Abstract
The multifunctional protein encoded by gene 4 of bacteriophage T7 (gp4) provides both helicase and primase activity at the replication fork. T7 DNA helicase preferentially utilizes dTTP to unwind duplex DNA in vitro but also hydrolyzes other nucleotides, some of which do not support helicase activity. Very little is known regarding the architecture of the nucleotide binding site in determining nucleotide specificity. Crystal structures of the T7 helicase domain with bound dATP or dTTP identified Arg-363 and Arg-504 as potential determinants of the specificity for dATP and dTTP. Arg-363 is in close proximity to the sugar of the bound dATP, whereas Arg-504 makes a hydrogen bridge with the base of bound dTTP. T7 helicase has a serine at position 319, whereas bacterial helicases that use rATP have a threonine in the comparable position. Therefore, in the present study we have examined the role of these residues (Arg-363, Arg-504, and Ser-319) in determining nucleotide specificity. Our results show that Arg-363 is responsible for dATP, dCTP, and dGTP hydrolysis, whereas Arg-504 and Ser-319 confer dTTP specificity. Helicase-R504A hydrolyzes dCTP far better than wild-type helicase, and the hydrolysis of dCTP fuels unwinding of DNA. Substitution of threonine for serine 319 reduces the rate of hydrolysis of dTTP without affecting the rate of dATP hydrolysis. We propose that different nucleotides bind to the nucleotide binding site of T7 helicase by an induced fit mechanism. We also present evidence that T7 helicase uses the energy derived from the hydrolysis of dATP in addition to dTTP for mediating DNA unwinding.
Helicases are molecular machines that translocate unidirectionally along single-stranded nucleic acids using the energy derived from nucleotide hydrolysis (1–3). The gene 4 protein encoded by bacteriophage T7 consists of a helicase domain and a primase domain, located in the C-terminal and N-terminal halves of the protein, respectively (4). The T7 helicase functions as a hexamer and has been used as a model to study ring-shaped replicative helicases. In the presence of dTTP, T7 helicase binds to single-stranded DNA (ssDNA)3 as a hexamer and translocates 5′ to 3′ along the DNA strand using the energy of hydrolysis of dTTP (5–7). T7 helicase hydrolyzes a variety of ribo and deoxyribonucleotides; however, dTTP hydrolysis is optimally coupled to DNA unwinding (5).
Most hexameric helicases use rATP to fuel translocation and unwind DNA (3). T7 helicase does hydrolyze rATP but with a 20-fold higher Km as compared with dTTP (5, 8). It has been suggested that T7 helicase actually uses rATP in vivo where the concentration of rATP is 20-fold that of dTTP in the Escherichia coli cell (8). However, hydrolysis of rATP, even at optimal concentrations, is poorly coupled to translocation and unwinding of DNA (9). Other ribonucleotides (rCTP, rGTP, and rUTP) are either not hydrolyzed or the poor hydrolysis observed is not coupled to DNA unwinding (8). Furthermore, Patel et al. (10) found that the form of T7 helicase found in vivo, an equimolar mixture of the full-length gp4 and a truncated form lacking the zinc binding domain of the primase, prefers dTTP and dATP. Therefore, in the present study we have restricted our examination of nucleotides to the deoxyribonucleotides.
The nucleotide binding site of the replicative DNA helicases, such as T7 gene 4 protein, bind nucleotides at the subunit interface (Fig. 1) located between two RecA-like subdomains that bind ATP (11, 12). The location of the nucleotide binding site at the subunit interface provides multiple interactions of residues with the bound NTP. A number of cis- and trans-acting amino acids stabilize the bound nucleotide in the nucleotide binding site and also provide for communication between subunits (13–15). Earlier reports revealed that the arginine finger (Arg-522) in T7 helicase is positioned to interact with the γ-phosphate of the bound nucleotide in the adjacent subunit (12, 16). However, His-465 (phosphate sensor), Glu-343 (catalytic base), and Asp-424 (Walker motif B) interacts with the γ-phosphate of the bound nucleotide in the same subunit (12, 17, 18). The arginine finger and the phosphate sensor have been proposed to couple NTP hydrolysis to DNA unwinding. Substitution of Glu-343, the catalytic base, eliminates dTTP hydrolysis (19), and substitution of Asp-424 with Asn leads to a severe reduction in dTTP hydrolysis (20). The conserved Lys-318 in Walker motif A interacts with the β-phosphate of the bound nucleotide and plays an important role in dTTP hydrolysis (21).
FIGURE 1.
Crystal structure of T7 helicase. A, crystal structure of the hexameric helicase C-terminal domain of gp4 (17). The structure reveals a ring-shaped molecule with a central core through which ssDNA passes. The inset shows the interface between two subunits of the helicase with adenosine 5′-{β,γ-imidol}-triphosphate in the nucleotide binding site. B, the nucleotide binding site of a monomer of the gp4 with the crucial amino acid residues reported earlier and in the present study is shown in sticks. The crystal structures of the T7 gene 4 helicase domain (12) with bound dTTP (C) and dATP (D). The structures shown are the nucleotide binding site of T7 helicase as viewed in Pymol by analyzing the PDB files 1cr1 and 1cr2 (12). Arg-504 and Tyr-535 sandwiches the base of the bound dNTP. Additionally, Arg-504 forms a hydrogen bridge with dTTP. Arg-363 interacts specifically with the 3-OH group of bound dATP. AMPPNP, adenosine 5′-(β,γ-imino)triphosphate.
Considering the wealth of information on the above residues that are involved in the hydrolysis of dTTP and the coupling of hydrolysis to unwinding, it is intriguing that little information is available on nucleotide specificity. Several crystal structures of T7 helicase in complex with a nucleotide triphosphate are available. However, most of structures were crystallized with a non-hydrolyzable analogue of dTTP or the nucleotide was diffused into the crystal. The crystal structure of the T7 helicase domain bound with dTTP or dATP was reported by Sawaya et al. (12). These structures assisted us in identifying two basic residues (Arg-363 and Arg-504) in close proximity to the sugar and base of the bound nucleotide whose orientation suggested that these residues could be involved in nucleotide selection. Arg-504 together with Tyr-535 sandwich the base of the bound nucleotide at the subunit interface of the hexameric helicase (Fig. 1). Arg-504 and Tyr-535 are structurally well conserved in various helicases (12). However, Arg-504 could make a hydrogen bridge with the OH group of thymidine, thus suggesting a role in dTTP specificity. On the other hand, Arg-363 is in close proximity (∼3.4 Å) to the sugar 3′-OH of bound dATP, whereas in the dTTP-bound structure this residue is displaced by 7.12 Å (Fig. 1) from the equivalent position. Consequently Arg-363 could play a role in dATP binding. The crystal structures do not provide any information on different interaction of residues with the phosphates of dATP and dTTP. However, alignment of the residues in the P-loops of different hexameric helicases reveals that the serine adjacent to the invariant lysine at position 319 (Ser-319) is conserved in bacteriophages, whereas bacterial helicases have a conserved threonine in the equivalent position (supplemental Fig. 1). Bacterial helicases use rATP in the DNA unwinding reactions. whereas T7 helicase preferentially uses dTTP, and bacteriophage T4 gene 41 uses rGTP or rATP (22).
Although considerable information is available on the role of residues in nucleotide binding and dTTP hydrolysis, very little is known on the determinants of nucleotide specificity. In the present study we made an attempt to address the role of a few selected residues (Arg-363, Arg-504, and Ser-319) in determining nucleotide specificity, especially dTTP and dATP, both of which are hydrolyzed and mediate DNA unwinding. We show that under physiological conditions T7 helicase uses the energy derived from the hydrolysis of dATP in addition to dTTP for mediating DNA unwinding.
EXPERIMENTAL PROCEDURES
Materials
E. coli DH5α (Novagen) was used for in vivo complementation studies. E. coli HMS174 (DE3) (Novagen) was used as the host strain to express T7 gene 4 and to purify wild-type and variants of gp4. Wild-type and mutant gene 4 are expressed from pET11b plasmid (Novagen) containing a T7 RNA polymerase promoter. Wild-type and gene 4-deficient T7 phage (T7Δ4-1) were from the laboratory collection. Oligonucleotides used in the assay to measure DNA binding (50-mer), to prepare helicase substrates (75- and 95-mer), and for mutagenesis of gene 4 were purchased from Integrated DNA technologies. M13 ssDNA and T4 polynucleotide kinase were purchased from New England Biolabs. Site-directed mutagenesis kit was purchased from Stratagene. Masateru Takahashi (Harvard Medical School) supplied T7 DNA polymerase. T7 gene 5 protein (gp5) forms a 1:1 complex with its processivity factor, E. coli thioredoxin. All of the chemicals and reagents were from Sigma unless otherwise mentioned.
Methods
Site-directed Mutagenesis, Protein Overproduction, and Purification—Plasmid pET11gp4–63 was used for expression and overproduction of wild-type gp4 (23). This plasmid was also used to create point mutations by using QuikChange II mutagenesis kit (Stratagene) in accordance with the manufacturer's instructions. The sequence of primers used to construct various single and double mutations are available on request. Mutations were confirmed by DNA sequencing. Constructs harboring wild-type or mutated gene 4 were transferred to E. coli strain HMS-174 (DE3) for overproduction of the corresponding proteins. Wild-type and altered gene 4 proteins were purified as described earlier (24).
Phage Complementation Assay—E. coli Dh5α containing a plasmid (pET11gp4–63 or its derivatives constructed in this study) that express gene 4 protein under a T7 promoter was grown to an A600 of 1. Serially diluted T7 phage stocks were mixed with an aliquot of the E. coli culture in 0.7% soft agar and poured onto LB plates. After an incubation of 4–6 h at 37 °C, the number of plaques appearing on the plate was determined. The number of plaques formed by plasmids harboring the wild-type gene 4 protein is normalized to 1. The relative efficiency of plating obtained with the mutated gene 4 constructs were determined by the number of plaques (PFU) formed by the mutated gene 4 constructs divided by the PFU of wild-type gene 4.
Nucleotide Hydrolysis Assay—Nucleotide hydrolysis was carried out at 37 °C in a reaction mixture containing 40 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 10 mm DTT, 50 mm potassium glutamate, 100 nm wild-type or altered gp4, 1 nm M13 ssDNA and different concentrations of [α-32P]dTTP, [α-32P]dATP, [α-32P]dGTP, or [α-32P]dCTP. After incubation for 30 min at 37 °C, EDTA at a final concentration of 250 mm was added to stop the reaction, and the reaction sample was kept on ice until it was spotted on TLC. [α-32P]dNDP, the product of hydrolysis, was separated from [α-32P]dNTP on TLC plates coated with polyethylene imine in a solvent containing 0.5 m formic acid and 0.5 m lithium chloride. The TLC plates were scanned by phosphorimaging (Fuji) and measured radiolabeled spots by the help of image quant software (Fuji). The data were further analyzed in GraphPad Prism software.
Double-stranded DNA Unwinding Assay—Displacement of a 5′-32P-labeled 75-mer oligonucleotide that is partially annealed to a 95-mer oligonucleotide by gp4 was measured. The helicase substrate was prepared by annealing a 5′-32P end-labeled 75-mer oligo (5′-CGC CGG GTA CCG AGC TCG AAT TCA CTG GCC GTC GTT TTA CAA CGT CGT GAC ATG CCT19-3′) with an unlabeled 95-mer oligo (5′-39T GGC ATG TCA CGA CGT TGT AAA ACG ACG GCC AGT GAA TTC GAG CTC GGT ACC CGG CG-3′). Reaction mixtures containing 100 nm labeled DNA substrate, 40 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 10 mm DTT, 50 mm potassium glutamate, 5 mm indicated nucleotides, and 100 nm wild-type or altered gp4 were incubated at 37 °C for different time periods. An aliquot of the reaction mixture was mixed with 5× stop buffer containing 0.4% (w/v) SDS, 40 mm EDTA, 8% (v/v) glycerol, and 0.1% (w/v) bromphenol blue to the final concentration. Unwound single-stranded oligonucleotides were separated from the helicase substrate in a 10% nondenaturing polyacrylamide gel. Subsequently, the gel was scanned by phosphorimaging and analyzed in Image gauge and GraphPad prism software.
ssDNA Binding Assay—A nitrocellulose filter binding assay was used for determining the ssDNA binding ability of wild-type or altered gp4. The reaction mixture (20 μl) containing 1 nm 5′-32P-labeled oligonucleotide (5′-ATG ACC ATG ATT TCG ACG TTT TTT TTT TTG GGG ATC CTC TAA CCT GCG CA-3′), 40 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 10 mm DTT, 50 mm potassium glutamate, 5 mm β,γ-methylene dTTP, and different concentrations of wild-type or altered gp4 was incubated at 37 °C for 30 min. Then the reaction mixture was filtered through a nitrocellulose membrane laid above a zeta probe membrane in a dot-blot filtration apparatus. The quantity of protein bound single-stranded DNA and free single-stranded DNA was measured by scanning the nitrocellulose and zeta probe membrane, respectively, by phosphorimaging.
Protein Oligomerization Assay— gp4 variants were examined for their ability to form hexamers in presence of non-hydrolysable dTTP analog (β,γ-methylene dTTP). The reaction mixture (20 μl) containing 1 μm wild-type or altered gp4, 40 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 10 mm DTT, 50 mm potassium glutamate, 5 mm β,γ-methylene dTTP was incubated for 20 min at 37 °C followed by the addition of the glutaraldehyde up to 0.033%, v/v. The reaction sample was kept in 37 °C for another 5 min, and then the reaction products were analyzed on a non-denaturing 10% polyacrylamide gel with a running buffer of 1× Tris borate-EDTA. After staining with Coomassie Blue, oligomerization of gp4 was verified by protein bands corresponding to markers greater than 250 kDa (25).
Strand Displacement DNA Synthesis Assay—M13 circular double-stranded having a 5′ tail was used to monitor strand displacement DNA synthesis. A replication fork was constructed by annealing M13 ssDNA to an oligonucleotide (5′-36TAATTCGTAATCATCATGGTCATAGCTGTTTCCT-3′) having 30 bases complementary to the M13 ssDNA and 36 bases forming a 5′-tail. Then gp5/thioredoxin (trx) was used to convert the ssDNA circle to double-stranded DNA circle. Phenol/chloroform was used to remove gp5/trx. Strand-displacement synthesis assay was carried out at 37 °C in a reaction mixture containing 40 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 10 mm DTT, 50 mm potassium glutamate, 10 nm gp5/trx, 10 nm wild-type or altered gp4 (hexamer), 500 μm each of dATP, [α-32P]dTTP (10 Ci/mmol), dCTP, and dGTP, and 10 nm M13 double-stranded DNA. After 10 min of incubation, the reaction was stopped with EDTA at a final concentration of 25 mm, and then the mixture was spotted onto a DE 81 filter paper. After washing 3 times with 0.3 m ammonium formate and 100% ethanol, incorporated [32P]dTMP was measured in a liquid scintillating counter.
RESULTS
Arginine at Position 363 Effects Specificity for dATP—Arg-363 is located in the nucleotide binding site of T7 helicase (Fig. 1). A crystal structure of the T7 helicase domain bound to dATP reveals an interaction of the deoxyribose of dATP (Fig. 1) with Arg-363, whereas no interaction was observed in the structure with dTTP. To explore the role of this residue in nucleotide specificity, we replaced Arg-363 with alanine to create helicase-R363A. The construct was expressed in E. coli HMS174 (DE3), and the overproduced protein was purified as described earlier (24). The altered protein was assayed for its ability to hydrolyze nucleotides in the presence of ssDNA and to catalyze the unwinding of duplex DNA.
In contrast to wild-type helicase, helicase-R363A does not hydrolyze dATP in the presence of ssDNA (Table 1). However, it does retain 90% (Vmax 11.4 μm·S-1) of the wild-type helicase level (Vmax 12.5 μm·S-1) of dTTP hydrolysis. The measured Km (2 mm) for dTTP for the altered protein was comparable to that of wild-type helicase (Table 1). Helicase-R363A did not hydrolyze dCTP and dGTP. As shown in Fig. 2A, wild-type helicase unwinds DNA using either dTTP or dATP (10, 26). In this assay helicase activity was measured by the ability of the enzyme to displace a 5′-32P-labeled 75-mer oligonucleotide annealed to a 95-mer oligonucleotide. Only 55 nucleotides of the duplex are complementary, leaving two ssDNA tails at one end to provide a binding site for the helicase (see the inset to Fig. 2). Helicase-R363A unwound the duplex DNA only in the presence of dTTP (Fig. 2B). As expected, in view of its inability to hydrolyze dATP, dATP could not support DNA unwinding. Helicase-R363A also could not utilize dGTP or dCTP in the unwinding reactions. The results show that Arg-363 is essential for the hydrolysis of dATP, dGTP, and dCTP.
TABLE 1.
Kinetic parameters of nucleotide hydrolysis by wild-type helicase and the helicase variants The maximal rates (Vmax) of hydrolysis of each dNTP (dTTP, dATP, dGTP, and dCTP) in the presence of ssDNA by wild-type helicase and helicase variants are compared. The substrate affinity (Km) of the proteins for each of the dNTP during the half-maximal rate was determined by the Michaelis-Menten equation using Graphad Prism software. Nucleotide hydrolysis reactions were performed as described under “Experimental Procedures.” The error bars represent the results from three independent experiments. The Vmax for dATP, dGTP, or dCTP hydrolysis by helicase-R363A was measured to be less than 0.02 μm·s–1; thus, Km for the nucleotide substrates could not be determined.
|
Helicase variants
|
dTTP
|
dATP
|
dGTP
|
dCTP
|
||||
|---|---|---|---|---|---|---|---|---|
| Vmax | Km | Vmax | Km | Vmax | Km | Vmax | Km | |
| μm/s | mm | μm/s | mm | μm/s | mm | μm/s | mm | |
| Wild type | 12.5 ± 0.6 | 1.8 ± 0.2 | 6.9 ± 0.8 | 0.7 ± 0.2 | 1.5 ± 0.2 | 1.1 ± 0.5 | 1.4 ± 0.2 | 9.2 ± 2.3 |
| R363A | 11.4 ± 1.4 | 2.0 ± 0.7 | <0.02 | <0.02 | <0.02 | |||
| R504A | 6.1 ± 0.4 | 1.9 ± 0.4 | 5.3 ± 0.5 | 0.9 ± 0.2 | 0.7 ± 0.2 | 1.6 ± 1.2 | 7.9 ± 1.4 | 4.1 ± 1.6 |
| S319T | 5.4 ± 0.5 | 1.6 ± 0.4 | 8.2 ± 0.9 | 1.8 ± 0.6 | 3.7 ± 0.6 | 0.8 ± 0.4 | 3.4 ± 0.5 | 1.7 ± 0.7 |
| R504A/S319T | 1.6 ± 0.1 | 0.6 ± 0.2 | 1.4 ± 0.1 | 0.2 ± 0.1 | 0.2 ± 0.02 | 5.2 ± 0.8 | 1.9 ± 0.2 | 1.1 ± 0.4 |
FIGURE 2.
Substitution of Arg-363 with alanine effects dATP, dCTP, and dGTP usage in unwinding DNA. DNA unwinding activity of wild-type helicase (A) and helicase-R363A (B). Reactions for measuring the unwinding of DNA were carried out as described under “Experimental Procedures.” The reaction contained a 5′-32P-labeled 75-mer oligonucleotide annealed to a 95-mer oligonucleotide (see the inset), a 5 mm concentration of the indicated dNTP, and 100 nm helicase. After incubation at 37 °C, aliquots of the reaction were removed at the indicated time of incubation. The products were then separated on 10% non-denaturing polyacrylamide gels in 1× Tris borate-EDTA buffer. The amount of labeled single-stranded oligonucleotide separated from the duplex DNA substrate in each case was calculated and expressed as percent of total helicase substrate. Results are the average of three experiments.
Arginine at Position 504 Couples dCTP Hydrolysis to DNA Unwinding—Arg-504 in T7 helicase is a conserved residue in the nucleotide binding sites of hexameric helicases (12). A crystal structure of the T7 helicase domain bound to dTTP shows Arg-504 forming a hydrogen bond with the hydroxyl group of the base. No interaction of Arg-504 with dATP is observed in a structure with dATP (Fig. 1). To explore the role of Arg-504 in dTTP specificity, we replaced Arg-504 with alanine to create helicase-R504A. The altered protein was overexpressed and purified as described for helicase-R363A.
Helicase-R504A hydrolyzes dTTP at only 50% (6 μm·S-1) that of the rate of wild-type helicase but retains 75% (5.3 μm·S-1) that of the wild-type level of dATP hydrolysis. The measured Km values for dTTP (1.9 mm) or dATP (0.9 mm) for the altered protein are comparable with that of wild-type helicase (Table 1). Surprisingly, helicase-R504A hydrolyzes dCTP (Vmax 7.9 μm·S-1) far better than wild-type helicase with a 2-fold reduction in Km (4 mm); it has a comparable level of dGTP hydrolysis. The ability of the altered helicase to unwind duplex DNA was measured using the DNA substrate and assay conditions described in Fig. 2. Helicase-R504A can couple dTTP or dATP hydrolysis to DNA unwinding (Fig. 3). In contrast to wild-type helicase, where optimal unwinding is observed with dTTP, helicase-R504A uses dATP and dTTP equally well. This result most likely is reflected in its almost equal rates of ssDNA-dependent hydrolysis of these two nucleotides (Table 1). Interestingly, helicase-R504A efficiently couples dCTP hydrolysis to unwinding of the duplex DNA, unlike wild-type helicase, where no unwinding is seen with dCTP. The results suggest that Arg-504 prevents the helicase from coupling the dCTP hydrolysis to DNA unwinding. We have also replaced both of the arginines at position 363 and 504 with alanine. The altered helicase-R363A/R504A cannot hydrolyze any of the four dNTPs. Not surprisingly, helicase-R363A/R504A did not catalyze DNA unwinding activity with any of the dNTPs (data not shown).
FIGURE 3.
Coupling of nucleotide hydrolysis and DNA unwinding by helicase-R504A. The ability of the helicase-R504A to unwind duplex DNA was measured with the indicated dNTPs (5 mm) as described in Fig. 2. The error bars represent the S.D. of three independent experiments.
Replacement of the T7 Unique Serine 319 in the P-loop with Threonine—Ser-319 in the T7 helicase is a conserved amino acid found in the phosphate binding loop (P-loop) of many bacteriophage helicases (supplemental Fig. 1). In the T7 helicase the hydroxyl group of Ser-319 interacts with the β-phosphate of the bound nucleotide with a hydrogen bridge. Ser-319 also contacts the γ-phosphate by coordinating the bound magnesium ion in the nucleotide binding site (12, 17). Substitution of Ser-319 with alanine inactivates ssDNA-dependent nucleotide hydrolysis and DNA unwinding activity (data not shown). Because most bacterial helicases have a threonine in the equivalent position of Ser-319 in the T7 helicase, we replaced the serine with a threonine. The altered helicase (helicase-S319T) hydrolyzes dATP (Vmax-8.2 μm·S-1) at levels comparable with wild-type T7 helicase (Vmax-6.9 μm·S-1). However, helicase-S319T hydrolyzes dTTP at 40% (Vmax-5.4 μm·S-1) that observed with wild-type helicase (Vmax 12.5 μm·S-1); the Km for dTTP (1.6 mm) was unchanged from that observed with wild-type helicase (Table 1). Helicase-S319T hydrolyzed dGTP and dCTP nearly 2-fold faster as compared with wild-type helicase. Helicase-S319T also differed from wild-type helicase in that the affinity (Km-1.7 mm) for dCTP was greater than that observed with the wild-type helicase (Km 9.2 mm). As shown in Fig. 2A wild-type helicase unwinds DNA using either dTTP or dATP but not dGTP or dCTP. Helicase-S319T also unwinds DNA in the presence of dTTP and dATP, but unlike wild-type helicase, it is also active, albeit less so, with dGTP or dCTP (Fig. 4A). Our results show that changing the hydroxyl bearing residue in the P-loop of T7 helicase from a serine to a threonine enables it to use dCTP and dGTP to mediate the unwinding of duplex DNA.
FIGURE 4.
DNA unwinding activity of helicase-S319T (A) and helicase-R504A/S319T (B). Reactions for measuring the unwinding of DNA by helicase-S319T and helicase-R504A/S319T in the presence of the indicated dNTPs (5 mm) were carried out as described for Fig. 2.
Helicase-S319T and helicase-R504A both hydrolyze dTTP 50% as efficiently as does wild-type helicase, with the ability to hydrolyze dATP unchanged. The results suggest that both Arg-504 and Ser-319 play a role in dTTP specificity. Consequently, we constructed a helicase, helicase-R504A/S319T, in which both of these residues were altered. Helicase-R504A/S319T hydrolyzes dTTP ∼ 10-fold less (Vmax 1.6 μm·S-1) than that observed with the wild-type protein (Table 1). The DNA unwinding activity with dTTP is also 10–15% that found with wild-type helicase (Fig. 4B). Like helicase-R504A and helicase-S319T, helicase-R504A/S319T can also use dCTP to mediate unwinding (Fig. 4B). The rate of dATP hydrolysis (Vmax 1.4 μm·S-1) is reduced up to 5-fold as compared with wild-type helicase. However, helicase-R504A/S319T unwinds duplex DNA 3-fold more efficiently with dATP than with dTTP or dCTP. dGTP is hydrolyzed poorly by helicase-R504A/S319T, and this nucleotide cannot support unwinding of DNA.
Effect of dATP, dGTP, and dCTP on the dTTP-coupled Unwinding of DNA—In vivo the four dNTPs are present at comparable levels in a range of concentrations from 0.1 to 0.3 mm (27) and consequently are available to T7 DNA helicase, which is capable of catalyzing the hydrolysis of all four. dTTP, however, is far more efficient at mediating the unwinding of duplex DNA as shown here and previously (23, 26). The hexameric structure of the helicase and the sequential activation of nucleotide binding sites as the DNA is transferred from one subunit to another raises a number of questions (19). In the absence of ssDNA all six nucleotide binding sites are randomly, but simultaneously, hydrolyzing nucleotides, albeit at a greatly reduced rate. In that all four dNTPs are available to the helicase in vivo, it seems likely that some of these sites will be occupied by nucleotides other than dTTP. The identity of the nucleotides accommodated in the six nucleotide binding sites is difficult to address, but the availability of helicase-R363A, helicase-R504A, and helicase-R504A/S319T provides an approach to dissecting this question. dATP is not hydrolyzed by helicase-R363A yet dTTP is hydrolyzed as well as that observed with wild-type helicase; hydrolysis of dTTP is coupled to the unwinding of DNA (Fig. 2B). On the other hand, helicase-R504A and helicase-R504A/S319T utilize dTTP poorly to fuel DNA unwinding as compared with dATP (Figs. 3 and 4B).
In the experiment shown in Fig. 5, the effect of adding dATP, dGTP, or dCTP to helicases using dTTP to unwind DNA was monitored. The DNA unwinding activity of wild-type helicase, helicase-R363A, helicase-R504A, and helicase-R504A/S319T was monitored for 5 min in the presence of dTTP with or without the addition of different concentrations of dNTP. The addition of dATP to reactions mediated by helicase-R363A inhibited unwinding by amounts proportional to the dATP present in the reaction (Fig. 5, A and B). In striking contrast, the addition of dATP to reactions with helicase-R504A or helicase-R504A/S319T markedly stimulated the reaction. Wild-type helicase was also stimulated by dATP but only by 20%. These results show that dTTP and dATP both engage one or more of the six-nucleotide binding sites of the hexameric helicase.
FIGURE 5.
Effect of dATP, dGTP, and dCTP on dTTP-dependent DNA unwinding activity. A, autoradiogram of the products of unwinding assays mediated by the indicated helicase. The indicated gene 4 proteins (100 nm) were incubated with the DNA helicase substrate described in Fig. 2 in a reaction containing 1 mm dTTP and the indicated amounts of dATP (0, 0.5, 1, 2, and 5 mm). In each case the reaction was first carried out with 1 mm dTTP with the indicated proteins (100 nm) for 30 s followed by the addition of the different concentrations of dATP. The reaction was further incubated for 5 min at 37 °C. Reaction products were separated on 10% non-denaturing polyacrylamide gel using 1× Tris borate-EDTA buffer. ds, double-stranded; ss, single-stranded. B–D, the graphs in B, C, and D show the quantitative analyses of the DNA unwinding activities of wild-type helicase, helicase-R363A, helicase-R504A, and helicase-R504A/S319T in the presence of 1 mm dTTP and the indicated amounts (0, 0.5, 1, 2, and 5 mm) of dATP, dGTP, and dCTP. The percentage of duplex DNA (100 nm) unwound by wild-type helicase (65%), helicase-R363A (30%), helicase-R504A (20%), and helicase-R504A/S319T (16%) in the presence of 1 mm dTTP after 5 min of incubation at 37 °C was normalized to 1 for the corresponding protein. The relative activity for each of the proteins upon the addition of dATP, dGTP, or dCTP was calculated. The reactions were carried out as described above for panel A. The results were analyzed by using Image gauge and GraphPad Prism software. The error bars represent the S.D. from three independent experiments.
Interestingly, the addition of dGTP or dCTP also increases DNA unwinding by ∼20% (Fig. 5, C and D), suggesting that dGTP and dCTP hydrolysis also can be coupled to DNA unwinding when dTTP is present. The addition of dGTP and dCTP marginally inhibits the dTTP-dependent DNA unwinding activity of helicase-R363A. However, this inhibition most likely derives from the observation that helicase-R363A does not hydrolyze dGTP and dCTP. Helicase-R504A, which does hydrolyze dGTP and dCTP, is stimulated by these nucleotides (Fig. 5, C and D). Helicase-R504A/S319T hydrolyzes dGTP at a very low rate, and dGTP has essentially no effect on unwinding of DNA by this helicase in the presence of dTTP. These results suggest that dNTPs freely exchange in the nucleotide binding sites of the hexameric T7 helicase. Our results also suggest that nucleotides that can be hydrolyzed in vitro by the helicase can be used by the protein for DNA unwinding in the presence of the preferred nucleotide such as dTTP for wild-type helicase.
Both dATP and dTTP Hydrolysis by T7 Helicase Are Required for T7 Phage Growth—T7Δ4, a phage in which the gene 4 coding region has been deleted, can grow only in E. coli cells harboring a plasmid that produces a functional gene 4 protein (28). T7Δ4, thus, provides a means to determine whether a genetically altered gene 4 protein can substitute for the wild-type protein to provide for T7 DNA replication and phage growth (28). Plasmids expressing the altered gene 4 were transformed into E. coli DH5-α. The host cells were then infected with T7Δ4, and the efficiency of plating was determined as described under “Experimental Procedures” (Table 2). Helicase-R363A, devoid of DNA-dependent dATP hydrolysis but with normal dTTP hydrolysis in vitro, supports the growth of T7Δ4 poorly (2.5% that observed with wild-type helicase) with smaller plaques. Plasmids encoding either helicase-S319T or helicase-R504A support the growth of T7Δ4 phage nearly as well, 60 and 100% efficiency, respectively, as does a plasmid encoding the wild-type helicase. Helicase-R504A/S319T, which lost up to 90% of its ability to couple dTTP hydrolysis to DNA unwinding, has an efficiency of plating of 0.1 with comparatively smaller plaque size. Helicase-S319A and -R363A/R504A, both devoid of dNTP hydrolysis and unwinding activities, do not support T7Δ4 phage growth (Table 2). These plasmids do not influence the growth of wild-type T7 phage. The results, taken together with the biochemical results, show that proteins devoid of either ssDNA-dependent dATP or dTTP hydrolysis activity do not support the growth of T7 phage.
TABLE 2.
Ability of plasmids encoding gene 4 proteins to complement T7Δ4 phage for growth The ability of the plasmids encoding gene 4 variants to support the growth of T7Δ4 in E. coli was examined. The number of plaques formed by plasmids harboring the wild-type gene 4 protein is normalized to 1. The relative efficiency of plating obtained with the mutated gene 4 constructs were determined by the number of plaques (PFU) formed by the mutated gene 4 constructs divided by the PFU of wild type gene 4. Efficiency of plating of ≤10–9 corresponds to the gene 4 construct unable to complement the T7Δ4 growths in the host bacteria.
| pET11b:gp4 construct | Efficiency of plating T7Δ4 |
|---|---|
| Wild type | 1 |
| R363A | 0.023 |
| R504A | 1 |
| R363A/R504A | ≤10–9 |
| S319T | 0.6 |
| S319A | ≤10–9 |
| R504A/S319T | 0.1 |
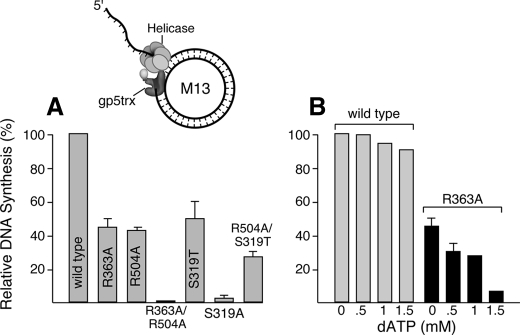
DNA Synthesis Mediated by DNA Polymerase and Helicase—The T7 gene 5 DNA polymerase (gp5) functions in vivo in a 1 to 1 complex with its processivity factor, E. coli trx. Gp5-trx catalyzes processive DNA synthesis on ssDNA templates but is unable to catalyze strand-displacement synthesis of duplex DNA (29). In the presence of T7 gp4, the DNA helicase unwinds the duplex ahead of the polymerase, exposing ssDNA template for the advancing gp5-trx. The two proteins together mediate extensive strand-displacement synthesis (30). In the experiment presented in Fig. 6 the amount of DNA synthesis catalyzed by gp5-trx and wild-type helicase in the strand-displacement DNA synthesis assay described under “Experimental Procedures” was assigned a relative value of 100%. In these reactions all four dNTPs (0.5 mm each) are present by necessity to support DNA synthesis. Helicase-R363A, helicase-R504A, helicase-S319T, and helicase-R504A/S319T were 2–4-fold less effective than wild-type helicase in mediating strand-displacement DNA synthesis with gp5-trx (Fig. 6A). Predictably, helicase-S319A and helicase-R363A/R504A, devoid of helicase activity, were ineffective in this reaction.
FIGURE 6.
Strand-displacement DNA synthesis mediated by T7 DNA polymerase and helicase. A, strand-displacement DNA synthesis mediated by gp5-trx and helicase was measured in an assay containing 10 nm M13 double-stranded DNA with a 5′-ssDNA tail on the interrupted strand, 0.5 mm dATP, dCTP, dGTP, [α-32P]dTTP (0.1 μCi), 10 nm gp5-trx, and 10 nm hexamer of the indicated T7 helicase (29). After incubation for 10 min at 37 °C the reactions were stopped by the addition of EDTA to 25 mm final concentration. DNA synthesis (7 picomole [32P]dTMP incorporated) mediated by gp5-trx and helicase was considered as 100% activity. The relative rate of DNA synthesis was calculated and plotted as a histogram using GraphPad prism software. Results are the average of three experiments. B, effect of increasing concentrations of dATP on DNA synthesis mediated by wild-type helicase or helicase-R363A and gp5-trx. The reaction was carried out with 0.5 mm each of dATP, dTTP, dGTP, and dCTP as described above for 30 s at 37 °C followed by the addition of different concentrations (0, 0.5, 1, and 1.5 mm) of dATP. The reaction was then incubated for 10 min at 37 °C. The reactions were stopped by the addition of EDTA to 25 mm final concentration. The DNA synthesis mediated by wild-type helicase and gp5-trx in the presence of 0.5 mm each of the dNTPs in the above reaction was considered as 100% activity. The error bars represent the S.D. from three different experiments.
The ability of helicase-R363A to support strand-displacement synthesis is somewhat surprising. The efficiency of plating T7Δ4 on the host, E. coli, is drastically reduced when the cells are complemented with plasmids encoding helicase-R363A. Furthermore, our results have shown that increased concentrations of dATP inhibit dTTP-dependent DNA unwinding of helicase-R363A. We measured the effect of an increased concentration of dATP on the ability of helicase-R363A to mediate strand-displacement synthesis with gp5-trx (Fig. 6B). Upon increasing the dATP concentrations 4-fold in the reaction, the ability of helicase-R363A to function in the strand-displacement synthesis assay decreased drastically (∼90% reduction). The increased concentration of dATP had essentially no effect on wild-type helicase in this reaction. Helicase-R504A and helicase-R504A/S319T were also relatively unaffected by the increased concentration of dATP (data not shown).
DNA Binding and Protein Oligomerization—Wild-type T7 helicase binds ssDNA with high affinity in the presence of the non-hydrolysable analog of dTTP, β,γ-methylene dTTP (23). In the presence of dTTP, DNA binding is markedly reduced, most likely because of the translocation of the helicase and its eventual dissociation. For this latter reason it was not possible to measure DNA binding of the variant helicases with dTTP or dATP. However, using a nitrocellulose filter binding assay helicase-R504A, -S319T, and -S319T/R504A bound 50-mer oligonucleotides quite similar to wild-type helicase with dissociation constants (Kd) in the range of 27–72 nm in the presence of β,γ-methylene dTTP (Fig. 7A). Helicase-R363A also bound the ssDNA but 4–5 times weaker than wild-type helicase (Kd-120 nm). Helicase-S319A and -R363A/R504A, both of which were inefficient in the hydrolysis of dTTP and in DNA unwinding, did not bind to ssDNA under these conditions.
FIGURE 7.
A, binding of helicases to ssDNA. Binding of T7 helicase to ssDNA was measured in a nitrocellulose filter assay. Reactions contained 1 nm 5′-32P-labeled 50-mer oligonucleotide, 5 mm β,γ-methylene dTTP, 10 mm MgCl2, 40 mm Tris-HCl (pH 7.5), 10 mm DTT, 50 mm potassium glutamate, and the indicated concentration of either wild-type helicase or one of the helicase variants. After incubation in 37 °C for 30 min the reaction mixture was filtered through a nitrocellulose membrane laid atop a charged zeta probe membrane with the help of a dot blot filtration apparatus. Protein-bound DNA on the nitrocellulose membrane and unbound free DNA on the zeta probe membrane were measured by radioactive intensity by phosphorimaging. The % oligonucleotide bound by each of the proteins is plotted against the corresponding protein concentration. The dissociation constant (Kd) calculated for each of the proteins using GraphPad prism software is presented. B, oligomerization of helicases. To determine the oligomeric state of wild-type and the variant helicases (indicated above the gel lanes), helicases were incubated with β,γ-methylene dTTP and a 50-mer oligoribonucleotide as described under “Experimental Procedures.” After incubation glutaraldehyde was added to stabilize the oligomeric forms as described under “Experimental Procedures.” The proteins were analyzed by electrophoresis on a 10% non-denaturing polyacrylamide gel. Each lane contained 25 pmol of the indicated helicase. After electrophoresis on a 10% non-denaturing polyacrylamide gel, the proteins were stained with Coomassie Blue. The molecular masses (kDa) of marker proteins are shown on the left.
T7 gene 4 protein forms higher order oligomers, predominantly hexamers in the reaction containing β,γ-methylene dTTP and ssDNA (6, 18). We have checked the ability of all the helicase variants to oligomerize as described under “Experimental Procedures.” All the altered gene 4 proteins form hexameric structures (Fig. 7B). The proteins that were defective in DNA binding, helicase-S319A and helicase-R363A/R504A, also formed higher order oligomers, suggesting that the DNA binding ability of gene 4 protein is not a prerequisite to form higher order oligomers.
DISCUSSION
DNA helicases play an essential role in DNA replication. In this communication we have extended the study of the DNA helicase encoded by bacteriophage T7. Not only is there considerable structural and biochemical information available on the helicase itself but there is also an understanding of its interactions of the other proteins of the T7 replisome. The replisome of phage T7 can be reconstituted with four purified proteins whose three-dimensional structures are known. DNA synthesis is carried out by T7 DNA polymerase, a complex of gene 5 DNA polymerase (gp5) bound tightly to its processivity factor, E. coli thioredoxin. Gene 4 protein (gp4) provides the helicase activity, the subject of this study, to unwind the parental DNA strands ahead of the polymerase. As the helicase unwinds the DNA, the polymerase synthesizes the leading strand continuously in a 5′-3′ direction. gp4 functions not only as helicase but also as a primase, synthesizing RNA primers that are required to initiate lagging strand synthesis. A dynamic interaction between the helicase, primase, and polymerase is crucial for timely separation of double-stranded, synthesis of primers, and initiation of lagging strand synthesis, and the coordination of synthesis of the two strands (31). Gene 2.5 protein (gp2.5) is a single-stranded DNA binding protein that coats the lagging strand extruded behind the helicase. gp2.5 also interacts with both gp5 and gp4 to stimulate their polymerase and primase activities, respectively.
gp4, as mentioned above, is a multifunctional protein in that it functions both as a helicase and as a primase. The helicase domain resides within the C-terminal half of the polypeptide and is covalently attached to the primase domain in the N-terminal half of the protein through a flexible linker. The oligomerization to form a hexamer involves residues at the N terminus of the helicase domain and within the linker. The DNA sequence encoding each domain can be cloned and the helicase and primase expressed separately. Although both the primase and helicase domains have activities corresponding to that found in their respective domains within the full-length protein, the helicase domain does, under certain conditions, impose dramatic changes in primase activity. In the current study we have, however, used the full-length gp4 to examine helicase activity.
T7 DNA helicase is unique among replicative hexameric helicases in that it preferentially uses dTTP as the source of energy for its translocation on ssDNA and its unwinding of duplex DNA (3). In addition, early in the characterization of the T7 helicase, its ability to hydrolyze most dNTPs and rNTPs was observed (8, 29). Not only is the enzyme capable of hydrolyzing all of the four dNTPs in the presence of ssDNA, but the hydrolysis of all four nucleotides by the helicase is observed during strand-displacement synthesis mediated by the polymerase and helicase on duplex DNA (29). Whereas dTTP and dATP will both function in the unwinding of duplex DNA by the helicase, dGTP and dCTP, although hydrolyzed by the helicase, cannot support unwinding of DNA. Here we show that the addition of dGTP, dCTP, or dATP to a reaction in which the helicase unwinds DNA in the presence of dTTP results in an enhancement of unwinding (Fig. 5). The results suggest that T7 helicase can couple the hydrolysis of dGTP, dCTP, and dATP to the unwinding of double-stranded DNA under conditions when it hydrolyzes dTTP; the helicase is likely to use all four dNTPs for DNA unwinding under physiological conditions.
Architecture of the Nucleotide Binding Site of T7 DNA Helicase—Hexameric helicases bind nucleotides at nucleotide-binding sites located at the interface of adjacent subunits (12, 17). A number of amino acids located in each of the subunits that create the nucleotide binding site play crucial roles in stabilizing and hydrolyzing the bound nucleotide. Amino acid residues conserved as the arginine finger, phosphate sensor, and catalytic base and a few residues in the Walker Motif A and Walker Motif B important for NTP hydrolysis are located around the phosphate groups of the NTP (Fig. 1, B–D) (11, 21). However, amino acid residues positioned nearer to the base of the bound nucleotide are more likely to determine nucleotide specificity. Consequently, in the present study we have focused on residues located near the base and sugar of the bound dNTP. Arg-363 and Arg-504 in T7 helicase are located in the nucleotide binding site. Arg-363 is positioned to interact with the 3′-OH of the bound dATP (Fig. 1). Arg-504 is conserved in the DnaB family of helicases, whereas a phenylalanine is positioned in DnaB family of helicases in place of Tyr-535 (3, 12). Both Arg-504 and Tyr-535 sandwich the base of the dNTP (12). However, Arg-504 can build a hydrogen bridge with the OH group of thymidine of the bound dTTP (Fig. 1). Such an interaction may be sufficient to create the observed preference for dTTP during DNA-dependent hydrolysis and unwinding of DNA.
To examine the role of Arg-363 and Arg-504 in accommodating different nucleotides for hydrolysis and for unwinding DNA, we have substituted alanine for these amino acids. The genetically altered helicases were then examined biochemically for their ability to use the various dNTPs. Arg-363 plays an important role in the hydrolysis of dATP, dGTP, and dCTP and in the ability of these nucleotides to fuel the unwinding of duplex DNA. However, it has little, if any effect on the utilization of dTTP for these reactions as measured by the ability of helicase-R363A to function in these reactions. Arg-504, on the other hand, does play a role in dTTP hydrolysis, as reflected in the 2-fold reduced level of dTTP hydrolysis and the change to a preference for dATP in the unwinding reaction with helicase-R504A. The rate of hydrolysis of dATP by helicase-R504A is not affected by the amino acid substitution. Thus, unlike wild-type helicase, helicase-R504A does not prefer dTTP. Taken together the results indicate that Arg-363 is responsible for dATP, dGTP, and dCTP specificity, whereas Arg-504 plays a role in determining specificity for dTTP. Most interesting is the finding that elimination of both of these arginines leads to an enzyme, helicase-R363A/R504A, that is unable to hydrolyze any of the four dNTPs. Clearly, these two arginines are critical for the accommodation of nucleotides in the active site. Structures of the helicase with dTTP or dATP (PDB files, 1cr1 and 1cr2) suggest that the architecture of the nucleotide binding sites is essentially identical with the bound nucleotide (12). However, Arg-363 is positioned to interact with dATP and not dTTP. Similarly, to accommodate dGTP and dCTP, Arg-363 might play a structural role. On the other hand helicase-R504A hydrolyzes dCTP and can couple its hydrolysis to unwinding of DNA, suggesting that Arg-504 does not allow wild-type helicase to couple dCTP hydrolysis to unwinding of DNA. These results taken together suggest that Arg-363 and Arg-504 play a disparate role in different dNTP binding and hydrolysis in the nucleotide binding site of T7 DNA helicase.
Ser-319 is an essential residue in the P-loop of the nucleotide binding site. Its replacement with alanine renders the helicase incapable of hydrolyzing any dNTPs. Bacterial helicases have a threonine at the equivalent position and, interestingly, prefer rATP for hydrolysis and unwinding of DNA. Indeed we find that substituting threonine for serine in the T7 helicase leads to a preference for dATP for DNA-dependent dNTP hydrolysis (Table 1). In addition, this altered protein, helicase-S319T, also uses dCTP and dGTP better than does wild-type helicase for DNA unwinding. Thus, simply changing the size and shape of the P-loop hydroxyl-bearing residue can alter a key catalytic mechanism.
In view of the reduced specificity for dTTP seen with helicase-R504A and helicase-S319T, we constructed helicase-R504A/S319T. These two amino acid changes together lead to a 90% reduction in the use of dTTP in the hydrolysis reaction and a strong preference for dATP in the unwinding of duplex DNA. These results suggest that Ser-319 and Arg-504 are collectively responsible for the preference for dTTP by the T7 helicase. In both instances, when the residue was altered (S319T or R504A) there was a loss of specificity for dTTP with no significant change in the preference for dATP in the nucleotide hydrolysis reaction. We propose that in the selection of a nucleotide some of the crucial amino acids are rearranged to provide for stability of the incoming nucleotide probably by an induced fit mechanism. However, the decisive factors behind the nucleotide specificity of helicase enzymes in general might be a consequence of physiological conditions.
Utilization of Different Nucleotides by T7 DNA Helicase—T7 DNA helicase hydrolyzes all four dNTPs (8), with dTTP preferentially used for DNA unwinding. Our results show that helicase-R363A devoid of dATP, dGTP, and dCTP hydrolysis was limited (2% of wild-type helicase) in its ability to support the growth of T7Δ4 phage lacking gene 4. Helicase-R504A/S319T can support the growth of T7Δ4 somewhat better (10% of wild-type helicase). Helicase-R504A/S319T hydrolyzes dATP and dCTP but is defective in dTTP and dGTP hydrolysis. Because dCTP and dGTP are hydrolyzed poorly by the wild-type helicase, the requirement of these activities might be less important than their ability to hydrolyze dATP and dTTP. Thus, in vivo complementation analyses of the helicase variants suggest that hydrolysis of both dATP and dTTP is required for bacteriophage growth.
The sequential model of NTP hydrolysis of hexameric helicases as revealed biochemically (18, 32) and structurally (13) depends on full occupancy of the subunits for DNA-dependent NTP hydrolysis. In this model, NTP hydrolysis only occurs on a single subunit at a time; the subunit most strongly interacting with the DNA strand. Upon completion of hydrolysis/release of NDP, the DNA is passed to the adjacent subunit next in line. Importantly, a hexamer containing a single subunit that cannot hydrolyze NTP or cannot bind DNA ruins the ability of the enzyme to hydrolyze NTP at a fast rate (18, 32). Thus, any unoccupied subunits would not allow DNA-dependent NTP hydrolysis. Similarly, in vitro the addition of a non-hydrolysable dTTP analog drastically reduces the hydrolysis of dTTP and the unwinding of DNA. The addition of dATP to reactions containing helicase-R363A also reduces these activities. Because helicase-R363A is unable to hydrolyze dATP, it may function in a manner similar to that observed with β,γ-methylene dTTP and wild-type helicase.
The nucleotide mixing studies with helicase-R363A, helicase-R504A, and helicase-R504A/S319T suggest that hydrolyzable dNTPs can increase the overall rate of DNA unwinding by T7 helicase under conditions where the preferred dNTP such as dTTP provides the energy. By contrast, nonhydrolysable dNTPs inhibits helicase activity. In fact, the Km for dATP (Km 0.7 mm) and dTTP (Km-1.8 mm) in the wild-type helicase reaction do not differ significantly (Table 1). However, the presence of dGTP (Km 1.1 mm) or dCTP (Km 9.2 mm) does not inhibit the dTTP- or dATP-dependent DNA unwinding activity of wild-type helicase, but rather increases it ∼20%. It, therefore, appears that the hydrolysis of nucleotides other than dTTP is a requirement for T7 DNA helicase to properly function in vivo.
Supplementary Material
Acknowledgments
We thank Steven Moskowitz (Advanced Medical Graphics) for illustrations and Samir Hamdan for reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM54397 (to C. C. R.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: ssDNA, single-stranded DNA; DTT, dithiothreitol; trx, thioredoxin.
References
- 1.Lohman, T. M., and Bjornson, K. P. (1996) Annu. Rev. Biochem. 65 169-214 [DOI] [PubMed] [Google Scholar]
- 2.Matson, S. W., and Kaiser-Rogers, K. A. (1990) Annu. Rev. Biochem. 59 289-329 [DOI] [PubMed] [Google Scholar]
- 3.Patel, S. S., and Picha, K. M. (2000) Annu. Rev. Biochem. 69 651-697 [DOI] [PubMed] [Google Scholar]
- 4.Frick, D. N., and Richardson, C. C. (2001) Annu. Rev. Biochem. 70 39-80 [DOI] [PubMed] [Google Scholar]
- 5.Tabor, S., and Richardson, C. C. (1981) Proc. Natl. Acad. Sci. U. S. A. 78 205-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egelman, E. H., Yu, X., Wild, R., Hingorani, M. M., and Patel, S. S. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 3869-3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim, D. E., Narayan, M., and Patel, S. S. (2002) J. Mol. Biol. 321 807-819 [DOI] [PubMed] [Google Scholar]
- 8.Matson, S. W., and Richardson, C. C. (1983) J. Biol. Chem. 258 14009-14016 [PubMed] [Google Scholar]
- 9.Hingorani, M. M., and Patel, S. S. (1996) Biochemistry 35 2218-2228 [DOI] [PubMed] [Google Scholar]
- 10.Patel, S. S., Rosenberg, A. H., Studier, W., and Johnson, K. A. (1992) J. Biol. Chem. 267 15013-15021 [PubMed] [Google Scholar]
- 11.Subramanya, H. S., Bird, L. E., Brannigan, J. A., and Wigley, D. B. (1996) Nature 28 379-383 [DOI] [PubMed] [Google Scholar]
- 12.Sawaya, M. R., Guo, S., Tabor, S., Richardson, C. C., and Ellenberger, T. (1999) Cell 99 167-177 [DOI] [PubMed] [Google Scholar]
- 13.Enemark, E. J., and Joshua-Tor, L. (2006) Nature 442 270-275 [DOI] [PubMed] [Google Scholar]
- 14.Moreau, M. J., McGeoch, A. T., Lowe, A. R., Itzhaki, L. S., and Bell, S. D. (2007) Mol. Cell 28 304-314 [DOI] [PubMed] [Google Scholar]
- 15.Lee, S. J., Qimron, U., and Richardson, C. C. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 8908-8913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crampton, D. J., Guo, S., Johnson, D. E., and Richardson, C. C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 4373-4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singleton, M. R., Sawaya, M. R., Ellenberger, T., and Wigley, D. B. (2000) Cell 101 589-600 [DOI] [PubMed] [Google Scholar]
- 18.Crampton, D. J., Ohi, M., Qimron, U., Walz, T., and Richardson, C. C. (2006)) J. Mol. Biol. 360 667-677 [DOI] [PubMed] [Google Scholar]
- 19.Crampton, D. J., Mukherjee, S., and Richardson, C. C. (2006) Mol. Cell. 21 165-174 [DOI] [PubMed] [Google Scholar]
- 20.Washington, M. T., Rosenberg, A. H., Grifin, K., Studier, F. W., and Patel, S. S. (1996) J. Biol. Chem. 271 26825-26834 [DOI] [PubMed] [Google Scholar]
- 21.Mendelman, L. V., Notarnicola, S. M., and Richardson, C. C. (1993) J. Biol. Chem. 268 27208-27213 [PubMed] [Google Scholar]
- 22.Venkatesan, M., Silver, L. L., and Nosal, N. G. (1982) J. Biol. Chem. 257 12426-12434 [PubMed] [Google Scholar]
- 23.Matson, S. W., and Richardson, C. C. (1985) J. Biol. Chem. 260 2281-2287 [PubMed] [Google Scholar]
- 24.Lee, S. J., and Richardson, C. C. (2001) J. Biol. Chem. 276 49419-49426 [DOI] [PubMed] [Google Scholar]
- 25.Notarnicola, S. M., Park, K., Griffith, J. D., and Richardson, C. C. (1995) J. Biol. Chem. 270 20215-20224 [DOI] [PubMed] [Google Scholar]
- 26.Matson, S. W., Tabor, S., and Richardson, C. C. (1983) J. Biol. Chem. 258 14017-14024 [PubMed] [Google Scholar]
- 27.Mathews, C. K. (1972) J. Biol. Chem. 247 7430-7438 [PubMed] [Google Scholar]
- 28.Mendelman, L. V., Notarnicola, S. M., and Richardson, C. C. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 10638-10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolodner, R., and Richardson, C. C. (1977) Proc. Natl. Acad. Sci. U. S. A. 74 1525-1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lechner, R. L., and Richardson, C. C. (1983) J. Biol. Chem. 258 11185-11196 [PubMed] [Google Scholar]
- 31.Hamdan, S. M., Johnson, D. E., Tanner, N. A., Lee, J. B., Qimron, U., Tabor, S., van Oijen, A. M., and Richardson, C. C. (2007) Mol. Cell 27 539-549 [DOI] [PubMed] [Google Scholar]
- 32.Liao, J. C., Jeong, Y. J., Kim, D. E., Patel, S. S., and Oster, G. (2005) J. Mol. Biol. 350 452-475 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.