Abstract
The bacterium Streptomyces anulatus 9663, isolated from the intestine of different arthropods, produces prenylated derivatives of phenazine 1-carboxylic acid. From this organism, we have identified the prenyltransferase gene ppzP. ppzP resides in a gene cluster containing orthologs of all genes known to be involved in phenazine 1-carboxylic acid biosynthesis in Pseudomonas strains as well as genes for the six enzymes required to generate dimethylallyl diphosphate via the mevalonate pathway. This is the first complete gene cluster of a phenazine natural compound from streptomycetes. Heterologous expression of this cluster in Streptomyces coelicolor M512 resulted in the formation of prenylated derivatives of phenazine 1-carboxylic acid. After inactivation of ppzP, only nonprenylated phenazine 1-carboxylic acid was formed. Cloning, overexpression, and purification of PpzP resulted in a 37-kDa soluble protein, which was identified as a 5,10-dihydrophenazine 1-carboxylate dimethylallyltransferase, forming a C–C bond between C-1 of the isoprenoid substrate and C-9 of the aromatic substrate. In contrast to many other prenyltransferases, the reaction of PpzP is independent of the presence of magnesium or other divalent cations. The Km value for dimethylallyl diphosphate was determined as 116 μm. For dihydro-PCA, half-maximal velocity was observed at 35 μm. Kcat was calculated as 0.435 s-1. PpzP shows obvious sequence similarity to a recently discovered family of prenyltransferases with aromatic substrates, the ABBA prenyltransferases. The present finding extends the substrate range of this family, previously limited to phenolic compounds, to include also phenazine derivatives.
The transfer of isoprenyl moieties to aromatic acceptor molecules gives rise to an astounding diversity of secondary metabolites in bacteria, fungi, and plants, including many compounds that are important in pharmacotherapy. However, surprisingly little biochemical and genetic data are available on the enzymes catalyzing the C-prenylation of aromatic substrates. Recently, a new family of aromatic prenyltransferases was discovered in streptomycetes (1), Gram-positive soil bacteria that are prolific producers of antibiotics and other biologically active compounds (2). The members of this enzyme family show a new type of protein fold with a unique α-β-β-α architecture (3) and were therefore termed ABBA prenyltransferases (1). Only 13 members of this family can be identified by sequence similarity searches in the data base at present, and only four of them have been investigated biochemically (3–6). Up to now, only phenolic compounds have been identified as aromatic substrates of ABBA prenyltransferases. We now report the discovery of a new member of the ABBA prenyltransferase family, catalyzing the transfer of a dimethylallyl moiety to C-9 of 5,10-dihydrophenazine 1-carboxylate (dihydro-PCA).2 Streptomyces strains produce many of prenylated phenazines as natural products. For the first time, the present paper reports the identification of a prenyltransferase involved in their biosynthesis.
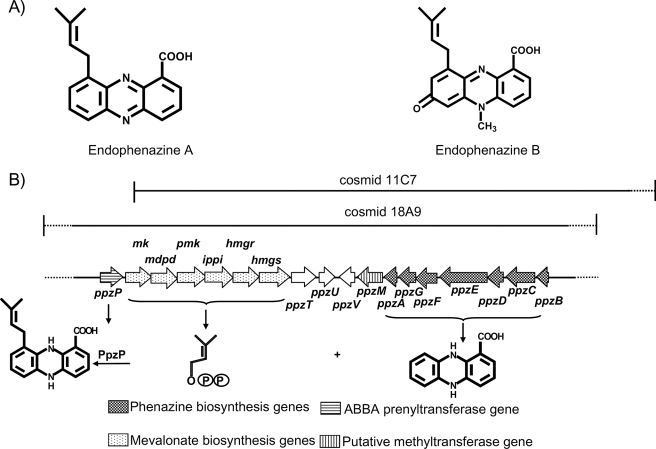
Streptomyces anulatus 9663, isolated from the intestine of different arthropods, produces several prenylated phenazines, among them endophenazine A and B (Fig. 1A) (7). We wanted to investigate which type of prenyltransferase might catalyze the prenylation reaction in endophenazine biosynthesis. In streptomycetes and other microorganisms, genes involved in the biosynthesis of a secondary metabolite are nearly always clustered in a contiguous DNA region. Therefore, the prenyltransferase of endophenazine biosynthesis was expected to be localized in the vicinity of the genes for the biosynthesis of the phenazine core (i.e. of PCA).
FIGURE 1.
A, prenylated phenazines from S. anulatus 9663. B, biosynthetic gene cluster of endophenazine A.
In Pseudomonas, an operon of seven genes named phzABCDEFG is responsible for the biosynthesis of PCA (8). The enzyme PhzC catalyzes the condensation of phosphoenolpyruvate and erythrose-4-phosphate (i.e. the first step of the shikimate pathway), and further enzymes of this pathway lead to the intermediate chorismate. PhzD and PhzE catalyze the conversion of chorismate to 2-amino-2-deoxyisochorismate and the subsequent conversion to 2,3-dihydro-3-hydroxyanthranilic acid, respectively. These reactions are well established biochemically. Fewer data are available about the following steps (i.e. dimerization of 2,3-dihydro-3-hydroxyanthranilic acid, several oxidation reactions, and a decarboxylation, ultimately leading to PCA via several instable intermediates). From Pseudomonas, experimental data on the role of PhzF and PhzA/B have been published (8, 9), whereas the role of PhzG is yet unclear. Surprisingly, the only gene cluster for phenazine biosynthesis described so far from streptomycetes (10) was found not to contain a phzF orthologue, raising the question of whether there may be differences in the biosynthesis of phenazines between Pseudomonas and Streptomyces.
Screening of a genomic library of the endophenazine producer strain S. anulatus now allowed the identification of the first complete gene cluster of a prenylated phenazine, including the structural gene of dihydro-PCA dimethylallyltransferase.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Culture Conditions—S. anulatus 9663 has been isolated previously from the intestine of different arthropods (7, 11). It was grown in liquid YMG medium or on solid MS medium. For production of secondary metabolites, the medium described by Sedmera et al. (12) was used.
Escherichia coli XL1 Blue MRF, E. coli SURE (Stratagene, Heidelberg, Germany), E. coli BW 25113, and E. coli ET 12567 (pUB307) were used for cloning and were grown in liquid or on solid (1.5% agar) Luria-Bertani or SOB medium at 37 °C. The REDIRECT technology kit for PCR targeting was obtained from Plant Bioscience Limited (Norwich, UK). For inactivation experiments, the aac(3)IV/oriT (apramycin resistance) cassette from pIJ773 (13) was used. Carbenicillin (50–100 μg ml-1), apramycin (50 μg ml-1), kanamycin (50 μg ml-1), chloramphenicol (25 μg ml-1), and nalidixic acid (20 μg ml-1) were used for selection of recombinant strains.
Chemicals—Kanamycin and carbenicillin were purchased from Genaxxon BioSciences GmbH (Biberach, Germany); apramycin and nalidixic acid were from Sigma; chloramphenicol was from Merck; and phenazine 1-carboxylic acid was from InFormatik. Dimethyllallyl diphosphate was synthesized as described in Ref. 14. Endophenazine A was isolated from cultures of Streptomyces cinnamonensis DSM 1024 as described in Ref. 7.
Genetic Procedures—Standard methods for DNA isolation and manipulation were performed as described by Kieser et al. (15) and Sambrook et al. (16). DNA fragments were isolated from agarose gels by using a PCR purification kit (Amersham Biosciences). Genomic DNA was isolated from Streptomyces strains by lysozyme treatment and phenol/chloroform extraction as described by Kieser et al. (15).
Construction and Screening of the Cosmid Library—Chromosomal DNA from S. anulatus was partially digested with Sau3AI, dephosphorylated and then ligated into the BamHI sites of SuperCos 1 (Stratagene) according to the manufacturer's instructions. The ligation products were packaged with Gigapack III XL (Stratagene) and transduced into E. coli SURE. Colony hybridization was performed on Hybond-N membranes (Amersham Biosciences). ephzA from S. cinnamonensis (10) was used as hybridization probe for the first screening of the library. The digoxigenin-labeled ephzA was generated using the PCR digoxigenin labeling mix (Roche Applied Science) with the primers ephzA_for (5′-ATG AGC ACC CCC CTG ACC ACC-3′) and ephzA_rev (5′-TCA GGA GGG GAT CCA GTC CCG-3′).
A second screening was performed by PCR for the identification of the following genes: phzD, phzF, hmgr (3-hydroxy-3-methyl-glutaryl-CoA reductase), hmgs (3-hydroxy-3-methylglutaryl-CoA synthase), and mdpd (mevalonate diphosphate decarboxylase). The following primers were used: PhzD_for (5′-CGC GCC GTC CTG (A/G)TN CA(C/T) GA(C/T) (A/C/T)T-3′) and phzD_rev (5′-CGG TGG TGG TCC CGG (G/C)(A/T)(A/G) AA(A/G) TCN (G/C)-3′); phzF_for (5′-CAT CCG GAT CTT GAC CCC NGT NAA (C/T)GA-3′) and phzF_rev (5′-GAG GGG CGC CCC AT(C/T) TCN CAN CC-3′); HMGR_for (5′-GGG CAT CGC CGC GAC CCT CGT GGA GGA GGG-3′) and HMGR_rev (5′-GCG ATG ACG GGG AGG CGC CGG GCG TTC TC-3′); HMGS_for (5′-GCC AAG TCC GCC GGN GTN TA(C/T) GT-3′) and HMGS_rev (5′-AGC CGG AAG GGG CCN GTN GT(C/T) TG-3′); MDPD_for (5′-GAC CCT GGA CGT CTT CCC NAC NAC NAC-3′) and MDPD_rev (5′-GCG TTC CGC TCG GC(A/G/T) AT(C/T) TCN-3′). PCRs were carried out with Taq polymerase.
Heterologous Expression of Cosmids ppzOS04 and ppzOS02— The plasmid pIJ787 (17) was first digested with DraI and BsaI, and the 4990-bp fragment containing the integrase cassette was used to replace the bla gene in the SuperCos 1 backbone in cosmids 11C7 and 18A9, using λ RED-mediated recombination (18, 19), generating ppzOS02 and ppzOS04, respectively. Both cosmids were first transformed into the nonmethylating host E. coli ET12567, and the nonmethylated DNA was introduced into Streptomyces coelicolor M512 via triparental conjugation.
Inactivation of the Gene ppzP—An apramycin resistance cassette (aac(3)IV) was amplified from plasmid pUG019 (17) using the following primers: ppzP_F (5′-CGC CCC AAG GGT GTC TTG TCG ACC TGT GGA GGA AAA ATG TCT AGA ATT CCG GGG ATC CGT CGA CC-3′) and ppzP_R (5′-CAA GCC CTT GTC CTT CAC ATG CCG ACG GGT GAG GCG CTA ACT AGT TGT AGG CTG GAG CTG CTT C-3′). Underlined are restriction sites for XbaI and SpeI, used for later removal of the cassette. The resulting 1077-bp PCR product was used to replace the ppzP gene on cosmid ppzOS04 by λ RED-mediated recombination, resulting in cosmid ppzOS05. Deletion of the aac(3)IV cassette from ppzOS05 was carried out by digestion with XbaI and SpeI and religation, resulting in cosmid ppzOS09. The resulting construct was introduced into S. coelicolor M512 via triparental conjugation (15).
Production and Analysis of Secondary Metabolites—Exconjugants of all mutants as well as wild type S. anulatus were precultured for 48 h in liquid YMG medium (50 ml). 50 ml of production medium (20) was then inoculated with 2.5 ml of the precultures. The flasks were agitated on a rotary shaker at 30 °C and 200 rpm for 120 h. For cultivation of mutants, all liquid media contained kanamycin (50 μg ml-1).
For isolation of endophenazine A, mycelia from 50-ml cultures were centrifuged at 3500 × g for 10 min. The supernatant was discarded, and the cells were extracted with methanol (10 ml) by vortexing. The extract was mixed with sodium acetate buffer (10 ml; 1 m, pH 4.0) and extracted with dichloromethane (5 ml). After separation of the organic phase, the solvent was evaporated, and the residue was redissolved in methanol (0.5 ml).
Extracts were analyzed with HPLC (Agilent 1100 series; Waldbronn, Germany) by using an Eclipse XDB-C18 column (4.6 × 150 mm, 5 μm; Agilent) at a flow rate of 1 ml min-1 with a linear gradient from 10 to 100% of solvent B in 20 min (solvent A: water/phosphoric acid (999:1); solvent B, acetonitrile) and detection at 252 and 365 nm. Additionally, a UV spectrum from 200 to 400 nm was logged by a photodiode array detector. The absorbance at 365 nm was used for quantitative analysis, employing authentic reference samples of PCA and endophenazine A as external standards.
Analysis by LC-MS—The extracts were examined with LC-MS and LC-MS2 analysis using a Nucleosil 100-C18 column (3 μm, 100 × 2 mm) coupled to an ESI mass spectrometer (LC/MSD Ultra Trap System XCT 6330; Agilent Technology). Analysis was carried out at a flow rate of 0.4 ml min-1 with a linear gradient from 10 to 100% of solvent B in 15 min (solvent A: water/formic acid (999:1); solvent B: acetonitrile/formic acid (999.4:0.6)). Detection was carried out at 230, 260, 280, 360, and 435 nm. Electrospray ionization (positive and negative ionization) in Ultra Scan mode with capillary voltage of 3.5 kV and heated capillary temperature of 350 °C was used for LC-MS analysis. For LC-MS2 and LC-MS3, the analysis was carried out in positive ionization mode with capillary voltage of 3.5 kV at 350 °C. For LC-MS2 identification of the enzymatic product endophenazine A, the mass 293 ± 0.5 Da was selected for fragmentation. In LC-MS3, the mass 275 ± 1 Da was selected for fragmentation.
Overexpression and Purification of PpzP Protein—ppzP was amplified by using the primers ppzP_pHis_F (5′-ACC TGT GGA GAA TTC ATG TCA GAA TCC GCT GAG CT-3′) and ppzP_pHis_R (5′-CCG GAC GGG CTC GAG GCT ATC CGG CAT CGG CGG TCA-3′). The underlined letters represent EcoRI and XhoI restriction sites, respectively. The resulting PCR fragment was digested with EcoRI and XhoI and ligated into plasmid pHis8 (21) digested with the same restriction enzymes. The resulting plasmid, pHis8-OS01, was verified by restriction mapping and sequencing.
E. coli BL21(DE3)pLysS cells harboring plasmid pHis8-OS01 were cultivated in 2 liters of liquid TB medium containing 50 μg ml-1 kanamycin and grown at 37 °C to an A600 of 0.6. The temperature was lowered to 20 °C, and isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 0.5 mm. The cells were cultured for a further 10 h at 20 °C and harvested. 30 ml of lysis buffer (50 mm Tris-HCl, pH 8.0, 1 m NaCl, 10% glycerol, 10 mm β-mercaptoethanol, 20 mm imidazole, 0.5 mg ml-1 lysozyme, 0.5 mm phenylmethylsulfonyl fluoride) were added to the pellet (40 g). After stirring at 4 °C for 30 min, cells were ruptured with a Branson sonifier by 4 °C. The lysate was centrifuged (55,000 × g, 45 min). Affinity chromatography with 4 ml of Ni2+-nitrilotriacetic acid-agarose resin (Qiagen, Hilden, Germany) was carried out according to the manufacturer's instructions, using 2 × 2.5 ml of 250 mm imidazole (in 50 mm Tris-HCl, pH 8.0, 1 m NaCl, 10% glycerol, 10 mm β-mercaptoethanol) for elution. Subsequently, a buffer exchange was carried out by PD10 columns (Amersham Biosciences), which were eluted with 50 mm Tris-HCl, pH 8.0, 1 m NaCl, 10% glycerol, and 2 mm 1,4-dithiothreitol. Approximately 30 mg of purified PpzP were obtained from 2 liters of cultures.
Assay for Prenyltransferase Activity—The reaction mixture (100 μl) contained 80 mm Na-TAPS (pH 7.5) (Sigma), 0.8 mm freshly prepared dihydro-PCA, 0.4 mm dimethylallyl diphosphate (DMAPP), 500 mm NaCl, and 5 μg/ml PpzP. For the preparation of dihydro-PCA, 90 μl of 100 mm freshly dissolved sodium dithionite (Merck) were mixed with 10 μl of 100 mm PCA dissolved in Tris-HCl, pH 8.0. 8 μl of this mixture was added to the incubation mixture. After incubation for 20 or 30 min at 30 °C, 15 μl of 100 mm sodium persulfate (Sigma) were added to oxidize dihydro-PCA and dihydroendophenazine A into PCA and endophenazine A, respectively. The mixture was immediately extracted with 100 μl of ethyl acetate/formic acid (975:25). After vortexing and centrifugation, 75 μl of the organic phase were evaporated. The residue was dissolved in 100 μl of methanol, and 35 μl thereof were analyzed by HPLC using an Eclipse XDB-C18 column (4.6 × 150 mm, 5 μm; Agilent) with the same mobile phase and gradient described above for secondary metabolite analysis. Detection was carried out at 252 and 365 nm. Quantitative analysis was carried out using the absorbance at 365 nm, as described under “Production and Analysis of Secondary Metabolites.”
Calculation of Kinetic Constants—Km and Kcat were calculated using the GraphPad Prism software, version 5.01 for Windows (GraphPad Software Inc., La Jolla, CA).
RESULTS
Cloning of a Biosynthetic Gene Cluster for Prenylated Phenazines—A genomic library of the phenazine producer strain S. anulatus was constructed in cosmid vector SuperCos 1 (Stratagene). 2000 independent cosmid clones were subjected to a colony blot screening with a labeled probe of the phenazine biosynthesis gene ephzA from S. cinnamonensis (10). From the genome size of streptomycetes (∼8.5 megabases) and the average insert size of SuperCos 1 (∼38 kb), it could be estimated that a single genome locus would be represented, on average, in nine different cosmid clones. However, the screening revealed 26 positive clones, indicating that more than one genomic locus hybridized with ephzA. In order to identify those cosmids that contained the correct locus and that were likely to contain the entire gene cluster, these 26 cosmids were screened with degenerate primers for two further phenazine biosynthetic genes, phzD and phzF, and for three genes of the mevalonate pathway for isoprenoid biosynthesis (see “Experimental Procedures”). In another Streptomyces strain, it had been shown that the prenyl moiety of endophenazine A is formed via the mevalonate pathway (22), whereas in streptomycetes, the methyl erythritol phosphate pathway is used for the formation of isoprenoids of primary metabolism (23). Two cosmids, 18A9 and 11C7, were identified that gave PCR products with all five primer pairs. Sequencing of the PCR products confirmed that they represented phzD, phzF, and the three mevalonate pathway genes. Sequencing of cosmid 18A9 revealed a DNA region of 17.5 kb comprising 18 putative coding sequences (Fig. 1B), which together code for all enzymatic functions expected to be involved in endophenazine formation. The results of data base comparisons for these genes are listed in Table 1. Seven of the putative coding sequences, designated as ppzA, ppzC, ppzD, ppzE, ppzF, ppzG, and ppzB, showed obvious similarities to the seven genes that direct the biosynthetic pathway from phosphoenolpyruvate and erythrose-4-phosphate to PCA in Pseudomonas strains (8, 9). In addition, the gene ppzM was identified, showing striking similarity to phzM from P. aeroginosa. phzM is proposed to be responsible for the N-methylation in the biosynthesis of pyocyanine (1-hydroxy-5-N-methylphenazine) (24). ppzM may therefore be responsible for the N-methylation reaction in the biosynthesis of endophenazine B (Fig. 1A).
TABLE 1.
Genes in the endophenazine biosynthetic gene cluster of S. anulatus 9663
| Gene | Amino acid | Proposed function | Ortholog identified by BLAST search | Identity/Similarity |
|---|---|---|---|---|
| ppzP | 299 | Prenyltransferase | Fnq26, S. cinnamonensis | 44/64 |
| mk | 345 | Mevalonate kinase | MEVK, Streptomyces sp. KO-3988 | 67/79 |
| mdpd | 351 | Diphosphomevalonate decarboxylase | MDPD, Streptomyces sp. KO-3988 | 73/80 |
| pmk | 371 | Phosphomevalonate kinase | PMK, Streptomyces sp. KO-3988 | 68/75 |
| ippi | 363 | Isopentenyldiphosphate δ-isomerase | Fni, Streptomyces sp. CL190 | 79/88 |
| hmgr | 353 | 3-Hydroxy-3-methylglutaryl coenzyme A reductase | HMGR, Streptomyces sp. CL190 | 86/92 |
| hmgs | 391 | 3-Hydroxy-3-methylglutaryl CoA synthase | HMGS, Streptomyces sp. CL190 | 78/87 |
| ppzT | 327 | 3-Oxoacyl-[acyl-carrier-protein] synthase | 3-Oxoacyl-[acyl-carrier-protein] synthase, Streptomyces sp. KO-3988 | 79/86 |
| ppzU | 221 | Flavodoxin | SGR_4078, Streptomyces griseus | 59/75 |
| ppzV | 206 | Hypothetical protein | Fnq22, S. cinnamonensis | 62/77 |
| ppzM | 340 | N-Methyltransferase | PhzM, Pseudomonas aeruginosa | 44/58 |
| ppzB | 168 | Phenazine biosynthesis | PhzB, Pseudomonas fluorescens | 51/67 |
| ppzG | 233 | FMN-dependent oxidase | PhzG, P. fluorescens | 54/71 |
| ppzF | 279 | trans-2,3-Dihydro 3-hydroxyanthranilate isomerase | PhzF, P. fluorescens | 68/77 |
| ppzE | 646 | 2-Amino-2-desoxy-isochorismate synthase | PhzE, P. fluorescens | 60/76 |
| ppzD | 207 | 2,3-Dihydro-3-hydroxy-anthranilate (DHHA) Synthase | PhzD, P. fluorescens | 60/76 |
| ppzC | 392 | 3-Deoxy-d-arabino-heptulosonic acid 7-phosphate synthase | PhzC, P. fluorescens | 59/73 |
| ppzA | 162 | Phenazine biosynthesis | PhzA, P. fluorescens | 78/87 |
Sequence analysis further revealed a group of six coding sequences with striking similarity to genes of the mevalonate pathway (23), leading from acetyl-CoA and acetoacetyl-CoA to isopentyl diphosphate and DMAPP (see Table 1). Orthologs of these six genes, arranged in the exact same order, are found in the biosynthetic gene cluster of furaquinocin (25), naphterpin (26), terepenticin (27), and BE 40644 (28). S. anulatus, therefore, is one of the rare Streptomyces species that possesses the genes of the mevalonate pathway, a feature limited to ∼1% of the strains of this genus (29).
Directly adjacent to the genes of the mevalonate pathway, a gene designated ppzP was identified (Fig. 1B). Its predicted gene product showed 44% identity to Fnq26, a prenyltransferase of the ABBA family that is involved in the biosynthesis of the meroterpenoid furanonaphtoquinone I (5).
Heterologous Expression of the Endophenazine Cluster in S. coelicolor M512—λ RED-mediated recombination in E. coli was used to replace the β-lactamase gene within the SuperCos 1 backbone of cosmid 18A9 with the cassette pIJ787 containing a tetracycline resistance gene and the integration function of the phage ΦC31 (13). The resulting cosmid ppzOS04 was introduced into the genome of S. coelicolor M512 using triparental conjugation (15).
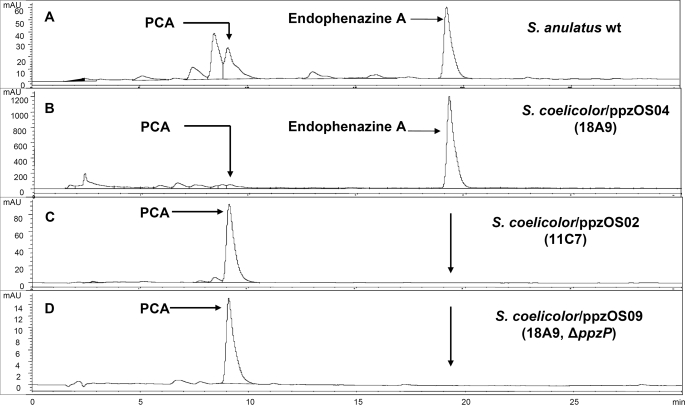
As control, the same pIJ787 cassette was introduced in an empty SuperCos 1 vector, and the resulting construct, ppzOS30, was also integrated into the genome of S. coelicolor M512. The secondary metabolite production of the resulting integration mutants was analyzed by HPLC-UV and LC-MS, using the wild type strain S. anulatus as comparison. In accordance with previous results (7), S. anulatus was found to produce PCA and endophenazine A (Fig. 2A). The polar compound PCA was predominantly found in the culture medium, but some amount was also present in the mycelial extract. The more lipophilic, prenylated compounds endophenazine A and B were associated with the cells (i.e. were mainly found in the pellet after centrifugation of the cultures). The heterologous expression strain S. coelicolor(ppzOS04) produced both phenazine 1-carboxylic acid (most of which was found in the medium) and endophenazine A (most of which was associated with the cells) (Fig. 2B). The presence of both compounds was also shown by LC-MS at m/z = 225 [M + H]+ and m/z = 293 [M + H]+ for PCA and endophenazine A, respectively. LC-MS2 showed that both compounds had the same fragmentation pattern as the authentic reference samples (data not shown). In contrast, the control strain S. coelicolor M512(ppzOS30), which had integrated the empty vector, did not produce any phenazines (data not shown).
FIGURE 2.
HPLC analysis of cell extracts of different Streptomyces strains (detection: UV, 365 nm). A, wild-type S. anulatus. B, S. coelicolor M512 containing the complete endophenazine cluster from cosmid 18A9. C, S. coelicolor M512 containing a partial endophenazine cluster (cosmid 11C7; see Fig. 1). D, S. coelicolor M512 containing the complete endophenazine cluster (cosmid 18A9) but with deletion of the putative prenyltransferase gene ppzP. PCA, phenazine 1-carboxylic acid. The polar compound PCA is predominantly found in the culture medium, but varying amounts are also present in the mycelial extracts, as shown here. The cultures also produced varying amounts of PCA-methyl ester, which is not separated from PCA in this HPLC system but could be identified in the LC-MS analysis (data not shown). The more lipophilic, prenylated compound endophenazin A is predominantly found in cell extracts, not in the culture medium.
LC-MS analysis showed that both S. anulatus and S. coelicolor(ppzOS04) also produced a small amount of endophenazine B as well as phenazine 1-carboxylic acid methyl ester (data not shown). Both compounds were identified by LC-MS2 in comparison with authentic reference samples.
As mentioned above, the screening of the cosmid library had identified a second cosmid, 11C7. Sequencing of the termini of its insert and comparison with the sequence obtained from 18A9 showed that 11C7 lacked the gene ppzP (the putative prenyltransferase) and 744 nucleotides of the mevalonate kinase gene but contained all other genes of the putative endophenazine cluster (see Fig. 1B). Cosmid 11C7 was expressed heterologously in S. coelicolor M512 using the same method described above. The resulting strain S. coelicolor(ppzOS02) produced phenazine 1-carboxylic acid and its methyl ester but no prenylated phenazines (Fig. 2C).
The results of the heterologous expression experiment with 18A9 prove that this cosmid contains all of the genes required for endophenazine biosynthesis. The experiment with 11C7 indicates that ppzP may be responsible for the prenylation reaction in this pathway. However, 11C7 lacked not only ppzP but also the promoter of the putative operon of mevalonate biosynthesis genes (see Fig. 1B) as well as part of the mevalonate kinase gene. Therefore, the heterologous expression of 18A9 and 11C7 did not provide conclusive evidence for the function of ppzP.
Inactivation of the Prenyltransferase Gene ppzP and Heterologous Expression of the Resulting Construct in S. coelicolor M512—λ RED-mediated recombination was used to create an in-frame deletion of the gene ppzP within the integrative cosmid ppzOS04. For this purpose, the cassette pIJ773, containing the apramycin-resistant gene acc(3)IV and flanked with XbaI and SpeI restriction sites, was used to replace the coding sequence of the ppzP gene. Subsequently, the cassette was excised by XbaI and SpeI digestion and religation of the compatible overhangs, leaving only 6 bp as a “scar” sequence between the start and the stop codon of ppzP. The resulting integrative cosmid ppzOS09 was introduced into the genome of S. coelicolor M512 as described above. HPLC and LC-MS analysis of the cultures (Fig. 2D) showed phenazine 1-carboxylic acid and its methyl ester but no endophenazine A or B. This strongly supported the hypothesis that ppzP is responsible for the prenylation reaction in endophenazine biosynthesis in the heterologous expression experiment. However, a polar effect of the deletion on the expression of the genes downstream of ppzP cannot be excluded with certainty.
Overexpression and Purification of PpzP—For the biochemical investigation of the putative prenyltransferase, the gene ppzP was cloned into a plasmid for expression as an N-terminally His-tagged protein (see “Experimental Procedures”). The resulting construct was introduced into E. coli BL21 (DE3)pLysS. Induction with isopropyl 1-thio-β-d-galactopyranoside resulted in expression of a protein of ∼37 kDa, as determined by SDS-PAGE (Fig. S1), coinciding with the calculated molecular mass of 37.138 kDa. Ni2+ affinity chromatography resulted in a protein of ∼90% purity. 30 mg of purified soluble PpzP were obtained from 2 liters of culture. This protein was used for the biochemical investigations described below.
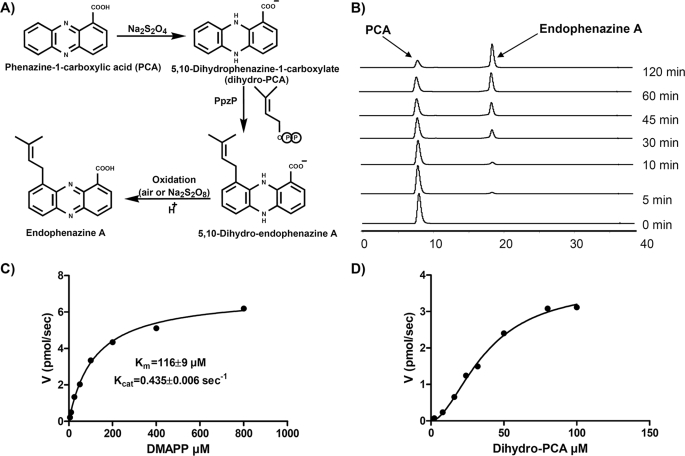
Identification of PpzP as 5,10-Dihydrophenazine 1-Carboxylate 9-Dimethylallyltransferase—Phenazine 1-carboxylic acid has been suggested as the product of the core pathway of phenazine biosynthesis from which the differently substituted or dimerized phenazines are derived by various tailoring reactions (8). In different Pseudomonas strains, PCA is converted by the monooxygenase PhzO to 2-hydroxy-PCA, by the amide synthethase PhzH to phenazine 1-carboxamide, and by the consecutive action of PhzM (methyltransferase) and PhzS (oxidoreductase) to pyocyanine (1-hydroxy-5-N-methyl-phenazine). However, when we incubated PCA with PpzP and DMAPP under various conditions, we did not observe product formation in HPLC analysis. From chemical reasoning, the reduced form of PCA (i.e. dihydro-PCA) (Fig. 3A), appeared to be an attractive candidate for the prenylation substrate, since it is much more reactive than PCA for an electrophilic attack at C-9. Dihydro-PCA is presumed to be the direct biosynthetic precursor of PCA (8). We therefore generated dihydro-PCA by reduction of PCA with sodium dithionite (Fig. 3A). When dihydro-PCA was incubated with PpzP and DMAPP, the time-dependent formation of an enzymatic product was readily observed by HPLC (Fig. 3B). Due to the very rapid oxidation of dihydro-PCA and its derivatives, they could not be quantified in the reduced form. After incubation, the reaction mixture was therefore oxidized with sodium persulfate (Na2S2O8), and substrate and product were analyzed in the oxidized form. The enzymatic product was thereafter identified as endophenazine A by LC-MS analysis showing the same retention time, UV spectrum, molecular ion (m/z = 293 in positive ionization), and fragmentation pattern in ESI-MS2 and ESI-MS3 as an authentic reference sample of endophenazine A.
FIGURE 3.
A, reaction scheme of the assay for dihydrophenazine 1-carboxylate dimethylallyltransferase activity. B, HPLC analysis of the dihydrophenazine 1-carboxylate prenyltransferase assay. C and D, product formation at different concentrations of DMAPP and dihydrophenazine 1-carboxylic acid. In C, dihydrophenazine 1-carboxylic acid was kept constant at 0.8 mm.In D, DMAPP was kept constant at 0.4 mm.
Biochemical Properties of PpzP—In the enzymatic assay described under “Experimental Procedures,” the formation of endophenazine A showed a linear dependence on the amount of PpzP (up to 1 μg) and on the reaction time (up to 45 min). The reaction was strictly dependent on the presence of active PpzP, DMAPP, and dihydro-PCA. Maximal product formation was observed at pH 7.5, with half-maximal values at pH 9.5 and pH 5.0. In sharp contrast to the trans-prenyldiphosphate synthases like FPP synthase (30) and to the aromatic prenyltransferase NphB of naphterpin biosynthesis (3), the catalytic activity of PpzP was independent of the presence of Mg2+ or any other divalent metal ions. The addition of EDTA (10 mm) even increased reaction velocity 1.5-fold. Similarly, the addition of 500 mm NaCl increased product formation 3-fold, whereas the addition of 50 mm MgCl2 and 100 mm CaCl2 increased product formation ∼1.5-fold. Therefore, 500 mm NaCl was routinely included in all assays (see “Experimental Procedures”). Although 10 mm Zn2+ had no effect on the reaction, the addition of 10 mm FeSO4 completely abolished the formation of endophenazine A.
PpzP was found to be specific for both DMAPP and dihydro-PCA. When geranyldiphosphate was used instead of DMAPP, no product formation was observed. Likewise, no prenylated products could be observed when PpzP was incubated with other phenazine substrates, such as phenazine, phenazine 1-carboxylic acid methyl ester, N-methyl-phenazine (as methyl sulfate salt), or pyocyanine (1-hydroxy-5-N-methyl-phenazine). These phenazine substrates were reduced with dithionite to their dihydro analogs in the same way as described for PCA. Since N-methyl-phenazine and pyocyanine are compounds with quaternary nitrogens, these reaction mixtures were analyzed directly without prior extraction with ethyl acetate.
We also tested the aromatic substrates of previously examined ABBA prenyltransferases (i.e. 4-hydroxyphenylpyruvate, flaviolin (2,5,7-trihydroxynaphthoquinone), 1,3-dihydroxynaphthalene, and 1,6-dihydroxynaphthalene). Of these, only flaviolin was prenylated by PpzP in the presence of DMAPP. LC-MS confirmed that the product was a monoprenylated flaviolin derivative. However, the reaction velocity was only 0.5% of that obtained with dihydro-PCA. A reaction mechanism of the C-prenylation of flaviolin under catalysis of Fnq26 was suggested by Haagen et al. (5). A similar mechanism may be expected for the prenylation of flaviolin by PpzP. However, the amount of the prenylated product was too low for a structural identification by NMR spectroscopy.
Using a constant concentration of dihydro-PCA (0.8 mm) and varying concentrations of DMAPP, a typical hyperbolic curve of product formation over substrate concentration was obtained (Fig. 3C), indicating that the reaction followed Michaelis-Menten kinetics. Nonlinear regression analysis resulted in a Km value of 116 ± 9 μm and a Kcat of 0.435 ± 0.006 s-1. Using different concentrations of dihydro-PCA in the presence of 0.4 mm DMAPP, a sigmoidal dependence of product formation on substrate concentration was observed (Fig. 3D). The half-maximal reaction velocity was obtained at ∼35 μm dihydro-PCA. Using this value (35 μm) as an estimate for the Km value for dihydro-PCA, the catalytic efficiency (Kcat/Km) of PpzP was calculated as 12,400 m-1 s-1. This is significantly higher than the value of 7.7 m-1 s-1 reported for the conversion of the (artificial) substrate 1,3-dihydroxynaphthalene by NphB (6) but is comparable with the value of 5280 m-1 s-1 calculated for the prenylation of the (genuine) substrate 4-HPP by CloQ (4). We therefore suggested that dihydro-PCA is the genuine substrate of PpzP.
Investigation of Fnq28 from S. cinnamonensis DSM 1042 for Dihydro-PCA Dimethylallyltransferase Activity—Our group recently reported a gene cluster of prenylated naphthoquinone and prenylated phenazine biosynthesis from S. cinnamonensis DSM 1042 (10). This strain produces, besides the prenylated naphthoquinone derivative furanonaphthoquinone I, the same prenylated phenazines as S. anulatus (10, 12, 20). The gene cluster from S. cinnamonensis contains genes assigned to furanonaphthoquinone I biosynthesis as well as a contiguous group of six genes (i.e. ephzBCDEGA) assigned to phenazine biosynthesis. By gene inactivation (10) and biochemical investigation (5), the prenyltransferase Fnq26 had been identified as the aromatic prenyltransferase of furanonaphthoquinone I biosynthesis. A second gene with similarity to ABBA prenyltransferases, designated as fnq28, was found immediately adjacent to the genes of the phenazine biosynthesis. Fnq28 shows 36% sequence identity with PpzP on the amino acid level. We tested whether Fnq28 also prenylates dihydro-PCA. For this purpose, Fnq28 was expressed as an N-terminally His-tagged protein, using the same method as employed for PpzP. Ni2+ affinity chromatography readily yielded a soluble protein of apparent homogeneity (data not shown). However, no prenylation of dihydro-PCA was observed with this protein, in clear contrast to the results with PpzP.
Fnq26 has been shown previously to transfer a geranyl moiety to flaviolin (2,5,7-trihydroxynaphthoquinone) or to the artificial substrate 1,3-dihydroxynaphthalene. We now tested whether Fnq26 could also use dihydro-PCA and either DMAPP or geranyldiphosphate as substrates. However, again no prenylation products were detected. These observations are consistent with the results from inactivation experiments, which had shown that endophenazine A is still formed in S. cinnamonensis after inactivation of both fnq26 and fnq28 (10). Therefore, neither Fnq26 nor Fnq28 catalyzes the prenylation reaction of endophenazine biosynthesis in S. cinnamonensis. In this organism, the enzyme that functionally corresponds to PpzP of S. anulatus is yet to be identified.
DISCUSSION
In this study, we have identified, for the first time, a prenyltransferase involved in the biosynthesis of prenylated phenazines. Sequence similarities and biochemical properties suggest that PpzP belongs to the recently discovered family of ABBA prenyltransferases (1). The present functional characterization of PpzP as 5,10-dihydrophenazine 1-carboxylate 9-dimethylallyltransferase now extends the substrate range of this family, previously limited to phenolic compounds, to include also phenazine derivatives.
At present, 13 genes with sequence similarity to ABBA prenyltransferases can be identified in the data base. A phylogenetic analysis of these genes (1) separates them into two clades. One of them comprises the 4-hydroxyphenylpyruvate 3-dimethylallyltransferases CloQ and NovQ from Streptomyces strains as well as four genes of unknown function from fungal genomes. The other clade comprises genes involved in the biosynthesis of prenylated naphthoquinones in different streptomycetes. A ClustalX analysis (data not shown) places PpzP into this second clade. Its closest ortholog is Fnq26, the prenyltransferase of furanonaphthoquinone I biosynthesis (5).
The x-ray structural analysis of the ABBA prenyltransferase NphB (formerly designated as Orf2) had revealed a novel protein fold, characterized by a β/α-barrel with 10 antiparallel β-strands. A structural model of PpzP (Fig. S2), generated using NphB as template, suggests a close similarity of the three-dimensional structure of the two proteins. The active center of NphB is localized within the spacious central cavity of the barrel, which has been suggested to explain the promiscuity of this enzyme for different aromatic substrates (3).
NphB requires the presence of Mg2+ for catalytic activity. X-ray crystallography has shown that the Mg2+ ion is coordinated by a single aspartate residue, by four water molecules, and by one of the oxygen atoms of the α-phosphate group of the isoprenoid diphosphate (3). In contrast, PpzP does not require Mg2+ ions for its activity, and the same has been reported for the ABBA prenyltransferases CloQ (4), SCO7190 (3) and Fnq26 (5). Modeling studies suggested that positively charged residues (Lys54 in CloQ, Arg51 in SCO7190, and Arg50 in Fnq26) may functionally substitute for Mg2+ in the binding of the α-phosphate group of the isoprenyl diphosphate (1, 3). An alignment of PpzP with the enzymes named above (Fig. S3) shows that also PpzP contains an arginine residue (Arg49) in the respective position of the first β strand. This may explain the Mg2+ independence of the PpzP reaction. Like the other ABBA prenyltransferases, PpzP does not contain the DDXXD motif typical for trans-prenyldiphosphate synthases (1).
In the biosynthesis of phenazines in different microorganisms, a common pathway leads to PCA, and this compound was believed to represent the branching point from which the pathways to differently substituted phenazines diverge (8). Our study now suggests that, at least for the prenylation of phenazines, the branching point is dihydro-PCA rather than PCA. This compound has been suggested (8) to be the product of the last enzyme-catalyzed step of the biosynthesis of the phenazine core, whereas its oxidation to PCA may occur nonenzymatically. Therefore, dihydro-PCA is likely to be available as substrate for PpzP in vivo.
Our study reports the first complete gene cluster of phenazine biosynthesis from streptomycetes, after a previously identified cluster turned out to be incomplete (10). Natural phenazines have important biological activities, including antibacterial, antitumor, antioxidant, and testosterone 5α-reductase-inhibiting activity (31). The present discovery of the endophenazine gene cluster from S. anulatus and the functional identification of PpzP as dihydro-PCA prenyltransferase may pave the way to the generation of new prenylated phenazines with improved biological activities by metabolic engineering and chemo-enzymatic synthesis.
Supplementary Material
Acknowledgments
We thank Kerstin Seeger for samples of Fnq26 and Fnq28, Ute Metzger and Elisa Haug-Schifferdecker for the synthesis of DMAPP, Andreas Kulik (Faculty of Biology, Tübingen University, Germany) for the LC-MS analysis, Christian Peifer (Medicinal Chemistry, Tübingen University, Germany) for the chemical synthesis of phenazine 1-carboxylic acid methyl ester, Joseph P. Noel (Salk Institute, La Jolla, CA) for plasmid pHis8, and Wulf Blankenfeldt (Max Planck Institute of Molecular Physiology, Düsseldorf, Germany) for helpful discussions.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) FN178498.
This work was supported by Deutsche Forschungsgemeinschaft Grant SPP 1152 EVOMET (to L. H.) and by a scholarship from Tishrin University, Syria (to O. S.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: PCA, phenazine 1-carboxylic acid; LC, liquid chromatography; MS, mass spectrometry; ESI, electrospray ionization; DMAPP, dimethylallyldiphosphate; TAPS, 3-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}-1-propanesulfonic acid; HPLC, high pressure liquid chromatography; MS2, second level tandem mass spectrometry; MS3, third level tandem mass spectrometry.
References
- 1.Tello, M., Kuzuyama, T., Heide, L., Noel, J. P., and Richard, S. B. (2008) Cell. Mol. Life Sci. 65 1459-1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hopwood, D. A. (2007) Streptomyces in Nature and Medicine: The Antibiotic Makers, Oxford University Press, New York
- 3.Kuzuyama, T., Noel, J. P., and Richard, S. B. (2005) Nature 435 983-987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pojer, F., Wemakor, E., Kammerer, B., Chen, H., Walsh, C. T., Li, S. M., and Heide, L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2316-2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haagen, Y., Unsöld, I., Westrich, L., Gust, B., Richard, S. B., Noel, J. P., and Heide, L. (2007) FEBS Lett. 581 2889-2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumano, T., Richard, S. B., Noel, J. P., Nishiyama, M., and Kuzuyama, T. (2008) Bioorg. Med. Chem. 16 8117-8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gebhardt, K., Schimana, J., Krastel, P., Dettner, K., Rheinheimer, J., Zeeck, A., and Fiedler, H. P. (2002) J. Antibiot. (Tokyo) 55 794-800 [DOI] [PubMed] [Google Scholar]
- 8.Mavrodi, D. V., Thomashow, L. S., and Blankenfeldt, W. (2008) in Pseudomonas: Model Organism, Pathogen, Cell Factory (Rehm, B. H. A., ed.) pp. 331-351, Wiley-VCH, Weinheim, Germany
- 9.Ahuja, E. G., Janning, P., Mentel, M., Graebsch, A., Breinbauer, R., Hiller, W., Costisella, B., Thomashow, L. S., Mavrodi, D. V., and Blankenfeldt, W. (2008) J. Am. Chem. Soc. 130 17053-17061 [DOI] [PubMed] [Google Scholar]
- 10.Haagen, Y., Glück, K., Fay, K., Kammerer, B., Gust, B., and Heide, L. (2006) ChemBioChem 7 2016-2027 [DOI] [PubMed] [Google Scholar]
- 11.Krastel, P., Zeeck, A., Gebhardt, K., Fiedler, H. P., and Rheinheimer, J. (2002) J. Antibiot. (Tokyo) 55 801-806 [DOI] [PubMed] [Google Scholar]
- 12.Sedmera, P., Pospíšil, S., and Novák, J. (1991) J. Nat. Prod. 54 870-872 [Google Scholar]
- 13.Gust, B., Chandra, G., Jakimowicz, D., Yuqing, T., Bruton, C. J., and Chater, K. F. (2004) Adv. Appl. Microbiol. 54 107-128 [DOI] [PubMed] [Google Scholar]
- 14.Woodside, A. B., Huang, Z., and Poulter, C. D. (1993) Org. Synth. 66 211-211 [Google Scholar]
- 15.Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000) Practical Streptomyces Genetics, John Innes Foundation, Norwich, UK
- 16.Sambrook, J., and Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 17.Eustáquio, A. S., Gust, B., Galm, U., Li, S. M., Chater, K. F., and Heide, L. (2005) Appl. Environ. Microbiol. 71 2452-2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gust, B., Challis, G. L., Fowler, K., Kieser, T., and Chater, K. F. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1541-1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datsenko, K. A., and Wanner, B. L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6640-6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tax, J., Sedmera, P., Vokoun, J., Urban, J., Karnetová, J., Stajner, K., Van[caron]ek, Z., and Krumphanzl, V. (1983) Collect. Czech. Chem. Commun. 48 527-532 [Google Scholar]
- 21.Jez, J. M., Ferrer, J. L., Bowman, M. E., Dixon, R. A., and Noel, J. P. (2000) Biochemistry 39 890-902 [DOI] [PubMed] [Google Scholar]
- 22.Bringmann, G., Haagen, Y., Gulder, T. A., Gulder, T., and Heide, L. (2007) J. Org. Chem. 72 4198-4204 [DOI] [PubMed] [Google Scholar]
- 23.Kuzuyama, T., and Seto, H. (2003) Nat. Prod. Rep. 20 171-183 [DOI] [PubMed] [Google Scholar]
- 24.Mavrodi, D. V., Bonsall, R. F., Delaney, S. M., Soule, M. J., Phillips, G., and Thomashow, L. S. (2001) J. Bacteriol. 183 6454-6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawasaki, T., Hayashi, Y., Kuzuyama, T., Furihata, K., Itoh, N., Seto, H., and Dairi, T. (2006) J. Bacteriol. 188 1236-1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takagi, M., Kuzuyama, T., Takahashi, S., and Seto, H. (2000) J. Bacteriol. 182 4153-4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isshiki, K., Tamamura, T., Sawa, T., Naganawa, H., Takeuchi, T., and Umezawa, H. (1986) J. Antibiot. (Tokyo) 39 1634-1635 [DOI] [PubMed] [Google Scholar]
- 28.Dairi, T. (2005) J. Antibiot. (Tokyo) 58 227-243 [DOI] [PubMed] [Google Scholar]
- 29.Sigmund, J. M., Clark, D. C., Rainey, F. A., and Anderson, A. S. (2003) Microb. Ecol. 46 106-112 [DOI] [PubMed] [Google Scholar]
- 30.Poulter, C. D. (2006) Phytochem. Rev. 5 17-26 [Google Scholar]
- 31.Laursen, J. B., and Nielsen, J. (2004) Chem. Rev. 104 1663-1685 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.