Abstract
A key structural component of amyloid fibrils is a highly ordered, crystalline-like cross-β-sheet core. Conformationally different amyloid structures can be formed within the same amino acid sequence. It is generally assumed that individual fibrils consist of conformationally uniform cross-β-structures. Using mammalian recombinant prion protein (PrP), we showed that, contrary to common perception, amyloid is capable of accommodating a significant conformational switching within individual fibrils. The conformational switch occurred when the amino acid sequence of a PrP variant used as a precursor substrate in a fibrillation reaction was not compatible with the strain-specific conformation of the fibrillar template. Despite the mismatch in amino acid sequences between the substrate and template, individual fibrils recruited the heterologous PrP variant; however, the fibril elongation proceeded through a conformational adaptation, resulting in a change in amyloid strain within individual fibrils. This study illustrates the high adaptation potential of amyloid structures and suggests that conformational switching within individual fibrils may account for adaptation of amyloid strains to a heterologous substrate. This work proposes a new mechanistic explanation for the phenomenon of strain conversion and illustrates the direction in evolution of amyloid structures. This study also provides a direct illustration that catalytic activity of self-replicating amyloid structures is not ultimately coupled with their templating effect.
The ability to form amyloid structures is considered to be one of the most general properties of a polypeptide backbone (1). Regardless of the specific peptides or proteins involved in fibril formation, all types of amyloid fibrils share a common structural motif that consists of a cross-β-structure (2). Cross-β-structures are comprised of highly ordered, nearly anhydrous, crystalline-like β-sheets stabilized by hydrogen bonding and densely packed side chains (3, 4). Growing evidence indicates that multiple amyloid structures referred to as amyloid strains could be formed within the same amino acid sequence (5–7).
Amyloids are capable of self-replicating (8). Self-replicating properties of amyloid fibrils are attributed to the unique arrangement of cross-β-strands that are assembled perpendicular to the fibrillar axis, where β-strands at the growing edge provide a template for recruiting and converting a monomeric precursor. The self-replicating property of the amyloid cross-β-structure consists of two activities: catalytic (i.e. the ability to convert a monomeric precursor into an amyloid state) and templating (i.e. the ability to accurately imprint the strain-specific conformation onto a newly recruited polypeptide). The templating activity is believed to be intimately coupled to the catalytic activity and accounts for the high fidelity of amyloid replication. High fidelity of replication requires identity or high homology between the amino acid sequences of a fibrillar template and a precursor substrate. The species specificity of a template-substrate interaction is believed to account for the species barrier in prion transmission and species specificity of in vitro cross-seeded fibrillation reactions. Local perturbations arising due to mismatches in packing of amino acid side chains within the crystalline-like cross-β-structures could prevent efficient replication of amyloid fibrils.
It is generally assumed that individual fibrils are structurally uniform, i.e. maintain the same structure of a cross-β-core throughout the fibrillar length. In the current study, we showed that, contrary to the common perception, amyloid fibrils are capable of accommodating significant conformational switching within individual fibrils. The conformational switch occurred when the amino acid sequence of the precursor substrate was not compatible with the conformation of the template. Despite mismatched amino acid sequences, individual fibrils were able to recruit the heterologous recombinant prion protein (PrP)2 variant; however, fibril elongation proceeded through switching to a new conformational state. The implications of these studies are multifold. First, our work illustrates the high adaptation potential of amyloid structures and suggests that the conformational switch accounts for adaptation of amyloid strains to the heterologous substrate. Second, the current studies propose a new molecular explanation for the phenomenon referred to as convergence of strains. Third, this work illustrates the directionality in evolution of amyloid structures, showing that the species-specific amyloid structures (i.e. structures that exist only within a single PrP sequence) can give rise to promiscuous or indiscriminative structures (structures compatible with several PrP variants), but not vice versa. Finally, our studies provide direct illustration that catalytic activity of self-replicating amyloid structures is not ultimately coupled with their templating effect.
EXPERIMENTAL PROCEDURES
Mouse and hamster full-length PrPs encompassing residues 23–230 and 23–231, respectively, were expressed and purified as described (9, 10) with modifications (11). To form amyloid fibrils, stock solutions of PrP were prepared immediately before use by resuspending lyophilized PrP powder in 5 mm MES (pH 6.0).
Formation of Amyloid Fibrils in 96-well Plates—Stock solutions of mouse or hamster PrPs were diluted with MES (pH 6.0) and guanidine hydrochloride to final concentrations of 50 mm and 2 m, respectively, and a final protein concentration of 2 μm. Thioflavin T was added to the reaction mixture to a final concentration of 10 μm. The fibrillation reactions were carried out in 96-well plates with a total reaction volume of 0.2 ml/well. To prepare fibrillar seeds for seeded reactions, fibrils were formed in manual format as described below and then sonicated for 10 s using a Bransonic-2510 bath sonicator (Branson Ultrasonics) and added to the reaction mixtures to a final amount of 0.1% (w/w) of PrP substrate. 96-Well plates were incubated at 37 °C with constant shaking at 900 rpm in a Fluoroskan Ascent CF microplate reader (Thermo Labsystems) as described (12). Multiple experiments were performed using mouse and hamster recombinant PrPs purified in separate batches.
Formation of Fibrils in Manual Format—Stock solutions of mouse or hamster PrPs were diluted with MES (pH 6.0) and guanidine hydrochloride to final concentrations of 50 mm and 2 m, respectively, and to a final protein concentration of 0.25–0.5 mg/ml. The fibrillation reaction was carried out in 1.5-ml conical plastic tubes (Fisher) in a total reaction volume of 0.42–0.6 ml at 37 °C with continuous agitation; a Clay Adams Nutator (Model 1105) was used for rotation (24 rpm), whereas the shaking was performed using a DELFIA plate shaker (Wallac) set on high speed (1150 rpm). For the seeded reactions, preformed fibrils were sonicated for 10 s using Bransonic-2510 bath sonicator and added to the reaction mixture to a final amount of 5% as calculated per total amount of PrP substrate. A series of control seeding experiments conducted with hamster PrP as a substrate revealed that the sonication (for up to 1 min) does not impair the ability of hamster S-fibrils (for “shaking”) or R-fibrils (for “rotation”) to replicate their individual conformations.
For the experiments in which composition of individual fibrils was analyzed by immunofluorescence microscopy or atomic force fluorescence microscopy (AFFM), the seeding reactions were carried out in the presence of 30% preformed fibrillar seeds. Seeds were not subjected to sonication prior to seeding.
Immunostaining and Fluorescence Microscopy—PrP fibrils (2 μg/ml) were deposited onto Permanox 8-well Lab-Tek chamber slides and double-stained with antibody as described previously (13) with minor modifications omitting formaldehyde fixation. To analyze fibril composition, the double staining was performed in the following order: 1) anti-PrP human antibody (Ab) D13 (1:3,000; recognizes epitope 96–104), 2) mouse Ab 3F4 (1:3,000; hamster-specific and recognizes epitope 109–112), and 3) a mixture of secondary Abs: goat anti-human and goat anti-mouse labeled with Alexa Fluor 488 and Alexa Fluor 546, respectively (Invitrogen/Molecular Probes; 1:1,000 for both Abs).
To analyze fibril conformation (i.e. the C-terminal epitope exposure), the following procedure for double staining was employed. Fab R2 (1:500; recognizes epitope 225–230) was used instead of Fab D13, and mouse Ab AG4 (1:1,000; recognizes epitope 37–50) was used instead of Ab 3F4. Fluorescence microscopy was carried out on an inverted microscope (Nikon Eclipse TE2000-U) using a 1.3 aperture Plan Fluor ×100 numerical aperture objective. The exposure times were 300 ms for Fab D13, 900 ms for Ab 3F4, 300 ms for Fab R2, and 100 ms for Ab AG4. Collected images were processed with WCIF ImageJ software (National Institutes of Health) as described previously (13). For both experimental settings, i.e. probing of fibrillar composition and conformation, four independent experiments were performed, and five fields of view were analyzed for each sample within each experiment.
AFFM—AFFM studies were performed using a PicoSPM LE AFM scanner (Molecular Imaging, Phoenix, AZ) mounted on the sample holder of the Eclipse TE2000-U inverted microscope equipped with a Photometrics CoolSnap HQ camera (Roper Scientific). Glass coverslips (25 × 25 mm, 0.13–0.17 mm thick; Fisher) were cleaned in two steps: (i) with organic solvents in a sonication bath, followed by (ii) incubation in 10:20:70 H2O2:H2O:H2SO4 (hydrogen peroxide:water:sulfuric acid) solution. After this, coverslips were rinsed extensively with deionized water (MilliQ, Millipore) and stored in it until sample application at +4 °C. The double immunostaining procedure was done as described above for Ab D13/Ab 3F4 with minor changes. Particularly, fibrils were diluted to a higher concentration of 5 μg/ml and deposited on freshly dried coverslips. After the final wash step of immunostaining, samples were covered by Tris-buffered saline with 0.02% sodium azide and stored at +4 °C. Before AFFM imaging, coverslips were rinsed gently but extensively in deionized water and dried with the slightest stream of nitrogen. After mounting the sample and AFM scanner onto the inverted microscope, the AFM tip position was aligned with the center of the shooting field of the camera using a laser diffraction pattern. Upon taking fluorescence pictures, the microscope lens was withdrawn, and AFM scanning was carried out with sample shifts controlled visually by an inverted microscope. The AFM scanner was used in acoustic alternating current mode with a silicon cantilever PPP-NCH (NANOSENSORS) with a tip radius of <7 nm and a spring constant of ∼42 newtons/m. The images (512 × 512 pixel scans) were collected at a scan rate of 0.7–1 lines/s. The resulting fluorescence and AFM amplitude variation images were aligned manually.
Fourier Transform Infrared Spectroscopy (FTIR)—FTIR spectra were measured with a Bruker Tensor 27 FTIR instrument (Bruker Optics, Billerica, MA) equipped with an MCT detector cooled with liquid nitrogen. Fibrils were dialyzed against 10 mm sodium acetate buffer (pH 5.0), concentrated up to 1–2 mg/ml using Nanosep 3K centrifugal devices (Pall, East Hills, NY), and 15 μl of each sample were loaded into a BioATR II cell. Three sets of 512 scans each were collected for each sample at 2 cm-1 resolution under constant purging with nitrogen. Spectra were corrected for water vapor, and background spectra of the same buffer were subtracted. The bands were resolved by Fourier self-deconvolution in the Opus 4.2 software package using a Lorentzian line shape and parameters equivalent to 20 cm-1 bandwidth at half-height and a noise suppression factor of 0.3.
RESULTS
In previous studies, two amyloid strains referred to as S- and R-fibrils were produced in vitro using full-length hamster PrP (7). These two strains were formed using the same stock of highly pure recombinant PrP under identical solvent conditions but different agitation modes: the S-strain was produced under shaking, whereas the R-strain was produced under rotation. As judged from FTIR, hydrogen-deuterium exchange Raman spectroscopy, electron microscopy, atomic force microscopy, and immunoconformational assay, the S- and R-fibrils displayed substantial differences with respect to the structure of the cross-β-cores, epitope exposure, and morphology (7).3
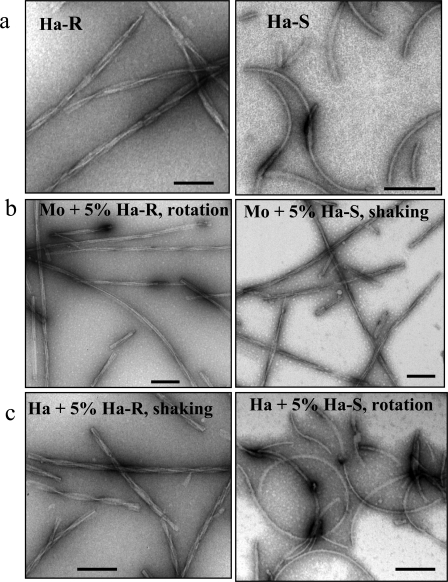
S-fibrils Fail to Replicate Using Mouse PrP as a Substrate—To test whether mouse PrP can adopt S- or R-conformations, we performed cross-seeding experiments, where 5% (w/w) of hamster S- or R-fibrils (Fig. 1a) were added to the conversion reactions containing mouse PrP. To make sure that agitation conditions did not undermine the effect of the seeds, the reactions seeded with R-fibrils were performed under rotation mode, whereas the reactions seeded with S-fibrils were performed under shaking mode. As judged from electron microscopy, the morphology of mouse daughter fibrils resembled that of hamster R-fibrils regardless of whether R- or S-seeds were used for seeding (Fig. 1b). The characteristic features of R-fibrils (mouse or hamster) were straight shape, polymorphous (twisted and non-twisted) morphology, and complex substructure arising from the assembling of multiple filaments, whereas the typical features of S-fibrils were curvy shape and relatively homogeneous smooth morphology. The cross-seeding experiments using mouse PrP as a substrate were repeated multiple times and always produced consistent results, illustrating that mouse PrP failed to acquire S-conformation.
FIGURE 1.
Electron microscopy images of R- and S-fibrils. a, hamster R-fibrils (Ha-R; left panel) and S-fibrils (Ha-S; right panel). b, mouse (Mo) fibrils formed in reactions seeded with 5% hamster R-fibrils under rotation (left panel) and with 5% hamster S-fibrils under shaking (right panel). c, hamster (Ha) fibrils formed in reactions seeded with 5% hamster R-fibrils under shaking (left panel) and with 5% hamster S-fibrils under rotation (right panel). Scale bars, 0.1 μm.
To make sure that failure of mouse PrP to inherit S-conformation was not due to the intrinsically low seeding activity of hamster S-seeds, we performed homologous seeding experiments in which we tested the ability of hamster S- and R-fibrils to transfer their S- or R-specific conformations to daughter hamster fibrils under agitation modes that favored opposite conformation. S-seeds were found to give rise to hamster S-fibrils under rotation, an agitation mode that produces R-fibrils in spontaneous reactions (Fig. 1c). Vice versa, the R-seeds produced hamster R-fibrils under shaking, a mode that gives rise to S-fibrils in non-seeded reactions. These experiments confirmed that both hamster S- and R-seeds replicate their individual conformations, despite unfavorable agitation modes when hamster PrP is used as a substrate.
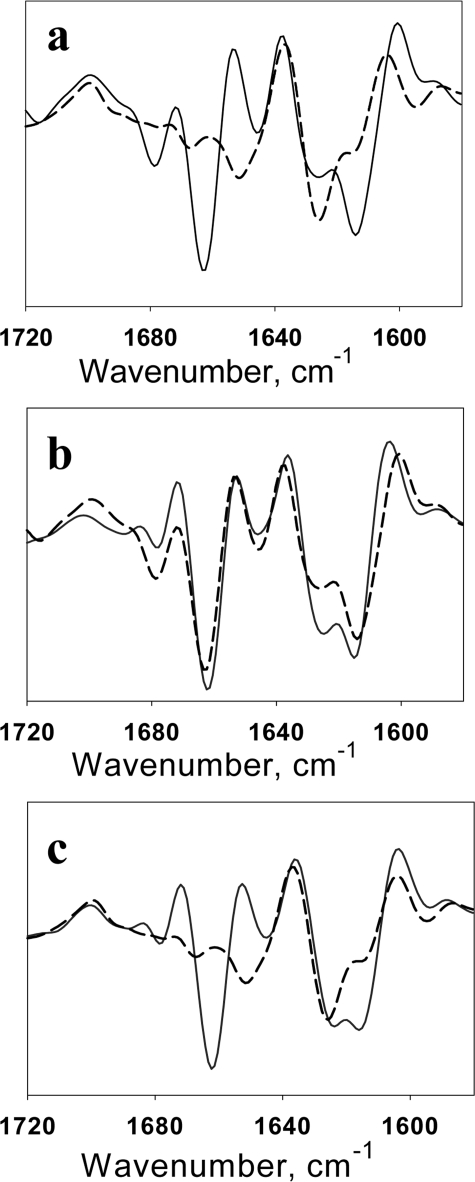
As an alternative method for analysis of cross-seeding reactions, we employed FTIR spectroscopy that revealed previously substantial differences in the secondary structure of hamster S- and R-fibrils (7). Consistent with previous data, a strong peak at 1662 cm-1 was present in the FTIR spectrum of hamster R-fibrils but absent in S-fibrils (Fig. 2a). Furthermore, in the region that accounts for the cross-β-sheet structure, the R-fibrils showed a characteristic double peak (at 1626 and 1613 cm-1), whereas the S-fibrils displayed only a single peak at 1625 cm-1 with a shoulder at 1616 cm-1. The FTIR spectra of daughter mouse fibrils produced in cross-seeded reactions with hamster R- or S-seeds mimicked the FTIR spectra of hamster R-fibrils in both conversion assays (Fig. 2, b and c). Both electron microscopy and FTIR spectroscopy demonstrated that mouse PrP did not acquire S-conformation in cross-seeding experiments with hamster S-fibrils.
FIGURE 2.
Second derivatives of the FTIR spectra. a, spectra of hamster R-fibrils (solid line) and S-fibrils (dashed line). b, spectrum of daughter mouse fibrils formed in reactions seeded with 5% hamster R-fibrils under rotation (solid line) and the spectrum of parent hamster R-fibrils (dashed line). c, spectrum of daughter mouse fibrils formed in reactions seeded with 5% hamster S-fibrils under shaking (solid line) and the spectrum of parent hamster S-fibrils (dashed line).
Mouse PrP Is Not Compatible with the S-conformation—The above experiments suggested that the S-conformation appeared to be hamster-specific, whereas the R-conformation can be accessed by both hamster and mouse PrPs. To test whether mouse PrPs can adopt an S-conformation, we employed an alternative format where the fibrillation reactions were carried out in mixtures of hamster and mouse PrPs under shaking, which is known to favor S-fibrils. As expected, in the absence of mouse PrP, hamster PrP produced S-fibrils as judged by electron microscopy and FTIR (supplemental Fig. S1, a and b). Surprisingly, adding as little as 0.1% of mouse PrP to the reaction mixture with hamster PrP was sufficient to abolish formation of S-fibrils and promote R-fibrils (supplemental Fig. S1a). The FTIR spectrum of fibrils formed in the mixture of hamster and mouse PrPs at a 99.9:0.1 molar ratio was remarkably different from that of hamster S-fibrils but similar to the spectrum of hamster R-fibrils (supplemental Fig. S2b). This experiment provided further support for the idea that mouse PrP is not compatible with the S-structure.
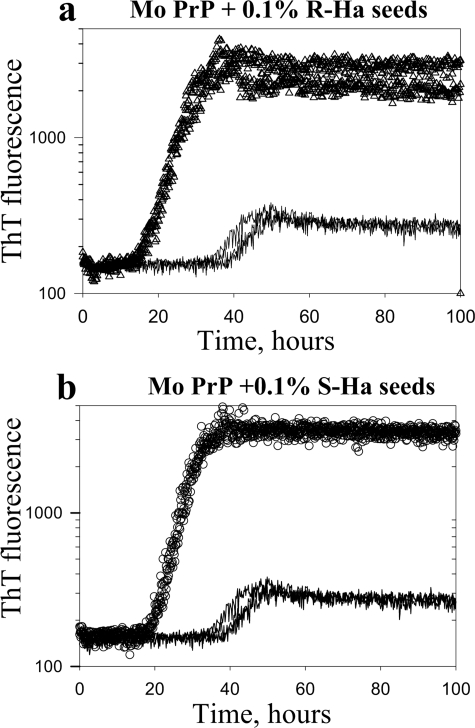
Both S- and R-fibrils Cross-seed Fibrillation of Mouse PrP— To test a possible correlation between the templating effect (the ability to transfer R- or S-specific conformations to daughter fibrils) and the seeding activity, we examined the kinetics of cross-seeded fibrillation. Considering that the S-conformation was not compatible with mouse PrP, we expected that only R-fibrils should exhibit a seeding effect in cross-seeding reactions. Contrary to our expectations, the S- and R-seeds both reduced the lag phase of fibrillation of mouse PrP to similar extent (Fig. 3, a and b). This experiment illustrated that both R- and S-fibrils showed similar efficacy in cross-seeding despite the lack of a templating effect in S-fibril-seeded fibrillation (i.e. the failure of mouse PrP to acquire S-conformation).
FIGURE 3.
Kinetics of cross-seeded fibrillation. a, the kinetics of fibrillation of mouse (Mo) PrP (2 μm) seeded with 0.1% (w/w) of hamster R-fibrils (Ha-R; triangles) and non-seeded controls (solid lines). b, the kinetics of fibrillation of mouse PrP (2 μm) seeded with 0.1% (w/w) of hamster S-fibrils (circles) and non-seeded controls (solid lines). The kinetic curves are shown in duplicates. The kinetics was monitored in 96-well plate as described under “Experimental Procedures.” The remaining lag phase observed in these seeded reactions should not be attributed to the species specificity of seeding between hamster seeds and mouse PrP. Similar lag phases were also observed in homologous seeding assays (9, 14).
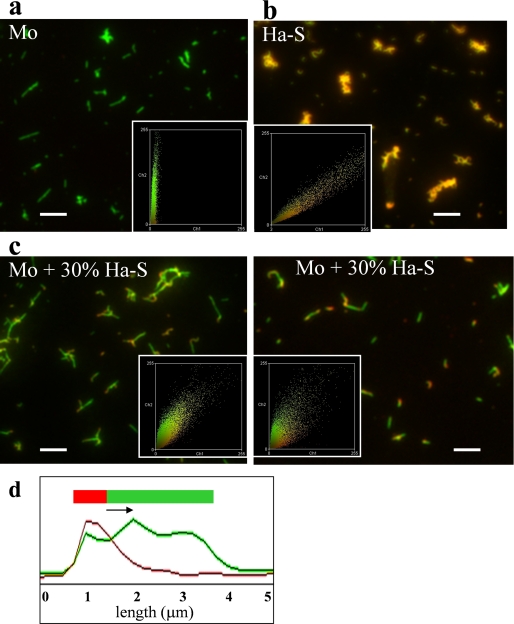
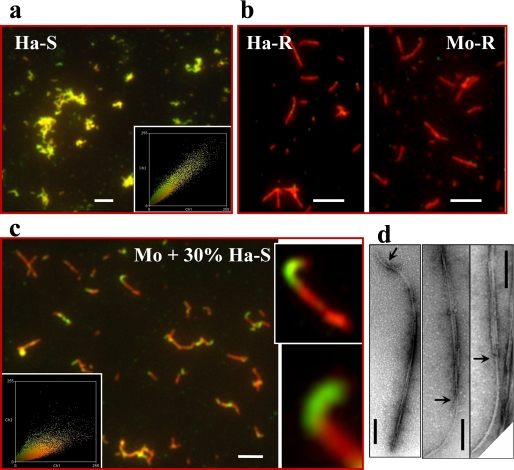
Cross-seeding with S-fibrils Produce “Hybrid” Hamster-Mouse Fibrils—The apparent discrepancy between the observation of seeding activity and the lack of templating effect was puzzling. These findings could be explained by two alternative mechanisms. (i) Cross-seeding did not involve elongation of a pre-existing seed; or (ii) cross-seeding proceeded through elongation of seeds but accommodated a switch from S- to R-conformation within individual fibrils. In contrast to the first mechanism, the last postulates formation of hybrid hamster-mouse fibrils. To answer this question, the composition of individual fibrils formed from S-seeded mouse PrP were examined using a double-staining immunofluorescence microscopy assay (13, 15). Because we wanted to test whether mouse PrP elongates individual S-fibrils, 30% of non-sonicated hamster S-fibrils were used in this experiment instead of 5% sonicated fibrils in the previous experiments. To determine the species of PrP within individual fibrils, Ab 3F4 that reacts with hamster PrP (the secondary Ab to 3F4 was labeled with Alexa Fluor 546; red) was used in combination with Ab D13, which detects both hamster and mouse PrPs (the secondary Ab to D13 was labeled with Alexa Fluor 488; green). Consistent with the color-coding design of this experiment, the fibrils produced in non-seeded reactions from mouse PrP appeared green (Fig. 4a), whereas the S-fibrils of hamster PrP were orange (Fig. 4b). The individual fibrils formed from S-seeded mouse PrP consisted of heterogeneous fragments (orange and green) indicating that these fibrils contained hamster and mouse sections (Fig. 4c). Fluorescence intensity profiles recorded along individual fibrils confirmed formation of hybrid hamster-mouse fibrils (Fig. 4d).
FIGURE 4.
Analysis of fibril composition. Microscopy images of fibrils produced in non-seeded reactions from mouse (Mo) PrP (a), hamster (Ha) PrP (b), or from mouse PrP in reactions seeded with 30% hamster S-fibrils (two panels in c). Fibrils were double-stained with Fab D13 (green) and Ab 3F4 (red). The microscopy images were transformed into two-dimensional fluorescence intensity scattering plots (insets) (13). Red fluorescence intensities are plotted on the horizontal axis, and the green intensities are plotted on the vertical axis. Scale bars, 5 μm. d, the fluorescence intensity profile of hamster S-seeded mouse fibril. The fluorescence profiles was measured along individual fibrils and recorded in both red and green channels.
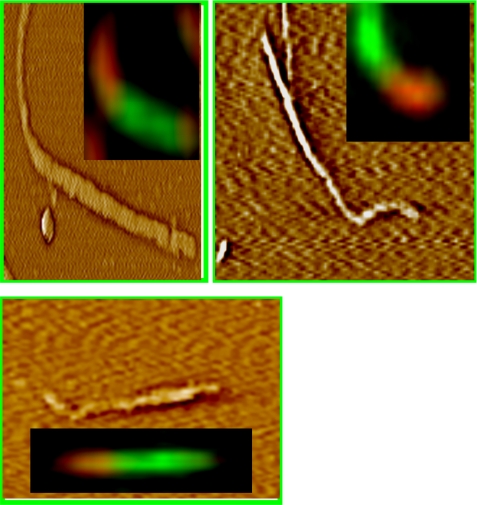
To make sure that hybrid hamster-mouse fibrils were not an artifact of lateral association of preformed hamster and mouse fibrils but were produced as a result of elongation of S-seeds, we employed atomic force fluorescence microscopy (AFFM). AFFM consisted of simultaneous measurements of the topological profile of individual fibrils by AFFM and fibril composition by double immunostaining (15). As judged from AFFM, the individual fibrils consisted of green and orange sections (Fig. 5). The orange sections composed of hamster PrP were curvy, reflecting curvy morphology of S-seeds, whereas the green sections composed of mouse PrP were straight, reflecting the rigid shape of the R-structure.
FIGURE 5.
Atomic force fluorescence microscopy imaging of hamster-mouse fibrils. Fibrils were formed from mouse PrP in the reactions seeded with 30% hamster S-fibrils and stained with Ab 3F4 (red) and Fab D13 (green). Approximately 100 individual hybrid fibrils were examined by AFFM; no signs of the lateral association between preformed individual hamster and mouse fibrils were found. Representative AFFM images shows that individual fibrils consisted of two sections: the sections made of hamster PrP had a curvy S-like shape and displayed red fluorescence, whereas the sections made of mouse PrP had a straight R-like shape and showed green fluorescence. Because fibrils were decorated by primary and secondary antibody, fibril dimensions and morphological details cannot be revealed from AFFM imaging.
The results of the fluorescence microscopy imaging and AFFM provided strong support for the second mechanism of cross-seeding, which implies that S-hamster seeds recruit mouse PrP that participates in elongation of S-fibrils. The alteration in shape within individual fibrils from curvy to straight suggested that elongation of S-seeds involves a switch to R-conformation.
Hybrid Hamster-Mouse Fibrils Show a Switch from S- to R-conformation—To test whether a conformational switch from S- to R-structure indeed occurred within individual fibrils, we probed solvent accessibility of the epitope 225–231 using a double-staining immunoconformational assay. In previous studies, the epitope 225–231 was found to be solvent exposed in S-fibrils but buried in R-fibrils (7, 13). To assess the solvent accessibility, Ab R2 (specific to the epitope 225–231) was used in a pair with reference Ab AG4, that binds to the epitope 37–59, which is known to be solvent-exposed in both R- and S-conformations. In this format, the immunofluorescence assay probes the immunoreactivity of the epitope 225–231, but not the species composition of fibrils. Because the secondary Ab to R2 was labeled with Alexa Fluor 488 (green) and the secondary Ab to AG4 was labeled with Alexa Fluor 546 (red), the S-conformation was expected to yield yellow or green colors in double-staining imaging, whereas the R-conformation was expected to be red. Consistent with this color-coding design, hamster S-fibrils that were used as seeds showed variations in color from yellow to green (Fig. 6a), whereas mouse or hamster R-fibrils were found to be red (Fig. 6b). Individual fibrils produced from mouse PrP in the presence of 30% hamster S-seeds were composed of yellow/green and red parts illustrating a switch from S-conformation (yellow or green parts) to R-conformation (red parts) within individual fibrils (Fig. 6c). Because shaking that was employed for fibrillation caused continuous fibril fragmentation, not all fibrils showed an S-R hybrid nature. Further examination of mouse fibrils produced in hamster S-seeded reactions by electron microscopy confirmed a switch in fibril morphology within individual fibrils from the curvy shape typical for S-conformation to the rigid shape typical for R-conformation (Fig. 6d).
FIGURE 6.
Analysis of fibril conformation. Immunofluorescence microscopy imaging of hamster S-fibrils (Ha-S; a), hamster R-fibrils (Ha-R; b, left), mouse R-fibrils (Mo; b, right), and fibrils produced from mouse PrP in reactions seeded with 30% hamster S-fibrils (c). Two enlarged images of fibrils are included in c. Fibrils were double-stained with Fab R2 (green) and Ab AG4 (red). The exposed conformation of epitope 225–230 is evident by green or yellow colors (a and c). The color variation from green to yellow in a and c arises as a result of stochastic variation in the ratio of R2 and AG4 bound within individual S-fibrils and across the fibrillar population and should not be viewed as an indication of structural polymorphism or heterogeneity within the S-population. The microscopy images were transformed into two-dimensional fluorescence intensity scattering plots (insets) as described previously (13). Red fluorescence intensities are plotted on the horizontal axis, and the green intensities are plotted on the vertical axis. Some structures showed an apparent switch back and forth between S- and R-conformations that was due to artifacts related to co-aggregation or overlap between several individual fibrils. Scale bars, 5 μm. d, electron microscopy images of fibrils produced from mouse PrP in the reactions seeded with 30% hamster S-fibrils. Arrows point to the transition region between S- and R-structures. Scale bars, 0.2 μm.
DISCUSSION
The current studies provided a direct illustration that amyloid structures are capable of accommodating conformational switching within individual fibrils. These findings are very surprising, considering that fibrils comprise crystalline-like structures consisting of highly ordered, densely packed cross-β-sheets (3, 4). Although three-dimensional structures of R- and S-fibrils are not available, several techniques including FTIR and Raman spectroscopy argue that conformational differences between these two amyloid strains do not only involve the fibril periphery but are encrypted in the structures of their cross-β-cores (7).3 Hydrogen-deuterium deep-UV Raman spectroscopy revealed a noticeable disparity in the strength of the cross-β-hydrogen bonds emphasizing differences in atomic structures of S- and R-specific cross-β-cores.3 These structural differences could be steadily propagated under the agitation conditions that favor opposite conformations, illustrating high fidelity of replication of S- or R-specific cross-β-structures (7).
Templating Versus Catalytic Activities of Amyloid Fibrils— The catalytic and templating activities are two key attributes of the amyloid self-replicating process. The ability to convert a monomeric precursor to an amyloid state is a manifestation of fibril catalytic activity, whereas transfer of seed-specific conformations to the daughter fibrils can be considered a manifestation of the templating activity. The catalytic and templating activities of amyloid structures are believed to be intimately coupled because of the self-replicating nature of cross-β-sheets (8).
The current studies showed that amyloid structures could display catalytic activity in the absence of a templating effect. Hamster S-seeds reduced the lag phase in cross-seeded reactions showing catalytic activity in converting mouse PrP. Unexpectedly, the daughter mouse fibrils did not inherit the S-specific conformation of seeds but instead acquired an alternative R-specific conformation. These experiments provide a direct demonstration that the catalytic activity of amyloid fibrils is not always accompanied by the templating effect. The absence of the templating effect is likely to be attributed to the lack of compatibility between the primary structure of a precursor substrate and the conformation of a seed. The newly described phenomenon of conformational switching within individual fibrils reconciles the apparent discrepancy between the ability to recruit/convert mouse PrP and the failure to imprint the S-specific conformation in cross-seeding assays.
Promiscuous or Indiscriminative Versus Species-specific Amyloid Structures—Our current and previous studies showed that the R-fibrils can be produced from hamster PrP, mouse PrP, or even from the mixtures of hamster and mouse PrPs (15). In contrast to the R-fibrils, formation of the S-fibrils appeared to be limited to hamster PrP. Strikingly, adding as little as 0.1% of mouse PrP to the reaction mixture containing hamster PrP was sufficient to abolish formation of S-fibrils in non-seeded reactions. In the cross-seeding assays that utilized mouse PrP as a substrate, the R-fibrils preserved their R-specific conformation (15), whereas the S-fibrils did not (Figs. 1 and 2). Unexpectedly, both R- and S-seeds were able to recruit mouse PrP and to form hybrid hamster-mouse fibrils (Fig. 6) (15). However, to accommodate mouse PrP, hybrid hamster-mouse fibrils formed in the presence of S-seeds displayed a conformational switch from the S- to R-structure within individual fibrils. These results show that two PrP amyloid strains formed within the same amino acid sequence display different compatibility to the heterologous PrP variant.
Considering that the R-structure is compatible with at least two PrP variants, this strain can be referred to as promiscuous or indiscriminative. In contrast, the S-structure appears to be hamster-specific or selective with respect to the PrP amino acid sequence that it can utilize for self-replicating. Therefore, the key difference between the R- and S-structures is in their ability to accommodate mismatches in PrP amino acid sequence without notable changes in individual structures. Consistent with this hypothesis, our previous studies showed that minor modifications in the PrP primary structure such as deletion of a few N-terminal residues or spontaneous oxidation of methionines were sufficient to switch the fibrillation pathway from S- to R-specific, even if such modifications were present in a very minor fraction of PrP (11).
The results of cross-seeding experiments with S- and R-fibrils showed remarkable parallels with the data on interspecies prion transmission. In recent studies, two bovine prion strains, BSE and BASE (bovine spongiform encephalopathy and bovine amyloidotic spongiform encephalopathy, respectively) showed strikingly different behavior upon serial passages in wild type mice (16). BSE was easily transmitted upon primary passage and preserved its individual strain-specific characteristics despite replication in a new host. Serial transmission of the BASE strain to wild type mice, however, showed substantial species barrier and, most surprisingly, induced a disease phenotype indistinguishable from that of BSE-infected mice, including BSE-specific biochemical and neuropathological features (16). In a manner similar to the conversion of the S- to R-structure described in the current study, BASE converted into the classical BSE strain upon replication using mouse PrPC (supplemental Fig. S2). A similar phenomena was described in the classical studies of Bruce and Dickinson (17), in which the scrapie agent 87A originally isolated from sheep was found to produce the ME7 strain upon serial transmission in wild type mice. Notably, 87A converted into ME7 at the dilutions well beyond the limiting dilution for ME7 (this procedure eliminated possible contamination of the original isolate with minor amounts of ME7), arguing that ME7 was generated de novo from 87A. The changes in strain properties observed during interspecies passages were referred to as “breakdown” of a strain and attributed to “mutations in the informational molecule of scrapie agent” (17). Subsequent studies introduced a “strain conversion” model that aimed to explain the strain-switching phenomena within the protein-only hypothesis (8, 18). The current studies could provide a mechanistic explanation for this effect and propose that a conformational switching within individual cross-β-structures underlies the strain conversion phenomenon.
Hierarchy of Amyloid Strains—The current and previous studies suggest that amyloid strains are not equally selective with respect to the amino acid sequence of a precursor substrate that they recruit for elongation. R-strain could be maintained within at least two sequences and showed no selectivity in recruiting mouse versus hamster substrates (this study and Ref. 15). However, the S-strain was able to replicate using only hamster PrP.
This difference in selectivity observed for two amyloid strains indicate a direction in which adaptation or evolution of self-propagating amyloid structures or prion strains could proceed upon interspecies transmission. One can predict that conformational adaptation could proceed only from selective or species-specific structures toward indiscriminative ones but not vice versa. A conversion from a species-specific to a less discriminative or promiscuous strain happens when a species-specific strain faces a heterologous substrate that is not compatible with the conformation of the original strain. Because indiscriminative strains are capable of accommodating mismatches in primary sequence, interspecies transmission will favor indiscriminative strains. The current results support the model that outlines a hierarchical relationship between promiscuous and species-specific strains presented in the recent review article (19).
Adaptation Potential of Amyloid Structure—The observation that amyloid states are capable of conformational switches within individual fibrils illustrates a surprisingly high adaptation potential of amyloid structures. The current work revealed that the elongation of S-fibrils proceeded through adaptive conformational switching, allowing recruitment of the PrP variant that otherwise was not compatible with the existing structure. Considering substantial differences between S- and R-specific cross-β-structures seen by FTIR and deep-UV Raman spectroscopy, conformational switching within individual fibrils should involve a significant change in atomic structure or global fold.
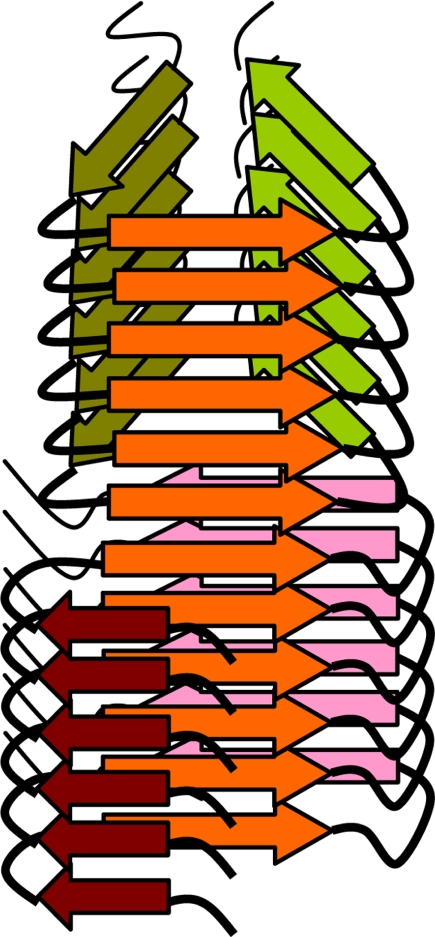
The question of how fibrillar conformation changes without interrupting fibril continuity remains to be addressed in future studies. One can assume that although two structures have substantially different global folds at the same time, they share common local structural motifs that account for the integrity of a hybrid fibril. As shown by a schematic diagram in Fig. 7, the same polypeptide region acquires the identical β-strand conformation within two fundamentally different folding structures. Because the region that acquires the common β-strand conformation is connected by hydrogen bonds to the identical region in the polypeptide molecules above and below, the β-sheet propagates along the whole length of fibril despite being part of two different global folds within individual molecules. Columns of hydrogen bonds running up and down the common β-sheet provide conformational stability for the whole hybrid structure and a template for recruiting new molecules at the fibrillar edges.
FIGURE 7.
A schematic diagram illustrating conformational switch within individual fibrils. The hybrid fibril consists of two segments with different global folds, both of which share β-strands (orange) with similar conformation that form parallel β-sheets. This diagram does not intend to model PrP structure within amyloid fibrils.
The implications of this newly observed phenomenon are manifold. An adaptive conformational switching within individual fibrils may provide a mechanistic explanation for modification or emergence of new prion strains (18, 20) or for the cross-seeding effect observed between non-homologous amyloidogenic proteins (21). Recent studies showed that amyloidosis of one protein can be initiated via seeding with fibrils of a non-related protein in the complex environment of a cell (22). Cross-talk between several yeast prion proteins provides other examples of direct interactions between newly forming and preexisting heterologous fibrils (23, 24).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant NS045585 (to I. V. B.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S2.
Footnotes
The abbreviations used are: PrP, recombinant prion protein; MES, 4-morpholineethanesulfonic acid; Ab, antibody; AFFM, atomic force fluorescence microscopy.
V. Shashilov, M. Xu, N. Makarava, R. Savtchenko, I. V. Baskakov, and I. K. Lednev, submitted for publication.
References
- 1.Dobson, C. M. (2002) Trends Biochem. Sci. 24 329-332 [DOI] [PubMed] [Google Scholar]
- 2.Tycko, R. (2006) Q. Rev. Biophys. 1-55 [DOI] [PubMed]
- 3.Diaz-Avalos, R., Long, C., Fontano, E., Balbirnie, M., Grothe, R., Eisenberg, D., and Caspar, D. L. D. (2003) J. Mol. Biol. 330 1165-1175 [DOI] [PubMed] [Google Scholar]
- 4.Nelson, R., Sawaya, M. R., Balbirnie, M., Madsen, A. O., Riekel, C., Grothe, R., and Eisenberg, D. (2005) Nature 435 773-778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petkova, A. T., Leapman, R. D., Gua, Z., Yau, W.-M., Mattson, M. P., and Tycko, R. (2005) Science 307 262-265 [DOI] [PubMed] [Google Scholar]
- 6.Tanaka, M., Chien, P., Naber, N., Cooke, R., and Weissman, J. S. (2004) Nature 428 323-328 [DOI] [PubMed] [Google Scholar]
- 7.Makarava, N., and Baskakov, I. V. (2008) J. Biol. Chem. 283 15988-15996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien, P., Weissman, J. S., and DePace, A. H. (2004) Annu. Rev. Biochem. 73 617-656 [DOI] [PubMed] [Google Scholar]
- 9.Bocharova, O. V., Breydo, L., Parfenov, A. S., Salnikov, V. V., and Baskakov, I. V. (2005) J. Mol. Biol. 346 645-659 [DOI] [PubMed] [Google Scholar]
- 10.Breydo, L., Bocharova, O. V., Makarava, N., Salnikov, V. V., Anderson, M., and Baskakov, I. V. (2005) Biochemistry 44 15534-15543 [DOI] [PubMed] [Google Scholar]
- 11.Ostapchenko, V. G., Makarava, N., Savtchenko, R., and Baskakov, I. V. (2008) J. Mol. Biol. 383 1210-1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bocharova, O. V., Breydo, L., Salnikov, V. V., and Baskakov, I. V. (2005) Biochemistry 44 6776-6787 [DOI] [PubMed] [Google Scholar]
- 13.Novitskaya, V., Makarava, N., Bellon, A., Bocharova, O. V., Bronstein, I. B., Williamson, R. A., and Baskakov, I. V. (2006) J. Biol. Chem. 281 15536-15545 [DOI] [PubMed] [Google Scholar]
- 14.Bocharova, O. V., Breydo, L., Salnikov, V. V., Gill, A. C., and Baskakov, I. V. (2005) Protein Sci. 14 1222-1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makarava, N., Lee, C. I., Ostapchenko, V. G., and Baskakov, I. V. (2007) J. Biol. Chem. 282 36704-36713 [DOI] [PubMed] [Google Scholar]
- 16.Capobianco, R., Casalone, C., Suardi, S., Mangieri, M., Miccolo, C., Limido, L., Catania, M., Rossi, G., Di Fede, G., Giaccone, G., Bruzzone, M. G., Minati, L., Corona, C., Acutis, P., Gelmetti, D., Lombardi, G., Groschup, M. H., Buschmann, A., Zanusso, G., Monaco, S., Caramelli, M., and Tagliavini, F. (2007) PLOS Pathog. 3 e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruce, M. E., and Dickinson, A. G. (1987) J. Gen. Virol. 68 79-89 [DOI] [PubMed] [Google Scholar]
- 18.Peretz, D., Williamson, R. A., Legname, G., Matsunaga, Y., Vergara, J., Burton, D., DeArmond, S., Prusiner, S., and Scott, M. R. (2002) Neuron 34 921-932 [DOI] [PubMed] [Google Scholar]
- 19.Baskakov, I. V., and Breydo, L. (2007) Biochim. Biophys. Acta 1772 692-703 [DOI] [PubMed] [Google Scholar]
- 20.Castilla, J., Gonzalez-Romero, D., Saa, P., Morales, R., De Castro, J., and Soto, C. (2008) Cell 134 757-768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Nuallain, B., Williams, A. D., Westermark, P., and Wetzel, R. (2004) J. Biol. Chem. 279 17490-17494 [DOI] [PubMed] [Google Scholar]
- 22.Yan, J., Fu, X., Ge, F., Zhang, B., Yao, J., Zhang, H., Qian, J., Tomozawa, H., Naiki, H., Sawashita, J., Mori, M., and Higuchi, K. (2007) Am. J. Pathol. 171 172-180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derkatch, I. L., Bradley, M. E., Hong, J. Y., and Liebman, S. W. (2001) Cell 106 171-182 [DOI] [PubMed] [Google Scholar]
- 24.Derkatch, I. L., Uptain, S. M., Quteiro, T. F., Krishnan, R., Lindquist, S. L., and Liebman, S. W. (2004) Proc. Acad. Natl. Sci. U. S. A. 101 12934-12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.