Abstract
Stenotrophomonas maltophilia is an opportunistic pathogen characterized for its intrinsic low susceptibility to several antibiotics. Part of this low susceptibility relies on the expression of chromosomally encoded multidrug efflux pumps, with SmeDEF being the most relevant antibiotic resistance efflux pump so far studied in this bacterial species. Expression of smeDEF is down-regulated by the SmeT repressor, encoded upstream smeDEF, in its complementary DNA strand. In the present article we present the crystal structure of SmeT and analyze its interactions with its cognate operator. Like other members of the TetR family of transcriptional repressors, SmeT behaves as a dimer and presents some common structural features with other TetR proteins like TtgR, QacR, and TetR. Differing from other TetR proteins for which the structure is available, SmeT turned out to have two extensions at the N and C termini that might be relevant for its function. Besides, SmeT presents the smallest binding pocket so far described in the TetR family of transcriptional repressors, which may correlate with a specific type and range of effectors. In vitro studies revealed that SmeT binds to a 28-bp pseudopalindromic region, forming two complexes. This operator region was found to overlap the promoters of smeT and smeDEF. This finding is consistent with a role for SmeT simultaneously down-regulating smeT and smeDEF transcription, likely by steric hindrance on RNA polymerase binding to DNA.
Opportunistic pathogens intrinsically resistant to antibiotics are currently a relevant health problem (1). Although several elements contribute to the intrinsic resistance of these bacteria (2, 3), the active efflux of antibiotics is a common mechanism relevant for their phenotype. Antibiotic efflux is because of the activity of multidrug (MDR)5 efflux pumps (4–7). Those elements are universally distributed among all living systems (8). In Gram-negative bacteria, the best characterized MDR pumps belong to the resistance nodulation division family. In most cases MDR pumps are down-regulated by specific transcription factors located upstream of the operon coding for the pump (9). Stable expression and, thus, a higher level of resistance is achieved by mutations in these regulatory elements. It is worth noting here that the constitutive expression of MDR pumps in these mutants is frequently linked to fitness costs (10–12). This indicates the need of a precise and stringent regulation of the system to avoid the nonspecific expression of these elements.
Stenotrophomonas maltophilia is an opportunistic pathogen that is considered as a prototype of intrinsically resistant bacteria (13). The first MDR pump described in this bacterial species was SmeDEF (14–16), an MDR determinant that contributes to intrinsic (17) and acquired (14, 16) resistance to several antibiotics and to biocides (18) in S. maltophilia. Expression of smeDEF is down-regulated by the transcriptional repressor SmeT (19, 20), located upstream, and in the complementary strand of smeDEF. Mutations leading to stable overexpression of smeDEF usually originate from SmeT, interfering with its binding to the smeT-smeDEF intergenic region (19, 20). It has been previously suggested that SmeT belongs to the TetR family of transcriptional regulators (19). This family, named from its first member, the tetracycline repressor, is widely distributed among bacteria (21). Members of this family characteristically present an N-terminal helix-turn-helix (HTH) DNA binding motif that exhibits a high degree of primary sequence identity among all of them. In contrast, the C-terminal domain, which is involved in both dimerization and in effector binding, does not seem to be conserved. The variation in sequence and structure of this ligand binding pocket probably accounts for the effector specificity. Structural and biochemical analyses to evaluate the response of some TetR proteins to the presence of their natural effectors have been performed, and it has been shown that the binding of the effectors to their binding pockets induces conformational changes in the conserved DNA binding domain. Under these circumstances, the repressor can no longer bind to the operator and is released, thus allowing the transcription of the repressed gene(s) from their cognate promoters.
Some members of the TetR family have been characterized at the structural level, among them Escherichia coli TetR (22–24), Staphylococcus aureus QacR (25), and Pseudomonas putida TtgR (26). Although sequence conservation is low, the three-dimensional structures of these proteins show a rather similar folding. However, despite these similarities, binding to their respective operators is different for some members of the family. TetR (27) binds to a palindromic sequence with a 2:1 stoichiometry (two subunits per each palindrome), whereas QacR binds to a pseudopalindromic region with a 4:1 stoichiometry (25), and EthR binds cooperatively to its operator with an 8:1 stoichiometry (28).
An important structural feature that impacts the activity of TetR proteins is their flexibility. For instance, for the TetR repressor, the mutation of some residues in the interface between the N- and C-terminal domains generates protein variants (revTetR mutants) that exhibit an N-terminal domain that is unfolded in the absence of the TetR effector, tetracycline (29, 30). After tetracycline binding, these domains are stabilized but remain flexible enough to adjust their DNA binding heads so that they can bind to the DNA. This extra flexibility clearly influences the way the protein interacts with the DNA and regulates transcription, as these mutations allow the activity of the TetR repressor even in the presence of its effector (31).
Similar to other members of the TetR family, SmeT is a transcriptional repressor (19). Because the promoters of smeDEF and smeT overlap, it has been suggested that the binding of SmeT to this DNA region would simultaneously repress smeDEF and smeT transcription by steric hindrance on RNA polymerase binding to DNA (19). Expression of the smeT and smeDEF genes would take place when SmeT is not bound to this DNA region, either due to mutations selected by antibiotic therapy (16, 20) or as the consequence of the binding of potential effectors to the C-terminal non-conserved region of the protein, followed by conformational changes that impede binding of the protein to its operator.
In the present study we have analyzed the structure of the SmeT protein and its DNA binding properties. Although the protein displays some common features with other members of the TetR family such as TtgR (26), IcaR (32), and QacR (25), SmeT was found to have some specific structural and functional characteristics.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions—The bacterial strains and plasmids used are shown in Table 1. The strains were grown in Luria-Bertani (LB) broth (33) at 37 °C unless otherwise specified.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strain | ||
| E. coli ER2566 | F– λ–fhuA2 [lon] ompT lacZ::T7 gene1 gal sulA11 Δ(mcrC-mrr)114::IS10 R(mcr-73::miniTn10-TetS)2 R(zgb-210::Tn10)(TetS) endA1 [dcm] | New England Biolabs and Refs. 34 and 35 |
| S. maltophilia D457 | Clinical isolate | 14 |
| Plasmids | ||
| pPS6 | pQE31 derivative that carries a KpnI-PstI restriction fragment containing smeT | 19 |
| pTYB1 | Cloning and expression vector used to fuse the C terminus of the target protein to an intein tag | New England Biolabs and Refs. 34 and 35 |
| pAHF1 | pTYB1 derivative used to express smeT fused to an intein tag | This study |
| pAHF2 | pGEMT-Easy derivative containing the smeD-SmeT intergenic region | 19 |
Plasmids Construction and Cloning—The smeT gene was PCR-amplified from the pPS6 plasmid (19) by PCR using the PCR Master Mix (Promega) with the primers smet NdeI forward (5′-GGAATTCCATATGGCCCGCAAGACCAAAGAGGA-3′) and smet SapI reverse (5′-GGTGGTTGCTCTTCCGCACGCCTCGGGCAGCGG-3′) to introduce restriction sites for NdeI and SapI (underlined). The conditions used were 95 °C for 5 min followed by 30 cycles of 95 °C for 60 s, 55 °C for 30 s, and 72 °C for 2 min and a final extension of 72 °C for 10 min. The PCR product was isolated from an agarose gel with Illustra ™ DNA and Gel Band Purification Kit (GE Healthcare), digested with NdeI (New England Biolabs), and SapI (New England Biolabs) and inserted in-frame into the pTYB1 vector (IMPACT-CN system, New England Biolabs) previously digested with the same restriction enzymes. The new plasmid, henceforth named pAHF1, encoding for the SmeT-Intein-CBD fusion protein, was transformed into E. coli ER2566 (IMPACT-CN system, New England Biolabs) for overexpression of the recombinant protein (34, 35).
Protein Expression and Purification—E. coli ER2566 strain harboring the plasmid pAHF1 was grown at 37 °C in LB broth supplemented with 100 μg/ml ampicillin to late-log phase (A600 = 0.8) and then induced with 0.5 mm isopropyl-β-d-thiogalactopyranoside overnight at 15 °C. The culture was harvested by centrifugation at 5000 × g for 15 min at 4 °C, and the pellet was resuspended in cold Tris-buffered saline (20 mm Tris-HCl, 0.5 m NaCl, 1 mm EDTA, pH 8). Cells were frozen at -20 °C.
The frozen cell suspensions were thawed, and 1 mm phenylmethylsulfonyl fluoride was added. Cells were disrupted by sonication, and Triton X-100 was added to a final concentration of 1%. The lysate was incubated for 1 h on ice and spun down at 20,000 × g for 30 min at 4 °C. The supernatant with the SmeT-Intein-CBD fusion protein was filtered through a 0.45-μm filter (Millipore) and loaded onto a Poly-Prep® chromatography column (Bio-Rad) containing 10 ml of chitin beads (New England Biolabs). The column was washed with 20 volumes of Tris-buffered saline followed by 3 beads volumes of cleavage buffer (20 mm Tris-HCl, 0.5 m NaCl, 100 mm dithiothreitol, 1 mm EDTA, pH 8), and the flow was stopped. After overnight incubation at 4 °C, the target protein was eluted with Tris-buffered saline. Dithiothreitol was removed from the sample by dialysis against Tris-buffered saline.
SmeT purification was assessed by SDS-PAGE. Samples were boiled in SDS sample buffer with β-mercaptoethanol and separated in a polyacrylamide gel containing 12% acrylamide and 0.1% SDS. The gel was stained with 0.025% Coomassie Brilliant Blue R. Protein concentrations were determined from absorbance at 280 nm, assuming a extinction coefficient of ε280 nm = 9970 m-1 cm-1 and by bicinchonic acid protein assay (Pierce) using bovine serum albumin as standard. For protein crystallization, a last purification step was carried out using a Sephacryl 100 gel filtration column in a buffer containing 300 mm NaCl and 20 mm Tris, pH 8.0.
Protein Crystallization—The protein was concentrated to 5 mg/ml, and crystals were obtained by the vapor diffusion method in drops containing 1 μl of protein and 1 μl of the crystallization agent, a solution of 200 mm Li2SO4, 28% MME2000, and 100 mm Tris, pH 8.5. The crystals displayed a plate morphology and belonged to the space group P21 with unit cell parameters a = 56.7 Å, b = 58.6 Å, c = 83.3 Å, and β = 103.1°. A dimer in the asymmetric unit gives a Matthews coefficient of 2.8 Å3 Da-1 and a solvent content of 55.8%. The crystals were soaked in a cryoprotectant solution containing 200 mm Li2SO4, 32% polyethylene glycol 2000 monomethyl ether, and 100 mm Tris, pH 8.5, and flash-cooled in liquid nitrogen.
Crystal Structure Determination and Refinement—Data were collected at the European Synchrotron Radiation Facility beamline BM14 (Grenoble, France). A native data set diffracting to 2.0 Å resolution and a mercury derivative diffracting to 2.1 Å were obtained. Both data sets were reduced using MOS-FLM (36) and SCALA (37) (Table 2). The structure of SmeT was solved by the SIRAS (Single Isomorphous Replacement with Anomalous Signal) method using these data with the HKL2MAP (38) graphic interface for the Shelxc/d/e programs (39). The model was refined against the native data set to final Rfree of 23.6% and Rcryst of 19.6%; see the statistics in Table 2. The phases provided by Shelxe (40) were good enough to determine the handedness, with some of the secondary structure elements clearly resolved in the electron density map. The figure of merit for the solution was 0.3. These phases were introduced in RESOLVE (41) using the PHENIX interface (42), and the program built automatically 167 residues with a figure of merit of 0.59. Subsequent building and refinement was done against the native data set; the first building/refinement cycles were done using ARP/wARP (43), and the successive refinement and manual building were done using REFMAC (44) and COOT (45). The translation-libration-screw (TLS) groups for REFMAC were as defined by the TLMSD server (46).
TABLE 2.
Data collection and refinement statistics
| Native | Mercury derivative | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Cell dimensions (Å,°) | a = 56.7, b = 58.6, c = 83.3, ß = 103.1 | a = 56.9, b = 58.5, c = 83.4, ß = 103.1 |
| Resolution (Å) | 2.0 | 2.1 |
| No. of observations | ||
| Measured | 465,623 | 480,417 |
| Unique | 35,083 | 31,381 |
| Completeness (%)a | 97.2 (96.3) | 100 (100) |
| Rmeasa (%) | 5.8 (21.9) | 9.5 (50.9) |
| Multiplicity | 7.4 (7.3) | 5.5 (5.5) |
| I/σ(I) | 25.4 (9.0) | 14.6 (3.5) |
| Phasing, figure of meritb | 0.30 (0.59) | |
| Refinement | ||
| Rcryst/Rfreec (%) | 19.6/23.6 | |
| No. of atoms protein/water | 2,963/269 | |
| r.m.s.d. | ||
| Bonds/angles (Å,°) | 0.015/1.453 | |
| B factor (Å2) main/side chain | 1.549/2.895 |
Values in parentheses are for the highest resolution shell
The first value is after Shelxe, and the value in parentheses is after Resolve
Rfree is Rcryst calculated for a test set of randomly selected 5% of the data
Structure Validation and Analysis—The stereochemistry of the final model was validated using Molprobity (47). The backbone dihedral angles of 99.7% of the residues fall in the most favored regions of the Ramachandran plot, and the remaining fall in the allowed regions. The PARVATI (48) server was used to analyze the anisotropy of the atomic displacement parameters calculated from the TLS approximation used in the refinement of the structure. The estimated mean anisotropy was 0.403 with a S.D. of 0.137.
A sequence alignment containing 288 sequences of different members of the TetR family was calculated using the program M-Coffee (49). We used the program Homolmapper (50) to map different properties of the alignment, such as conservation of sequence, charge, or hydrophobicity, on the surface of the structure. The similarity was calculated using the matrix BLO-SUM62. Electrostatic calculations were carried out with the program APBS (51), and the electrostatic distribution was represented using Chimera (52). The figures were done using Chimera or Pymol (53).
Homology Analysis—The Uniref100 data base was searched with the sequence of SmeT using the PSI-Blast algorithm (www.ncbi.nlm.nih.gov). The CD-HIT program (54) allowed us to retrieve the 288 most representative sequences (sharing less than 70% identity) out from the 487 sequences gathered by PSI-Blast at convergence. Finally, M-Coffee (49) produced a consensus alignment of the selected sequences by merging the alignments obtained through the ClustalW, t_coffee, poa, muscle, mafft, dialignt, pcma, and probcons algorithms. M-Coffee reported a quality score of 80 for this consensus alignment.
DNA Labeling—For the electrophoretic mobility shift assay (EMSA) experiments, the 30-bp oligonucleotide, 5′-GTTTACAAACAAACAAGCATGTATGTATAT-3′, containing the hypothetical operator site was 5′ end-labeled with [γ-32P]dATP (GE Healthcare) using T4 polynucleotide kinase (New England Biolabs) and later hybridized with its complementary non-labeled DNA strand by heating the samples at 90 °C for 3 min and cooling down at room temperature.
For the footprinting and missing nucleoside assays, DNA probes were generated by PCR with PCR Master Mix (Promega) using the plasmid pAHF2 as template. To amplify the whole 223-bp smeD-smeT intergenic region, the primers sme 46 (5′-GGGTGTGGGTACGAGTGC-3′) and sme 47 (5′-GACGGAAAGGCTCTTGGAG-3′) were used under the conditions described in Sanchez et al. (19). A 158-bp fragment containing the hypothetical operator site was amplified by using primers sme 135 (5′-AAAGCCCGCAGATCGCGCCCA-3′) and sme 46. To distinguish the binding on each DNA strand, one of the oligonucleotides was 5′ end-labeled with [γ-32P]dATP as described above. PCR conditions were 95 °C for 5 min followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min and a final extension of 72 °C for 10 min. The PCR products were purified by 8% (w/v) denaturing urea-PAGE and extracted by the crush and soak method (55).
EMSA—EMSA experiments were performed incubating the end-labeled 30-bp double-stranded oligonucleotide with increasing concentrations (0, 0.2, 0.4, 0.8, 1.6, and 3.2 μm) of purified SmeT in binding buffer (10 mm Tris-HCl, 50 mm KCl, 10 mm MgCl2, 1 mm EDTA, pH 7.2, 50 μg/ml bovine serum albumin, 1 mm dithiothreitol, 5% (v/v) glycerol, and 100 μg/ml poly(dI-dC) as nonspecific competitor DNA) for 30 min at room temperature. For competitions assays, increasing concentrations of non-labeled 30-bp double-stranded oligonucleotide were added. Retarded complexes were separated on a 6% nondenaturing polyacrylamide gel (37.5:1 acrylamide:bisacrylamide). Electrophoresis was performed at room temperature with 89 mm Tris borate and 2 mm EDTA buffer for 90 min at 100 V and dried before autoradiography.
DNase I Footprinting Assays—DNase I footprinting assays were performed as described (55). For the smeD coding strand, the oligonucleotides used were sme 46 (5′ end-labeled) and non-labeled sme 47. The PCR conditions were as previously stated, and the binding conditions were as described for EMSA. After incubation, the samples were digested with 0.05 units of DNase I (Roche Applied Science) for 3 min at room temperature. Reactions were stopped by the addition of EDTA (20 mm final concentration), and DNA was precipitated with 0.3 m potassium acetate and 2.5 volumes of ethanol. DNA fragments were resolved by electrophoresis on an 8% (w/v) polyacrylamide, 7 m urea denaturing gel and visualized by autoradiography. For the smeT coding strand the conditions used were as above, but the 5′ end-labeled oligonucleotide was sme 47, and DNA fragments were separated on a 6% (w/v) polyacrylamide, 7 m urea sequencing gel. For each labeled strand, a Maxam and Gilbert sequencing reference ladder was prepared and run in parallel.
Missing Nucleoside Assay—Missing nucleoside assays were performed as previously described (55) with 5′ end-labeled double-stranded DNA produced by PCR. To visualize the smeD coding strand, the smeD-smeT intergenic region was amplified using 5′ end-labeled sme 46 and non-labeled sme 47. For the smeT coding strand, the primers used were 5′ end-labeled sme 135 and non-labeled sme 46, amplifying the 158-bp fragment containing the hypothetical SmeT binding site. Once purified, the PCR products were dissolved in 10 mm Tris-HCl, 50 mm KCl, 10 mm MgCl2, 1 mm EDTA, pH 7.2, and 100 μg/ml poly(dI-dC) and subjected to hydroxyl radical cleavage by adding 3 μl of a freshly prepared solution containing 4 mm EDTA, 2 mm FeSO4·7H2O, 16 mm sodium ascorbate, and 7% (v/v) H2O2. After 4 min the reaction was stopped with 2 μl of 100 mm thiourea and 2 μl of 0.5 m EDTA. DNA fragments were precipitated, resuspended in 20 μl of EMSA binding buffer, and incubated with SmeT for 40 min at room temperature. Protein-bound DNA was separated from unbound DNA on a 6% (w/v) polyacrylamide nondenaturing gel with 89 mm Tris borate and 2 mm EDTA and extracted by the crush and soak method (55). After DNA precipitation, samples were resuspended in sequencing loading buffer and analyzed in 8% (w/v) urea-polyacrylamide gels. Samples were run in parallel to a Maxam and Gilbert sequencing reference ladder.
RESULTS AND DISCUSSION
SmeT Structure Overview—To avoid structural distortions that could be produced as the consequence of the presence of tags, SmeT was purified as an intein-fusion protein, and the tag used for purification by affinity chromatography was removed as described under “Experimental Procedures.” This allowed the purification of SmeT in its native form. This purified SmeT protein was used for all functional and structural assays.
The structure of SmeT was solved by the SIRAS method using a mercury derivative and refined against a native data set diffracting to 2 Å with final Rfree of 23.6% and Rcryst of 19.6%. The final model includes residues 7–116 and 126–217 for subunit A and residues 13–20, 35–44, 56–116, and 123–216 for subunit B. That means that in subunit B several of the residues in the N-terminal domain were not modeled because the density in that region was not good enough.
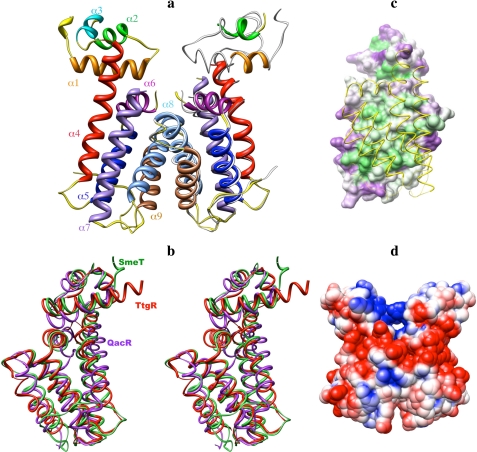
The crystal structure of SmeT, illustrated in Fig. 1a, appeared to be a homodimer, as found in most members of the TetR family. Gel filtration and ultracentrifugation data (data not shown) showed that the dimer is the relevant species in solution. Each subunit consists of 9 helices organized in two domains arranged sequentially from the N to the C terminus. The r.m.s.d. between both subunits is 0.811 Å, with the biggest differences corresponding to the N-terminal domain and the residues close to the missing loop in the C-terminal domain. The r.m.s.d. for the C-terminal domain is 0.706 Å.
FIGURE 1.
Structure of SmeT. a, ribbon representation of SmeT. The helices 1–9 are displayed in different colors and have been labeled only in subunit A. Subunit A has been superposed onto subunit B and is shown in gray. b, stereo view of the structural comparison between SmeT (green), QacR (magenta, ID code 1JUP) and TtgR (red, ID code 2UXU). The figure shows the extended N and C termini of SmeT. c, conservation of the hydrophobic character of the residues in the interface of SmeT dimer. The figure has been prepared using HOMOLMAPPER based on an alignment of 288 sequences from members of the TetR family prepared by M-Coffee. Residues in green have a conserved hydrophobic character, and residues in magenta do not. Subunit B has been represented as a yellow backbone tube to show the dimerization area. d, electrostatic surface potential of SmeT dimer. Because the model is incomplete, subunit A was superposed onto subunit B, and the dimer generated in that way was used for the calculations. The electrostatic potential was calculated by APBS and is mapped on the solvent-accessible surface by CHIMERA. Electrostatic potential values range from positive (blue, 5 kT/e) to negative (red, -5 kT/e), where k is Boltzmann's constant, T is the absolute temperature, and e is the proton charge.
The N-terminal domain of SmeT comprises the first three helices, α1 (residues 14–27), α2 (34–41), α3 (45–49), and the beginning of helix α4, which as in other members of the TetR family, participates in the stabilization of this domain (22). Helices α2 and α3 form the HTH motif involved in DNA binding. The C-terminal domain is composed by the remaining six helices, α4 (54–76), α5 (85–102), α6 (104–114), α7 (127–149), α8 (159–179), and α9 (186–201), connected by loop regions. The N-terminal domain contacts with the C-terminal domain through a number of interactions that are relevant for its stabilization. First, the beginning of helix α4 participates in the formation of the hydrophobic core that stabilizes the N-terminal domain; Val-58, Leu-59, and Ile-62 are anchored in a hydrophobic pocket formed by residues from helices α1, α2, and α3. Apart from these hydrophobic interactions, there is a network of salt bridges/hydrogen bonds; Arg-65 (from helix α4) establishes ionic interactions with the negatively charged residues Glu-14, Asp-18, and Glu-21 (from helix α1), and some residues from helix α6 make hydrogen bonds with residues from helix α1: Glu-21—Arg-105, Glu-21—Thr-109, Thr-109 —His-25, and His-25—Lys-108. These interactions have been reported to be crucial for the mechanism of activation-deactivation in QacR and TetR (9). In both proteins the binding of an effector into the C-terminal domain induces structural changes in this domain, one of them being the displacement of helix α6. Because this helix is anchored to the DNA binding domain through the interactions described above, the translocation of this helix provokes a change in the orientation of the DNA binding domain of both subunits, generating a ligand-bound conformation of the repressor that is no longer able to bind DNA.
The N-terminal Domain of SmeT Is Partly Disordered in the Crystal—The loop between helices α6 and α7, residues 117–125 in subunit A and 117–122 in subunit B, could not be solved in the crystal. Furthermore, most of the residues in the N-terminal domain of subunit B were not visible in the electron density. A detailed inspection of the crystal packing revealed a lack of crystal contacts along the N-terminal domain of subunit B. The more extensive crystallographic contacts of subunit A may have contributed to its stabilization in the crystal. A comparison of the principal axes of the libration tensors of the TLS correction showed that the TLS group corresponding to the subunit B N-terminal domain has larger and more anisotropic values than those of subunit A. Altogether, these facts could explain the poor definition of this region in the electron density map of subunit B. The only residues with clear density in this domain belong to secondary structure elements, namely, to the helices α2 and α3. Because the ability of intrinsically unfolded proteins and domains to fold upon interaction with functional partners has been recognized (56), this observation argues for an intrinsic instability of the domain, which could be only partly folded in solution, a situation that might play a functional role in the recognition by SmeT of its cognate DNA operator.
The capability of the N-terminal domains to fit into the DNA major grooves is essential for the mechanism of SmeT, so it can be important for the protein to keep a certain degree of flexibility. This assumption is supported by finding of the revTetR variants, which present a disordered N-terminal domain that is folded and stabilized when the tetracycline is bound to the protein (29, 30). The stabilization of the N-terminal domain due to the binding of the ligand has also been described for the native TetR. We hypothesize that the DNA binding domains of SmeT would be partly unfolded in solution, folding only upon ligand or DNA binding. In the structure of SmeT, the N-terminal domain of subunit A is visible and seems to be perfectly ordered. It is important to stress the fact that the ligand pocket in subunit A is occupied by an extra electron density that indicates the presence of an unknown ligand. We may speculate that this ligand might stabilize the N-terminal domain of that subunit. In subunit B, we do not see any indication of a putative ligand. This fact along with the smaller number of crystallographic contacts established by this subunit could explain the disorder of its N-terminal domain.
Structural Comparison with Other Members of the TetR Family—A structural similarity search using the DALI server with the whole subunit A of SmeT as probe gives the best score (Z = 19.5 r.m.s.d. = 2.5 Å for 187 residues) for the structure of TtgR (26). TtgR is also found to be the most similar one when only the N-terminal or C-terminal domain is used as the probe. A good score is also obtained for the two members of the family that have been more extensively characterized, TetR from E. coli (22) and QacR from S. aureus (25, 57).
Despite these similarities, the structure of SmeT displays a number of interesting differences. Particularly relevant is the presence of extended N and C termini (Fig. 1b). The extra residues in the N terminus have an extended conformation and fold over the HTH motif. Among the members of the TetR family for which structural data are available, only TtgR presents an extended N terminus. However, in this case the extra residues form part of the helix α1 and remain far away from the HTH motif (Fig. 1b). Besides this extension in the N-terminal domain, SmeT also presents an extension in the C-terminal domain, namely, residues 202–218, that is not present in other members of the family. These residues are in an extended conformation without secondary structure, being stabilized through some few interactions with residues from helices α5 and α9 of the same subunit.
To further analyze the characteristic features of SmeT, a sequence alignment containing 288 sequences of different members of the TetR family was calculated using the program M-Coffee. The program Homolmapper was used then to map different properties of the alignment, such as conservation of sequence, charge, or hydrophobicity, on the surface of the structure. The sequence conservation is reasonable for the DNA binding domain (32.7% overall similarity for the 288 sequences aligned, residues 8–49) and very low for the C-terminal domain (12.3% overall similarity, residues 54–201). The similarity was calculated using the matrix BLOSUM62. Not surprisingly, helix α3 in the HTH, the helix that recognizes the DNA, is the most conserved region. Other residues that display a high conservation among the different members of the TetR family (higher than 75%) are Ile-16, Gly-28, Ile-37, Tyr-43, Tyr-49, Phe-52, all in the N-terminal domain, and Gly-151 in the loop between helices α7 and α8, in the base of the ligand binding domain, at the opposite side of the DNA binding domain.
When the hydrophobicity was analyzed in the same way, we found several conserved hydrophobic patches on the surface. Although there is no sequence conservation in the dimerization region, the interacting surface of the dimerization domain displays a clear and conserved hydrophobic character (Fig. 1c, green). Besides, the residues lining the ligand pocket also exhibit a conserved hydrophobic character. There is another hydrophobic patch in the DNA binding domain involved in the anchoring of the N-terminal into the C-terminal domain. Finally, three of the residues likely involved in DNA binding, Tyr-49, Tyr-43, and Trp-50, have a hydrophobic character conserved across the sequence alignment.
Dimerization Domain of SmeT—Dimerization is mainly mediated by the C-terminal domain. Despite its low degree of sequence homology within the members of the TetR family, the dimerization domain exhibits a very clear topological similarity (Fig. 1b) and includes several regions with a markedly conserved hydrophobic character (see above). The formation of the dimer buries a 1328 Å2 surface area for subunit A and 1273 Å2 for subunit B. The dimerization interface is mostly formed by helices α8 and α9 from both subunits via van der Waals contacts established by the lateral chains of non-polar residues. This four-helical dimerization region accounts for most of the buried surface. Besides, residues from helices α6 and α7 contribute to the interface, some in the loop connecting these helices. Part of this loop (residues 117–125 in subunit A and 117–122 in subunit B) is not visible in the electron density map for any of the subunits, due probably to its flexibility. In addition to the hydrophobic interactions, a network of hydrogen bonds and four strong salt bridges between Arg-134 and Glu-180 and between Arg-164 and Asp-189 from both subunits were also found.
DNA Binding Domain of SmeT—As stated above, the HTH DNA binding domain of SmeT (residues 1–54) is more conserved than the dimerization domain among the members of the TetR family. When we represented the electrostatic distribution on the surface of the protein, we saw that, with the exception of the N-terminal domain, the charge in the surface of the protein is mostly negative (Fig. 1d). The N-terminal domain is positively charged, although the charge is not very strong. In the recognition helix α3, only Arg-45 contributes to this positive charge; the rest of the positive residues are His-27 in helix α1, Arg-31 in the loop between helices α1 and α2, His-51 in the loop between helices α3 and α4, and Lys-53 and Lys-55, both, in helix α4. SmeT has some extra residues in the N terminus as compared with other members of the TetR family (Fig. 1b). Notably, these extra residues display a marked negative charge (arising from residues Glu-7 and Asp-8) that stays close to the positive charge of the HTH motif. Moreover, these residues protrude from the HTH motif and would overlap with SmeT DNA operator. Therefore, to allow DNA binding, their conformation should change. Because TtgR, the other member of the family that presents an extended N terminus (Fig. 1b), also has some acidic residues (three glutamic acids) in its extension, it is tempting to speculate that such N-terminal extensions might modulate the binding of SmeT and TtgR to their cognate DNA operators.
Ligand Binding Pocket—Members of the TetR family of repressors have effector binding pockets mostly lined by hydrophobic residues along with some residues, usually charged, that determine their specificity. These proteins can recognize a wide variety of molecules including antibiotics, flavonoids, and aromatic compounds (7). The common feature of these molecules is the presence of at least one aromatic ring. We have analyzed the potential ligand binding pockets of SmeT using the programs PASS (58), LIGSITE (59), and CASTp (60). All of them found the same cavity in the ligand binding domain of each subunit.
The volume of this cavity (as calculated by CASTp) is 630 Å3, which is smaller than previously calculated values for other TetR family members, ranging from 800 Å3 in ActR (61) to 1500 Å3 in TtgR (26). However, the volume of the cavity may be subjected to variations depending on whether a ligand is bound or not and on the nature of the ligand(s). The cavity (Fig. 2) is lined by residues His-67, Phe-70, Met-71, Leu-74, Val-92, Met-93, Ser-96, Phe-97, Leu-100, Met-110, Met-113, Leu-114, Gly-132, Phe-133, Ala-136, Met-140, Leu-166, His-167, Val-170, Leu-171, and Leu-174. Strikingly, six among them adopt a dual conformation, namely, Phe-70, Met-93, Met-110, Met-113, Phe-133, and Met-140. Residues with multiple side-chain conformations have been described for other multidrug-binding proteins, including members of the TetR family (26, 62). Their multiple conformations facilitate the interaction with different drugs binding in different configurations.
FIGURE 2.
Effector binding pocket of SmeT. a, the figure shows the secondary structure of subunit A of SmeT in pale brown, and the binding pocket is represented in magenta. Pathways leading to this cavity were calculated using CAVER and are represented using PYMOL. b, the surface of SmeT subunit A is in green, and subunit B is in blue, showing the entrance between helices 4, 5, and 6. The binding pocket for subunit A is represented in magenta. c, stereo view of the residues lining the hydrophobic cavity of SmeT.
Only three of the residues lining the cavity, His-67, His-167, and Ser-96, are polar. In contrast, QacR presents in this cavity four negatively charged residues (Glu-57, Glu-58, Glu-90, and Glu-120) that allow this protein to recognize cationic drugs (63, 64). Conversely, the binding pocket of TtgR can accommodate negatively charged ligands due to the presence of positively charged residues, such as Arg-176 (26). Because the binding pocket of SmeT is much smaller than the one of TtgR, we may speculate that SmeT should accommodate a different range of effectors than TtgR. Although the architecture of SmeT binding pocket could give a hint about its possible ligand(s), such guesses are hampered by the fact that ligand charges can be shielded not only by free carboxylate and amino groups but also by the dipoles of several oxygen or nitrogen atoms.
The extra electron density in the binding pocket of subunit A suggests the presence of a ligand molecule that was present during protein production or crystallization. Such has been the case for other TetR proteins like EthR from Mycobacterium tuberculosis (65). The shape of the extra density found in the ligand pocket of SmeT suggests that the putative ligand(s) may engage in a stacking interaction with Phe-70 and establish hydrogen bonds to His-67 and His-167. To find possible pathways connecting the ligand binding site and the outside of the protein, the structure of SmeT was analyzed using the program CAVER (66). Two paths were found (Fig. 2a); the entrance for the first one is at a triangle formed by helices α4 (at the level where this helix slight bends), α5, and α6. This entrance would be equivalent to the one proposed for TtgR (26) near the point where helix α4 exhibits a 65° inward bending. In SmeT, this bending exists but is not so pronounced. Interestingly, residue Phe-70 is just at the entrance of this path. This residue displays a double conformation; when only one of the conformations is included (the so-called open one), we clearly see a cleft in the surface that connects with the ligand binding site (Fig. 2b). This observation supports the notion that Phe-70 may be the gate of the ligand binding pocket and could act as a regulating switch. The structurally equivalent residue of SmeT Phe-70 in TetR, His-64, is also important for the mechanism of action of the protein. TetR His-64 is the pivotal residue in the rotational rearrangement found between the ligand and the DNA-bound conformations of the protein. The second path connecting the cavity and the outside of the SmeT is opened to the surface between helix α6, the N terminus of helix α5, the C terminus of helix α8, the N terminus of helix α9, and the loop between helix α8 and α9.
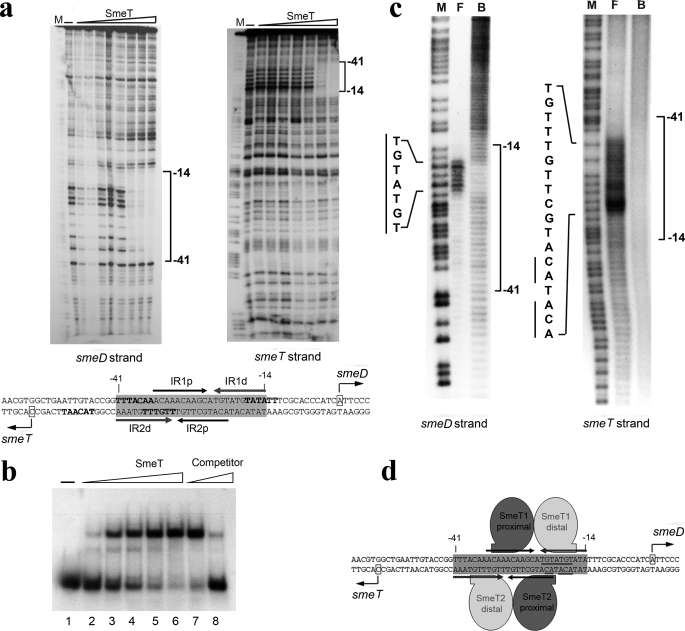
The SmeT Operator Has Two Binding Sites and Includes the Promoter Regions for smeD and smeT—It had been previously demonstrated that His-tagged SmeT is able to partly shift the migration of the smeT-smeD intergenic region in an EMSA, suggesting that this region likely contains the SmeT operator (19). To ascertain that native SmeT binds this DNA region, we used purified SmeT protein free of any affinity tag and the labeled intergenic smeT-smeD region. DNase I footprinting assays were performed to identify the operator sequence of SmeT. As shown in Fig. 3a, SmeT protected from DNase I digestion a 30-bp region that contains a pseudopalindromic sequence of 28 bp that includes the -10 and -35 regions of PsmeT promoter and the -35 region of PsmeD promoter. EMSA assays using a 30-bp DNA that contains the SmeT putative operator showed that SmeT binds to this region forming two complexes (Fig. 3b). The appearance of the two shifted bands was dependent on the protein concentration, and the addition of non-labeled specific DNA (30-bp DNA) competed the binding, indicating the specificity of the interactions. Altogether, these results strongly suggest that the binding of SmeT to its operator will simultaneously inhibit not just the expression of the efflux pump operon smeDEF, but also that of its own gene smeT, by hindering the binding of the RNA polymerase. This negative feedback mechanism provides a means to limit the amount of SmeT protein in the bacterial cell and, therefore, to control the fast response of the system to external inputs.
FIGURE 3.
Binding of SmeT to its operator DNA. a, identification of the SmeT binding site by a DNase I-footprinting assay. An end-labeled 223-bp DNA segment containing the smeD-smeT intergenic region was incubated with increasing concentrations of SmeT and later digested by DNase I. SmeT protects in both DNA strands a 28-bp sequence located between positions -14 and -41 from the smeD transcription start site. Two inverted repeats located within the protected region (IR1 and IR2, indicated by arrows) may represent the binding sequences for SmeT. IR1p, inverted repeat 1 proximal arm (relative to the dyad that relates the two dimers); IR1d, inverted repeat 1 distal arm; IR2p, inverted repeat 2 proximal arm; IR2d, inverted repeat 2 distal arm. smeD and smeT transcription start sites are indicated with a box. The -35 and -10 regions of both promoters are highlighted in bold. b, binding to a 30-bp DNA that contains the putative SmeT operator. The end-labeled 30-bp DNA was incubated with increasing concentrations (0, 0.2, 0.4, 0.8, 1.6, and 3.2 μm) of SmeT (lanes 1–6, respectively). Complexes were resolved in a 6% (w/v) nondenaturating polyacrylamide gel. With SmeT at 3.2 μm, the binding was competed by the unlabeled probe (1 and 10 μm, lanes 7 and 8. c, critical nucleosides for SmeT binding to DNA. Missing nucleoside assays were performed with the whole end-labeled 223-bp intergenic region for the smeD strand and a 158-bp fragment containing the hypothetical SmeT binding site for the smeT strand. Both DNA fragments were subjected to hydroxyl radical cleavage and were later incubated with SmeT. Bound and free DNA were separated on a 6% (w/v) polyacrylamide nondenaturing gel, purified from the gel, and analyzed in 8% (w/v) urea-polyacrylamide gels. Nucleosides critical for SmeT binding in both strands are indicated with bars. M, DNA sequence ladder; F, free DNA; B, bound DNA. d, SmeT-Operator DNA binding model. SmeT binds to a 28-bp region placed between positions -14 and -41 from the smeD transcriptional start site. This operator DNA contains a pseudopalindromic sequence with two overlapping inverted repeats (IR1 and IR2) that can accommodate a pair of SmeT dimers each. Our results suggest that the strongest interaction is carried out by the SmeT dimer that binds to the IR1 in the smeD coding strand through binding to the sequence TGTATGT. Once this first dimer is bound, the second dimer binds cooperatively. Critical nucleosides for SmeT binding are underlined. smeD and smeT transcription start sites are indicated with a box.
Two main different modes of binding to DNA have been described for the TetR family. Whereas TetR binds to a palindromic, 15-bp site with a 2:1 stoichiometry (two subunits per dsDNA site), QacR binds to a pseudopalindromic region with stoichiometry 4:1; that is, four subunits bind to opposite sites of a 28-bp DNA sequence, each HTH motif of a given dimer making a different set of contacts. We have shown that SmeT behaves as a dimer in solution and that it recognizes a pseudomalindromic 28-bp DNA region. EMSA assays and the size of the protected region determined with the footprinting assays are consistent with the formation of two complexes (likely cooperatively) between SmeT and its operator sequence. This hypothesis is further supported by the presence of two inverted repeats (Fig. 3a) in the opposite strands of the SmeT-protected DNA defined by the footprinting assays. Our data are consistent with a model in which SmeT binds DNA similarly as QacR. This is in agreement with common structural features of these two proteins, although the DNA sequences recognized either by one or another of the proteins are different (see below). The residues that in QacR interact with the DNA are conserved in SmeT, mainly those entering the major groove. In QacR these are Tyr-40 and Tyr-41 (Tyr-49 and Trp-50 in SmeT) via hydrophobic interactions, Lys-36 (Arg-45 in SmeT) making a hydrogen bond, and Gly-37 (Gly46 in SmeT), whose amide nitrogen is hydrogen-bonded to one of the DNA bases. The presence of a Gly in this position (a proline in TetR) seems to be crucial for the specificity because it makes possible a very close fit between the protein and the DNA (9).
The distance between two consecutive major grooves on a canonical B-DNA is ∼34 Å. In QacR, the center-to-center distance between the helices α3 of each subunit of a dimer is of 37 Å (measured between Tyr-40 Cα atoms). The 3 Å difference results in a curvature being induced in the DNA upon repressor binding. However, upon binding to its effector, QacR undergoes a large conformational change so that the helix α3-helix α3′ distance increases to 47 Å, and the protein cannot bind DNA (64). Because the model for the N-terminal domain of SmeT in subunit B is incomplete, to estimate the distance between the DNA binding domains, we have superposed the subunit A onto subunit B and reconstructed a complete dimer (Fig. 1a, virtual subunit B in gray). The distance between Tyr-49 and Tyr-49′ (residue equivalent to QacR Tyr-40) is 42 Å. Because electron density for a small molecule is clearly defined in the effector binding pocket of subunit A of SmeT (see above), this distance may correspond to an effector-bound form of SmeT, suggesting that this protein might undergo conformational rearrangements. These conformational changes will be facilitated by the flexibility displayed by subunit B, which does not contain any molecule in its binding pocket.
Critical Nucleosides for the Binding of SmeT to DNA—The nucleotides critical for SmeT binding to its operator were identified by missing nucleoside assays (Fig. 3c). The SmeT-DNA complex formation was not possible when the smeDEF coding strand lacked any of the nucleosides of the sequence TGTATGT located between positions -18 and -24 from the smeD transcription start. In the smeT coding strand, a greater number of nucleosides was important for DNA binding, including those complementary to the sequence found crucial for the smeD coding strand. We must note that with this method we cannot ascertain whether the binding is prevented because the missing nucleosides impede specific amino acid-DNA interactions or because they alter the DNA conformation required for SmeT binding.
Our data support that SmeT binds to its operator, forming two complexes, and that the binding protects a 28-bp region containing two pseudopalindromic sequences (Fig. 3d). One half pseudopalindrome, TGTATGT, in the strand encoding for the efflux pump, is crucial for the binding of SmeT to its operator. Our results are consistent with a model in which a first SmeT homodimer binds to the operator, establishing a critical interaction between one of the subunits (SmeT1 distal) and the TGTATGT zone. For the second homodimer, the distal subunit (SmeT2 distal) would recognize a similar zone, and its binding might be facilitated by the previously bound dimer. The overall structure of this DNA region, with two pseudopalindromic sequences, is similar for those members of the TetR family that bind DNA in a 4:1 stoichiometry. However, there are clear differences between the DNA regions recognized by proteins from the TetR family from either Gram-positive (QacR and IcaR) or Gram-negative (SmeT, TtgR and MtrR) bacteria. Whereas the sequence TGTATGTA is highly conserved in the operators of the aforementioned Gram-negative bacteria, it is absent in the case of Gram-positive microorganisms. Furthermore, Clustal analysis based in the identity of these DNA binding sites rendered a sharp separation between the operators of Gram-positive and Gram-negative bacteria. This indicates that, although the members of the TetR family share rather similar structural features, their operators have evolved divergently.
Conclusions—The structure of the SmeT repressor shows several similarities to TtgR and QacR. However, it has clear differences that might account for its specificity. Differing from all members of the TetR family for which a structural analysis is available, SmeT presents two extensions at both its termini that might modulate its interaction with DNA. Our data are consistent with the binding of SmeT to a pseudopalindromic region of 28 bp that is well conserved in the operators of members of the TetR family from Gram-negative bacteria but is not conserved in Gram-positives. Although the mechanism of binding (two dimers per operator) and the protein domains involved are rather similar, the operator differences reflect the distinct specificity of the members of the TetR family. The specificity is further highlighted by the small size of the effector binding pocket SmeT as compared with other members of the family. Notably, SmeT has the smallest size, whereas TtgR, which is its closest relative, presents the largest. This suggests that the type and range of effectors recognized by these two repressors might be different.
Acknowledgments
We acknowledge the EMBL Grenoble Outstation for providing support for measurements at the European Synchrotron Radiation Facility under the European Research Infrastructure Action FP6 program “Structuring the European Research Area'I3 Programme.” We thank Hassan Belrhali and Martin Walsh for assistance in using beamline BM14 and Miguel Ortiz-Lombardía for discussion and comments on this manuscript.
The atomic coordinates and structure factors (code 2w53) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by Grants BIO2005-04278, BFU2005-05055, BFU2006-00767/BMC, and BIO-2008-00090 from the Spanish Ministry of Science and Innovation, LSHM-CT-2005-518152 and LSHM-CT-2005-018705 from the European Union, and REIPI RD06/0008 from the Spanish Ministry of Health.
Footnotes
The abbreviations used are: MDR, multidrug resistance; HTH, helix-turn-helix; TLS, translation-libration-screw; EMSA, electrophoretic mobility shift assay; r.m.s.d., root mean square deviations; CBD, chitin binding domain.
References
- 1.WHO (2000) in World Health Organization Report in Infectious Diseases, Geneva
- 2.Fajardo, A., Martinez-Martin, N., Mercadillo, M., Galan, J. C., Ghysels, B., Matthijs, S., Cornelis, P., Wiehlmann, L., Tummler, B., Baquero, F., and Martinez, J. L. (2008) PLoS ONE 3 e1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez, J. L., Fajardo, A., Garmendia, L., Hernandez, A., Linares, J. F., Martinez-Solano, L., and Sanchez, M. B. (2009) FEMS Microbiol. Rev. 33 44-65 [DOI] [PubMed] [Google Scholar]
- 4.Vila, J., and Martinez, J. L. (2008) Curr. Drug Targets 9 797-807 [DOI] [PubMed] [Google Scholar]
- 5.Piddock, L. J. (2006) Clin. Microbiol. Rev. 19 382-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poole, K. (2005) J. Antimicrob. Chemother. 56 20-51 [DOI] [PubMed] [Google Scholar]
- 7.Li, X. Z., and Nikaido, H. (2004) Drugs 64 159-204 [DOI] [PubMed] [Google Scholar]
- 8.Martinez, J. L., Sanchez, M., Martinez-Solano, L., Hernandez, A., Garmendia, L., Fajardo, A., and Alvarez-Ortega, C. (2009) FEMS Microbiol. Rev. 33 430-449 [DOI] [PubMed] [Google Scholar]
- 9.Grkovic, S., Brown, M. H., and Skurray, R. A. (2002) Microbiol. Mol. Biol. Rev. 66 671-701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alonso, A., Morales, G., Escalante, R., Campanario, E., Sastre, L., and Martinez, J. L. (2004) J. Antimicrob. Chemother. 53 432-434 [DOI] [PubMed] [Google Scholar]
- 11.Linares, J. F., Lopez, J. A., Camafeita, E., Albar, J. P., Rojo, F., and Martinez, J. L. (2005) J. Bacteriol. 187 1384-1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez, P., Linares, J. F., Ruiz-Diez, B., Campanario, E., Navas, A., Baquero, F., and Martinez, J. L. (2002) J. Antimicrob. Chemother. 50 657-664 [DOI] [PubMed] [Google Scholar]
- 13.Crossman, L. C., Gould, V. C., Dow, J. M., Vernikos, G. S., Okazaki, A., Sebaihia, M., Saunders, D., Arrowsmith, C., Carver, T., Peters, N., Adlem, E., Kerhornou, A., Lord, A., Murphy, L., Seeger, K., Squares, R., Rutter, S., Quail, M. A., Rajandream, M. A., Harris, D., Churcher, C., Bentley, S. D., Parkhill, J., Thomson, N. R., and Avison, M. B. (2008) Genome Biology 9 http://genomebiology.com/2008/9/4/R74 [DOI] [PMC free article] [PubMed]
- 14.Alonso, A., and Martinez, J. L. (1997) Antimicrob. Agents Chemother. 41 1140-1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alonso, A., and Martinez, J. L. (2000) Antimicrob. Agents Chemother. 44 3079-3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alonso, A., and Martinez, J. L. (2001) Antimicrob. Agents Chemother. 45 1879-1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang, L., Li, X. Z., and Poole, K. (2001) Antimicrob. Agents Chemother. 45 3497-3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez, P., Moreno, E., and Martinez, J. L. (2005) Antimicrob. Agents Chemother. 49 781-782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez, P., Alonso, A., and Martinez, J. L. (2002) Antimicrob. Agents Chemother. 46 3386-3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez, P., Alonso, A., and Martinez, J. L. (2004) Antimicrob. Agents Chemother. 48 2274-2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramos, J. L., Martinez-Bueno, M., Molina-Henares, A. J., Teran, W., Watanabe, K., Zhang, X., Gallegos, M. T., Brennan, R., and Tobes, R. (2005) Microbiol. Mol. Biol. Rev. 69 326-356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orth, P., Schnappinger, D., Hillen, W., Saenger, W., and Hinrichs, W. (2000) Nat. Struct. Biol. 7 215-219 [DOI] [PubMed] [Google Scholar]
- 23.Hinrichs, W., Kisker, C., Duvel, M., Muller, A., Tovar, K., Hillen, W., and Saenger, W. (1994) Science 264 418-420 [DOI] [PubMed] [Google Scholar]
- 24.Orth, P., Cordes, F., Schnappinger, D., Hillen, W., Saenger, W., and Hinrichs, W. (1998) J. Mol. Biol. 279 439-447 [DOI] [PubMed] [Google Scholar]
- 25.Schumacher, M. A., Miller, M. C., Grkovic, S., Brown, M. H., Skurray, R. A., and Brennan, R. G. (2002) EMBO J. 21 1210-1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alguel, Y., Meng, C., Teran, W., Krell, T., Ramos, J. L., Gallegos, M. T., and Zhang, X. (2007) J. Mol. Biol. 369 829-840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hillen, W., and Berens, C. (1994) Annu. Rev. Microbiol. 48 345-369 [DOI] [PubMed] [Google Scholar]
- 28.Engohang-Ndong, J., Baillat, D., Aumercier, M., Bellefontaine, F., Besra, G. S., Locht, C., and Baulard, A. R. (2004) Mol. Microbiol. 51 175-188 [DOI] [PubMed] [Google Scholar]
- 29.Resch, M., Striegl, H., Henssler, E. M., Sevvana, M., Egerer-Sieber, C., Schiltz, E., Hillen, W., and Muller, Y. A. (2008) Nucleic Acids Res. 36 4390-4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reichheld, S. E., and Davidson, A. R. (2006) J. Mol. Biol. 361 382-389 [DOI] [PubMed] [Google Scholar]
- 31.Kamionka, A., Bogdanska-Urbaniak, J., Scholz, O., and Hillen, W. (2004) Nucleic Acids Res. 32 842-847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeng, W. Y., Ko, T. P., Liu, C. I., Guo, R. T., Liu, C. L., Shr, H. L., and Wang, A. H. (2008) Nucleic Acids Res. 36 1567-1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atlas, R. M. (1993) Handbook of Microbiological Media, p. 492, CRC Press, Boca Raton, FL
- 34.Chong, S., Mersha, F. B., Comb, D. G., Scott, M. E., Landry, D., Vence, L. M., Perler, F. B., Benner, J., Kucera, R. B., Hirvonen, C. A., Pelletier, J. J., Paulus, H., and Xu, M. Q. (1997) Gene (Amst.) 192 271-281 [DOI] [PubMed] [Google Scholar]
- 35.Chong, S., Montello, G. E., Zhang, A., Cantor, E. J., Liao, W., Xu, M. Q., and Benner, J. (1998) Nucleic Acids Res. 26 5109-5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leslie, A. G. (2006) Acta Crystallogr. D Biol. Crystallogr. 62 48-57 [DOI] [PubMed] [Google Scholar]
- 37.CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50 760-763 [DOI] [PubMed] [Google Scholar]
- 38.Pape, T., and Schneider, T. R. (2004) J. Appl. Crystallogr. 37 843-844 [Google Scholar]
- 39.Schneider, T. R., and Sheldrick, G. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58 1772-1779 [DOI] [PubMed] [Google Scholar]
- 40.Sheldrick, G. M. (2002) Z. Kristallogr. 217 644-650 [Google Scholar]
- 41.Terwilliger, T. C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59 1688-1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams, P. D., Grosse-Kunstleve, R. W., Hung, L. W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K., and Terwilliger, T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58 1948-1954 [DOI] [PubMed] [Google Scholar]
- 43.Perrakis, A., Morris, R., and Lamzin, V. S. (1999) Nat. Struct. Biol. 6 458-463 [DOI] [PubMed] [Google Scholar]
- 44.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 45.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 46.Pinter, J., and Merritt, E. A. (2006) J. Appl. Crystallogr. 39 109-111 [Google Scholar]
- 47.Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B., III, Snoeyink, J., Richardson, J. S., and Richardson, D. C. (2007) Nucleic Acids Res. 35 375-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merritt, E. A. (1999) Acta Crystallogr. D Biol. Crystallogr. 55 1109-1117 [DOI] [PubMed] [Google Scholar]
- 49.Moretti, S., Armougom, F., Wallace, I. M., Higgins, D. G., Jongeneel, C. V., and Notredame, C. (2007) Nucleic Acids Res. 35 645-648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rockwell, N. C., and Lagarias, J. C. (2007) BMC Bioinformatics 8 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker, N. A., Sept, D., Joseph, S., Holst, M. J., and McCammon, J. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10037-10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., and Ferrin, T. E. (2004) J. Comput. Chem. 25 1605-1612 [DOI] [PubMed] [Google Scholar]
- 53.DeLano, W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientifics, Palo Alto, CA
- 54.Li, W., and Godzik, A. (2006) Bioinformatics 22 1658-1659 [DOI] [PubMed] [Google Scholar]
- 55.Rojo, F., and Alonso, J. C. (1995) Nucleic Acids Res. 23 3181-3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dyson, H. J., and Wright, P. E. (2005) Nat. Rev. Mol. Cell Biol. 6 197-208 [DOI] [PubMed] [Google Scholar]
- 57.Murray, D. S., Schumacher, M. A., and Brennan, R. G. (2004) J. Biol. Chem. 279 14365-14371 [DOI] [PubMed] [Google Scholar]
- 58.Brady, G. P., Jr., and Stouten, P. F. (2000) J. Comput. Aided Mol. Des. 14 383-401 [DOI] [PubMed] [Google Scholar]
- 59.Huang, B., and Schroeder, M. (2006) BMC Struct. Biol. 6 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dundas, J., Ouyang, Z., Tseng, J., Binkowski, A., Turpaz, Y., and Liang, J. (2006) Nucleic Acids Res. 34 116-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willems, A. R., Tahlan, K., Taguchi, T., Zhang, K., Lee, Z. Z., Ichinose, K., Junop, M. S., and Nodwell, J. R. (2008) J. Mol. Biol. 376 1377-1387 [DOI] [PubMed] [Google Scholar]
- 62.Brooks, B. E., Piro, K. M., and Brennan, R. G. (2007) J. Am. Chem. Soc. 129 8389-8395 [DOI] [PubMed] [Google Scholar]
- 63.Peters, K. M., Schuman, J. T., Skurray, R. A., Brown, M. H., Brennan, R. G., and Schumacher, M. A. (2008) Biochemistry 47 8122-8129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schumacher, M. A., Miller, M. C., Grkovic, S., Brown, M. H., Skurray, R. A., and Brennan, R. G. (2001) Science 294 2158-2163 [DOI] [PubMed] [Google Scholar]
- 65.Dover, L. G., Corsino, P. E., Daniels, I. R., Cocklin, S. L., Tatituri, V., Besra, G. S., and Futterer, K. (2004) J. Mol. Biol. 340 1095-1105 [DOI] [PubMed] [Google Scholar]
- 66.Petrek, M., Otyepka, M., Banas, P., Kosinova, P., Koca, J., and Damborsky, J. (2006) BMC Bioinformatics 7 316. [DOI] [PMC free article] [PubMed] [Google Scholar]