Abstract
Phosphatidylinositol (PI) 3-kinase mediates multiple pathways that regulate many aspects of the cell including metabolism, survival, migration, and proliferation. Both Akt and cytokine-independent survival kinase (CISK)/SGK3 are known AGC family protein kinases that function downstream of PI 3-kinase. Although the Akt signaling pathway has been studied extensively, the specific signaling cascades that are modulated by CISK remain to be elucidated. To understand CISK function, we affinity-purified the CISK protein complex and identified Flightless-I (FLII) as a novel downstream target of CISK. Here we show that FLII is an in vivo substrate of CISK that functions downstream of PI 3-kinase. CISK can associate with FLII and phosphorylate FLII at residues Ser436 and Thr818. FLII has been shown to act as a co-activator for nuclear hormone receptors such as estrogen receptor (ER). We demonstrate here that CISK can enhance ER transcription, which is dependent on its kinase activity, and mutation of CISK phosphorylation sites on FLII attenuates its activity as an ER co-activator. Furthermore, FLII knockdown by RNA interference renders 32D cells more sensitive to interleukin-3 withdrawal-induced apoptosis, suggesting that FLII itself is also a survival factor. These findings support the model that CISK phosphorylates FLII and activates nuclear receptor transcription and suggest a new cell survival signaling pathway mediated by PI 3-kinase and CISK.
Cell death and survival are tightly regulated throughout development, through the action of numerous factors and pathways (1–6). Of these, PI2 3-kinase and its downstream effectors are among the most widely studied. PI 3-kinase pathway is essential for survival and proliferation of mammalian cells and has been implicated in cancer (7–10). Through the regulation of D3-phosphoinositol levels in cells, PI 3-kinases control the activity of 3-phosphoinositide-dependent kinase and members of the AGC (cAMP-dependent protein kinase/protein kinase G/protein kinase C) extended superfamily of kinases, including Akt and SGK isoforms (11–16). Akt1 has been shown to promote cell survival, by activating NF-κB (17–19), phosphorylating, and inhibiting pro-apoptotic proteins such as Bad and forkhead transcription factors (20–25).
As in the case of Akt and SGK1, CISK/SGK3 also functions downstream of PI 3-kinase, and its in vivo kinase activity can be inhibited by PI 3-kinase inhibitors (11). Originally cloned from an enhanced retroviral mutagen-mediated cell survival genetic screen (11), CISK overexpression allows IL-3-dependent cells to survive in the absence of IL-3. CISK exhibits high homology in the kinase domain to other SGK family proteins and all three isoforms of Akt and is capable of phosphorylating Akt substrates such as Bad and forkhead transcription factor FKHRL1 (11). Interestingly, unlike other SGK family members, CISK mRNA levels do not change in response to serum or glucocorticoid stimulation (26).
CISK is the only member of the SGK family kinases that contains a Phox homology (PX) domain (11). Similar to the pleckstrin homology domain of Akt, the PX domain of CISK can also bind phosphoinositides (27). CISK PX domain preferentially binds phosphatidylinositol 3-phosphate, phosphatidylinositol 3,5-bisphosphate, and phosphatidylinositol 3,4,5-trisphosphate and targets CISK to early endosomes (27, 28). In contrast, Akt exhibits diffuse staining in the cytosol with low amount of nuclear staining (29). Endosomes are important vesicles for protein sorting and trafficking. Growth factor receptors are usually sorted in endosomes for recycling or degradation (30). The endosomal localization of CISK suggests that CISK may regulate distinct pathways from Akt.
CISK is known to up-regulate a variety of transport systems when overexpressed in Xenopus oocyte (31–33). CISK knock-out mice have impaired intestinal sodium-dependent glucose transport but normal intestinal transport of phenylalanine, cysteine, glutamine, and proline (34). These mice also display defects in hair follicle development with delayed hair development and abnormal hair follicles in adulthood (35). Decreased cell proliferation and fewer hair bulb progenitors appear to contribute to these defects. Interestingly, CISK null mice show striking resemblance to epidermal growth factor receptor null mice in their hair development phenotypes (36). Epidermal growth factor is known to translocate from the cell surface through endosomes and has been shown to co-localize with CISK in the same vesicles during its translocation process (27). Therefore, CISK inactivation in mice might impair epidermal growth factor signaling via endosome trafficking. Akt levels are increased in CISK knock-out mice, suggesting possible functional overlap between CISK and Akt (37). Wnt signaling pathway has been proposed to mediate the impaired hair development in CISK knock-out mice (35). In CISK null mice, reduced nuclear localization of β-catenin was observed, and overexpression of CISK can increase lymphoid enhancer factor-1 transcription in vitro. However, the in vivo lymphoid enhancer factor-1 transcription activity in hair follicle cells remains unchanged in CISK null mice (37).
The CISK knock-out mouse model has provided valuable information regarding the physiological function of CISK (34, 35, 37–39) but still left unanswered questions regarding the molecular mechanism of CISK signaling or the physiological substrates of CISK. One potential CISK target may be the E3 ubiquitin ligase AIP4. AIP4 can interact with CISK and be phosphorylated by CISK in vitro, which in turn may be critical to CISK-mediated inhibition of the degradation of the chemokine receptor CXCR4 (40). To identify potential CISK substrates in the CISK pathway, we purified the CISK complex from IL-3-dependent 32D cells overexpressing FLAG-tagged CISK. Here we report the identification of the mammalian homologue of Drosophila flightless I (FLII) as a novel in vivo substrate of CISK. Furthermore, we demonstrate that CISK and FLII regulate cell survival as well as the transcriptional activity of nuclear receptors.
MATERIALS AND METHODS
Constructs and Cell Lines—Full-length or mutant CISK cDNAs were cloned into the pcDNA or pBabe expression vector. The CISK construct used for large scale immunoprecipitation is tagged with HA at the N terminus and FLAG at the C terminus. Activated CISK (myrCISK) was tagged with the myristylation sequence at the N terminus and the FLAG epitope at the C terminus. The CISK K191A mutant was generated by mutating Lys191 in the ATP-binding site of the kinase domain to Ala. Full-length or mutant FLII cDNAs were cloned into the pCL vector and tagged with FLAG at both termini. Ser436 and Thr818 mutations were generated by site-directed mutagenesis. 293T and HeLa cells were used for kinase assays and luciferase assays. Retroviruses encoding various CISK and FLII proteins were generated by transfecting BOSC23 cells with various CISK and FLII constructs and then used to infect 32D cells. These cells were selected in puromycin (1 μg/ml) for 3 days after infection to generate stable lines. Puromycin-selected cells were allowed to recover for 24 h before further analysis.
RNAi Vectors and Sequences—The sequences were as follows: FLII shRNA1, gatccccGTTCTACGAGGCTGACTGCttcaagagaGCAGTCAGCCTCGTAGAACtttttgga; mouse FLII mFLII-SR3-1, gatccccACTGAAGAAACTCGTCCTGAA-ttcaagagaTTCAGGACGAGTTTCTCAGTtttttgga; human CISK shRNA3, gatccccGGCAGCTTTGGCAAGGTTC-ttcaagagaGAACCTTGCCAAAGCTGCCtttttgga; human CISK shRNA41, gatccccGAGAACGGTCCTTTCCTGA-ttcaagagaTCAGGAAAGGACCGTTCTCtttttgga; and mouse CISK shRNA43, gatccccGCGAGTGGTTTGTCTTCAG-ttcaagagaCTGAAGACAAACCACTCGCtttttgga. These DNA fragments were cloned into the pcl-mU6 shRNA retroviral vector between the BamHI and HindIII sites as previously described (41). siRNA oligonucleotides for vimentin, hFLII (GUGGUACAACAUCGACUUC), and human CISK (GUUCUGGUUUCAGTGGGUU) were ordered from Dharmacon.
Preparation of S100 Fraction—32D cells overexpressing CISK were grown in suspension to 1 × 106 cells/ml. All of the steps were carried out at 4 °C unless otherwise noted. More than 109 cells were collected and washed in cold phosphate-buffered saline and hypotonic buffer (10 mm Tris, pH 7.3, 10 mm KCl, 1.5 mm MgCl2, 0.2 mm phenylmethylsulfonyl fluoride, and 10 mm 2-mercaptoethanol). The cells were then allowed to swell for 15 min in hypotonic buffer, homogenized until percentage of cell lysis reached ∼80%, and centrifuged at 14,000 × g for 15 min. High salt buffer (30 mm MgCl2, 300 mm Tris, pH 7.3, and 1.4 m KCl) was added to the supernatant (at 1/11 of v/v ratio), followed by centrifugation at 44,000 × g for ≥1 h. Cleared supernatant was then dialyzed in BC100 buffer (20 mm Tris, pH 7.3, 20% glycerol, 0.2 mm EDTA, 100 mm KCl, 0.2 mm phenylmethylsulfonyl fluoride, and 10 mm 2-mercaptoethanol) for 3 h and aliquotted for storage at -80 °C. The aliquots were centrifuged again at 44,000 × g for ≥20 min just before immunoprecipitation experiments.
Immunoprecipitation, Western Blotting, and Antibodies— For large scale immunoprecipitation of the CISK complex, anti-FLAG M2 antibody-conjugated agarose beads (100 μl) (Sigma) were incubated with 50 mg of S100 lysate and incubated for 3 h at 4 °C. The beads were then washed four times with 1× NETN (1 mm EDTA, 40 mm Tris, pH 8, 100 mm NaCl, and 0.5% Non-idet P-40). The associated proteins were subsequently eluted from the beads using 200 μg/ml FLAG peptides (Sigma).
For small scale immunoprecipitation, the cells were freshly lysed in lysis buffer (20 mm Tris, pH 8.0, 150 mm NaCl, 10% glycerol, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 0.15 unit/ml aprotinin, 20 μm leupeptin, 1 mm sodium vanadate, 1 μm β-glycerol phosphate, and 1 μm dithiothreitol), followed by a 2-h incubation with appropriate primary antibodies and an additional hour of incubation with protein A/G-agarose beads (Santa Cruz) at 4 °C. The beads were then washed four times with lysis buffer and then boiled in 2× SDS Lamini buffer. For Western blotting analysis, the immunoprecipitates or cell lysates were resolved by 8% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Bio-Rad).
Rabbit polyclonal antibodies for CISK (against the PX domain) and phosphorylated FLII (against sequences surrounding Ser436) were generated by Bethyl Laboratories. The monoclonal FLII antibody was purchased from Santa Cruz Laboratories. Anti-Akt substrate antibodies (from Cell Signaling) were raised against RXRXX(S/T) sequences. Anti-FLAG and anti-HA antibodies were from Sigma-Aldrich.
In Vitro Kinase Assay—Lysates from 1 × 107 293T cells cotransfected with FLAG-tagged CISK and FLAG-tagged FLII were immunoprecipitated with 15 μl of anti-FLAG conjugated agarose beads (Sigma). The beads were then used for in vitro kinase assays as previously described (42). Briefly, the reaction was carried out in 20 μl of reaction volume for 30 min at 30 °C in kinase buffer (50 mm Tris, pH 7.5, 10 mm MgCl2, 1 mm dithiothreitol) plus 40 μm ATP, 10 μCi of [γ-32P]ATP (3,000 cpm/pmol), and the immunoprecipitates. The reaction mixtures were subsequently resolved by SDS-PAGE, blotted onto polyvinylidene difluoride membranes, and exposed for autoradiography or Western blotted with appropriate antibodies.
Cell Survival Assay—Cell survival was measured by trypan blue exclusion assay (11) or propidium iodide staining followed by flow cytometry. 32D cells stably expressing different CISK or FLII shRNA sequences were plated in the absence of IL-3 and collected at different time points after IL-3 withdrawal to assay for cell survival.
Luciferase Assays—Luciferase assays were carried out as described (11). HeLa cells were transiently transfected with various constructs at different concentrations using Lipofectamine 2000 (Invitrogen). The total amount of transfected DNA was normalized using pcDNA3. RNAi oligonucleotides were transfected using TransIt TKO transfection reagent (Mirus). The ER-dependent reporter construct (50 ng/transfection) and the ERα construct (5 ng/transfection) were described previously (43). β-Galactosidase (50 ng/transfection) was used as a control to normalize transfection efficiency. 17β-Estradiol (1 ng/ml) was added on the second day post-transfection. The cells were subsequently incubated for one more day before being harvested to assay for luciferase activity as described in the manufacturer's manual (Promega).
RT-CES Assay—MCF-7 cells expressing various CISK and FLII constructs were generated by retroviral transduction. The cells were then selected in puromycin for 3 days and allowed to recover for at least 24 h before further analysis. These cells were subsequently cultured in estrogen-free medium for 3 days, seeded onto E-plates in the absence or presence of 10-10 m 17β-estradiol (E2) and monitored using the RT-CES® system from ACEA Biosciences. Cell index curves were obtained using the RT-CES® SP software. The coefficients (slopes) of the curves were then calculated and graphed. The experiments were repeated at least twice for each cell line, and a similar trend was observed for each cell line.
RESULTS
Identification of FLII in the CISK Protein Complex—SGK family kinases including CISK and Akt isoforms share high sequence homology and a number of downstream targets (10, 16). To determine the pathways unique to CISK, we performed large scale immunoprecipitations to affinity purify CISK and its associated proteins from the cytosolic fractions of 32D cells expressing FLAG-tagged CISK. The precipitated proteins were then analyzed by mass spectrometry sequencing.
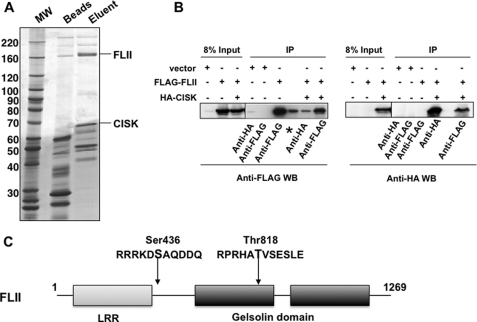
Among the CISK-associated proteins was the mammalian homologue of Drosophila melanogaster FLII (Fig. 1A) (44). FLII belongs to the gelsolin family of actin-binding proteins (Fig. 1C). Homozygous knock-out of FLII led to embryonic lethality in mice, highlighting the importance of FLII for early embryonic development (45). To confirm the physical interaction between FLII and CISK in vivo, we performed co-immunoprecipitation experiments using 32D cells expressing FLAG-tagged FLII alone or together with HA-tagged CISK. As shown in Fig. 1B, immunoprecipitation of HA-CISK could bring down FLAG-FLII (left panel), and immunoprecipitation of FLAG-FLII could pull down HA-CISK (right panel). These data support the large scale immunoprecipitation data and suggest that FLII can indeed interact with CISK.
FIGURE 1.
Mammalian FLII associates with CISK. A, large scale immunoprecipitation (IP) of FLAG-tagged CISK was performed using the S100 fraction from FLAG-CISK expressing mouse 32D cells with anti-FLAG antibody-conjugated agarose beads. The immunoprecipitates were eluted with FLAG peptides, resolved by SDS-PAGE, and stained with Coomassie Blue. The visible bands were excised and sequenced by mass spectrometry. B, co-immunoprecipitation of FLII and CISK. Cell extracts from 32D cells stably expressing vector alone, FLAG-tagged FLII, or FLAG-FLII and HA-tagged CISK together were immunoprecipitated using either anti-FLAG or anti-HA antibodies. The immunoprecipitates were resolved by SDS-PAGE and analyzed by Western blotting using the indicated antibodies. *, nonrelevant lane. C, diagram of the domain structure of FLII. The putative CISK phosphorylation sites are indicated. LRR, leucine-rich repeat.MW, molecular mass.
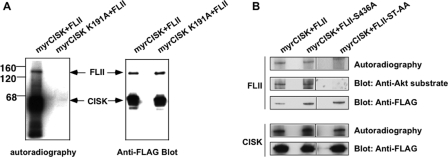
CISK Phosphorylates FLII at Ser436 and Thr818 in Vitro—Next, we examined whether FLII could be a kinase substrate for CISK. To this end, FLII was co-transfected into 293T cells with either myristylated CISK (an active form of CISK) or kinase-dead CISK (CISK K191A mutant) (11). All three constructs were FLAG tagged so that anti-FLAG immunoprecipitation would bring down CISK and FLII together for in vitro kinase assays (42). As shown in Fig. 2A, we could detect phosphorylated FLII in immunoprecipitates of active but not kinase-dead CISK, suggesting that FLII could be phosphorylated by CISK.
FIGURE 2.
CISK phosphorylates FLII at Ser436 and Thr818in vitro. 293T cells co-expressing FLAG-tagged activated CISK (myrCISK, with an N-terminal myristylation signal) or kinase-dead myrCISK mutant (myrCISK K191A) in combination with FLAG-tagged wild type FLII or FLII S436A or the FLII S436A/T818A mutant (FLII ST-AA) were generated. CISK and FLII proteins were co-immunoprecipitated with anti-FLAG antibody-conjugated agarose beads and subjected to in vitro kinase assays in the presence of [32P]ATP. A, the kinase reaction mixtures were resolved by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and analyzed by autoradiography (left) or Western blotting using anti-FLAG antibodies (right). B, CISK can phosphorylate FLII on both Ser436 and Thr818, and phosphorylated FLII can be recognized by anti-Akt substrate antibodies. In vitro kinase reactions were similarly carried out and analyzed as in A.
Akt is known to phosphorylate Ser/Thr within the RXRXX(S/T) motif (46). The high sequence homology between CISK and Akt kinase domains suggests shared substrate recognition sequences between these two kinases. Indeed, we have found CISK to phosphorylate a number of Akt substrates in vitro (11). Sequence analysis revealed two potential CISK phosphorylation sites on FLII, Ser436 and Thr818, that also contain the RXRXX(S/T) motif (Fig. 1C); we therefore reasoned that antibodies against phosphorylated Akt substrates should recognize phosphorylated FLII as well. Indeed, Western blotting with an anti-Akt substrate antibody revealed a band that corresponded to phosphorylated FLII (Fig. 2B). To investigate whether Ser436 and Thr818 could be phosphorylated by CISK, we generated FLII mutants with Ala mutations of either Ser436 (S436A; Fig. 2B) or Thr818 (data not shown). Such single mutations did not affect CISK phosphorylation of FLII, as determined by in vitro kinase assays. However, FLII could no longer be phosphorylated in vitro or recognized by anti-Akt substrate antibodies when both Ser436 and Thr818 were mutated (FLII ST-AA) (Fig. 2B). These data indicate that CISK can phosphorylate FLII at both Ser436 and Thr818 in vitro.
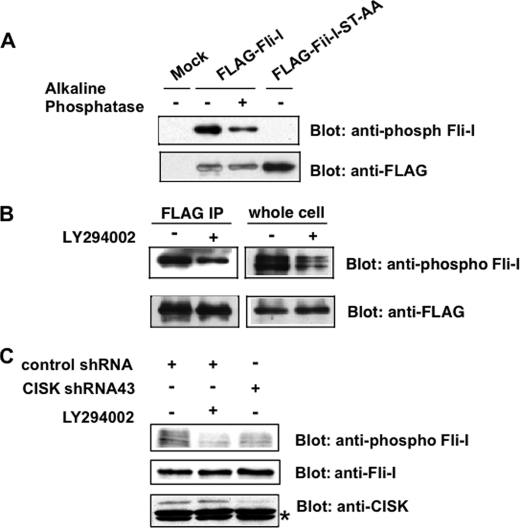
CISK Phosphorylates FLII at Ser436 in Vivo—To examine the in vivo phosphorylation state of FLII, we generated an antibody against phosphorylated Ser436 and its flanking sequences on FLII. 32D cells expressing FLAG-tagged wild type FLII or FLII phosphorylation site mutants were generated. FLII proteins were then immunoprecipitated with anti-FLAG antibodies and Western blotted using the phospho-specific antibody. The anti-Ser(P)436 FLII antibody was able to recognize wild type FLII, but not the S436A/T818A double mutant FLII ST-AA (Fig. 3A) or FLAG-tagged FLII S436A (data not shown). Furthermore, phosphatase treatment also led to a reduction of phospho-antibody reactive signal toward wild type FLII (Fig. 3A). These results suggest that the Ser(P)436-FLII antibody specifically recognizes the phosphorylated Ser436 site on FLII.
FIGURE 3.
CISK phosphorylates FLII at Ser436in vivo. A, generation of a phospho-specific antibody for Ser436 of FLII. FLAG-tagged wild type FLII or the S436A/T818A double mutant (FLII-ST-AA) was expressed in 32D cells. The FLAG-tagged proteins were immunoprecipitated (IP) with anti-FLAG antibody-conjugated agarose beads and eluted using FLAG peptides. For phosphatase treatment, alkaline phosphatase was added to the eluted proteins and incubated for 30 min at 37 °C. The samples were then resolved by SDS-PAGE and Western blotted using the indicated antibodies. B, Ser436 phosphorylation may be regulated by PI 3-kinase. 32D cells expressing FLAG-tagged FLII were treated with the PI 3-kinase inhibitor LY294002 in the absence of IL-3 for 2 h. The cells were then collected and analyzed as described in A. whole cell, whole cell lysate. C, CISK knockdown led to reduced Ser436 phosphorylation. The lysates from 32D cells expressing control shRNA (CISK shRNA41) or CISK shRNA, or LY294002-treated control shRNA expressing 32D cells were Western blotted using the indicated antibodies. The asterisk indicates cross-reactive bands that served as loading controls.
We have shown previously that CISK functions downstream of the PI 3-kinase pathway and that its kinase activity can be inhibited by PI 3-kinase inhibitors (11). If FLII is a substrate of CISK, inhibition of PI 3-kinase activity should also attenuate FLII phosphorylation. To test this, FLAG-FLII expressing 32D cells were first treated with the PI 3-kinase inhibitor LY294002, and total lysates or anti-FLAG immunoprecipitates were then prepared to examine FLII phosphorylation. We were able to observe a ≥50% decrease in FLII phosphorylation (presumably at Ser436) in both total cell lysates and anti-FLAG immunoprecipitates following LY294002 treatment, as determined by the anti-Ser(P)436 FLII antibody (Fig. 3B), indicating that the phosphorylation of FLII at Ser436 is at least partially dependent on the PI 3-kinase pathway. Taken together with our results from immunoprecipitation and in vitro assays, these data point to CISK as the candidate kinase for FLII phosphorylation at Ser436 in vivo.
We went on to investigate FLII phosphorylation in CISK knockdown cells. CISK protein levels can be stably knocked down by ≥80%, in 32D cells stably expressing a small hairpin RNA (shRNA43) against CISK (Fig. 3C). Importantly, FLII phosphorylation at Ser436 (as indicated by the anti-phospho-FLII antibody) was reduced by >50% in these cells (Fig. 3C), lending further support to the hypothesis that phosphorylation of FLII at Ser436 depends on CISK in vivo.
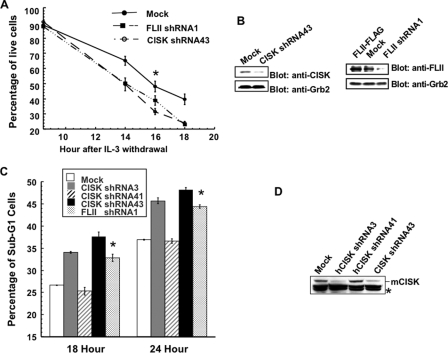
CISK and FLII Promote IL-3-mediated Survival in 32D Cells— CISK was originally identified through an enhanced retroviral mutagen-mediated gain-of-function genetic screen as a survival kinase in IL-3-mediated pathways (47). We found that CISK overexpression could protect 32D cells from IL-3 withdrawal-induced apoptosis (11). To more definitively demonstrate that CISK is indeed required for IL-3-mediated survival, we examined the survival of CISK shRNA43 cells (in which CISK was stably knocked down) in the absence of IL-3. Consistent with our previous findings, RNAi inhibition of CISK rendered the cells more susceptible to IL-3 withdrawal and resulted in ∼20% more cell death at 18 h after IL-3 withdrawal (Fig. 4, A and B). To further confirm this observation, we generated additional CISK shRNA constructs that targeted different regions of CISK for expression in 32D cells. These cells were cultured in the absence of IL-3 and analyzed by propidium iodide staining and flow cytometry. Consistent with our results obtained by trypan blue exclusion assay, the percentage of sub-G1 cells (indicative of apoptotic populations) following IL-3 withdrawal was reversely correlated with the protein level of CISK (Fig. 4, C and D). Expression of the constructs that more efficiently knocked down CISK levels (shRNA3 and shRNA43) resulted in higher percentages of sub-G1 cells, providing strong proof that CISK is indeed a survival factor.
FIGURE 4.
CISK and FLII are required for IL-3 dependent survival. A, 32D cells stably expressing shRNA sequences against CISK or FLII were collected at different time points after IL-3 withdrawal. Percentages of survival cells were determined by trypan blue exclusion. The error bars indicate standard error (≥3 independent experiments). The p value for the time point indicated by the asterisk is <0.03 for FLII and <0.002 for CISK. B, RNAi knockdown of CISK or FLII by shRNA expression. Extracts prepared from 32D cells stably expressing CISK or FLII shRNA sequences were analyzed by Western blotting with the indicated antibodies. Grb2 was used as a loading control. C, 32D cells stably expressing different CISK shRNA sequences or FLII shRNA were collected at 18 or 24 h after IL-3 withdrawal and processed for propidium iodide staining and flow cytometry analysis. The percentages of sub-G1 cells were then calculated and plotted. The error bars indicate standard error (≥3 independent experiments). The p values for the time points indicated by the asterisks are ≤0.001. D, Western blot analysis of CISK expression in 32D cells stably expressing various CISK shRNA constructs. The asterisk indicates a cross-reactive band that was also used as a loading control.
The kinase activity appears critical for CISK survival function (11), indicating that CISK likely mediates survival through phosphorylation of its substrates. FLII was recently reported to be able to inhibit caspase activity and prevent cell death induced by caspase expression (48). In addition, FLII was shown to be a co-activator for nuclear receptors and β-catenin (49, 50), which have also been implicated in cell survival and growth regulation (51–53). We therefore speculated that FLII might be one of the downstream targets in CISK-mediated survival, and reducing FLII expression might compromise the cell survival of IL-3-dependent cells. Indeed, cells with FLII level stably knocked down by shRNA expression were more susceptible to IL-3 withdrawal-induced apoptosis, at a level similar to CISK shRNA43 cells (Fig. 4, A and B). Additionally, FLII knockdown led to an increase in the percentage of sub-G1 cells (Fig. 4C). Taken together with the data that FLII interacts with CISK, these observations indicated that the two proteins likely function in the same pathway to control survival.
CISK/FLII Complex Activates Nuclear Receptor Pathway—Recently, FLII was reported to be a co-activator of nuclear receptors such as ER and thyroid receptor (49). FLII can interact with the nuclear receptor co-activator SRC-2 complex and synergistically increase its transcriptional co-activation activity. Our findings that FLII is a substrate of CISK suggest that CISK-FLII interaction may regulate the transcription activity of nuclear receptors such as ER.
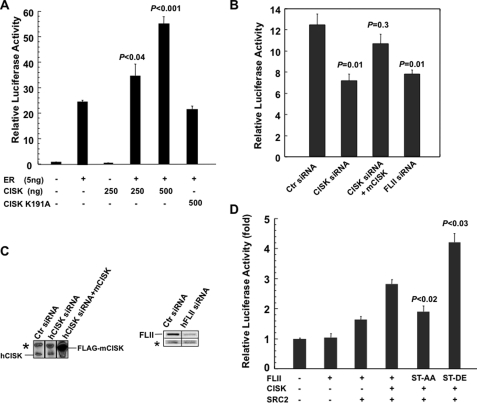
To test this idea, we carried out reporter assays in HeLa cells using a luciferase construct containing 4× ERE (54). As shown in Fig. 5A, co-expressing CISK increased ER-mediated transcriptional activation in a dose-dependent manner. In contrast, expressing kinase-dead CISK failed to achieve the same result, indicating that the kinase activity of CISK is required for enhancing ER transcriptional activity. In addition, knocking down endogenous CISK and FLII expression by siRNA led to decreased luciferase activity in the same reporter assay (Fig. 5, B and C), and the effect of knocking down human CISK on ER activity could be rescued by the co-expression of mouse CISK (mCISK; Fig. 5, B and C). These data suggest that CISK and FLII may activate transcription activity of the estrogen receptor.
FIGURE 5.
CISK positively regulates ER transcription activity through phosphorylation of FLII. A, CISK expression activates ER transcription activity. HeLa cells were transiently transfected with the ER-dependent luciferase reporter ERE-Luc (50 ng), ERα (5 ng), and wild type CISK (in varying amounts) or kinase-dead CISK (K191A). 17β-Estrodial was added to the cells 24 h after transfection. Cells were collected to assay luciferase activities at 40 h after transfection. The error bars indicate standard error (n ≥ 3). The p values were calculated using Student's t test (n ≥ 3). B, RNAi-mediated suppression of CISK and FLII inhibits ER activity. HeLa cells were transiently transfected with ERE-Luc (50 ng), ERα (5 ng), and the indicated siRNA oligonucleotides (100 nm). Mouse CISK (mCISK, 75 ng) was also included for rescue. Ctr siRNA, control siRNA oligonucleotide against vimentin. The error bars indicate standard error (n = 3). The p values, which indicate unpaired probability with control RNAi data, were calculated using Student's t test (n = 3). C, Western blot analysis of cell extracts from B using anti-CISK and FLII antibodies. The asterisk indicates cross-reactive bands that also served as loading controls. D, luciferase assays were performed as described in A. ERE-Luc and ERα were transfected into HeLa cells in combination with CISK, SRC-2, and wild type or mutated FLII. The error bars indicate standard error (n ≥ 3). The p values were calculated using Student's t test (n ≥ 3).
To investigate how CISK may regulate ER activity, we first tested whether CISK could enhance the co-activator function of FLII. As shown in Fig. 5D, co-expressing FLII and SRC-2 increased ER transcriptional activity more than FLII alone, consistent with published data that FLII couples with SRC-2 to activate ER (49). Interestingly, CISK expression further augmented co-activator activity of SRC-2 and FLII on ER. To further determine whether CISK regulates ER activity through phosphorylating FLII, we utilized FLII phosphorylation site mutants. Potential CISK phosphorylation sites on FLII were mutated to either Ala to block phosphorylation (ST-AA for S436A/T818A) or Asp/Glu (ST-DE for S436D/T818E) to mimic phosphorylation. These mutations did not change the interaction between FLII and SRC-2 (data not shown). FLII ST-AA exhibited decreased activity as an ER co-activator, whereas FLII ST-DE exhibited higher co-activation activity for ER (Fig. 5D). Collectively, our data indicate that CISK can up-regulate ER activity by phosphorylating its co-activator FLII.
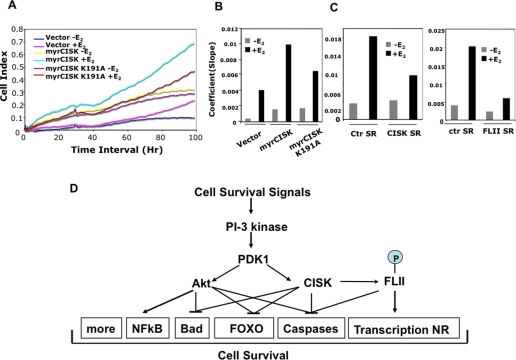
The human breast adenocarcinoma cell line MCF-7 is ER-positive, and estrogen has been shown to protect MCF-7 cells from apoptosis (55). Our results imply that CISK and FLII may regulate estrogen-dependent cell survival. To test this idea, we went on to generate estrogen-responsive MCF-7 cells that stably expressed the various wild type and mutant CISK proteins, as well as shRNA sequences against CISK and FLII. These cells were first cultured in estrogen-free medium for 3 days and then seeded onto special E-plates in the absence or presence of E2. Cell growth was then monitored using RT-CES system (56). In this assay, expression of activated CISK (myrCISK) led to increased cell growth in the presence of E2, whereas mutation in the kinase domain (myrCISK K191A) diminished such an effect (Fig. 6, A and B). In addition, CISK or FLII knockdown by shRNA expression resulted in reduced E2-dependent cell growth (Fig. 6C). These results are consistent with our finding that CISK regulates ER activity through FLII and suggest a new cell survival signaling pathway mediated by PI 3-kinase and CISK.
FIGURE 6.
A, real time measurement of MCF-7 cell survival. MCF-7 cells that stably expressed vector alone, myristylated CISK (myrCISK), or kinase-dead myrCISK (K191A) were seeded onto E-plates in the absence (-) or presence (+) (10-10 m) of E2 and monitored using the RT-CES system to derive the Cell Index (arbitrary units). A representative cell index plot is presented here. B, the coefficients (slopes) from curves described in A were calculated and graphed. C, MCF-7 cells expressing shRNA sequences against human CISK (CISK SR, CISK shRNA43) or human FLII (FLII SR, FLII shRNA1) were similarly examined. The coefficients (slopes) from the curves were calculated and plotted. CISK shRNA41 and a mouse FLII shRNA sequence (mFLII-SR3-1) were used as controls (ctr SR). D, a diagram of the proposed CISK survival signaling pathway.
DISCUSSION
Post-translational modifications such as protein phosphorylation by kinases are pivotal to development and cellular function. Unfortunately, identification of the authentic in vivo substrate(s) of a particular kinase often constitutes the bottle-neck step in studying kinase-mediated pathways. Further complicating this process is the presence of numerous isoforms or homologues that may share substrate sequence recognition. Great progress has been made in more accurately predicating substrate specificities of a kinase and in fishing out potential targets genome wide. The challenge still remains, however, to sift through these findings and locate the true physiological targets.
The availability of a substrate to a kinase is determined by multiple factors including subcellular compartmentalization and formation of protein complexes that may sequester a particular protein. In some cases, the substrates can form relatively stable complexes with their upstream kinases, thereby enabling their identification through affinity purification. In this study, we have successfully identified FLII as a CISK-associated protein and an in vivo substrate of CISK, providing important clues to understanding the signaling pathways controlled by CISK.
With a Ser/Thr kinase domain that is highly homologous to Akt (26) and a PX domain that can interact with phospholipids (27), the structural organization of CISK may afford CISK unique functions compared with other members of the AGC family of kinases. We identified the mammalian homologue of Drosophila FLII in the CISK complex. FLII is a gelsolin family actin-binding protein, containing a leucine-rich repeat domain in its N terminus (57). We present evidence here that supports the notion that the Ser436 and Thr818 residues of FLII are potential CISK phosphorylation sites (Figs. 2 and 3). For example, CISK can phosphorylate these sites in vitro, and in vivo Ser436 phosphorylation could be detected using a phospho-specific antibody. Furthermore, phosphorylation of FLII at Ser436 can be attenuated by CISK knockdown or treatment with PI 3-kinase inhibitors. These data collectively provide strong proof that FLII is an in vivo substrate of CISK.
The CISK-FLII connection provides new insight into the mechanisms of CISK and FLII activity. CISK was originally identified as a survival kinase (11). This survival activity may be important for hair development in mice, as evidenced in CISK knock-out mice that exhibit impaired proliferation in hair follicle development and reduced hair bulb progenitor cells (35). Both CISK inhibition and FLII inhibition by RNAi-mediated knockdown could sensitize 32D cells to apoptosis induced by IL-3 withdrawal (Fig. 4), suggesting that FLII functions in the same survival pathway downstream of CISK (Fig. 6D). Previous studies have also linked CISK to the regulation of endosomal trafficking of survival signal molecules such as growth factor and chemokine receptors (35, 37, 40). FLII is an actin-binding protein whose phosphorylation by CISK might impact actin reorganization and endosome trafficking. Interestingly, CISK may also attenuate degradation of receptors through its interaction with the E3 ubiquitin ligase AIP4 (40). The molecular relationships among different CISK-associated proteins warrant further exploration.
Our studies have placed FLII as a downstream mediator of PI 3-kinase/CISK signaling pathway. FLII may prevent apoptosis by directly inhibiting caspase activity following cytokine depletion (Fig. 6D) (48). Additionally, CISK and FLII may modulate cell survival through the pathway presented here. Nuclear receptors including ER are known to control the expression of several genes that are involved in cell survival and growth (58). FLII has recently been identified to complex with the nuclear receptor co-activator SRC-2 and CARM1 (49). Overexpressing FLII could increase the transcription activity of ER that is dependent on SRC-2. We demonstrate here that CISK and its kinase activity are required for full ER activation (Fig. 5, A and B). Interestingly, mutations of CISK phosphorylation sites of FLII affected its ability to regulate ER activity (Fig. 5D). These data suggest that one important function of CISK is regulating nuclear receptor (in this case ER) pathways through phosphorylating FLII and that CISK may modulate the activity of ER and/or other nuclear receptors to execute its cell survival function.
Estrogen is important throughout development, not only for regulating female reproductive systems but also bone formation (59) and brain development (60). Increased ER activity tightly correlates with breast cancer and ovarian cancer (61). ER-positive breast cancer can be effectively treated with the anti-estrogen drug tamoxifen but tamoxifen-resistant tumors eventually develop. This resistance has been correlated with increased levels of growth factors such as IGF-1 and heregulin that can activate PI 3-kinase (62). PI 3-kinase can increase both estrogen-dependent and independent ER activity in vivo. Estrogen-independent ER activation can be mediated through Akt (63, 64), where Akt may activate ER by directly phosphorylating Ser167 (65). However, PI 3-kinase can also activate ER through pathways other than Akt. Our study suggests CISK as an alternative kinase that may function in Akt-independent ER activation downstream of PI 3-kinase.
CISK regulation of ER most likely occurs via phosphorylating FLII. Although phosphorylation of FLII by CISK does not affect its interaction with SRC-2 (data not shown), it might still affect FLII interaction with other proteins in the transcription activation complex. FLII can translocate between the nucleus and cytoplasm (66). The endosome localized CISK might regulate the trafficking of FLII and other proteins required for ER-dependent transcription. We did not detect any gross changes in the overall localization of FLII with Ser436 and Thr818 mutations (data not shown). However, such observations could not rule out the possibility that CISK regulates the dynamic translocation of FLII upon estrogen stimulation. Further investigation is needed to detail how CISK phosphorylation of its target may affect ER-dependent transcription and to offer new insight into the development and progression of diseases such as breast cancer.
Acknowledgments
We thank A. Safari, Paola Mussi, and Liuh-Yow Chen for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants CA84208 and GM069572 (to Z. S.) and CA119689 (to J. M. X.). This work was also supported by funds from the Welch Foundation (to Z. S.).
Footnotes
The abbreviations used are: PI, phosphatidylinositol; CISK, cytokine-independent survival kinase; SGK, serum and glucocorticoid-regulated kinase; FLII, Flightless-I; ER, estrogen receptor; ERE, estrogen-responsive element; shRNA, small hairpin RNA; RNAi, RNA interference; IL, interleukin; PX, Phox homology; E2, 17β-estradiol; E3, ubiquitin-protein isopeptide ligase; HA, hemagglutinin; siRNA, small interfering RNA.
References
- 1.Wyllie, A. H. (1997) Eur. J. Cell Biol. 73 189-197 [PubMed] [Google Scholar]
- 2.Nagata, S. (1997) Cell 88 355-365 [DOI] [PubMed] [Google Scholar]
- 3.Thompson, C. B. (1995) Science 267 1456-1462 [DOI] [PubMed] [Google Scholar]
- 4.Evan, G. I., and Vousden, K. H. (2001) Nature 411 342-348 [DOI] [PubMed] [Google Scholar]
- 5.Green, D. R. (2003) Immunol. Rev. 193 5-9 [DOI] [PubMed] [Google Scholar]
- 6.Lowe, S. W., Cepero, E., and Evan, G. (2004) Nature 432 307-315 [DOI] [PubMed] [Google Scholar]
- 7.Bellacosa, A., Testa, J. R., Staal, S. P., and Tsichlis, P. N. (1991) Science 254 274-277 [DOI] [PubMed] [Google Scholar]
- 8.Chang, H. W., Aoki, M., Fruman, D., Auger, K. R., Bellacosa, A., Tsichlis, P. N., Cantley, L. C., Roberts, T. M., and Vogt, P. K. (1997) Science 276 1848-1850 [DOI] [PubMed] [Google Scholar]
- 9.Carracedo, A., and Pandolfi, P. P. (2008) Oncogene 27 5527-5541 [DOI] [PubMed] [Google Scholar]
- 10.Manning, B. D., and Cantley, L. C. (2007) Cell 129 1261-1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, D., Yang, X., and Songyang, Z. (2000) Curr. Biol. 10 1233-1236 [DOI] [PubMed] [Google Scholar]
- 12.Brunet, A., Park, J., Tran, H., Hu, L. S., Hemmings, B. A., and Greenberg, M. E. (2001) Mol. Cell. Biol. 21 952-965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alessi, D. R., James, S. R., Downes, C. P., Holmes, A. B., Gaffney, P. R., Reese, C. B., and Cohen, P. (1997) Curr. Biol. 7 261-269 [DOI] [PubMed] [Google Scholar]
- 14.Stokoe, D., Stephens, L. R., Copeland, T., Gaffney, P. R., Reese, C. B., Painter, G. F., Holmes, A. B., McCormick, F., and Hawkins, P. T. (1997) Science 277 567-570 [DOI] [PubMed] [Google Scholar]
- 15.Stephens, L., Anderson, K., Stokoe, D., Erdjument-Bromage, H., Painter, G. F., Holmes, A. B., Gaffney, P. R., Reese, C. B., McCormick, F., Tempst, P., Coadwell, J., and Hawkins, P. T. (1998) Science 279 710-714 [DOI] [PubMed] [Google Scholar]
- 16.Tessier, M., and Woodgett, J. R. (2006) J. Cell. Biochem. 98 1391-1407 [DOI] [PubMed] [Google Scholar]
- 17.Ozes, O. N., Mayo, L. D., Gustin, J. A., Pfeffer, S. R., Pfeffer, L. M., and Donner, D. B. (1999) Nature 401 82-85 [DOI] [PubMed] [Google Scholar]
- 18.Romashkova, J. A., and Makarov, S. S. (1999) Nature 401 86-90 [DOI] [PubMed] [Google Scholar]
- 19.Kane, L. P., Shapiro, V. S., Stokoe, D., and Weiss, A. (1999) Curr. Biol. 9 601-604 [DOI] [PubMed] [Google Scholar]
- 20.Datta, S. R., Dudek, H., Tao, X., Masters, S., Fu, H., Gotoh, Y., and Greenberg, M. E. (1997) Cell 91 231-241 [DOI] [PubMed] [Google Scholar]
- 21.del Peso, L., Gonzalez-Garcia, M., Page, C., Herrera, R., and Nunez, G. (1997) Science 278 687-689 [DOI] [PubMed] [Google Scholar]
- 22.Cardone, M. H., Roy, N., Stennicke, H. R., Salvesen, G. S., Franke, T. F., Stanbridge, E., Frisch, S., and Reed, J. C. (1998) Science 282 1318-1321 [DOI] [PubMed] [Google Scholar]
- 23.Brunet, A., Bonni, A., Zigmond, M. J., Lin, M. Z., Juo, P., Hu, L. S., Anderson, M. J., Arden, K. C., Blenis, J., and Greenberg, M. E. (1999) Cell 96 857-868 [DOI] [PubMed] [Google Scholar]
- 24.Franke, T. F., Hornik, C. P., Segev, L., Shostak, G. A., and Sugimoto, C. (2003) Oncogene 22 8983-8998 [DOI] [PubMed] [Google Scholar]
- 25.Toker, A., and Yoeli-Lerner, M. (2006) Cancer Res. 66 3963-3966 [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi, T., Deak, M., Morrice, N., and Cohen, P. (1999) Biochem. J. 344 189-197 [PMC free article] [PubMed] [Google Scholar]
- 27.Xu, J., Liu, D., Gill, G., and Songyang, Z. (2001) J. Cell Biol. 154 699-705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing, Y., Liu, D., Zhang, R., Joachimiak, A., Songyang, Z., and Xu, W. (2004) J. Biol. Chem. 279 30662-30669 [DOI] [PubMed] [Google Scholar]
- 29.Xu, J., Liu, D., and Songyang, Z. (2002) J. Biol. Chem. 277 35561-35566 [DOI] [PubMed] [Google Scholar]
- 30.Komada, M., and Kitamura, N. (2005) J. Biochem. (Tokyo) 137 1-8 [DOI] [PubMed] [Google Scholar]
- 31.Dieter, M., Palmada, M., Rajamanickam, J., Aydin, A., Busjahn, A., Boehmer, C., Luft, F. C., and Lang, F. (2004) Obes. Res. 12 862-870 [DOI] [PubMed] [Google Scholar]
- 32.Embark, H. M., Bohmer, C., Palmada, M., Rajamanickam, J., Wyatt, A. W., Wallisch, S., Capasso, G., Waldegger, P., Seyberth, H. W., Waldegger, S., and Lang, F. (2004) Kidney Int. 66 1918-1925 [DOI] [PubMed] [Google Scholar]
- 33.Baltaev, R., Strutz-Seebohm, N., Korniychuk, G., Myssina, S., Lang, F., and Seebohm, G. (2005) Pfluegers Arch. 450 26-33 [DOI] [PubMed] [Google Scholar]
- 34.Sandu, C., Rexhepaj, R., Grahammer, F., McCormick, J. A., Henke, G., Palmada, M., Nammi, S., Lang, U., Metzger, M., Just, L., Skutella, T., Dawson, K., Wang, J., Pearce, D., and Lang, F. (2005) Pfluegers Arch 451 437-444 [DOI] [PubMed] [Google Scholar]
- 35.McCormick, J. A., Feng, Y., Dawson, K., Behne, M. J., Yu, B., Wang, J., Wyatt, A. W., Henke, G., Grahammer, F., Mauro, T. M., Lang, F., and Pearce, D. (2004) Mol. Biol. Cell 15 4278-4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Threadgill, D. W., Dlugosz, A. A., Hansen, L. A., Tennenbaum, T., Lichti, U., Yee, D., LaMantia, C., Mourton, T., Herrup, K., Harris, R. C., et al. (1995) Science 269 230-234 [DOI] [PubMed] [Google Scholar]
- 37.Alonso, L., Okada, H., Pasolli, H. A., Wakeham, A., You-Ten, A. I., Mak, T. W., and Fuchs, E. (2005) J. Cell Biol. 170 559-570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grahammer, F., Artunc, F., Sandulache, D., Rexhepaj, R., Friedrich, B., Risler, T., McCormick, J. A., Dawson, K., Wang, J., Pearce, D., Wulff, P., Kuhl, D., and Lang, F. (2006) Am. J. Physiol. 290 R945-R950 [DOI] [PubMed] [Google Scholar]
- 39.Lang, U. E., Wolfer, D. P., Grahammer, F., Strutz-Seebohm, N., Seebohm, G., Lipp, H. P., McCormick, J. A., Hellweg, R., Dawson, K., Wang, J., Pearce, D., and Lang, F. (2006) Behav. Brain Res. 167 75-86 [DOI] [PubMed] [Google Scholar]
- 40.Slagsvold, T., Marchese, A., Brech, A., and Stenmark, H. (2006) EMBO J. 25 3738-3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang, J., Wan, M., Zhang, Y., Gu, P., Xin, H., Jung, S. Y., Qin, J., Wong, J., Cooney, A. J., Liu, D., and Songyang, Z. (2008) Nat. Cell Biol. 10 731-739 [DOI] [PubMed] [Google Scholar]
- 42.Eckhart, W., Hutchinson, M. A., and Hunter, T. (1979) Cell 18 925-933 [DOI] [PubMed] [Google Scholar]
- 43.Li, Q., Chu, M. J., and Xu, J. (2007) Mol. Cell. Biol. 27 8073-8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell, H. D., Schimansky, T., Claudianos, C., Ozsarac, N., Kasprzak, A. B., Cotsell, J. N., Young, I. G., de Couet, H. G., and Miklos, G. L. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 11386-11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campbell, H. D., Fountain, S., McLennan, I. S., Berven, L. A., Crouch, M. F., Davy, D. A., Hooper, J. A., Waterford, K., Chen, K. S., Lupski, J. R., Ledermann, B., Young, I. G., and Matthaei, K. I. (2002) Mol. Cell. Biol. 22 3518-3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawlor, M. A., and Alessi, D. R. (2001) J. Cell Sci. 114 2903-2910 [DOI] [PubMed] [Google Scholar]
- 47.Deng, C. X., and Scott, F. (2000) Oncogene 19 1059-1064 [DOI] [PubMed] [Google Scholar]
- 48.Li, J., Yin, H. L., and Yuan, J. (2008) J. Cell Biol. 181 321-333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Archer, S. K., Behm, C. A., Claudianos, C., and Campbell, H. D. (2004) Biochem. Soc Trans 32 940-942 [DOI] [PubMed] [Google Scholar]
- 50.Lee, Y. H., and Stallcup, M. R. (2006) Nucleic Acids Res. 34 5052-5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kucharova, S., and Farkas, R. (2002) Endocr. Regul. 36 37-60 [PubMed] [Google Scholar]
- 52.McDonnell, D. P., and Norris, J. D. (2002) Science 296 1642-1644 [DOI] [PubMed] [Google Scholar]
- 53.Moon, R. T., Kohn, A. D., De Ferrari, G. V., and Kaykas, A. (2004) Nat. Rev. Genet. 5 691-701 [DOI] [PubMed] [Google Scholar]
- 54.Mussi, P., Liao, L., Park, S. E., Ciana, P., Maggi, A., Katzenellenbogen, B. S., Xu, J., and O'Malley, B. W. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 16716-16721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, T. T., and Phang, J. M. (1995) Cancer Res. 55 2487-2489 [PubMed] [Google Scholar]
- 56.Xing, J. Z., Zhu, L., Jackson, J. A., Gabos, S., Sun, X. J., Wang, X. B., and Xu, X. (2005) Chem. Res. Toxicol. 18 154-161 [DOI] [PubMed] [Google Scholar]
- 57.Campbell, H. D., Fountain, S., Young, I. G., Claudianos, C., Hoheisel, J. D., Chen, K. S., and Lupski, J. R. (1997) Genomics 42 46-54 [DOI] [PubMed] [Google Scholar]
- 58.Musgrove, E. A., Sergio, C. M., Loi, S., Inman, C. K., Anderson, L. R., Alles, M. C., Pinese, M., Caldon, C. E., Schutte, J., Gardiner-Garden, M., Ormandy, C. J., McArthur, G., Butt, A. J., and Sutherland, R. L. (2008) PLoS ONE 3 e2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Syed, F., and Khosla, S. (2005) Biochem. Biophys. Res. Commun. 328 688-696 [DOI] [PubMed] [Google Scholar]
- 60.Peri, A., Danza, G., and Serio, M. (2005) J. Endocrinol. Investig. 28 285-293 [DOI] [PubMed] [Google Scholar]
- 61.Castagnetta, L., Granata, O. M., Cocciadiferro, L., Saetta, A., Polito, L., Bronte, G., Rizzo, S., Campisi, I., Agostara, B., and Carruba, G. (2004) Ann. N. Y. Acad. Sci. 1028 233-246 [DOI] [PubMed] [Google Scholar]
- 62.Herynk, M. H., and Fuqua, S. A. (2007) Adv. Exp. Med. Biol. 608 130-143 [DOI] [PubMed] [Google Scholar]
- 63.Stoica, G. E., Franke, T. F., Wellstein, A., Czubayko, F., List, H. J., Reiter, R., Morgan, E., Martin, M. B., and Stoica, A. (2003) Mol. Endocrinol. 17 818-830 [DOI] [PubMed] [Google Scholar]
- 64.Lonard, D. M., and O'Malley, B. W. (2007) Mol. Cell 27 691-700 [DOI] [PubMed] [Google Scholar]
- 65.Campbell, R. A., Bhat-Nakshatri, P., Patel, N. M., Constantinidou, D., Ali, S., and Nakshatri, H. (2001) J. Biol. Chem. 276 9817-9824 [DOI] [PubMed] [Google Scholar]
- 66.Davy, D. A., Campbell, H. D., Fountain, S., de Jong, D., and Crouch, M. F. (2001) J. Cell Sci. 114 549-562 [DOI] [PubMed] [Google Scholar]