) | Non-X-link TnI (83-94) | Non-X-link TnI (49-60) | X-link TnC (47-59) | X-link TnC (77-83) | Non-X-link TnC (100-112) |
|---|---|---|---|---|---|---|
| 1 | P 42.2 | L 21.8 | L 21.8 | M 36.2 | F 26.3 | L 21.8 |
| 2 | A 69.4 | E 3.5 | NDa | L 58.3 | L 58.3 | F 10.5 |
| 3 | P 26.1 | L 6.8 | T 2.4 | G 19.3 | V 25.4 | R 5.9 |
| 4 | A 34.2 | ND | L 8.4 | Q 25.7 | M 6.8 | M 6.8 |
| 5 | P 16.4 | G 7.8 | L 6.3 | N 15.1 | M 11.8 | F 4.5 |
| 6 | ND | L 7.7 | L 7.7 | P 13.2 | V 10.7 | D 6.0 |
| 7 | R 6.2 | G 7.1 | Q 3.0 | T 8.8 | R 6.2 | ND |
| 8 | F 3.3 | I 2.3 | P 6.0 | N 4.7 | ||
| 9 | A 4.8 | A 4.8 | E 6.1 | A 4.8 | ||
| 10 | E 7.0 | ND | E 7.0 | P 3.0 | ||
| 11 | L 4.0 | Q 2.1 | L 4.0 | G 6.7 | ||
| 12 | Q 3.4 | E 4.9 | Q 3.4 | Y 2.1 | ||
| 13 | E 5.6 | I 2.4 |
ND, not determined quantitatively due to low yields.
Tandem mass spectrometry was used to determine specific sites of cross-linking from manually collected capillary HPLC fractions that were either directly infused in the case of the LTQ-FTICR or mixed with MALDI matrix and infused into an ABI 4700 MALDI TOF/TOF. The triply charged precursor ion 2608 was static sprayed into an LTQ-FTICR to determine the specific intramolecular cross-link of the S5C mutant specifically to aa Met-155 of hcTnI (Fig. 5A). The doubly charged precursor ion 2645 was static sprayed into an LTQ-FTICR to determine the specific intermolecular cross-link of the S5C mutant specifically to aa Met-47 of hcTnC (Fig. 5B). The doubly charged precursor ion 2055 was static sprayed into an LTQ-FTICR to determine the specific intermolecular cross-link of the S5C mutant specifically to amino acid Met-80 of TnC (Fig. 5C). The triply charged precursor ion 2454 was static sprayed into an LTQ-FTICR to determine the specific intramolecular cross-link of the I19C mutant specifically to aa Met-154 of hcTnI (Fig. 5D). The singly charged precursor ion 2491 was infused into an ABI 4700 MALDI TOF/TOF to determine the specific intermolecular cross-link of the I19C mutant specifically to aa Met-47 of TnC (Fig. 5E). The singly charged precursor ion 1901 was infused into an ABI 4700 MALDI TOF/TOF to determine the specific intermolecular cross-link of the I19C mutant specifically to amino acid Met-80 of hcTnC (Fig. 5F).
FIGURE 5.
Tandem mass spectrometry. BP-MAL, cross-linker mass 277.073 with hydrolyzed maleimide ring + 18 = 295.073 (30). Some spectra have two series of b/y ions represented by a 1 or 2 preceding ion type as shown in C, E, and F. A, spectra of the intramolecular cross-linked troponin I labeled at position 5 and cross-linked to position 155. Cross-linked peptide complex was direct infused into an LTQ-FTICR instrument and MS/MS ions are from the m/z 869.7264 precursor ion (z = 3), Δ0.153 ppm. B, spectra of the intermolecular cross-linked troponin I labeled at position 5 and cross-linked to position 47 of troponin C. Cross-linked peptide complex was directly infused into an LTQ-FTICR instrument, and MS/MS ions are from the m/z 1323.0645 precursor ion (z = 2), Δ3.25 ppm. * = precursor (z = 2) loss of H2O. C, spectra of the intermolecular cross-linked troponin I labeled at position 5 and cross-linked to position 80 of troponin C. Cross-linked peptide complex was directly infused into an LTQ-FTICR (precursor ion) instrument, and MS/MS (LTQ) ions are from the m/z 1027.9635 precursor ion (z = 2), Δ2.48 ppm. * = 1b6 (z = 1) loss of H2O. D, spectra of the intramolecular cross-linked troponin I labeled at position 19 and cross-linked to position 154. Cross-linked peptide complex was direct infused into an LTQ-FTICR instrument, and MS/MS ions are from the 818.3980 precursor ion (z = 3), Δ0.57 ppm. * = b11 (z = 2) loss of ammonia on Gln. E, spectra of the intermolecular cross-linked troponin I labeled at position 19 and cross-linked to position 47 of troponin C. Cross-linked peptide complex was analyzed by MALDI TOF/TOF instrument, and MS/MS ions are from the 2491.4072 precursor ion (z = 1), Δ117 ppm. The three spectra are zoomed in from the same spectra. * = cleavage of the BP-MAL from PAPAPCR. ** = loss of H2S from the cysteine of PAPAPCR. F, spectra of the intermolecular cross-linked troponin I labeled at position 19 and cross-linked to position 80 of troponin C. Cross-linked peptide complex was analyzed by MALDI TOF/TOF instrument, and MS/MS ions are from the 1901.1526 precursor ion (z = 1), Δ124 ppm. The two spectra are zoomed in from the same spectra. * = 2a2 (z = 1).
DISCUSSION
Our data are the first to identify specific sites of intramolecular interactions between the unique N terminus of hcTnI and the switch peptide (∼149–162) at Met-154 and Met-155. Under all conditions we also identified sites of intermolecular cross-linking between N-hcTnI at I19C and S5C and positions Met-47 and Met-80 of N-hcTnC. Moreover, measurements of Ca2+ regulation of ATPase rate revealed insights into the interaction of the N-hcTnI with hcTnI Met-154 and Met-155. As discussed below these new insights on the role of Met residues in the switch peptide comes from our data on the different effects of cross-linked products with BP-Mal located at position 19 of hcTnI (cross-linked to Met-154) versus BP-MAL located at position 5 of the N terminus (cross-linked to Met-155). In thin filaments reconstituted with troponin-containing hcTnI-I19C there was intramolecular cross-linking to Met-154, and Ca2+ was able to partially switch on myosin-S1 ATPase activity (Fig. 2B) whether the cross-linking was carried out in either the presence or absence of Ca2+. However, when thin filaments were regulated by hcTnI-S5C with an intramolecular cross-link to Met-155, Ca2+ regulation was completely lost, when the cross-linking was carried out in the absence of Ca2+.
There are several possibilities for the differential Ca2+ regulation with hcTnI-I19C compared with hcTnI-S5C mutant. One possibility as illustrated in Fig. 6, is that Met-154 (cross-linked from I19C) exists on a different side of the switch peptide from Met-155 (cross-linked to S5C) (28). Met-155 is more exposed and accessible compared with the Met 154 in the core crystal structure in the presence of Ca2+. The 10-Å distance of the cross-linker along with the flexibility of the N-hcTnI (11, 12) should allow for the interaction as depicted in Fig. 6. Thus we speculate when cross-linking occurred at Met-154 there was a more prominent effect on the closed state by reducing the movement of the complex not allowing hcTnC to fully open, or by stabilizing the closed state of hcTnC to a greater extent. Moreover, the N-hcTnI likely moves to a different position when interacting with either Met-155 or Met-154 allowing for different stabilizing effects of the switch peptide. The Ca2+ binding site of hcTnC may be affected by the interaction between N-hcTnI and Met-154 as depicted in Fig. 6A. Conversely when the N-hcTnI cross-links with Met-155 the N-hcTnI may move around to the other side of the N-hcTnC, which allows for normal Ca2+ regulation when cross-linked in the presence of Ca2+ Fig. 6B. When cross-linked in the absence of Ca2+ the conformation of hcTnC is likely different and this different interaction of the N-hcTnI with hcTnC may contribute to the ablation of Ca2+ regulation.
As proposed by Baryshnikova et al. (29) and Li et al. (28), there is also the possibility that cross-linking of the N-hcTnI to the switch peptide induces an allosteric effect influencing hcTnC conformation. In the case of Met-155 the N-hcTnI may hold the switch peptide in a conformation that inhibits normal allosteric effects on hcTnC conformation. The position of I19C relative to S5C in hcTnI structure may also be responsible for the different effects of the cross-linked products on Ca2+ control of ATPase rate. Position 19 is closer to the core structure of troponin than position 5 and, when cross-linked, may retard the interaction of the switch peptide with N-hcTnC thus stabilizing the closed state of hcTnC. Our demonstration of relatively abundant cross-linking for N-hcTnI-I19C to hcTnC is consistent with a previous report (10) and may contribute to the blunted response to Ca2+. Therefore, position hcTnI-I19C interacting with N-hcTnC (suspected Met-47 see “Discussion” below) may be the intermolecular site responsible for the blunted response to Ca2+ in the N-hcTnI-I19C mutant. It is also possible that differences in Ca2+ regulation of thin filaments regulated by hcTnI-S5C and hcTnI-I19C are due to increased cross-linked products in the S5C mutant involving troponin T.
Compared with results obtained with hcTnI-I19C, Ca2+ regulation was completely lost, when the cross-linking was carried out in the absence of Ca2+ in thin filaments regulated by hcTnI-S5C with an intramolecular cross-link to Met-155. This result indicates that the conformational state in the absence of Ca2+ is such that, when cross-linked at Met-155 and then reconstituted into the thin filament, the troponin complex is locked in a closed state. Once cross-linked in the absence of Ca2+ the closed state may interfere with Ca2+ binding to hcTnC, however, due to the flexibility of the N-hcTnI the Ca2+ binding site of hcTnC should only be partially blocked. Our data support the idea of flexibility in N-hcTnI based on multiple sites of interaction identified at the same point of time during cross-linking. In addition, the specific cross-linking sites of N-hcTnI (S5C mutant) to hcTnT may be different and cause the loss of Ca2+ regulation. We were not able to determine specific sites for hcTnT due to low yields from the HPLC (retention time 47.4 min in Fig. 3A).
The intermolecular cross-linking sites N-hcTnC from N-hcTnI were at positions Met-47 and Met-80. The Met-47 and Met-80 flank the switch peptide with Met-47 being more exposed to the surface and the Met-80 residue being more internally positioned in a similar fashion as the Met-155 and Met-154 cross-links (Fig. 6C). The relative abundance of site-specific intermolecular cross-linking could not be definitively determined with mass spectrometry or SDS-PAGE. However, the intramolecular cross-linking abundance was not greatly changed from hcTnI position 5 and 19 based on SDS-PAGE. We found two cross-linked protein bands migrating at different positions in SDS-PAGE corresponding to an I-C complex. We were unable to use in gel digests to identify specific sites due to low yield. Thus we used in-solution digests, which were much more efficient, and only then were we able to obtain enough cross-linked peptide to determine the specific sites of interaction. We were unable to determine specifically which band corresponded to which cross-link due to incomplete separation using HPLC. We think the SDS-PAGE band difference arises from the two sites identified as Met-47 and Met-80; one site is exposed in the Ca2+ saturated structure (Met-47), whereas the other is embedded (internally located) in the N-lobe. Based on the structure (Fig. 6C) and accessibility, the I-C2 is likely the cross-link with Met-47 and the I-C1 is from the Met-80. The intramolecular cross-linking efficiency did not change significantly in ±Ca2+ conditions nor did the intermolecular cross-linking of hcTnI to hcTnT suggesting the proximities or efficiency of cross-linking was not affected by ± Ca2+.
Our data have important implications with regard to advancing our understanding of the role of N-hcTnI. The N-terminal peptide of hcTnI is now known to be a critical factor in control of cardiac dynamics (13, 15). Although the last ∼12 amino acids of the N-terminal extension of cTnI are highly conserved, there are significant differences in the primary structure of the first 20 amino acids of cTnI in hearts of large and small animals. For example, the first 20 amino acids in both human and mouse cTnI are acidic; however, the mouse has more of a net negative charge than the human isoform due to an additional glutamic acid and a lack of two basic arginines. Our identification of the intramolecular sites of interaction of the hcTnI N-terminal region with the hcTnI switch peptide provides a basis for further testing hypotheses for the relation between primary structure of the N-terminal peptide of cTnI and tuning the contraction/relaxation dynamics of the heart to heart rate.
Acknowledgments
Special thanks to Chao Yuan for help with static spray on the LTQ-FTICR.
This work was supported, in whole or in part, by National Institutes of Health Grants PO1 HL 62426 (Project 1) (to R. J. S.) and RO1 HL 82923 (to T. K.). Use of LTQ-FT and Voyager De Pro was provided by the CBC-UIC research Resources Center Proteomics and Informatics Services Facility, which was established by a grant from The Searle Funds at the Chicago Community Trust to the Chicago Biomedical Consortium. The ABI 4700 was used at the Proteomics Core Facility at the University of Chicago.
Footnotes
The abbreviations used are: hcTnI, human cardiac troponin I; BP-MAL, benzophenone-4-maleimide; N-hcTnI, N terminus of human cardiac troponin I; hcTnC, human cardiac troponin C; hcTnT, human cardiac troponin T; N-hcTnC, N terminus of human cardiac troponin C; HPLC, high-performance liquid chromatography; MOPS, 4-morpholinepropanesulfonic acid; MALD-TOF, matrix-assisted laser desorption ionization time-of-flight; S1, myosin subfragment-1; LTQ, linear trap quadrupole; FTICR, Fourier transform ion cyclotron resonance.
W. L. DeLano (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palto Alto, CA.
References
- 1.Fentzke, R. C., Buck, S. H., Patel, J. R., Lin, H., Wolska, B. M., Stojanovic, M. O., Martin, A. F., Solaro, R. J., Moss, R. L., and Leiden, J. M. (1999) J. Physiol. 517 143–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arteaga, G. M., Palmiter, K. A., Leiden, J. M., and Solaro, R. J. (2000) J. Physiol. 526 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konhilas, J. P., Irving, T. C., Wolska, B. M., Jweied, E. E., Martin, A. F., Solaro, R. J., and de Tombe, P. P. (2003) J. Physiol. 547 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westfall, M. V., and Metzger, J. M. (2007) J. Mol. Cell Cardiol. 43 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolska, B. M., Vijayan, K., Arteaga, G. M., Konhilas, J. P., Phillips, R. M., Kim, R., Naya, T., Leiden, J. M., Martin, A. F., de Tombe, P. P., and Solaro, R. J. (2001) J. Physiol. 536 863–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moir, A. J., Solaro, R. J., and Perry, S. V. (1980) Biochem. J. 185 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ray, K. P., and England, P. J. (1976) FEBS Lett. 70 11–16 [DOI] [PubMed] [Google Scholar]
- 8.Solaro, R. J., Moir, A. J., and Perry, S. V. (1976) Nature 262 615–617 [DOI] [PubMed] [Google Scholar]
- 9.Ward, D. G., Brewer, S. M., Calvert, M. J., Gallon, C. E., Gao, Y., and Trayer, I. P. (2004) Biochemistry 43 4020–4027 [DOI] [PubMed] [Google Scholar]
- 10.Ward, D. G., Brewer, S. M., Cornes, M. P., and Trayer, I. P. (2003) Biochemistry 42 10324–10332 [DOI] [PubMed] [Google Scholar]
- 11.Takeda, S., Yamashita, A., Maeda, K., and Maeda, Y. (2003) Nature 424 35–41 [DOI] [PubMed] [Google Scholar]
- 12.Howarth, J. W., Meller, J., Solaro, R. J., Trewhella, J., and Rosevear, P. R. (2007) J. Mol. Biol. 373 706–722 [DOI] [PubMed] [Google Scholar]
- 13.Solaro, R. J., Rosevear, P., and Kobayashi, T. (2008) Biochem. Biophys. Res. Commun. 369 82–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorman, G., and Prestwich, G. D. (1994) Biochemistry 33 5661–5673 [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi, T., and Solaro, R. J. (2006) J. Biol. Chem. 281 13471–13477 [DOI] [PubMed] [Google Scholar]
- 16.Schenk, P. M., Baumann, S., Mattes, R., and Steinbiss, H. H. (1995) BioTechniques 19 196–198, 200 [PubMed] [Google Scholar]
- 17.Wang, Z. Y., Sarkar, S., Gergely, J., and Tao, T. (1990) J. Biol. Chem. 265 4953–4957 [PubMed] [Google Scholar]
- 18.Luo, Y., Wu, J. L., Li, B., Langsetmo, K., Gergely, J., and Tao, T. (2000) J. Mol. Biol. 296 899–910 [DOI] [PubMed] [Google Scholar]
- 19.Rarick, H. M., Opgenorth, T. J., von Geldern, T. W., Wu-Wong, J. R., and Solaro, R. J. (1996) J. Biol. Chem. 271 27039–27043 [DOI] [PubMed] [Google Scholar]
- 20.Swartz, D. R., and Moss, R. L. (1992) J. Biol. Chem. 267 20497–20506 [PubMed] [Google Scholar]
- 21.Webb, M. R. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 4884–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fritz, J. D., Swartz, D. R., and Greaser, M. L. (1989) Anal. Biochem. 180 205–210 [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. (1970) Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- 24.Layland, J., Cave, A. C., Warren, C., Grieve, D. J., Sparks, E., Kentish, J. C., Solaro, R. J., and Shah, A. M. (2005) FASEB J. 19 1137–1139 [DOI] [PubMed] [Google Scholar]
- 25.Parker, L., Engel-Hall, A., Drew, K., Steinhardt, G., Helseth, D. L., Jr., Jabon, D., McMurry, T., Angulo, D. S., and Kron, S. J. (2008) J. Mass Spectrom. 43 518–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan, C., Sheng, Q., Tang, H., Li, Y., Zeng, R., and Solaro, R. J. (2008) Am. J. Physiol. 295 H647–H656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu, E. T., Hawkins, A., Kuntz, I. D., Rahn, L. A., Rothfuss, A., Sale, K., Young, M. M., Yang, C. L., Pancerella, C. M., and Fabris, D. (2008) J. Proteome Res. 7 4848–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, M. X., Spyracopoulos, L., and Sykes, B. D. (1999) Biochemistry 38 8289–8298 [DOI] [PubMed] [Google Scholar]
- 29.Baryshnikova, O. K., Li, M. X., and Sykes, B. D. (2008) J. Mol. Biol. 375 735–751 [DOI] [PubMed] [Google Scholar]
- 30.Leszyk, J., Tao, T., Nuwaysir, L. M., and Gergely, J. (1998) J. Muscle Res. Cell Motil. 19 479–490 [DOI] [PubMed] [Google Scholar]