Abstract
Histatins, a family of salivary proteins, have antimicrobial activity. Candida albicans, which is killed by histatins, induces oral candidiasis in individuals with compromised immune systems. Although the functional significance of histatins has been documented, their biological and physiological functions against host cells have not been clarified. In this study, we found that histatin 3, a member of the histatin family, binds to heat shock cognate protein 70 (HSC70). These proteins were co-localized in the cytoplasm and nucleus in human gingival fibroblasts following non-heat and heat shock. Histatin 3 induced stimulation of DNA synthesis and cell survival in human gingival fibroblasts in a dose-dependent manner. This DNA synthesis was found to be dependent on HSC70 by knockdown experiments. The effect of heat shock on DNA synthesis induced by histatin 3 was ∼2-fold higher than that of non-heat shock. When the histatin 3 uptake into cells was inhibited by monodansylcadaverine or when histatin 3 binding to HSC70 was precluded by 15-deoxyspergualin, DNA synthesis by histatin 3 was ∼2-fold less than that without monodansylcadaverine or 15-deoxyspergualin. Although HSC70 directly bound to p27Kip1 (a cyclin-dependent kinase inhibitor), histatin 3 increased the binding between those proteins but not with a peptide capable of binding to HSC70. Moreover histatin 3 prevented ATP-dependent dissociation of HSC70-p27Kip1. ATP was unable to form a histatin 3-HSC70(D10N)-p27Kip1 complex (HSC70(D10N) is a mutant attenuating ATPase activity). These findings suggest that histatin 3 may be involved in cell proliferation through the regulation of HSC70 and p27Kip1 in oral cells.
Oral non-immune defense is associated with saliva. Some salivary proteins, such as histatins, have antibacterial and anti-fungal activities and protect oral tissues from pathogenic microorganisms (1). The histatin family of proteins, consisting of 12 members that are histidine-rich and consist of cationic 3–4-kDa proteins found in the saliva secreted by the salivary glands of humans and higher primates, are localized in human oral tissues (2–7). Histatins in saliva are also present in healthy adults at concentrations of 50–425 μg/ml (8). Histatins 1 and 3 are full-length proteins of 38 and 32 amino acids in length, respectively. Histatin 5 comprises 24 amino acids and is either a proteolytic product of histatin 3 or a protein translated from a post-transcriptionally modified histatin 3 mRNA (3). Other members of the histatin family are also generated by proteolytic degradation during secretion and have been characterized (9, 10). Histatins 3 and 5 exhibit antimicrobial activity against Candida albicans at physiological concentrations of 15–50 μm (2, 11–13). In addition, histatin 5 has been shown to inhibit a trypsin-like protease and the cysteine protease clostripain, which are produced by Bacteroides gingivalis (an oral bacterium suspected of being the pathogen of periodontal disease) and Clostridium histolyticum, respectively (14, 15).
A wide variety of stresses, including environmental, pathological, and physiological stimuli, induce the synthesis of heat shock proteins (HSPs)2 in prokaryotic and eukaryotic cells (16). However, low level expression of HSPs is observed under normal physiological conditions (17). HSPs play roles as chaperones in the correct folding of proteins and assembly of their subunits and in transporting certain proteins across the membrane into organelles (18). HSC70 has been identified as a constitutively expressed HSP in cells (19). The structure of HSC70 contains domains corresponding to an ATPase, substrate-binding domain, and a lid (trapping and untrapping substrates) from the N to C termini (20). A nuclear localization signal and a nuclear localization-related signal for HSC70 have been identified in amino acid regions 246–262 and 473–492, respectively (21, 22). An aspartate residue at amino acid 10 of HSC70 is essential for ATPase activity (23).
Cyclins and cyclin-dependent kinases (CDKs) regulate the progression of the cell cycle in eukaryotic cells (24). p27Kip1 is a cyclin-dependent kinase inhibitor and negatively regulates cell cycle progression. Phosphorylation and ubiquitination of p27Kip1 result in its degradation by the proteasome system, thereby inducing cell cycle progression from G1 to S phases (25–27). Therefore, p27Kip1 plays a pivotal role in the control of the cell cycle.
The functions of histatins in oral cells, such as constitutive periodontal cell HGFs, have not been clarified. In this study, we identified a strong association of histatin 3 with HSC70 and observed that this association progressed from the G1 to S phases and thereby induced DNA synthesis in HGFs. These findings provide insight into another physiological function of salivary proteins in oral cells.
EXPERIMENTAL PROCEDURES
Cell Culture—COS-7, HEK293, and HGF cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich) with 10% fetal bovine serum (FBS), 100 units/ml penicillin G, and 100 μg/ml streptomycin at 37 °C in a 5% CO2 and 95% air humidified incubator. HGFs used in this study were obtained from volunteers after appropriate informed consent was obtained. The Ethics Committee of Matsumoto Dental University approved the study protocol. HGFs isolated from adhering gingival tissue on extracted teeth were cultured on collagen-coated plates in medium.
Plasmid Constructions—pHybLex/Zeo-cBASH(1–62) and p2D11-6 plasmids have been described by Imamura et al. (28). All PCR experiments described below were carried out using 1.75 units of Expand High Fidelity polymerase (Roche Applied Science) and the following templates (1 ng each) and primers (100 pmol each). For pHybLex/Zeo-hHistatin 3, template primer 5′-GCAAAGAGACATCATGGGTATAAAAGAAAATTCCATGAAAAGCATCATTCACATCGAGGCTATAGATCAAATTATCTGTA-3′ and primers hHis3-1 (5′-GGGAATTCGGATCCGATTCACATGCAAAGAGACATCATGGG-3′ and 5′-GGGCGGCCGCTCAATTGTCATACAGATAATTTGATCT-3′) were used. The amplified human histatin 3 DNA was cloned into pCR-BluntII-TOPO (Invitrogen; named pCR-BluntII-hHistatin 3). The EcoRI-EcoRI fragment from pCR-BluntII-hHistatin 3 was then cloned into pHybLex/Zeo (Invitrogen). For pYESTrp-hHSP70, template pEF-hHSP70 and primers hHSP70-1 (5′-GGAAGCTTGAATTCATGGCCAAAGCCGCGGCGATCGGC-3′) and hHSP70-A (5′-GGGCGGCCGCCTAATCTACCTCCTCAATGGTGGG-3′) were used. The HindIII-NotI fragment from the amplified human HSP70 DNA was inserted into the same sites of pYESTrp (Invitrogen). For pYESTrp-hHSC70, template pTARGET-HSC73 and primers hHSC70-1 (5′-GGGGATCCGAATTCATGTCCAAGGGACCTGCAGTTGG-3′) and hHSC70-A (5′-GGGCGGCCGCTTAATCAACCTCTTCAATGGTGGG-3′) were used. The BamHI-HincII fragment from the amplified human HSC70 DNA and a HincII-NotI fragment from pTARGET-HSC73 were inserted into the BamHI and NotI sites of pYESTrp. For pGST-hHistatin 3, the EcoRI-EcoRI fragment from pHybLex/Zeo-hHistatin 3 was cloned into pGEX-5X-1 (GE Healthcare). For pGST-hHistatins 4 and 5, template pHybLex/Zeo-hHistatin 3 and primers 5′-GGGTCGACTCAATTGTCATACAGATAATTTGA-3′ and 5′-GGAATTCAGAAAATTCCATGAAAAG-3′ for histatin 4 and primers hHis3-1 and 5′-GGTCGACTCAATAGCCTCGATGTGAATG-3′ for histatin 5 were used. EcoRI-SalI fragments from amplified histatin 4 or 5 DNA were inserted into the same sites of pGEX-5X-1. For pCAT7-hHSC70, the EcoRI-XhoI fragment from pYESTrp-hHSC70 was inserted into the same sites of pCAT7-neo (28). For pCAT7-hHSC70(D10N), the KpnI-SmaI fragment from pDsRed2-hHSC70(D10N) was inserted into the same sites of pCAT7-neo. For pCAT7-hHSC70(Δ1–384), template pDsRed2-hHSC70 and primers hHSP70-2 (5′-GGCTCGAGCTTCTGAGAATGTTCAAGATTTGCTGC-3′) and hHSP70-B (5′-CAAGTGAATTCTTGGATGACACCTTGTCCC-3′) were used. The XhoI-EcoRI fragment from the amplified HSC70 DNA and an EcoRI-KpnI fragment from pCAT7-hHSC70 were inserted into the XhoI and KpnI sites of pCAT7-neo. For pCAT7-hHSC70(Δ385–543), two cycles of PCR were carried out. For one reaction of the first cycle, template pDsRed2-hHSC70 and primers hHSC70-3 (5′-CATCTTGTCTGGAGACAAGTCCTATGCCTTCAACATG-3′) and hHSC70-A were used. For another reaction, template pDsRed2-hHSC70 and primers hHSC70-C (5′-CATGTTGAAGGCATAGGACTTGTCTCCAGACAAGATG-3′) and hHSC70-4 (5′-GTATTGAGATCGATTCTCTCTATG-3′) were used. In the second cycle, for the template, the two fragments of the first cycle were annealed through their 3′-end homologies, and primers hHSC70-4 and hHSC70-A were used. The HindIII-KpnI fragment from the amplified HSC70 DNA and an EcoRI-HindIII fragment from pCAT7-hHSC70 were inserted into the EcoRI and KpnI sites of pCAT7-neo. For pCAT7-hHSC70(Δ542–646), an EcoRI-EcoRI fragment from pCAT7-hHSC70 was cloned into pCAT7-neo. For pECFP-hHistatin 3, an EcoRI-SalI fragment from pCR-BluntII-hHistatin 3 was inserted into the same sites of pECFP-C1 (Clontech). For pDsRed2-hHSC70, a NotI-KpnI fragment from pCAT7-hHSC70 was inserted into the same sites of pCR-Blunt (Invitrogen; named pCR-Blunt-hHSC70(N-K)). The XhoI-KpnI fragment from pCR-Blunt-hHSC70(N-K) was inserted into the same sites of pDsRed2-C1 (Clontech). For pDsRed2-hHSC70(D10N), a BamHI-NotI fragment from pGST6P-HSC70(D10N) was inserted into the same sites of pCR-Blunt (named pCR-Blunt-hHSC70(D10N)(B-N)). The KpnI-ApaI fragment from pCR-Blunt-hHSC70(D10N)(B-N) was inserted into the same sites of pDsRed2-C1. For pSINsi-HSC70 and pSINsi-HSC70-scr, the target sequence of HSC70 for small interfering RNA (siRNA) has been described by Kose et al. (29), and its scrambled (control) sequence was 5′-GACTAGTCACATATGTATGCTGTTA-3′. Oligonucleotides for the construction of plasmids were designed, and plasmid construction was performed as described according to instructions in the pSINsi-hU6 DNA user manual (Takara).
Yeast Two-hybrid System—The procedure was performed as described in the user manual of Hybrid Hunter (Invitrogen) (28). Plasmids for bait and prey were transformed into yeast strain L40. A β-galactosidase assay was performed thereafter.
Transfection and Infection—For immunoprecipitation, expression vectors of pECFP-hHistatin 3 and pCAT7-hHSC70 or pCAT7-hHSC70(D10N) (5 μg each) were mixed with TransIT-LT1 reagents (Mirus) before being co-transfected into 4 × 106 COS-7 cells and harvested 48 h later. For GST pulldown assays, 5 μg of each expression vector, pCAT7-hHSC70, pCAT7-hHSC70(D10N), pCAT7-hHSC70(Δ1–384), pCAT7-hHSC70(Δ385–543), or pCAT7-hHSC70(Δ542–646), were transfected into COS-7 or HEK293 cells. For confocal laser microscopy, 1 μg of each expression vector, pECFP-hHistatin 3, pDsRed2-hHSC70, and/or pDsRed2-hHSC70(D10N), was transfected into 2 × 105 HGFs. Preparations and infections of retroviruses (6 × 104 colony-forming units/ml) for siRNA were performed as described in the material accompanying the retrovirus constructive system Ampho (Takara).
Synthetic Peptides—Human histatins 3, 4, and 5; fluorescein isothiocyanate (FITC)-histatin 3 (Scrum Inc.); and P3a peptide (Biosynthesis Inc.) were chemically synthesized and purified by high performance liquid chromatography. The purity of the synthetic peptides was >85%. (Pro-Pro-Gly)10·9H2O (Peptide Institute Inc.) was used as a control.
Antibodies—Antiserum against human histatin 3 protein was produced by immunizing a rabbit with a synthetic peptide corresponding to the whole histatin 3 conjugated to keyhole limpet hemocyanin. The serum was purified using an affinity column carrying protein A (Invitrogen). The following antibodies were purchased: mouse monoclonal anti-T7-tag (Novagen), goat anti-GST (GE Healthcare), rabbit anti-GFP serum, Alexa Fluor 594 goat anti-mouse IgG (Molecular Probes), mouse monoclonal anti-human HSC70 (B-6), mouse monoclonal anti-cyclin D1 (DCS-6), mouse monoclonal anti-cyclin E (E-4), rabbit polyclonal anti-CDK2 (M2), rabbit polyclonal anti-p27Kip1 (C-19; for immunoprecipitation), mouse monoclonal anti-p27Kip1 (F-8; for Western blotting) (Santa Cruz Biotechnology), and mouse monoclonal anti-β-actin (Abcam).
Immunoprecipitation, GST Pulldown Assay, and Western Blotting—Immunoprecipitation, GST pulldown, and Western blotting analyses were performed as described previously (28, 30). For immunoprecipitation under physiological conditions, HGFs were cultured with 25 μm histatin 3 for 24 h and then heat-shocked at 42 °C for 5 h. Proteins extracted from HGFs were immunoprecipitated with anti-HSC70 antibody. Precipitates were analyzed by Western blotting. For immunoprecipitation of HSC70/p27Kip1, HGFs were cultured in DMEM containing 0.1% FBS over 24 h and stimulated with 10% FBS, 30 μm P3a, and histatin 3 in the presence or absence of 10 μg/ml 15-deoxyspergualin (15-DSG) (Spanidin® injection, Nippon Kayaku) for 24 h. Then proteins extracted from HGFs were immunoprecipitated with anti-p27Kip1 antibody, and precipitates were analyzed by Western blotting. For immunoprecipitation of ATP-dependent dissociation, the procedure was performed as described by Nakamura et al. (31). HGFs were cultured in DMEM containing 0.1% FBS over 24 h. Proteins extracted from HGFs were immunoprecipitated with anti-p27Kip1 antibody. Precipitates were incubated with 200 μm P3a and histatin 3 on ice for 15 min, and 1 mm ATP (GE Healthcare), ADP (Oriental Yeast), and ATPγS (Roche Applied Science) were added before Western blotting was conducted. For GST pulldown assays, GST and GST-histatin 3, 4, and 5 proteins were expressed in Escherichia coli BL21(DE3) (Invitrogen). The proteins that had been immobilized to glutathione-Sepharose 4B (GE Healthcare) were mixed with cell lysates from transfected cells. After washing thoroughly, precipitates were analyzed by Western blotting. For GST pulldown assays of ATP-dependent dissociation, proteins extracted from transfected cells were mixed with the immobilized GST-histatin 3 protein. Precipitates were incubated with 1 mm ATP, ADP, and ATPγS on ice for 30 min after which Western blotting was performed. For Western blotting of cyclin D1, cyclin E, and CDK2, HGFs were cultured in DMEM containing 0.1% FBS over 24 h and stimulated with 10% FBS, 3 and 30 μm bovine serum albumin (BSA; Nacalai Tesque), and peptides for 8 h. Cell lysates were then prepared, and Western blotting was carried out. For siRNA experiments, HGFs were infected with retroviruses for siRNAs targeting HSC70 and its control. Cells were cultured in DMEM containing 10% FBS for 24 h and subsequently cultured in DMEM containing 0.1% FBS over 24 h. Cell lysates were then prepared, and Western blotting was carried out.
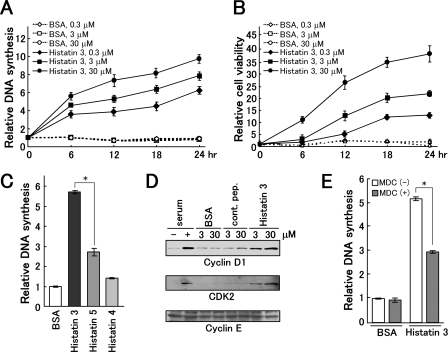
DNA Synthesis and 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Assays—HGFs (1 × 104) were cultured in DMEM containing 0.1% FBS over 24 h. For treatment with histatin 3, cells were cultured with histatin 3 and BSA at 0.3, 3, and 30 μm for the specified time periods (see Fig. 4, A and B). For treatment with various histatins, HGFs were cultured with 0.3 μm BSA and histatins 3, 4, and 5 for 24 h. For siRNA experiments, HGFs were cultured and infected with retroviruses for siRNA as described above. Cells were cultured with 0.3 μm histatin 3 for 8 h. For heat shock treatment, HGFs were cultured with 0.3 μm BSA and peptides at 37 °C for 3 h. Next cells were cultured at 42 °C for 2 h and then at 37 °C for 24 h. For monodansylcadaverine (MDC) or 15-DSG treatment, HGFs were cultured with 0.3 μm BSA and peptides in the presence or absence of 20 μm MDC or 10 μg/ml 15-DSG for 24 h. The level of DNA synthesis in cells was determined by measuring BrdUrd incorporation using the Frontier BrdU Cell Proliferation Assay kit (Exalpha Biologicals). For MTT assays, HGFs were cultured with 10 μl of 5 mg/ml MTT (Sigma-Aldrich) for 2–4 h. Then 100 μl of 0.04 n HCl, isopropanol was added to the medium and completely dissolved. Samples were measured by Microplate Reader model 550 (Bio-Rad) using a dual wavelength of 450/595 nm (test/reference) for DNA synthesis analyses and 595/655 nm for MTT assays. Figs. 4, 5, 6, 7 show representative examples of three identical experiments with essentially identical results.
FIGURE 4.
Effect of histatin 3 on DNA synthesis and cell survival. A and B, DNA synthesis and cell survival induced by histatin 3. After HGFs had been cultured in DMEM containing 0.1% FBS over 24 h, histatin 3 and BSA (control) were added to cells at the concentrations indicated, and cells were cultured for the indicated time periods. DNA synthesis in cells and cell viability were determined by measuring BrdUrd incorporation using the Frontier BrdU Cell Proliferation Assay kit and by an MTT assay, respectively. Each data point is the mean of duplicated samples with error bars showing standard deviations. C, DNA synthesis in the presence of histatins 4 and 5. Histatins and BSA at 0.3 μm were added to HGFs, and DNA synthesis was analyzed as described in A and B. Bars represent the means of duplicated samples with error bars showing standard deviations. *, p < 0.01 (Student's t test). D, examination of expression patterns for cyclin D1, cyclin E, and CDK2. HGFs were cultured in DMEM containing 0.1% FBS over 24 h. Cells were then stimulated with serum, BSA, control peptide (cont. pep.), and histatin 3 for 8 h. Expression levels of cyclin D1, cyclin E, and CDK2 were examined by Western blotting with the respective antibodies. E, DNA synthesis in cells after the addition of 0.3 μm histatin 3 with or without MDC. DNA synthesis was examined as described under “Experimental Procedures.” Bars represent the means of duplicated samples with error bars showing standard deviations. *, p < 0.01 (Student's t test).
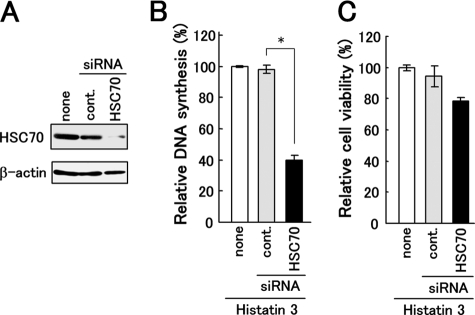
FIGURE 5.
Effect of knockdown of HSC70 in HGFs on DNA synthesis induced by histatin 3. A, HGFs were infected with retroviruses expressing siRNAs targeting HSC70 and its control (cont.). Proteins in cell extracts from HGFs were analyzed by Western blotting with anti-HSC70 (upper) and anti-β-actin (lower) antibodies. B and C, DNA synthesis and cell viability after the addition of histatin 3 to HSC70 knockdown cells were examined as described under “Experimental Procedures.” Bars represent the means of duplicated samples with error bars showing standard deviations. *, p < 0.01 (Student's t test).
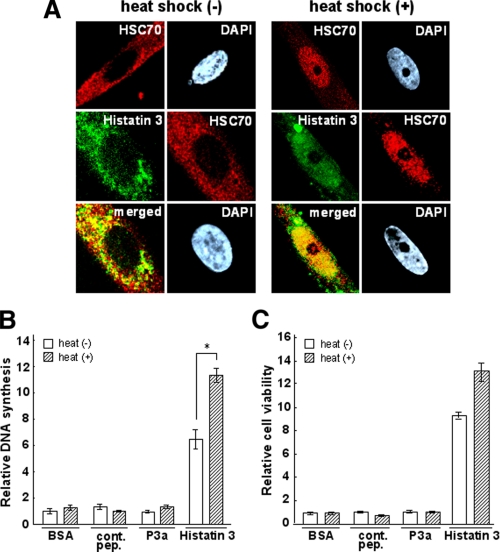
FIGURE 6.
Effect of heat shock on the localization of histatin 3 and HSC70 in living cells and on DNA synthesis and cell survival. A, HGFs were cultured with FITC-histatin 3 for 12 h and heat-shocked at 42 °C for 5 h (right) or not treated (left). Cells were stained with anti-HSC70 antibody and 4′,6-diamidino-2-phenylindol dihydrochloride (DAPI) (nuclear staining), and images were obtained using confocal laser microscopy. Upper panel, localization of HSC70; middle panel, localizations of histatin 3 and HSC70; lower panel, merged images of histatin 3 and HSC70. B and C, DNA synthesis and cell viability in the presence of histatin 3 under heat shock conditions were examined as described under “Experimental Procedures.” Bars represent the means of duplicated samples with error bars showing standard deviations. *, p < 0.01 (Student's t test). cont. pep., control peptide.
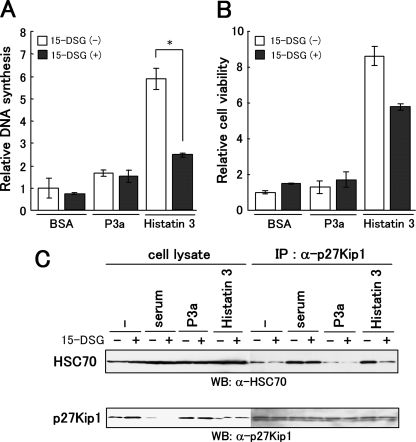
FIGURE 7.
Effect of 15-DSG on DNA synthesis, cell survival, and the binding of HSC70 to p27Kip1 after treatment of cells with histatin 3. A and B, DNA synthesis and cell viability in the presence of 15-DSG and histatin 3 were examined as described under “Experimental Procedures.” Bars represent the means of duplicated samples with error bars showing standard deviations. *, p < 0.01 (Student's t test). C, effect of 15-DSG on the binding of HSC70 to p27Kip1 after the addition of histatin 3 to cells. HGFs were cultured in DMEM containing 0.1% FBS over 24 h and stimulated with serum, P3a, and histatin 3 in the presence or absence of 15-DSG. At 24 h, proteins extracted from cells were immunoprecipitated (IP) with anti-p27Kip1 antibody. Precipitates were analyzed by Western blotting (WB) with anti-HSC70 (top) and anti-p27Kip1 (bottom) antibodies.
Confocal Laser Microscopy—An LSM510 microscope (Carl Zeiss) was used for confocal laser microscopy. The excitation and emission wavelengths of ECFP, DsRed2, FITC, Alexa Fluor 594, and 4′,6-diamidino-2-phenylindol dihydrochloride were 458 and 463–495 nm, 563 and 570–602 nm, 488 and 516 nm, 543 and 570–720 nm, and 358 and 461 nm, respectively. For heat shock analysis, HGFs were cultured with 1 μm FITC-histatin 3 for 12 h and then heat-shocked at 42 °C for 5 h. Cells were fixed with 4% paraformaldehyde at room temperature for 30 min and permeabilized with 0.5% Triton X-100 at 25 °C for 10 min. After blocking with 1% skim milk in Tris-buffered saline containing 0.1% Tween 20 (0.1% TBS-T) at 25 °C for 30 min, cells were incubated with anti-HSC70 antibody at 4 °C overnight. Then cells were washed with 0.1% TBS-T and incubated with Alexa Fluor 594 goat anti-mouse IgG at 25 °C for 1 h. After washing cells with phosphate-buffered saline, nuclei were stained with 0.3 mm 4′,6-diamidino-2-phenylindol dihydrochloride (Molecular Probes) at 25 °C for 5 min.
RESULTS
Association of Histatin 3 with Heat Shock Cognate Protein HSC70—The biological and physiological functions of histatin 3 against host cells are poorly understood. As a starting point, we looked for histatin 3-associated proteins in mammalian cells. A previous study reported that Ssa1p/Ssa2p proteins, members of the HSP family from C. albicans and Saccharomyces cerevisiae, bind to histatin 5 and induce fungicidal activity (32). We therefore examined the association of histatin 3 with human HSPs by a yeast two-hybrid analysis using histatin 3 as bait. As shown in Fig. 1A, histatin 3 bound to HSC70 as well as the positive control, BASH-(1–62)/BNAS2 (28). In contrast, neither combination of the empty bait or prey vectors showed binding. HSP70, an HSP whose expression is induced by stresses such as heat, did not bind to histatin 3, suggesting that histatin 3 preferentially binds to HSC70, an HSP constitutively expressed in the absence of stress in cells.
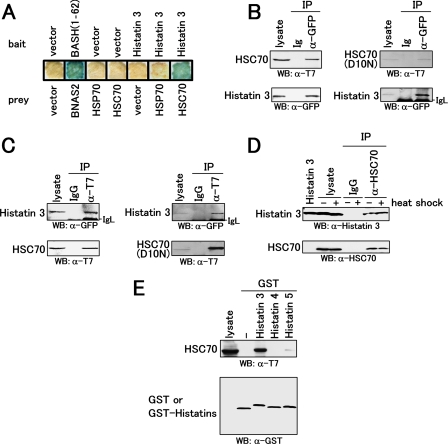
FIGURE 1.
Interaction of histatin 3 with heat shock protein HSC70. A, expression vectors encoding histatin 3 (bait) and heat shock proteins (prey) were transformed into yeast L40. Transformants were cultured on a membrane filter, and a β-galactosidase assay was performed. Vector and BASH-(1–62)/BNAS2 represent empty and positive control vectors, respectively. B, COS-7 cells were co-transfected with expression vectors encoding CFP-tagged histatin 3 and T7-tagged HSC70 or T7-tagged HSC70(D10N). Proteins extracted from COS-7 cells were immunoprecipitated (IP) with anti-GFP serum and control serum (Ig). Precipitates as well as cell lysates (lysate) were analyzed by Western blotting (WB) with anti-T7 antibody (top) and anti-GFP serum (bottom). C, proteins extracted from COS-7 cells were immunoprecipitated with anti-T7 antibody and control IgG. Precipitates were analyzed by Western blotting with anti-GFP serum (top) and anti-T7 antibody (bottom). D, HGFs were cultured with histatin 3 under non-heat (-) and heat (+) shock conditions. Proteins extracted from cells were immunoprecipitated with anti-HSC70 antibody. Precipitates were analyzed by Western blotting with anti-histatin 3 (top) and anti-HSC70 (bottom) antibodies. E, GST pulldown assays for members of the histatin family and HSC70. An expression vector encoding T7-tagged HSC70 was transfected into COS-7 cells. Proteins extracted from COS-7 cells were mixed with GST and GST-fused histatins 3, 4, and 5 that had been immobilized to glutathione-Sepharose beads. Precipitates were analyzed by Western blotting with anti-T7 (top) and anti-GST (bottom) antibodies.
To determine the binding of histatin 3 to HSC70 in mammalian cells, immunoprecipitation analyses were carried out. Expression vectors encoding CFP-tagged histatin 3 (CFP-histatin 3) and T7-tagged HSC70 (T7-HSC70) or T7-tagged HSC70(D10N) (T7-HSC70(D10N), a substituted mutant in which Asp at amino acid 10 is changed to Asn (23)), were co-transfected into COS-7 cells. CFP-histatin 3 in the cell lysate was immunoprecipitated with anti-GFP serum, and Western blotting was carried out using anti-T7 antibody. As shown in Fig. 1B, T7-HSC70 and T7-HSC70(D10N) were co-precipitated with CFP-histatin 3 but not with CFP alone (data not shown) and control Ig. Conversely for immunoprecipitation with anti-T7 antibody followed by Western blotting with anti-GFP serum, CFP-histatin 3 was co-precipitated with T7-HSC70 and T7-HSC70(D10N) (Fig. 1C). These results indicate that histatin 3 and HSC70 or HSC70(D10N) are associated with each other.
We then examined the interaction between histatin 3 and HSC70 under physiological and heat shock conditions. HGFs were cultured with histatin 3 for 24 h and then heated at 42 °C for 5 h. HSC70 in the cell lysate was immunoprecipitated, and Western blotting was carried out using anti-histatin 3 antibody. As shown in Fig. 1D, histatin 3 was co-precipitated with HSC70, indicating that histatin 3 is taken up by HGFs and thereafter binds to HSC70 in cells regardless of heat shock.
Next we tested whether other members of the histatin family are capable of binding to HSC70. T7-HSC70 was transiently expressed in COS-7 cells, and the cell lysate was subjected to binding with GST-fused histatins (GST-histatins) 3, 4, and 5. As shown in Fig. 1E, GST-histatin 3 bound to T7-HSC70 more strongly than did GST-histatin 5, and GST-histatin 4 and GST alone did not bind to T7-HSC70. These results suggest that some, but not all, histatins are associated with HSC70, and histatin 3 in particular is predominant. We thereafter focused on the interaction of histatin 3 with HSC70.
Determination of the Histatin 3-binding Domain of HSC70— To determine the histatin 3-binding domain of HSC70, expression vectors encoding various deletion mutants of T7-HSC70 were prepared (Fig. 2A) and transfected into COS-7 cells. GST pulldown assays were carried out using the cell lysate and GST-histatin 3. As shown in Fig. 2B, deletion of the 385–543 region of HSC70 resulted in the loss of histatin 3 binding. Similarly the GST protein alone could not bind to either wild-type or mutant HSC70s. Taken together, these results indicate that the 385–543 region (substrate-binding domain) of HSC70 is essential for binding with histatin 3.
FIGURE 2.
Determination of the histatin 3-binding domain of HSC70. A, schematic representation of HSC70 and its mutants. HSC70(Δ1–384), HSC70(Δ385–543), and HSC70(Δ542–646) indicate HSC70 with deleted ATPase, peptide (substrate)-binding, and variable (lid) domains, respectively. All cDNAs were inserted into the pCAT7-neo vector. B, GST pulldown assays for histatin 3 and various HSC70s. Expression vectors encoding various HSC70s were transfected into COS-7 cells, and binding experiments and Western blotting (WB) were carried out as described in Fig. 1E. WT, wild type.
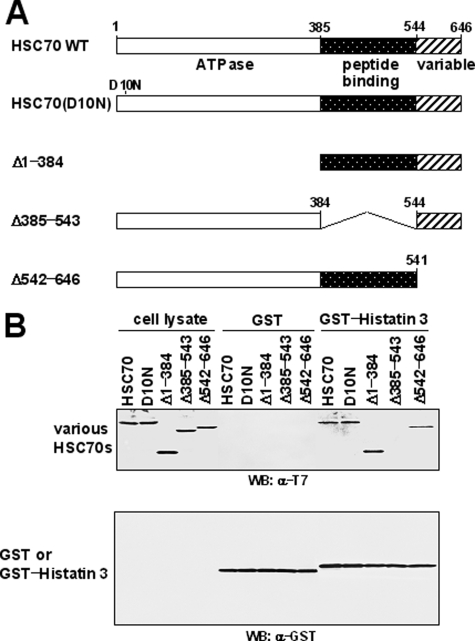
Co-localization of Histatin 3 with HSC70 in Living Cells—To examine the localization of histatin 3 and HSC70 in living cells, expression vectors encoding CFP-histatin 3, DsRed-HSC70, and DsRed-HSC70(D10N) were transfected into HGFs, and cells were examined by confocal laser microscopy. As shown in Fig. 3A, histatin 3 was diffusely distributed throughout cells, whereas HSC70 and HSC70(D10N) were localized in the cytoplasm (Fig. 3, B and C, respectively). When CFP-histatin 3 and DsRed-HSC70 were co-expressed, histatin 3 was diffusely distributed throughout cells (Fig. 3D), and HSC70 was mainly localized in the cytoplasm and slightly in the nucleus (Fig. 3E). The co-localization of these proteins can be observed in “merged” figures (Fig. 3F). Conversely HSC70(D10N) was found to be co-localized with histatin 3 in the cytoplasmic region (Fig. 3, G–I). This result is consistent with the observation that HSC70(D10N) was observed in the cytoplasm even under conditions in which HSC70 migrated into the nucleus (29). These results indicate that histatin 3 is associated with HSC70 in living cells.
FIGURE 3.
Localization of histatin 3 and HSC70 in living cells. Expression vectors encoding CFP-histatin 3, DsRed2-HSC70, and DsRed2-HSC70(D10N) were transfected alone (A–C) or co-transfected in various combinations (D–I) into HGFs. At 48 h after transfection, the localizations of proteins in cells were examined by confocal laser microscopy. F and I, merged images of D and E or G and H, respectively.
DNA Synthesis and Cell Survival Induced by Histatin 3— Histatin 3 was found to be associated with HSC70 in vitro and in vivo (Figs. 1, 2, 3). Because HSPs are involved in the regulation of the cell cycle (31, 33), DNA synthesis and cell survival were examined in HGFs in the presence of histatin 3. The levels of DNA synthesis in cells were determined by measuring BrdUrd incorporation into DNA, and cell viability was measured by an MTT assay. As shown in Fig. 4A, histatin 3, but not BSA, stimulated DNA synthesis in a dose-dependent manner; cell viability was also enhanced by the addition of histatin 3 but not BSA (Fig. 4B). These results suggest that histatin 3 is involved in the proliferation of HGFs.
To further examine whether the stimulation of DNA synthesis is induced by other histatins in HGFs, we conducted BrdUrd incorporation assays using histatins 4 and 5. As shown in Fig. 4C, the level of DNA synthesis after the addition of histatin 5 to cells was ∼2-fold lower than that after the addition of histatin 3. The stimulatory effect of histatin 4 was similar to that of BSA. These results suggest that the association of some, but not all, histatins with HSC70 (Fig. 1E) is required for stimulating DNA synthesis.
Because the progression of the cell cycle from G1 to the S phase is controlled by cyclins and CDKs (25–27), we examined whether the expression pattern of these proteins was affected by histatin 3. To do this, HGFs were cultured in DMEM containing 0.1% FBS over 24 h, and cells were stimulated with histatin 3 for 8 h. The expression levels of cyclin D1 and CDK2 in cells were analyzed by Western blotting with the respective antibodies. As shown in Fig. 4D, histatin 3, like serum (a positive control), induced the expression of cyclin D1 and CDK2 in a dose-dependent manner, but BSA and a nonspecific control peptide did not. Cyclin E was expressed at similar levels after the addition of histatin 3, serum, BSA, and a control peptide. These results suggest that histatin 3 is involved in inducing the expression of cell cycle regulators such as cyclin D1 and CDK2 during the G1/S transition.
We observed that, when cultured in the presence of histatin 3, HGFs took up histatin 3 (Figs. 1D and 6A). In an attempt to resolve the underlying mechanism, we supplemented the culture medium with MDC in addition to histatin 3. MDC is an inhibitor of membrane-bound transglutaminase and interferes with clathrin-mediated receptor trafficking as demonstrated for CD91 (34). The results indicated that the uptake of histatin 3 by HGFs depends on endocytosis (data not shown). We then examined whether the stimulatory effect of histatin 3 on DNA synthesis is affected by the addition of MDC to cells. As shown in Fig. 4E, the level of DNA synthesis induced by histatin 3 was significantly decreased in the presence of MDC compared with that observed in the absence of MDC (p < 0.01), suggesting that the internalization of histatin 3 by HGFs through endocytosis is necessary for stimulating DNA synthesis.
Effect of HSC70 on DNA Synthesis Induced by Histatin 3—We then examined whether DNA synthesis induced by histatin 3 is dependent upon HSC70. When the expression of HSC70 was knocked down by introducing an siRNA targeting HSC70 into HGFs (Fig. 5A), the level of DNA synthesis induced by histatin 3 was found to be ∼40% of that obtained in HGFs when introducing a control siRNA (p < 0.01; Fig. 5B). Cell viability in the presence of histatin 3 in HSC70 knocked down HGFs was ∼80% compared with the control (Fig. 5C, none). These results indicate that the stimulatory effect of histatin 3 on DNA synthesis is dependent upon HSC70, and it is possible that the association of these proteins is required for DNA synthesis in HGFs.
Effect of Heat Shock on the Localization of Histatin 3 and HSC70 in Living Cells, DNA Synthesis, and Cell Survival—It has been reported that HSC70 shuttles between the cytoplasm and nucleus (35) and that translocation into the nucleus occurs when cells are stimulated by heat shock (36). To determine whether histatin 3 comigrates with HSC70 into the nucleus in living cells following heat shock, HGFs were cultured with FITC-histatin 3 for 12 h and heat-shocked at 42 °C for 5 h followed by staining with anti-HSC70 antibody. Cells were inspected by confocal laser microscopy. As shown in Fig. 6A, as reported previously (36), HSC70 was predominantly translocated from the cytoplasm to the nucleus under heat shock conditions (upper panel of “heat shock (-)” versus “heat shock (+)”). When histatin 3 was added to HGFs, histatin 3 and HSC70 were mainly localized in the cytoplasm and partially in the nucleus under non-heat shock conditions (middle and lower panels of heat shock (-)). Following heat shock, these proteins translocated into the nucleus and co-localized (middle and lower panels of heat shock (+)). These results suggest that histatin 3 binds to HSC70 in living cells regardless of heat shock and that the complex is capable of translocating into the nucleus.
We then examined the effect of heat shock on DNA synthesis and cell survival after the addition of histatin 3 to cells. As shown in Fig. 6B, the level of DNA synthesis induced by histatin 3 increased significantly in response to heat shock (p < 0.01). In contrast, BSA, a control peptide, and P3a (a peptide from the clathrin light chain that binds to HSC70 (37)) did not affect DNA synthesis. Cell viability in the presence of histatin 3 was not affected by heat shock (Fig. 6C). These results suggest that histatin 3 plays an important role in cell proliferation through interaction with HSC70 even when cells are heat-shocked.
Effect of 15-DSG on DNA Synthesis and Cell Survival Induced by Histatin 3 and on Binding of HSC70 to p27Kip1—We further examined whether the specific interaction of histatin 3 and HSC70 is necessary for DNA synthesis and cell survival. BrdUrd incorporation and MTT assays were carried out using 15-DSG. 15-DSG, which has a peptidomimetic structure and is an immunosuppressive agent, binds specifically to HSC70 and is also thought to preclude peptide binding to HSC70 (38, 39). As shown in Fig. 7A, the level of DNA synthesis induced by histatin 3 decreased significantly in the presence of 15-DSG compared with that in the absence of 15-DSG (p < 0.01). Although cell survival was not affected by the addition of 15-DSG alone at this concentration (40), the level of cell viability by histatin 3 in the presence of 15-DSG was reduced to ∼70% of that obtained in the absence of 15-DSG (Fig. 7B). Neither BSA nor P3a affected DNA synthesis or cell viability. These results indicate that the association between histatin 3 and HSC70 is very important for cell proliferation in HGFs.
A previous study showed that HSC70 directly interacts with p27Kip1, a cell cycle regulator, during the G1/S transition (31). Because our results also showed that the binding of histatin 3 and HSC70 may be correlated with DNA synthesis through the G1/S transition in HGFs, we examined the effect of histatin 3 with or without 15-DSG on HSC70-p27Kip1 complex formation. HGFs were cultured in DMEM containing 0.1% FBS over 24 h, and cells were stimulated with histatin 3 in the presence or absence of 15-DSG. Then p27Kip1 in the cell lysate was immunoprecipitated, and precipitates were analyzed by Western blotting with anti-HSC70 antibody. As shown in Fig. 7C, after the addition of histatin 3 alone to cells, HSC70 bound as strongly to p27Kip1 as it did after the addition of serum with or without 15-DSG. Conversely decreased binding between HSC70 and p27Kip1 was observed in the presence of both histatin 3 and 15-DSG, and P3a had little effect on their binding activities. These results suggest that the binding of HSC70 to p27Kip1 is reinforced by the binding of histatin 3 to HSC70 during the G1/S transition.
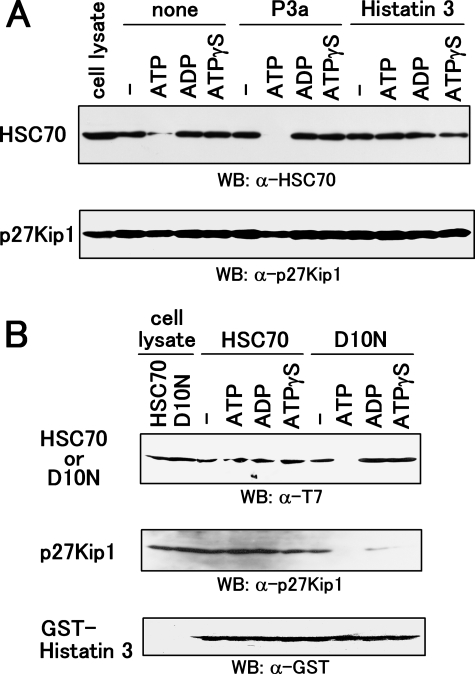
Prevention of ATP-dependent Dissociation of the HSC70-p27Kip1 Complex by Histatin 3—It has been shown previously that ATP induces the dissociation of the HSC70-p27Kip1 complex (31). The ATP (but not ADP)-bound form of HSPs is in a state of rapid flux between the binding and release of the substrate (20). We examined the effect of binding between HSC70 and p27Kip1 in the presence of histatin 3, ATP, ADP, and a non-hydrolyzable ATP analog, ATPγS. p27Kip1 in cell lysates from HGFs was immunoprecipitated, and precipitates were mixed with histatin 3 and nucleotides before Western blotting using anti-HSC70 antibody. As shown in Fig. 8A, HSC70 strongly bound to p27Kip1 in the presence of histatin 3 and ATP, whereas binding decreased dramatically with the addition of P3a and ATP. ADP and ATPγS did not affect binding in the presence or absence of histatin 3 and P3a. These results indicate that histatin 3 stabilizes the HSC70-p27Kip1 complex even when ATP is present.
FIGURE 8.
Effect of nucleotides on the complex formation of histatin 3-HSC70-p27Kip1. A, association of HSC70 with p27Kip1 in the presence or absence of histatin 3 and nucleotides. HGFs were cultured in DMEM containing 0.1% FBS over 24 h. Proteins extracted from cells were immunoprecipitated with anti-p27Kip1 antibody, and precipitates were treated with P3a and histatin 3 followed by incubation with ATP, ADP, and ATPγS. Western blotting (WB) was then carried out using anti-HSC70 (top) and anti-p27Kip1 (bottom) antibodies. B, effect of nucleotides on the association of HSC70-p27Kip1 with histatin 3. HEK293 cells were transfected with the expression vectors encoding T7-tagged HSC70 and T7-tagged HSC70(D10N). Proteins in cell extracts were mixed and precipitated with GST-histatin 3. Precipitates were incubated with ATP, ADP, and ATPγS, and Western blotting was carried out using anti-T7 (top), anti-p27Kip1 (middle), and anti-GST (bottom) antibodies.
Finally we examined the effect of nucleotides on the complex formation of histatin 3-HSC70-p27Kip1 or histatin 3-HSC70(D10N)-p27Kip1. T7-HSC70 and T7-HSC70(D10N) were expressed in HEK293 cells, and GST pulldown assays were carried out using proteins extracted from cells and GST-histatin 3. Precipitates were then incubated with nucleotides, and Western blotting was carried out using anti-T7 and anti-p27Kip1 antibodies. As shown in Fig. 8B, HSC70 binding to histatin 3 was associated with p27Kip1 in the presence of all nucleotides; however, ATP decreased the binding of HSC70(D10N)-p27Kip1 to histatin 3. In addition, although ADP and ATPγS both dramatically decreased the binding of p27Kip1 to HSC70(D10N), HSC70(D10N) bound to histatin 3. p27Kip1 did not bind directly to GST-histatin 3 (data not shown). These results suggest that the Asp-10 residue of HSC70 is important for HSC70 binding to histatin 3 when ATP is present and that neither ADP nor ATPγS appears to be essential for the binding of p27Kip1 to HSC70(D10N).
DISCUSSION
It has been reported previously that, together with epidermal growth factor, histatin 5 enhances cell proliferation in rabbit costal chondrocytes (41). However, the precise mechanism of this action is not understood, and it is essential to clarify how histatins affect the physiology of mammalian cells, especially oral cells. In this study, we found that histatin 3 interacts with HSC70 from mammalian cells but not with HSP70. Histatin 5 was also weakly associated with HSC70 compared with histatin 3. The binding of histatin 3 to HSC70 prevented ATP-dependent dissociation of the HSC70-p27Kip1 complex. Moreover histatin 3 did not bind with the HSC70(D10N)-p27Kip1 complex in the presence of ATP. The association of histatin 3 and HSC70 may correlate with cell proliferation through a cell cycle regulator, p27Kip1, in oral HGFs, which constitute the major cellular population of gingival tissue (42).
In general, although mammalian cell proliferation is primarily regulated by extracellular signals such as growth factors, other extracellular signals have been reported, including a peptide from the nuclear localization signal of fibroblast growth factor-1 that exhibits mitogenic activity and stimulates DNA synthesis in a fibroblast growth factor receptor-independent manner in NIH3T3 cells, enhancing the expression of cyclin D1 (43–45). In conjunction with our present study, these previous findings suggest that some, but not all, peptides from both native and partial proteins can function as effectors (factors) of cell proliferation.
The G1/S transition in the cell cycle of mammalian cells is one of the checkpoints at which the balance between activating and inhibitory molecules appears critical, and actions such as the overexpression of cyclins and the down-regulation of cyclin-dependent kinase inhibitors are all likely to be observed (46). Among the various cyclin-dependent kinase inhibitors identified, p27Kip1 regulates cell cycle progression through ubiquitin/proteasomal degradation (47–49). In addition, proteins that interact with p27Kip1, such as Jab1 and stress protein p8, control the degradation of p27Kip1 (50, 51). HSC70 is also a p27Kip1-binding protein and is involved in cell cycle progression (31, 52). In addition, HSC70 is ubiquitinated and degraded with its binding protein by the 26 S proteasome (53). Our findings show that p27Kip1 interacts with the histatin 3-HSC70 complex and that this ternary complex may be stable regardless of the presence of ATP (Fig. 8; see discussion below). Moreover histatin 3 induced the G1/S transition, thereby stimulating HSC70-dependent DNA synthesis in HGFs (Fig. 5). It is tempting to speculate that the binding of p27Kip1 to the histatin 3-HSC70 complex and/or the ternary complex might cause ubiquitination and degradation through the ubiquitin/proteasome system, leading to cell cycle progression. Further studies are required to clarify this issue.
The present findings indicated that histatin 3 was co-localized with HSC70 in the nucleus under non-heat shock conditions (even more so under heat shock conditions) (Figs. 3 and 6A). We also found that histatin 3 accumulated in the nucleus when HGFs were cultured with FITC-histatin 3 at 37 °C for 4 days (data not shown). This migration of histatin 3 appears to depend on the nuclear localization signal and nuclear localization-related signal-like sequences of HSC70 in living cells (Fig. 6A and Refs. 21 and 22) because no such sequences in histatin 3 were found in a search for motifs. The translocation of HSC70 to the nucleus under physiological conditions corroborates the findings of a study using microinjection of fluorescently labeled HSC70 into the cytoplasm of rat embryo fibroblasts (36). Our findings may therefore imply that the histatin 3-HSC70 complex shuttles easily between the cytoplasm and nucleus under physiological conditions, thereby associating with p27Kip1 even though p27Kip1 exists in the nucleus and/or cytoplasm. In fact, the translocation of p27Kip1 from the nucleus to the cytoplasm has been observed (50, 54).
In addition, our findings showed that the formation of the HSC70-p27Kip1 complex was reinforced by the addition of histatin 3 but not by the addition of P3a (a peptide that binds to HSC70 (37)) even when ATP was present (Fig. 8A). Although the HSC70-p27Kip1 complex bound to histatin 3 in the presence of ATP, the HSC70(D10N)-p27Kip1 complex did not. Moreover in the presence of ADP and ATPγS, the amount of p27Kip1 that bound to histatin 3-HSC70(D10N) was very low (Fig. 8B). These findings imply that binding may be associated with differences in affinities between histatin 3 and HSC70 or HSC70(D10N) in the presence of nucleotides and also by changes in the conformation (conformational stability) between histatin 3-HSC70 (or HSC70(D10N))-nucleotide complexes. It has been reported that HSC70 has a high peptide affinity when bound to ADP (20). However, in the presence of ATP, peptide-HSC70 is in a low affinity complex (55); in the case of DnaK, an E. coli homolog of HSC70, conformational changes in its substrate-binding region are induced by the binding of ATP (56). Another possibility is that ATPase activity of HSC70 may also affect the formation of the ternary complex. A previous study showed that HSC70(D10N) dramatically decreases ATPase activity, although the affinities between HSC70(D10N) and ATP (or the substrate) and the conformation of HSC70(D10N) are similar to those of HSC70 (23); the reason for this decrease in ATP activity is not yet understood. It therefore seems plausible that the binding of histatin 3 to HSC70 must induce conformational changes in HSC70, thereby leading to the formation of a stable complex even when ATP is present, especially toward p27Kip1. In such a case, the ATPase activity of HSC70 may also affect the formation of the ternary complex. Furthermore these presumed actions seem to imply that the ternary complex still exists in the stable state in cells even though the concentration of intracellular nucleotides dramatically changes during G1/S progression. In fact, total cellular ATP pools have been shown to increase during G1/S progression in 3T6 cells, whereas nuclear stores do not, and nuclear ATP/ADP ratios decrease once cells enter the S phase (57).
In this study, we identified an interaction between a salivary protein and a molecule from oral cells that may be related to cell proliferation, in particular to G1/S transition. The present findings help us to better understand the functions and mechanisms of salivary proteins against host cells in the oral cavity.
Acknowledgments
We are grateful to Dr. Hiroyoshi Ariga for pTARGET-HSC73, pEF-hHSP70, and reading this manuscript. We are also grateful to Drs. Naoko Imamoto and Shingo Kose for pGST6P-HSC70(D10N).
This work was supported by a grant-in-aid for scientific research (C).
Footnotes
The abbreviations used are: HSP, heat shock protein; HSC70, heat shock cognate protein 70; CDK, cyclin-dependent kinase; HGF, human gingival fibroblast; BASH, B cell adaptor containing SH2 domain; BNAS2, BASH N terminus-associated protein 2; GST, glutathione S-transferase; siRNA, small interfering RNA; 15-DSG, 15-deoxyspergualin; MDC, monodansylcadaverine; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; (E)CFP, (enhanced) cyan fluorescent protein; dansyl, 5-dimethylaminonaphthalene-1-sulfonyl; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; FITC, fluorescein isothiocyanate; GFP, green fluorescent protein; ATPγS, adenosine 5′-O-(thiotriphosphate); BSA, bovine serum albumin.
References
- 1.Tenovuo, J. (1998) Acta Odontol. Scand. 56 250-256 [DOI] [PubMed] [Google Scholar]
- 2.Oppenheim, F. G., Yang, Y. C., Diamond, R. D., Hyslop, D., Offner, G. D., and Troxler, R. F. (1986) J. Biol. Chem. 261 1177-1182 [PubMed] [Google Scholar]
- 3.Sabatini, L. M., and Azen, E. A. (1989) Biochem. Biophys. Res. Commun. 160 495-502 [DOI] [PubMed] [Google Scholar]
- 4.Oppenheim, F. G., Xu, T., McMillian, F. M., Levitz, S. M., Diamond, R. D., Offner, G. D., and Troxler, R. F. (1988) J. Biol. Chem. 263 7472-7477 [PubMed] [Google Scholar]
- 5.Pollock, J. J., Denepitiya, L., MacKay, B. J., and Ianoco, V. J. (1984) Infect. Immun. 44 702-707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu, T., Telser, E., Troxer, R. F., and Oppenheim, F. G. (1990) J. Dent. Res. 69 1717-1723 [DOI] [PubMed] [Google Scholar]
- 7.Wang, P.-L., Kanehira, T., Nagai, N., Nagatsuka, H., Imamura, Y., Tanaka, J., and Kuboki, Y. (2005) Dent. Jpn. (Tokyo) 41 39-41 [Google Scholar]
- 8.Lal, K., Pollock, J. J., Santarpia, R. P., III, Heller, H. M., Kaufman, H. W., Fuhrer, J., and Steigbigel, R. T. (1992) J. Acquired Immune Defic. Syndr. 5 904-914 [PubMed] [Google Scholar]
- 9.Troxler, R. F., Offner, G. D., Xu, T., Vanderspek, J. C., and Oppenheim, F. G. (1990) J. Dent. Res. 69 2-6 [DOI] [PubMed] [Google Scholar]
- 10.Perinpanayagam, H. E. R., Van Wuyckhuyse, B. C., Ji, Z. S., and Tabak, L. A. (1995) J. Dent. Res. 74 345-350 [DOI] [PubMed] [Google Scholar]
- 11.Raji, P. A., Edgerton, M., and Levin, M. J. (1990) J. Biol. Chem. 265 3898-3905 [PubMed] [Google Scholar]
- 12.Xu, T., Levitz, M., Diamond, R., and Oppenheim, F. G. (1991) Infect. Immun. 59 2549-2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzgerald, D. H., Coleman, D. C., and O'Connell, B. C. (2003) FEMS Microbiol. Lett. 220 247-253 [DOI] [PubMed] [Google Scholar]
- 14.Nishikata, M., Kanehira, T., Oh, H., Tani, H., Tazaki, M., and Kuboki, Y. (1991) Biochem. Biophys. Res. Commun. 174 625-630 [DOI] [PubMed] [Google Scholar]
- 15.Gusman, H., Grogan, J., Kagan, H. M., Troxler, R. F., and Oppenheim, F. G. (2001) FEBS Lett. 489 97-100 [DOI] [PubMed] [Google Scholar]
- 16.Lindquist, S., and Craig, E. A. (1988) Annu. Rev. Genet. 22 631-677 [DOI] [PubMed] [Google Scholar]
- 17.Craig, E. A., and Gross, C. A. (1991) Trends Biochem. Sci. 16 135-140 [DOI] [PubMed] [Google Scholar]
- 18.Ellis, R. J., and van der Vies, S. M. (1991) Annu. Rev. Biochem. 60 321-347 [DOI] [PubMed] [Google Scholar]
- 19.Chappell, T. G., Welch, W. J., Schlossman, D. M., Palter, K. B., Schlesinger, M. J., and Rothman, J. M. (1986) Cell 45 3-13 [DOI] [PubMed] [Google Scholar]
- 20.Sullivian, C. S., and Pipas, J. M. (2002) Microbiol. Mol. Biol. Rev. 66 179-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsukahara, F., and Maru, Y. (2004) J. Biol. Chem. 279 8867-8872 [DOI] [PubMed] [Google Scholar]
- 22.Lamian, V., Small, G. M., and Feldherr, C. M. (1996) Exp. Cell Res. 228 84-91 [DOI] [PubMed] [Google Scholar]
- 23.Huang, S.-P., Tsai, M.-Y., Tzou, Y.-M., Wu, W.-G., and Wang, C. (1993) J. Biol. Chem. 268 2063-2068 [PubMed] [Google Scholar]
- 24.Murray, A. W. (2004) Cell 116 221-234 [DOI] [PubMed] [Google Scholar]
- 25.Sheaff, R. J., Groudine, M., Gordon, M., Roberts, J. M., and Clurman, B. E. (1997) Genes Dev. 11 1464-1478 [DOI] [PubMed] [Google Scholar]
- 26.Vlach, J., Hennecke, S., and Amati, B. (1997) EMBO J. 16 5334-5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montagnoli, A., Fiore, F., Eytan, E., Carrano, A. C., Draetta, G. F., Hershko, A., and Pagano, M. (1999) Genes Dev. 13 1181-1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imamura, Y., Katahira, T., and Kitamura, D. (2004) J. Biol. Chem. 279 26425-26432 [DOI] [PubMed] [Google Scholar]
- 29.Kose, S., Furuta, M., Koike, M., Yoneda, Y., and Imamoto, N. (2005) J. Cell Biol. 171 19-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuji, S., Okamoto, M., Yamada, K., Okamoto, N., Goitsuka, R., Alnold, R., Kiefer, F., and Kitamura, D. (2001) J. Exp. Med. 194 529-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura, S., Tatuno, I., Noguchi, Y., Kitagawa, M., Kohn, L. D., Saito, Y., and Hirai, A. (1999) Biochem. Biophys. Res. Commun. 257 340-343 [DOI] [PubMed] [Google Scholar]
- 32.Xeuwei, S., Reddy, M. S., Baev, D., and Edgerton, M. (2003) J. Biol. Chem. 278 28553-28561 [DOI] [PubMed] [Google Scholar]
- 33.Miralski, K. L., Welch, W. J., and Morimoto, R. I. (1989) J. Cell Biol. 108 413-423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies, P. J., Davies, D. R., Levitzki, A., Maxfield, F. R., Milhaud, P., Willingham, M. C., and Pastan, I. H. (1980) Nature 283 162-167 [DOI] [PubMed] [Google Scholar]
- 35.Mandell, R. B., and Feldherr, C. M. (1990) J. Cell Biol. 111 1775-1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welch, W. J., and Mizzen, L. A. (1988) J. Cell Biol. 106 1117-1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deluca-Flaherty, C., McKay, D. B., Parham, P., and Hill, B. L. (1990) Cell 62 875-887 [DOI] [PubMed] [Google Scholar]
- 38.Nadler, S., Tepper, M., Schacter, B., and Mazzucco, C. (1992) Science 258 484-486 [DOI] [PubMed] [Google Scholar]
- 39.Nadeau, K., Nadler, S., Saulnier, M., Tepper, M., and Walsh, C. (1994) Biochemistry 33 2561-2567 [DOI] [PubMed] [Google Scholar]
- 40.Panjwani, N., Akbari, O., Garcia, S., Brazil, M., and Stockinger, B. (1999) J. Immunol. 163 1936-1942 [PubMed] [Google Scholar]
- 41.Murakami, Y., Nagata, H., Shizukuishi, S., Nakashima, K., Okawa, T., Takigawa, M., and Tsunemitsu, A. (1994) Biochem. Biophys. Res. Commun. 198 274-280 [DOI] [PubMed] [Google Scholar]
- 42.Lindhe, J., Karring, T., and Lang, P. N. (2003) Clinical Periodontology and Implant Dentistry, 4th Ed., p. 19, Blackwell Munksgaard, Copenhagen
- 43.Lin, Y.-Z., Yao, S. Y., and Hawinger, J. (1996) J. Biol. Chem. 271 5305-5308 [DOI] [PubMed] [Google Scholar]
- 44.Imamura, T., Engleka, K., Zhan, Z., Tokita, Y., Forough, R., Roeder, D., Jackson, A., Maier, J. A. M., Hla, T., and Maciag, T. (1990) Science 249 1567-1570 [DOI] [PubMed] [Google Scholar]
- 45.Komi, A., Suzuki, M., and Imamura, T. (1998) Exp. Cell Res. 243 408-414 [DOI] [PubMed] [Google Scholar]
- 46.Sherr, C. J., and Roberts, J. M. (1995) Genes Dev. 9 1149-1163 [DOI] [PubMed] [Google Scholar]
- 47.Nakayama, K., and Nakayama, K. I. (1998) BioEssays 20 1020-1029 [DOI] [PubMed] [Google Scholar]
- 48.Polyak, K., Lee, M. H., Erdjument-Bromage, H., Koff, A., Roberts, J. M., Tempst, P., and Massague, J. (1994) Cell 78 59-66 [DOI] [PubMed] [Google Scholar]
- 49.Toyoshima, H., and Hunter, T. (1994) Cell 78 67-74 [DOI] [PubMed] [Google Scholar]
- 50.Tomoda, K., Kubota, Y., and Kato, J. (1999) Nature 398 160-165 [DOI] [PubMed] [Google Scholar]
- 51.Malicet, C., Hoffmeister, A., Moreno, S., Closa, D., Dagorn, J. C., Vasseur, S., and Iovanna, J. L. (2006) Biochem. Biophys. Res. Commun. 339 284-289 [DOI] [PubMed] [Google Scholar]
- 52.Zeise, E., Kühl, N., Kunz, J., and Rensing, L. (1998) Cell Stress Chaperones 3 94-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urushitani, M., Kurisu, J., Tateno, M., Hatakeyama, S., Nakayama, K., Kato, S., and Takahashi, R. (2004) J. Neurochem. 90 231-244 [DOI] [PubMed] [Google Scholar]
- 54.Connor, M. K., Kotchetkov, R., Cariou, S., Resch, A., Lupetti, R., Beniston, R. G., Melchior, F., Hengst, L., and Slingerland, J. M. (2003) Mol. Biol. Cell 14 201-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takeda, S., and McKay, D. B. (1996) Biochemistry 35 4636-4644 [DOI] [PubMed] [Google Scholar]
- 56.Buchberger, A., Theyssen, H., Schröder, H., McCarty, J. S., Virgallita, G., Milkereit, P., Reinstein, J., and Bukau, B. (1995) J. Biol. Chem. 270 16903-16910 [DOI] [PubMed] [Google Scholar]
- 57.Rapaport, E., Garcia-Blanco, M. A., and Zamecnik, P. C. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 1643-1647 [DOI] [PMC free article] [PubMed] [Google Scholar]