Abstract
Abasic (AP) sites are very frequent and dangerous DNA lesions. Their ability to block the advancement of a replication fork has been always viewed as a consequence of their inhibitory effect on the DNA synthetic activity of replicative DNA polymerases (DNA pols). Here we show that AP sites can also affect the strand displacement activity of the lagging strand DNA pol δ, thus preventing proper Okazaki fragment maturation. This block can be overcome through a polymerase switch, involving the combined physical and functional interaction of DNA pol β and Flap endonuclease 1. Our data identify a previously unnoticed deleterious effect of the AP site lesion on normal cell metabolism and suggest the existence of a novel repair pathway that might be important in preventing replication fork stalling.
Loss of purine and pyrimidine bases is a significant source of DNA damage in prokaryotic and eukaryotic organisms. Abasic (apurinic and apyrimidinic) lesions occur spontaneously in DNA; in eukaryotes it has been estimated that about 104 depurination and 102 depyrimidation events occur per genome per day. An equally important source of abasic DNA lesions results from the action of DNA glycosylases, such as uracil glycosylase, which excises uracil arising primarily from spontaneous deamination of cytosines (1). Although most AP sites are removed by the base excision repair (BER)5 pathway, a small fraction of lesions persists, and DNA with AP lesions presents a strong block to DNA synthesis by replicative DNA polymerases (DNA pols) (2, 3). Several studies have been performed to address the effects of AP sites on the template DNA strand on the synthetic activity of a variety of DNA pols. The major replicative enzyme of eukaryotic cells, DNA pol δ, was shown to be able to bypass an AP lesion, but only in the presence of the auxiliary factor proliferating cell nuclear antigen (PCNA) and at a very reduced catalytic efficiency if compared with an undamaged DNA template (4). On the other hand, the family X DNA pols β and λ were shown to bypass an AP site but in a very mutagenic way (5). Recent genetic evidence in Saccharomyces cerevisiae cells showed that DNA pol δ is the enzyme replicating the lagging strand (6). According to the current model for Okazaki fragment synthesis (7–9), the action of DNA pol δ is not only critical for the extension of the newly synthesized Okazaki fragment but also for the displacement of an RNA/DNA segment of about 30 nucleotides on the pre-existing downstream Okazaki fragment to create an intermediate Flap structure that is the target for the subsequent action of the Dna2 endonuclease and the Flap endonuclease 1 (Fen-1). This process has the advantage of removing the entire RNA/DNA hybrid fragment synthesized by the DNA pol α/primase, potentially containing nucleotide misincorporations caused by the lack of a proofreading exonuclease activity of DNA pol α/primase. This results in a more accurate copy synthesized by DNA pol δ. The intrinsic strand displacement activity of DNA pol δ, in conjunction with Fen-1, PCNA, and replication protein A (RP-A), has been also proposed to be essential for the S phase-specific long patch BER pathway (10, 11). Although it is clear that an AP site on the template strand is a strong block for DNA pol δ-dependent synthesis on single-stranded DNA, the functional consequences of such a lesion on the ability of DNA pol δ to carry on strand displacement synthesis have never been investigated so far. Given the high frequency of spontaneous hydrolysis and/or cytidine deamination events, any detrimental effect of an AP site on the strand displacement activity of DNA pol δ might have important consequences both for lagging strand DNA synthesis and for long patch BER. In this work, we addressed this issue by constructing a series of synthetic gapped DNA templates with a single AP site at different positions with respect to the downstream primer to be displaced by DNA pol δ (see Fig. 1A). We show that an AP site immediately upstream of a single- to double-strand DNA junction constitutes a strong block to the strand displacement activity of DNA pol δ, even in the presence of RP-A and PCNA. Such a block could be resolved only through a “polymerase switch” involving the concerted physical and functional interaction of DNA pol β and Fen-1. The closely related DNA pol λ could only partially substitute for DNA pol β. Based on our data, we propose that stalling of a replication fork by an AP site not only is a consequence of its ability to inhibit nucleotide incorporation by the replicative DNA pols but can also stem from its effects on strand displacement during Okazaki fragment maturation. In summary, our data suggest the existence of a novel repair pathway that might be important in preventing replication fork stalling and identify a previously unnoticed deleterious effect of the AP site lesion on normal cell metabolism.
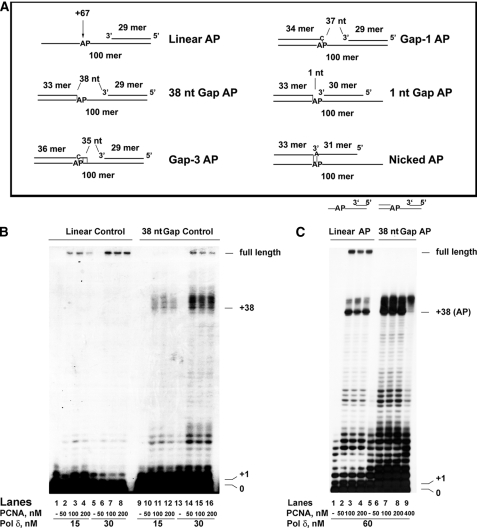
FIGURE 1.
An abasic site immediately upstream of a double-stranded DNA region inhibits the strand displacement activity of DNA polymerase δ. The reactions were performed as described under “Experimental Procedures.” A, schematic representation of the various DNA templates used. The size of the resulting gaps is indicated in nt. The position of the AP site on the 100-mer template strand is indicated relative to the 3′ end. Base pairs in the vicinity of the lesion are indicated by dashes. The size of the gaps (35–38 nt) is consistent with the size of ssDNA covered by a single RP-A molecule, which has to be released during Okazaki fragment synthesis when the DNA pol is approaching the 5′-end of the downstream fragment. When the AP site is covered by the downstream terminator oligonucleotide (Gap-3 and Gap-1 templates) the nucleotide placed on the opposite strand is C to mimic the situation generated by spontaneous loss of a guanine or excision of an oxidized guanine, whereas when the AP site is covered by the primer (nicked AP template), the nucleotide placed on the opposite strand is A to mimic the most frequent incorporation event occurring opposite an AP site. B, human PCNA was titrated in the presence of 15 nm (lanes 2–4 and 10–12) or 30 nm (lanes 6–8 and 14–16) recombinant human four subunit DNA pol δ, on a linear control (lanes 1–8) or a 38-nt gap control (lanes 9–16) template. Lanes 1, 5, 9, and 13, control reactions in the absence of PCNA. C, human PCNA was titrated in the presence of 60 nm DNA pol δ, on a linear AP (lanes 2–4) or 38-nt gap AP (lanes 6–9) template. Lanes 1 and 5, control reactions in the absence of PCNA.
EXPERIMENTAL PROCEDURES
Oligonucleotide DNA Substrates—All of the oligonucleotides both undamaged and containing a single synthetic AP site (tetrahydrofurane moiety) were from Eurogentec (Angers, France) and PAGE-purified according to the manufacturer's protocol. For detailed sequence description see supplemental “Experimental Procedures.” Primers were 5′-labeled with 3000 Ci/mmol [γ-32P]ATP with T4 polynucleotide kinase, according to the manufacturer's protocol.
Proteins—Recombinant human DNA pol β was from Trevigen Inc. (Gaithersburg, MD). Recombinant human DNA pols δ and λ, PCNA, RP-A, and Fen-1 were isolated as described (12–16).
Antibodies—Antibodies against DNA pol β were from our laboratory (13). Antibodies against Fen-1 were purchased form GeneTex, Inc. (San Antonio, TX).
Primer Elongation Assays—For denaturing gel analysis of DNA synthesis products, the reaction mixtures (10 μl) contained 50 mm Tris-HCl, pH 7.0, 0.25 mg/ml BSA, 1 mm dithiothreitol, 50 μm dNTPs, 1 mm MgCl2, and 10–20 nm (3′-OH ends) of the 5′ 32P-labeled primer/templates. Concentrations of DNA pols λ and β, PCNA, RP-A, and Fen-1 were as indicated in the figures and figure legends. The reactions were incubated for 5 min at 37 °C (unless otherwise stated) and then stopped by the addition of standard denaturing gel loading buffer (95% formamide, 10 mm EDTA, xylene cyanol, and bromphenol blue), heated at 95 °C for 3 min, and loaded on a 7 m urea, 10% polyacrylamide gel.
Far Western Blot Analysis—Purified recombinant human Fen-1 (500 ng) and BSA (500 ng) were mixed with Laemmli buffer and subjected to SDS-PAGE, and the proteins were transferred to a nitrocellulose membrane. All of the subsequent steps were preformed at 4 °C. The membrane was first incubated two times for 10 min in denaturing buffer (6 m guanidine hydrochloride in phosphate-buffered saline). To renature proteins the membrane was incubated in serial dilutions (1:1) of denaturing buffer and phosphate-buffered saline supplemented with 1 mm dithiothreitol, up to final concentration of 93.75 mm guanidine hydrochloride. The membrane was then blocked for 1 h in 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.3% Tween, and 10% milk powder, followed by washing for 10 min in wash buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.3% Tween, 0.25% milk powder, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride). Additionally the membrane was cut in half, followed by incubation for 2 h 30 min at 4 °C and then 30 min at room temperature, with one half in 1 μg/ml dilution of purified recombinant DNA pol β in wash buffer and the other half in wash buffer alone. The membranes were then washed four times for 10 min in wash buffer, with an exception that a second wash contained 0.0001% glutaraldehyde. From this point a conventional immunoblot analysis was performed, and the proteins were detected with the respective specific antibodies.
RESULTS
An Abasic Site Immediately Upstream of a Double-stranded DNA Region Inhibits the Strand Displacement Activity of DNA Polymerase δ—Human DNA pol δ was tested on linear or gapped DNA templates, either undamaged or bearing a single AP site (tetrahydrofuran) at position +38 from the 3′-OH of the primer (Fig. 1A). The gapped DNA substrate was constructed so that the size of the gap was 38 nt, and the abasic site was placed at the last single-stranded (ss) template position, immediately upstream the double-stranded (ds) region. In all cases, the downstream terminator oligonucleotide was 5′-phosphorylated with cold ATP, whereas the 5′-end of the primer was radioactively labeled. As shown in Fig. 1B, with the undamaged templates, DNA pol δ was expected to be able to synthesize full-length products in a strictly PCNA-dependent manner both on the linear (lanes 1–8) and on the gapped (lanes 9–16) DNA substrates. However, on gapped DNA, pol δ was effectively slowed down by the presence of a downstream dsDNA, as shown by the accumulation of products >38 residues long (lanes 10–12 and 14–16) and by the requirement of an higher DNA pol δ concentration than with the linear template to reach full-length products (compare lanes 2–4 with lanes 10–12 and 14–16). When DNA pol δ was tested on the same templates but in the presence of an AP site, a strong accumulation of products was noted at the position corresponding to the lesion both on the linear and on the gapped DNA templates (Fig. 1C, lanes 2–4 and 6–9). However, whereas on the linear template DNA pol δ was eventually able to overcome the lesion, reaching full-length products on the gapped DNA the synthesis was abortive, with accumulation of products between +38 and +45, and no full-length synthesis, even in the presence of high PCNA concentrations (lanes 6–9), was observed. Thus, the presence of an additional block, such as an AP site, immediately upstream of a dsDNA region was able to prevent strand displacement by DNA pol δ.
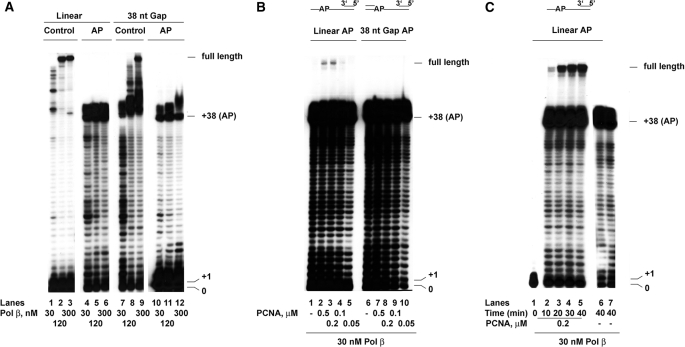
PCNA Promotes Efficient Bypass of an Abasic Site by DNA Polymerase β on Linear but Not on Gapped DNA Templates— Next, human DNA pol β was tested on the same templates. As shown in Fig. 2A, the ability of DNA pol β to synthesize full-length products on the linear (lanes 1–3) or gapped (lanes 7–9) undamaged templates increased as a function of enzyme concentration, according to the well known distributive mode of DNA synthesis by DNA pol β. DNA pol β was able to bypass an AP site both on linear (lanes 4–6) or gapped (lanes 10–12) DNA templates but with abortive synthesis of products slightly longer than +38, even at the highest concentration tested, suggesting that the lesion significantly increased the distributive DNA synthesis. DNA pol β has been reported to physically interact with PCNA, even though no functional role for this interaction has been shown so far. Thus, increasing amounts of PCNA were titrated in the reaction in the presence of low amounts of DNA pol β on both the linear and gapped DNA templates bearing an AP site. As shown in Fig. 2B, PCNA promoted the synthesis of full-length products past the lesion, but only on the linear template (lanes 1-5). If the mechanism of termination at the AP site was an increased dissociation rate of the enzyme from the DNA template, the bypass efficiency should be also influenced by the incubation time. To test this, a time course experiment was performed in the presence of fixed amounts of DNA pol β and PCNA, starting from 10 min (the same incubation time of the experiments shown in Fig. 2B) and up to 40 min. As shown in Fig. 2C, the amount of full-length products synthesized by DNA pol β increased with time (lanes 2–5). PCNA was essential for the stimulation of DNA synthesis, because in its absence no full-length products accumulated, even after 40 min (lanes 6 and 7). Thus, PCNA appears to be a stimulatory factor for DNA pol β bypass of an AP site. This is the first report of a functional interaction between these two proteins. However, no full-length synthesis could be detected on gapped DNA even with long incubation times (data not shown), suggesting that, similar to what has been observed with DNA pol δ (Fig. 1B), the presence of an AP site immediately upstream of a dsDNA region constitutes a strong block also to DNA pol β activity.
FIGURE 2.
PCNA promotes efficient bypass of an abasic site by DNA polymerase β on linear but not on gapped DNA templates. The reactions were performed as described under “Experimental Procedures.” A, recombinant human DNA pol β was incubated 10 min in the presence of linear control (lanes 1–3), linear AP (lanes 4–6), 38-nt gap control (lanes 7–9), or 38-nt gap AP (lanes 10–12) templates. B, 30 nm of DNA pol β were incubated 10 min in the absence (lanes 1 and 6) or in the presence (lanes 2–5 and 7–10) of increasing amounts of PCNA on a linear AP (lanes 1–5) or a 38 nt gap AP(lanes 6–10) template. C, 30 nm of DNA pol β were incubated for the indicated times on a linear AP substrate in the presence (lanes 1–5) or in the absence (lanes 6 and 7) of 200 nm PCNA.
The Presence of a Double-stranded DNA Region Upstream of an Abasic Site Facilitates Bypass by DNA Polymerase δ but Not β— Next, we investigated whether the position of the AP site relative to the dsDNA region might influence the ability of DNA pol δ to overcome the lesion. To this aim, a gapped substrate was constructed, where the AP site was placed 3 nt within the dsDNA, resulting in a ssDNA gap of 35 nt. As shown in Fig. 3A, DNA pol δ was able to fully displace the downstream terminator oligonucleotide, synthesizing full-length products (lanes 2–5). The addition of the ssDNA-binding protein RP-A further promoted full-length DNA synthesis (lanes 6–9). When DNA pol β was tested under the same conditions (Fig. 3B), however, no full-length synthesis was detected, irrespective of the presence or RP-A alone (lanes 9–12) or in combination with PCNA (lanes 17–20). Thus, the position of the AP lesion on gapped DNA substrates relative to the 5′-dsDNA region is important for DNA pol δ but not for DNA pol β. It has been suggested that DNA pol β interacts with a gapped DNA differently from DNA pol δ, by contacting first the 5′-downstream end of the gap (17). This different binding mode between DNA pols β and δ might explain the observed differences.
FIGURE 3.
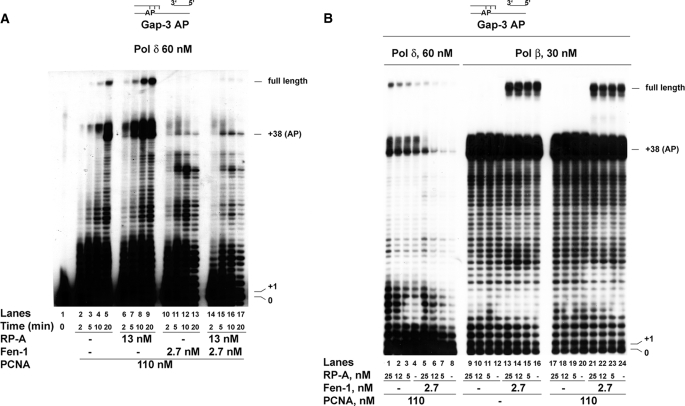
Fen-1 inhibits the synthesis of products beyond the abasic site by DNA polymerase δ but is essential for efficient strand displacement-dependent bypass of the abasic site by DNA polymerase β. The reactions were performed as described under “Experimental Procedures.” A, 60 nm DNA pol δ were incubated for the indicated times on the Gap-3 AP template in the presence of 110 nm PCNA alone (lanes 2–5) or in combination with 13 nm (as trimer) RP-A (lanes 6–9) or 2.7 nm recombinant human Fen-1 (lanes 10–13) or both (lanes 14–17). B, 60 nm DNA pol δ (lanes 1–8) or 30 nm DNA pol β (lanes 9–24), were incubated with 110 nm PCNA alone (lanes 1–8 and 17–24) or in combination with 2.7 nm Fen-1 (lanes 5–8 and 21–24), in the absence (lanes 4, 8, 20, and 24) or in the presence (lanes 1–3, 17–19, and 21–23) of increasing amounts of RP-A. Lanes 9–16, RP-A titration in the presence of 30 nm DNA pol β in the absence of PCNA and in the absence (lanes 9–12) or in the presence (lanes 13–16) of 2.7 nm Fen-1.
Fen-1 Inhibits the Synthesis of Products beyond the Abasic Site by DNA Polymerase δ but Is Essential for Efficient Strand Displacement-dependent Bypass of the Abasic Site by DNA Polymerase β—When the AP site is placed 3 nt within the dsDNA, both DNA pol δ and β would generate a short (3-nt) DNA Flap before encountering the AP site. In eukaryotic cells the endonuclease Fen-1 can specifically bind and cut Flap DNA structures. When Fen-1 was added in the reaction together with DNA pol δ, a strong reduction of DNA synthesis was observed (Fig. 3A), irrespective of the absence (lanes 10–13) or presence (lanes 14–17) of a fixed amount of RP-A. Because RP-A was able to promote full-length synthesis by DNA pol δ on this template (Fig. 3A, lanes 6–9), increasing amounts of RP-A were titrated into the reaction. As shown in Fig. 3B, in the absence of Fen-1, RP-A effectively promoted synthesis by DNA pol δ in a dose-dependent manner (lanes 1–4). However, the addition of Fen-1 (lanes 5–8) resulted in strong inhibition of DNA synthesis. Inhibition of DNA pol δ synthetic activity by Fen-1 on linear templates has been reported. However, on this gapped template, at the highest RP-A concentration, Fen-1 addition caused a strong reduction of bypass products (i.e. those of >38 residues, arising from strand displacement synthesis), whereas the amount of shorter products (i.e. those of <38 residues, synthesized during the gap filling step) was almost unaffected (compare lane 1 with lane 5), suggesting a mechanistic relationship between strand displacement by DNA pol δ and Fen-1 inhibition.
DNA pol β was unable to overcome the block imposed by an AP site on gapped DNA, resulting in abortive synthesis of short bypass products even in the presence of PCNA and RP-A (Figs. 2B and 3B). However, when Fen-1 was added into the reaction, efficient bypass of the AP site was achieved and resulted in the accumulation of full-length products, irrespective of whether PCNA and RP-A were absent (Fig. 3B, lanes 13–16) or present (lanes 21–24). When similar experiments were repeated in the presence of increasing salt concentrations, a stimulatory effect of PCNA on the strand displacement activity of DNA pol β on the 38-nt Gap AP template could be observed even in the presence of Fen-1, suggesting that both proteins were required for lesion bypass under physiological salt conditions (supplemental Fig. S1). Thus, Fen-1 and PCNA appear to be necessary to promote efficient strand displacement-dependent AP site bypass by DNA pol β.
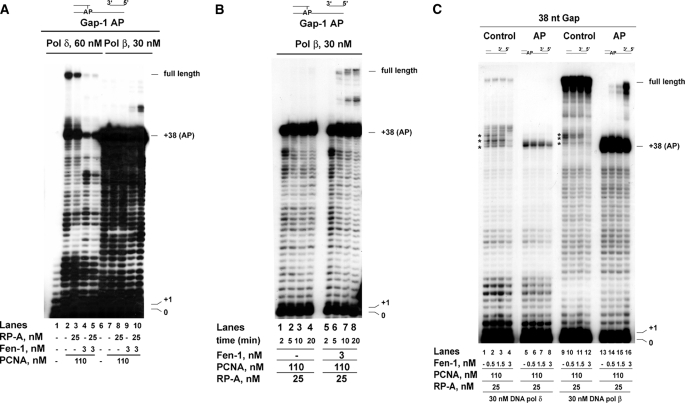
The Effect of Fen-1 on the Abasic Site Bypass by DNA Polymerases δ and β Is Not Dependent on the Position of the Lesion— Next, the influence of Fen-1 on the ability of DNA pol δ or β to bypass an AP site was tested on a gapped substrate where the AP site was placed 1 nt within the 5′-ds region, resulting in a ssDNA gap of 37 nt. On this template, an incoming DNA pol has to displace only 1 nt to face the AP site on the template. As shown in Fig. 4A, DNA pol δ was able to bypass the lesion by extending the products up to the full length (lanes 2 and 3). However, the addition of Fen-1 caused a strong reduction of the bypass products (lanes 4 and 5), suggesting that a 1-nt Flap was already sufficient to activate the inhibitory mechanism by Fen-1. RP-A did not show any effect on this reaction (compare lanes 2 and 4 with lanes 3 and 5). When these experiments were repeated with DNA pol β, extension of products beyond the lesion was observed again only in the presence of Fen-1 (compare lanes 7 and 8 with lanes 9 and 10) and RP-A stimulated the reaction (compare lane 9 with lane 10). However, the overall efficiency of Fen-1 stimulation was lower than with the 35-nt gapped DNA template (compare Fig. 4A with Fig. 3B). The experiments shown in Fig. 4A were incubated for 10 min. Increasing the incubation time up to 20 min (Fig. 4B) resulted in higher accumulation of full-length products by DNA pol β but always in a strictly Fen-1-dependent manner (compare lanes 1–4 with lanes 5–8). Thus, the effects of Fen-1 on either DNA pol δ or β were not influenced by the length of the Flap generated during strand displacement-dependent bypass of the AP site.
FIGURE 4.
The effect of Fen-1 on the abasic site bypasses by DNA polymerases δ and β are not dependent on the position of the lesion. The reactions were performed as described under “Experimental Procedures.” A, 60 nm DNA pol δ (lanes 1–5) or 30 nm DNA pol β (lanes 6–10) were incubated on the Gap-1 AP template, in the absence (lanes 1 and 6) or in the presence of 110 nm PCNA, either alone (lanes 2 and 7) or in combination with 25 nm RP-A (lanes 3 and 8) or 3 nm Fen-1 (lanes 4 and 9) or both (lanes 5 and 10). B, 30 nm DNA pol β were incubated for the indicated times on the Gap-1 AP template in the presence of PCNA and RP-A and in the absence (lanes 1–4) or in the presence (lanes 5–8) of 3 nm Fen-1. C, 30 nm DNA pol δ (lanes 1–8) or DNA pol β (lanes 9–16) were incubated on the 38 nt Gap control (lanes 1–4 and 9–12) or AP (lanes 5–8 and 13–16) template in the presence of PCNA and RP-A and in the absence (lanes 1, 5, 9, and 13) or in the presence (lanes 2–4, 6–8, 10–12, and 14–16) of increasing amounts of Fen-1. Asterisks mark the position of pausing sites during strand displacement DNA synthesis.
Fen-1 Can Promote Strand Displacement-dependent Bypass of an Abasic Site Immediately Upstream of a Double-stranded DNA Region by DNA Polymerase β but Not by δ—As shown in Figs. 1 and 2, the presence of an AP site immediately upstream of the 5′-dsDNA tract on a gapped template (38-nt Gap) constituted a strong block to strand displacement by both DNA pol δ and β. When increasing amounts of Fen-1 were titrated on this template in the presence of DNA pol δ, PCNA, and RP-A, (Fig. 4C), inhibition rather than stimulation of synthesis past the lesion was observed (lanes 5–8). In contrast, in the presence of DNA pol β, Fen-1 promoted efficient strand displacement beyond the lesion, with generation of full-length products (lanes 13–16). The effects of Fen-1 were also dependent on the presence of the lesion. In fact, on the undamaged 38-nt gapped template, no inhibition by Fen-1 of DNA pol δ was observed (Fig. 4C, lanes 1–4). On the contrary, Fen-1 facilitated strand displacement by both DNA pol δ and β, as shown by the reduction in the pausing sites corresponding to the first strand displacement events (Fig. 4C, compare lane 1 with lane 4 and lane 9 with lane 12; pausing sites are marked with asterisks at positions 38–42).
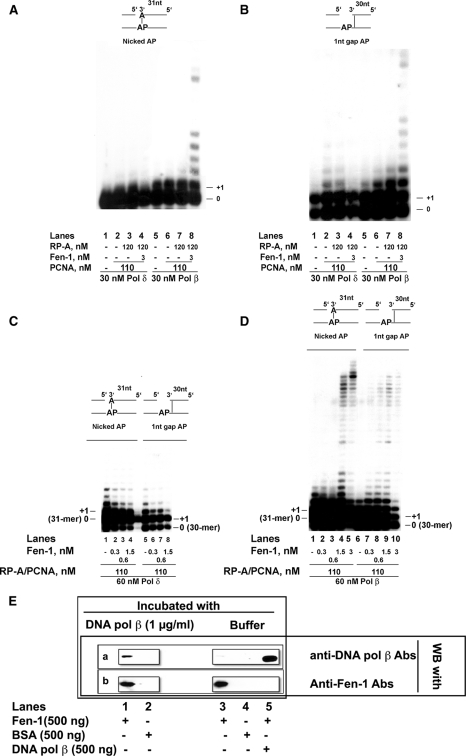
Fen-1 Can Act on Abasic Site Bypass by DNA Polymerases δ and β Either at the Incorporation or Elongation Steps—The data presented above show that Fen-1 can differentially influence the activities of DNA pol δ and β while they attempt to bypass an AP site during strand displacement DNA synthesis. However, a question still remains as to whether this action occurs at the level of incorporation or elongation. To verify this, Fen-1 was tested in the presence of either DNA pol δ or β on two different templates, mimicking the reaction intermediates generated during gap filling before and after incorporation opposite the lesion. To this aim, in a first construct a 30-mer primer was annealed to the template strand, which terminates immediately upstream of the lesion, together with a 5′-phosphorylated terminator oligonucleotide that was annealed immediately downstream the AP site. This generated a 1-nt gapped DNA, with the AP site as the only ssDNA template residue, mimicking the reaction intermediate at the pre-incorporation step. In a second construct, a 31-mer primer was used that annealed opposite the lesion. This, together with the downstream terminator oligonucleotide, generated a nicked DNA, with an A residue base-paired to the AP site, thus mimicking the most frequent post-incorporation product generated by AP site bypass. As shown in Fig. 5A, the nicked DNA was a poor substrate for DNA pol δ, which incorporated one nucleotide only when PCNA and RP-A were present (lanes 2 and 3) but not in their absence (lane 1). The addition of Fen-1, however, completely abolished nucleotide incorporation by DNA pol δ even in the presence of PCNA and RP-A (lane 4). On the opposite, DNA pol β efficiently incorporated one nucleotide on this substrate, either in the absence (lane 5) or in the presence of PCNA and RP-A (lanes 6 and 7). The addition of Fen-1 greatly stimulated strand displacement-coupled DNA synthesis by DNA pol β, allowing the incorporation of several nucleotides (lane 8). With the 1-nt gapped DNA substrate (Fig. 5B), DNA pol δ could efficiently fill the gap opposite the AP site even in the absence of PCNA and RP-A (lane 1). The addition of PCNA and RP-A allowed limited strand displacement activity, resulting in the incorporation of additional two nucleotides (lanes 2 and 3). Again, the addition of Fen-1 completely abolished strand displacement by DNA pol δ, resulting in the accumulation of +1 product only, even in the presence of PCNA and RP-A (lane 4). DNA pol β, as expected, was able to efficiently fill the gap and also to perform limited strand displacement, generating +2 products (lane 5). PCNA and RP-A further stimulated its activity, allowing +3 products to be synthesized (lanes 6 and 7). However, the addition of Fen-1 dramatically increased the strand displacement-dependent DNA synthesis by DNA pol β, with the generation of long elongation products (lane 8). Next, higher amounts of DNA pol δ (Fig. 5C) or β (Fig. 5D) were incubated on both substrates, in the presence of PCNA and RP-A, and in the absence or presence of increasing amounts of Fen-1. Under these conditions, in the absence of Fen-1, DNA pol δ showed limited strand displacement activity on both the nicked (lane 1) or the 1-nt gapped (Fig. 5C, lane 5) templates. The addition of Fen-1 caused a clear reduction of the strand displacement products (lanes 2–4 and 5–8). DNA pol β in the absence of Fen-1 also showed on both substrates limited strand displacement activity (Fig. 5D, lanes 1 and 6), which was, however, dramatically stimulated by Fen-1 in a dose-dependent manner (lanes 2–5 and 7–10). Thus, Fen-1 was able to inhibit DNA pol δ and to stimulate DNA pol β, during AP site bypass, both at the incorporation and elongation steps.
FIGURE 5.
Fen-1 influences both the incorporation and elongation steps by DNA polymerases δ and β during abasic site bypass and can physically interact with DNA polymerase β. The reactions were performed as described under “Experimental Procedures.” A, 30 nm DNA pol δ (lanes 1–4) or DNA pol β (lanes 5–8) were incubated on the nicked AP template in the absence (lanes 1 and 5) or in the presence of PCNA either alone (lanes 2 and 6) or in combination with RP-A (lanes 3 and 7) or 120 nm RP-A together with 3 nm Fen-1 (lanes 4–8). B, as in A but on the 1-nt gap AP template. C, 60 nm DNA pol δ were incubated on the nicked AP (lanes 1–4) or on the 1-nt Gap AP (lanes 5–8) templates, in the presence of a combination of equimolar amounts of PCNA and RP-A (110 nm), and in the absence (lanes 1 and 5) or in the presence (lanes 2–4 and 6–8) of increasing amounts of Fen-1. D, as in C but in the presence of 60 nm DNA pol β. E, 500 ng of Fen-1 (lanes 1, 3, and 5), BSA (lanes 2 and 4) or DNA pol β (lane 5) were incubated either in the presence (lanes 1 and 2) or in the absence (lanes 3–5) of DNA pol β. The membranes were developed either with anti-Fen-1 (membrane a) or anti-DNA pol β (membrane b) antibodies, and the signals were detected by chemiluminescence.
DNA Polymerase β Physically Interacts with Fen-1—It has been shown that both DNA pol β and Fen-1 interact with PCNA. To verify whether they also interact with each other, a Far Western blot analysis was performed. Fen-1 was immobilized on a nitrocellulose membrane, and after in situ renaturation, the membrane was incubated with a solution of 1 μg/ml DNA pol β. Development of the membrane with anti-DNA pol β antibodies revealed the presence of DNA pol β on the membrane in correspondence of the signal of Fen-1 (lane 1). Replacement of Fen-1 with the same amount of BSA (lane 2) or omission of DNA pol β from the incubation buffer (lane 3) abolished the signal of DNA pol β, suggesting that Fen-1 and DNA pol β can physically interact.
DNA Polymerase λ Cooperates Less Efficiently with Fen-1 in Abasic Site Bypass than DNA Polymerase β—DNA pol λ is another family X enzyme closely related to DNA pol β. As shown in supplemental Fig. S2 A, on both linear and gapped DNA substrates, DNA pol λ was able to synthesize full-length products in the absence (lanes 1–3 and 7–9) but not in the presence (lanes 4–6 and 10–12) of an AP site. Comparison with DNA pol β (Fig. 2A) also showed that DNA pol λ was generally less efficient on all the substrates tested (supplemental Fig. S2). Next, DNA pol λ was tested on undamaged or AP-containing 38-nt gapped DNA substrates in the absence or in the presence of Fen-1 and PCNA. As shown in supplemental Fig. S2, Fen-1 promoted strand displacement-coupled DNA synthesis on undamaged DNA (compare lane 1 with lanes 2–4). The addition of PCNA (lanes 8–11) further promoted strand displacement, leading to full-length products synthesis (compare lane 8 with lanes 9–11). In the presence of an AP site, however, DNA pol λ was stalled at the lesion (lanes 5 and 12), and Fen-1 could only promote very limited strand displacement synthesis beyond the AP site (lanes 6 and 7). The addition of PCNA enhanced the effect of Fen-1 (lanes 12–15) but did not increase the size of the elongation products made by strand displacement. Thus, DNA pol λ could cooperate with Fen-1 and PCNA in AP site bypass on gapped DNA substrates but with lower efficiency than DNA pol β (compare supplemental Fig. S2 with Fig. 4C).
DISCUSSION
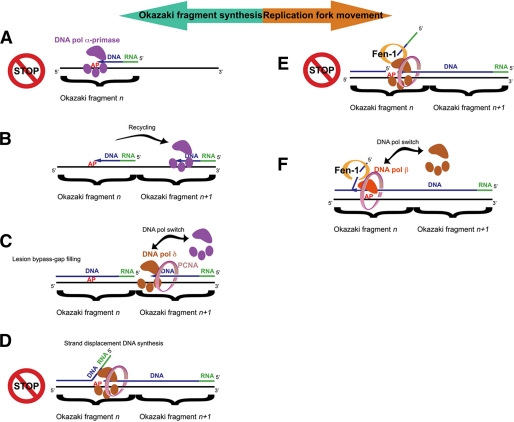
In eukaryotes, little is known on how DNA lesions differentially affect leading versus lagging strand replication and whether and how they are differentially handled during the lesion bypass (3, 18, 19). The effects of blocking lesions such as AP sites have been always viewed as the consequence of the inability of replicative DNA pols to perform translesion synthesis because of a dramatic drop of either the incorporation or the elongation rates when attempting to copy AP sites. This is certainly true in the case of leading strand replication. However, in the case of lagging strand synthesis, where about 2.5 × 107 Okazaki fragments have to be correctly replicated, the situation is much more complex (Fig. 6). The current model predicts that, after the synthesis of an RNA/DNA hybrid fragment of about 30 nt by DNA pol α/primase, DNA pol δ gets in and proceeds to synthesize the remaining 100–120 nt of a typical Okazaki fragment. When the 5′-end of the downstream fragment is reached, DNA pol δ initiates strand displacement DNA synthesis, removing the RNA/DNA hybrid synthesized by DNA pol α/primase. If an AP site is encountered by DNA pol α/primase, Okazaki fragment synthesis is impaired (Fig. 6A). Studies based on the analysis of replication products using SV40-origin containing DNA plasmids in UV-irradiated mammalian cells suggested that a blocking lesion on the lagging strand template inhibits the completion of the nascent Okazaki fragment, leaving a small gap in the lagging strand (20, 21). Synthesis of a new Okazaki fragment (fragment n+1 in Fig. 6B) is then initiated by DNA pol α/primase, whereas the gap on the damaged strand of the previous fragment (fragment n in Fig. 6C) is being filled presumably by DNA pol δ and/or β. After the polymerase switch between DNA pol α/primase and DNA pol δ has occurred, and DNA pol δ has reached the 5′-end of the downstream Okazaki fragment (fragment n in Fig. 6C), strand displacement synthesis is initiated. During this step, however, DNA pol δ will encounter the AP site still present on the template strand (Fig. 6D). Biochemical data have shown that DNA pol δ is considerably slowed down by the presence of an AP site on the template strand (4, 22). Thus, it is conceivable that strand displacement synthesis will be paused at the site of the lesion, enabling Fen-1 to process the generated Flap (Fig. 6E). The data presented in this work suggest that the resulting intermediate, where the AP site is placed at the ss/dsDNA junction, constitutes an impassable block for DNA pol δ, even in the presence of PCNA and RP-A. This potentially very dangerous situation can be resolved through a special polymerase switch, whereby the repair enzyme DNA pol β can replace DNA pol δ and, in coordination with Fen-1, continues the strand displacement synthesis, thus bypassing the lesion (Fig. 6F). Fen-1 appears to be absolutely required for this reaction, and the fact that Fen-1 and DNA pol β physically interact strengthen this. A stimulatory effect of Fen-1 on the activity of DNA pol β has been already shown to take place in the context of BER reactions (23). Because both of these enzymes also interact with PCNA (24), it is possible to envisage a coordinated trade-off mechanism, where pausing of DNA pol δ allows the recruitment of Fen-1 to the Flap through interaction with PCNA. Then DNA pol β is recruited by physical interaction with Fen-1 and PCNA, thus replacing DNA pol δ. As shown in Fig. 2 (B and C), PCNA promoted AP site bypass by DNA pol β. This is the first time that a functional effect for the physical interaction between DNA pol β and PCNA is reported (25, 26). In summary, our data suggest that, because the first 30 nt of each Okazaki fragment have to be copied twice, first by DNA pol α/primase and subsequently by DNA pol δ in the context of strand displacement synthesis, the presence of an AP site in this tract will cause a double block that must be resolved by two distinct mechanisms. Thus, our data identify a previously unnoticed deleterious effect of the AP site lesion on normal cell metabolism and suggest the existence of a novel repair pathway that might be important in preventing replication fork stalling.
FIGURE 6.
Model for the bypass of the replication block imposed by an abasic site on the lagging strand of the replication fork. Because of the antiparallel nature of the DNA double helix, the apparent direction of lagging strand synthesis (green arrow) is opposite to the direction of replication fork advancement (orange arrow). For simplicity only the relevant proteins are shown. For details see text.
Supplementary Material
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures“ and Figs. S1 and S2.
Footnotes
The abbreviations used are: BER, base excision repair; AP, abasic; pol, polymerase; PCNA, proliferating cell nuclear antigen; Fen-1, Flap endonuclease 1; RP-A, replication protein A; BSA, bovine serum albumin; nt, nucleotide(s); ss, single-stranded; ds, double-stranded.
References
- 1.Lindahl, T. (1993) Nature 362 709-715 [DOI] [PubMed] [Google Scholar]
- 2.Shibutani, S., Takeshita, M., and Grollman, A. P. (1997) J. Biol. Chem. 272 13916-13922 [DOI] [PubMed] [Google Scholar]
- 3.Higuchi, K., Katayama, T., Iwai, S., Hidaka, M., Horiuchi, T., and Maki, H. (2003) Genes Cells 8 437-449 [DOI] [PubMed] [Google Scholar]
- 4.Mozzherin, D. J., Shibutani, S., Tan, C. K., Downey, K. M., and Fisher, P. A. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 6126-6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanca, G., Villani, G., Shevelev, I., Ramadan, K., Spadari, S., Hubscher, U., and Maga, G. (2004) Biochemistry 43 11605-11615 [DOI] [PubMed] [Google Scholar]
- 6.Nick McElhinny, S. A., Gordenin, D. A., Stith, C. M., Burgers, P. M., and Kunkel, T. A. (2008) Mol. Cell 30 137-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bae, S. H., and Seo, Y. S. (2000) J. Biol. Chem. 275 38022-38031 [DOI] [PubMed] [Google Scholar]
- 8.Maga, G., Villani, G., Tillement, V., Stucki, M., Locatelli, G. A., Frouin, I., Spadari, S., and Hubscher, U. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14298-14303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossi, M. L., and Bambara, R. A. (2006) J. Biol. Chem. 281 26051-26061 [DOI] [PubMed] [Google Scholar]
- 10.Stucki, M., Pascucci, B., Parlanti, E., Fortini, P., Wilson, S. H., Hubscher, U., and Dogliotti, E. (1998) Oncogene 17 835-843 [DOI] [PubMed] [Google Scholar]
- 11.Pascucci, B., Stucki, M., Jónsson, Z. O., Dogliotti, E., and Hübscher, U. (1999) J. Biol. Chem. 274 3040-3046 [DOI] [PubMed] [Google Scholar]
- 12.Podust, V. N., Chang, L. S., Ott, R., Dianov, G. L., and Fanning, E. (2002) J. Biol. Chem. 277 3894-3901 [DOI] [PubMed] [Google Scholar]
- 13.Maga, G., Crespan E., Wimmer, U., van Loon, B., Amoroso, A., Mondello, C., Belgiovine, C., Ferrari, E., Locatelli, G., Villani, G., and Hübscher, U. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 20689-20694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonsson, Z. O., Hindges, R., and Hubscher, U. (1998) EMBO J. 17 2412-2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henricksen, L. A., Umbricht, C. B., and Wold, M. S. (1994) J. Biol. Chem. 269 11121-11132 [PubMed] [Google Scholar]
- 16.Stucki, M., Jonsson, Z. O., and Hübscher, U. (2001) J. Biol. Chem. 276 7843-7849 [DOI] [PubMed] [Google Scholar]
- 17.Beard, W. A., Prasad, R., and Wilson, S. H. (2006) Methods Enzymol. 408 91-107 [DOI] [PubMed] [Google Scholar]
- 18.Nikolaishvili-Feinberg, N., and Cordeiro-Stone, M. (2001) Biochemistry 40 15215-15223 [DOI] [PubMed] [Google Scholar]
- 19.Cordeiro-Stone, M., and Nikolaishvili-Feinberg, N. (2002) Mutat. Res. 510 91-106 [DOI] [PubMed] [Google Scholar]
- 20.Cordeiro-Stone, M., Zaritskaya, L. S., Price, L. K., and Kaufmann, W. K. (1997) J. Biol. Chem. 272 13945-13954 [DOI] [PubMed] [Google Scholar]
- 21.Mezzina, M., Menck, C. F., Courtin, P., and Sarasin, A. (1988) J. Virol. 62 4249-4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maga, G., Villani, G., Ramadan, K., Shevelev, I., Tanguy Le Gac, N., Blanco, L., Blanca, G., Spadari, S., and Hubscher, U. (2002) J. Biol. Chem. 277 48434-48440 [DOI] [PubMed] [Google Scholar]
- 23.Liu, Y., Beard, W. A., Shock, D. D., Prasad, R., Hou, E. W., and Wilson, S. H. (2005) J. Biol. Chem. 280 3665-3674 [DOI] [PubMed] [Google Scholar]
- 24.Maga, G., and Hubscher, U. (2003) J. Cell Sci. 116 3051-3060 [DOI] [PubMed] [Google Scholar]
- 25.Kedar, P. S., Kim, S. J., Robertson, A., Hou, E., Prasad, R., Horton, J. K., and Wilson, S. H. (2002) J. Biol. Chem. 277 31115-31123 [DOI] [PubMed] [Google Scholar]
- 26.Maga, G., Villani, G., Crespan, E., Wimmer, U., Ferrari, E., Bertocci, B., and Hübscher, U. (2007) Nature 447 606-608 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.