Abstract
PRIP (peroxisome proliferator-activator receptor interacting protein) is a nuclear receptor coactivator required for mammary gland development. To understand the function of PRIP in breast tumorigenesis, we established a mammary tumor cell line with the PRIPLoxp/Loxp genotype. By knocking out the PRIP gene in the tumor cell line, we demonstrated that PRIP deficiency led to inhibited tumor formation without affecting tumor cell proliferation. The PRIP deficiency was associated with decreased cell invasion and migration capabilities. We found that PRIP deficiency substantially reduced FOS gene expression. A chromatin immunoprecipitation assay revealed that PRIP was recruited to the FOS promoter. In addition, we demonstrated that PRIP also directly up-regulated the FOS gene expression in human breast cancer cells. Promoter analysis showed that PRIP acted through serum-responsive factor to regulate FOS gene expression. Finally, by re-expressing the FOS gene, we confirmed that the inhibited tumor formation of PRIP-deficient tumor cells was due to reduced expression of the FOS gene.
PRIP (ASC-2/RAP250/TRBP/NRC) is one of the nuclear receptor coactivators isolated by our laboratory and others (1–5). PRIP was shown to be a component of a large protein complex, ASCOM (MLL3 and MLL4 complex), including MLL3 or MLL4, ASH-2, Rbbp5, PTIP, WDR5, hDPY-30, and UTX (6–8). MLL3 and MLL4 carry a SET (Su(var)3–9, Enhancer of Zeste, Trithorax) domain with histone methyltransferase activity that methylates Lys4 on histone H3. UTX is a histone demethylase that demethylates trimethylated Lys27 on histone H3 (9, 10). Since the methylation of H3K4 and H3K27 are marks of active and silent states of gene expression, respectively, the association of PRIP with these enzymes indicates that it functions through modifying chromatin structure. In addition, we found that PRIP binds PIMT (PRIP-interacting protein with methyltransferase domain), and its function is stimulated by PIMT (11). PRIP also interacts with CAPER (coactivator of activating protein-1 and estrogen receptors) and COAA (coactivator activator) (12, 13), both of which contain RNA recognition motifs. In addition to nuclear receptors, PRIP was found to potentiate the transcriptional activity of NF-κB, Smad, Cre, AP-1, and serum-responsive factor (SRF)2 (3, 14).
We and others demonstrated that PRIP null mutation is embryonic lethal (15–17). To analyze the role of PRIP in mammary gland development, we generated mouse models with a null mutation of PRIP in mammary glands (18). Mammary glands with PRIP deficiency showed decreased ductal side branching and alveolar proliferation with relatively differentiated function intact. The ductal branching of mammary gland in response to estrogen treatment was attenuated in PRIP mutant glands. These studies demonstrated that PRIP plays critical roles in mammary gland development. In addition, the PRIP gene is amplified and overexpressed in a proportion of breast cancers (5), indicating that it may be involved in breast cancer development.
FOS is an immediate early gene belonging to a class of genes that are rapidly activated in response to intracellular signaling cascades (19). FOS and other members of the Fos family dimerize with Jun proteins to form AP-1 transcription factor complex (20). AP-1-regulated genes include critical modulators for invasion and metastasis; proliferation, differentiation, and survival; and angiogenesis (20). However, different cells often have a different set of genes controlled by AP-1, which leads to a different biological consequence due to increased AP-1 activity (20).
SRF is the transcription factor recognizing the serum-responsive element often found in the immediate early genes, including FOS (21). In addition to its role in cell proliferation, recent studies have revealed that SRF is a critical transcription factor regulating actin cytoskeleton and contractile genes (22).
To understand the function of PRIP in breast tumorigenesis, we established a mammary tumor cell line with the PRIPLoxp/Loxp genotype. By knocking out the PRIP gene in the tumor cell line, we demonstrated that PRIP promoted tumor formation by enhancing the SRF-mediated FOS gene expression.
EXPERIMENTAL PROCEDURES
Isolation of Mammary Epithelial Cells and Retroviral Infection—The inguinal glands were collected from PRIPLoxp/Loxp mice. Mammary epithelial cells were separated from stromal cells by collagenase digestion and Percoll gradient centrifugation (23). 5 × 106 Phoenix-Eco packaging cells were transfected with 15 μg of Pbabe-H-rasV12 (provided by Bob Weinberg, MIT) through the calcium phosphate precipitation method. Forty-eight hours after the transfection, viral supernatant was added to 5 × 105 mammary epithelial cells with 8 μg/ml Polybrene. After 5 h of incubation, the medium with retrovirus was replaced with fresh medium.
Knock-out of the PRIP Gene—Mammary tumor cells were infected with retroviruses expressing Cre-ERtam or control viruses. The cells were selected with 1 μg/ml puromycin. The cells were then treated with tamoxifen (1 μm) for 2 days.
Cell Proliferation Assay—Cells were seeded into 6-well plates at a density of 1 × 104 cells/well with fresh DMEM plus 10% FBS. Viable cells were counted after trypan blue staining once a day for 4 days.
Soft Agar Colony Formation Assay—Tumor cells (2 × 103) were suspended in a 1-ml mixture containing 0.35% (w/v) agar and DMEM supplemented with 10% FBS and layered on a 1-ml base of 0.7% (w/v) agar in DMEM with 10% FBS in a 6-well plate. Colonies were counted and photographed 2 weeks after plating. Each soft agar assay was performed in triplicate.
In Vivo Tumorigenicity Assay—1 × 106 mammary epithelial cells were subcutaneously injected into 8-week-old BALB/c athymic nude mice. The maximal tumor diameter was determined by caliber measurements once a week up to the 4th week.
Migration and Invasion Assays—For migration assays, transwell chamber membranes (Costar) were coated with 15 μg/ml collagen I. 1 × 105 cells suspended in DMEM/bovine serum albumin were added to the upper chamber, and DMEM containing 5% FBS was added to the lower chamber. After incubating for 14 h, nonmigrating cells were removed from the upper chamber with a cotton swab. Cells that had migrated to the lower surface of the membrane were stained with crystal violet. Migration was quantified by counting cells/mm2 using bright field optics. For invasion assays, chambers (8-μm pore size; Costar) were coated with Matrigel diluted 1:2 with α-minimum Eagle's medium. 1 × 105 cells were added to the chamber and allowed to invade for 14 h. Cells that had invaded to the lower surface of the filter were stained and counted. Each assay was repeated three times.
Microarray Hybridization—The tumor cells were used for the preparation of total RNA by the TRIzol (Invitrogen) method. Total RNA was used to make cRNA for hybridization with mouse genome (Affymetrix) as instructed. Microarray data analysis was performed with Affymetrix microarray software.
siRNA Transfection—siRNA smart pool oligonucleotides targeting human PRIP and control siRNAs were purchased from Dharmacon. Human breast cancer BT-474 cells were purchased from ATCC and maintained in DMEM containing 10% heat-inactivated FBS. The cells were transfected with control siRNA or PRIP siRNA using Lipofectamine 2000 reagent (Invitrogen) and cultured in phenol red-free minimum Eagle's medium containing 10% charcoal-stripped FBS. After 24 h, total RNA was then isolated using TRIzol reagent.
Quantitative Real Time RT-PCR—RT-PCR was performed with the SuperScript one-step RT-PCR kit from Invitrogen. Quantitative PCR was performed on ABI 7300 (Applied Biosystems) by using the SYBR Green Supermix (Applied Biosystems) according to the manufacturer's protocol. The expression levels of FOS or FOSB were normalized against β-actin RNA. The primers used were as follows: FOSB, 5′-ATAGCCTTGGCTTCCCGGC-3′ and 5′-AAGAGATGAGGGTGGGTTGC-3′; FOS, 5′-GCGTCATCCTCCCGCTGCA-3′ and 5′-GGCTGCACCAGCCACTGCA-3′; mouse β-actin, 5′-CCATCTACGAGGGCTATGCT-3′ and 5′-GCAAGTTAGGTTTTGTCAAAGA-3′; human β-actin, 5′-CGAGCACAGAGCCTCGCC-3′ and 5′-ACGATGGAGGGGAAGACGG-3′.
Chromatin Immunoprecipitation—Mammary tumor cells were maintained in phenol red-free modified Eagle's medium supplemented with 10% charcoal-dextran-stripped fetal bovine serum. Cells were cross-linked with 1% formaldehyde for 10 min at room temperature. After cells were collected, chromatin immunoprecipitation was performed as previously described (24), using antibodies against PRIP, SRF, or trimethylated H3K4 (Abcam, Cambridge, MA). The primer pair F1/R1 for the mouse FOS promoter is 5′-CGTCAATCCCTCCCTCCTTT-3′ and 5′-AGGATTTCGGAGATGGTCCC-3′. Sequences for control primer pair F2/R2 upstream of the mouse FOS promoter are 5′-CTTCTCTGCACTGATTTGGG-3′ and 5′-GGTCATTGTCCAGCAATCTG-3′. Sequences for the primer F1/R1 for the human FOS promoter are 5′-TCAATCCCTCCCCCCTTACA-3′ and 5′-TCTAAACGTCACGGGCTCAA-3′. Sequences for the primer F2/R2 located upstream of the human FOS promoter are 5′-ACTGCTACCCTGTAAGCTCG-3′ and 5′-CTTAACTCTTTCCTTCTGGC-3′. In all cases, PCR was performed with a serial dilution of input and various cycles (29–35 cycles) to ensure that amplification was maintained in the linear range. Each assay was repeated three times. Representative results were presented.
Promoter Analysis—The three FOS promoter fragments and the mutant FOS promoter fragment were amplified from mouse genomic DNA by PCR using forward primers 5′-AGCTAGGGTACCGCCGGCGAGCTGTTCCCGT-3′,5′-AGCTAGGGTACCTCCCTCCTTTACACAGGA-3′,5′-AGCTAGGGTACCTGCGTCAGCAGGTTTCCA-3′, and 5′-AGCTAGGGTACCTCCCTCCTTTACACAGGATGTGGATATTACCACATCTGC-3′ and the common reverse primer 5′-GCTAGCTAGATCTCCAGGGGTAGACACTGGT-3′. The PCR products were digested with KpnI and BglII and were inserted into pGL3-Basic KpnI/BglII sites (Promega, Madison, WI) to generate FOS1-LUC, FOS2-LUC, FOS3-LUC, and mutant FOS-LUC. PRIP-/- cells (1 × 105) were plated in DMEM containing 10% fetal calf serum in 6-well plates and were cultured for 24 h before transfection. Transfections were performed using Lipofectamine 2000 reagent (Invitrogen) with β-galactosidase expression vector pCMV-β as an internal control for transfection efficiency. Cell extracts were prepared 36 h after transfection and were assayed for luciferase and β-galactosidase activities.
Immunoprecipitation—Nuclear extracts from Ras-induced tumor cells (100 μg) were immunoprecipitated by anti-SRF (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or control IgG in the buffer (20 mm Tris-HCl, pH 7.9, 180 mm KCl, 0.2 mm EDTA, 0.05% Nonidet P-40, 0.5 mm phenylmethylsulfonyl fluoride, and 1 mm dithiothreitol). After extensive washing using the same buffer, the bound proteins were subjected to Western blot analysis using anti-PRIP.
Electrophoretic Mobility Shift Assay—The electrophoretic mobility shift assay was performed according to standard protocols. Nuclear extracts were prepared from wild type tumor cells. The double-stranded SRE oligonucleotide (5′-AGGATGTCCATATTAGGACATCTG-3′) was end-labeled using T4 polynucleotide kinase and [γ-32P]ATP. 5 μg of nuclear extracts were incubated for 20 min at room temperature with 32P-labeled SRE probe. DNA-protein complexes were resolved on a 6% TBE polyacrylamide gel. Gels were dried and autoradiographed at -80 °C. For supershift experiments, nuclear extracts were incubated with anti-SRF or anti-PRIP in binding buffer for 45 min at room temperature before 32P-labeled probe was added to the binding mixture.
Generation of Stable Cell Lines Expressing FOS and Tumor Formation—The PRIP-/- cells were transfected with 10 μg of pcDNA3-FLAG-Fos (Addgene) using Lipofectamine 2000. After selection with G418 (300 μg/ml), the individual clones were picked up, expanded, and examined for Fos expression. The PRIP+/+ cells, PRIP-/- cells not expressing exogenous Fos, and PRIP-/- cells expressing exogenous Fos were injected subcutaneously into nude mice. The maximal tumor diameter was determined by caliber measurements once a week up to the 4th week.
RESULTS
PRIP Is Not Required for the Proliferation of Ras-induced Mammary Tumor Cells—PRIPLoxP/LoxP mammary cells were isolated and infected with a retrovirus expressing an oncogenic form of the H-ras gene, H-rasV12. Two weeks later, individual clones were picked up and expanded. Many of the cells underwent senescence followed by apoptosis, whereas some of the cells proliferated. The process continued for about 4 months. Finally, a cell line emerged with vigorous growth. The cell line is estrogen receptor-negative and expresses keratin 18 but not smooth muscle actin, indicating that the cell line was derived from a luminal epithelial cell. The cells also formed colonies when cultured in soft agar. In addition, we found that the cells generated invasive tumors in nude mice.
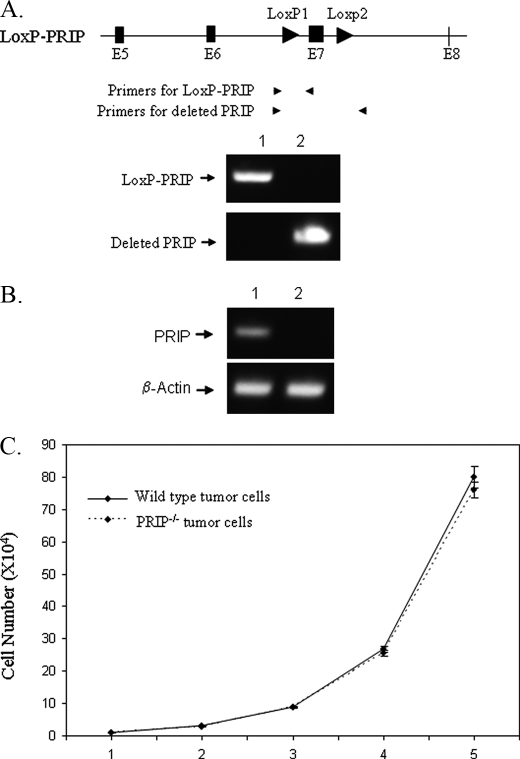
To mutate the PRIP gene, the Ras-induced tumor cells were infected with a retrovirus expressing Cre-ERtam or a control retrovirus. After selection with puromycin, the cells were treated with tamoxifen for 2 days to activate the Cre recombinase and induce the knock-out of the PRIP gene. The deletion of the PRIP gene in mammary epithelial cells was demonstrated by PCR using primers specific for the deleted and LoxP-PRIP gene (Fig. 1A). RT-PCR showed that PRIP mRNA from tumor cells with deleted PRIP gene was absent (Fig. 1B). The successful deletion indicated that PRIP is not essential for the growth of Ras-induced mammary tumor cells. To find if PRIP deficiency affects the cell proliferation, PRIP-/- and PRIP+/+ tumor cells were inoculated at equal densities, and their numbers were counted every day for 5 days. The PRIP-/- and wild type tumor cells showed a similar rate of proliferation (Fig. 1C).
FIGURE 1.
Loss of PRIP does not affect the proliferation of Ras-induced mammary tumor cells. A, generation of PRIP knock-out cells. Ras-induced mammary tumor cells with PRIPLoxP/LoxP genotype were infected with retroviruses expressing Cre-ERtam or control viruses. Genomic DNA was prepared after the cells were treated with tamoxifen for 2 days. PCR was performed with primers specific for the LoxP-PRIP transgene or the deleted PRIP gene. Lane 1, tumor cells infected with control retroviruses; lane 2, tumor cells infected with Cre-ERtam expressing retroviruses. B, loss of PRIP expression in PRIP knock-out cells. Total RNA was prepared from tumor cells treated with tamoxifen. RT-PCR was performed to examine PRIP expression. Lane 1, cells infected with a control. Lane 2, cells infected with retroviral-Cre-ERtam. C, viable cells were counted by trypan blue staining at different times after initial seeding of 2 × 104 cells. PRIPLoxP/LoxP cells infected with viruses expressing Cre-ERtam (PRIP-/-) and cells infected with control viruses (wild type) showed similar growth rates.
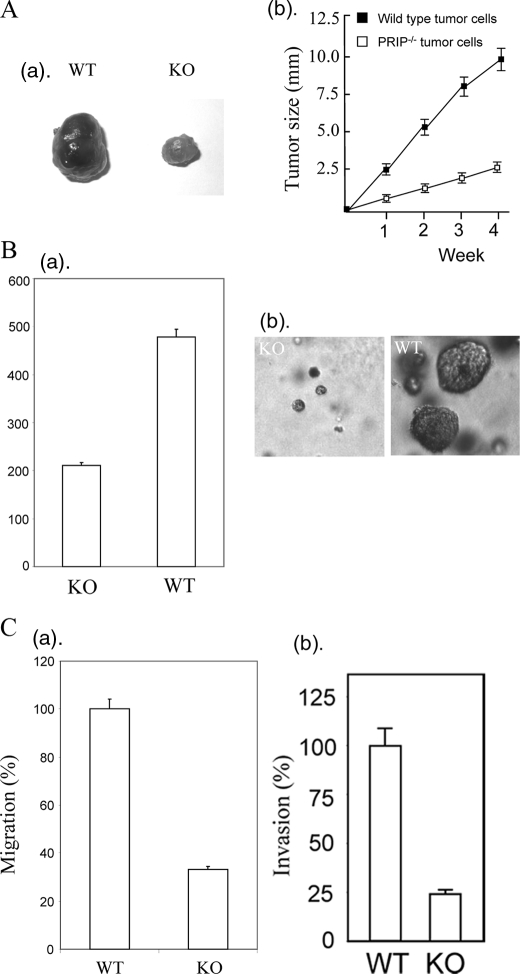
Loss of PRIP Inhibits Tumor Formation—We subcutaneously injected PRIP-/- or PRIPLoxP/LoxP Ras-induced tumor cells into nude mice for tumor formation. Surprisingly, the tumor formed by PRIP-deficient tumor cells grew far more slowly than wild type tumor cells (Fig. 2A). To determine the potential influence of PRIP on the anchorage-independent growth, we inoculated PRIP-deficient tumor cells and wild type tumor cells into soft agar. The PRIP-deficient tumor cells showed both reduced number and size of colonies compared with wild type tumor cells (Fig. 2B). Since PRIP expression had no significant effect on the growth rate of tumor cells on plastic, we tested if loss of PRIP affects the cell migration and invasion capability. PRIP-deficient cells and wild type cells were plated onto Matrigel-coated transwell chambers to measure invasion or onto collagen-coated transwell chambers to measure migration. The loss of PRIP significantly decreased migration and invasion by mammary tumor cells (Fig. 2C).
FIGURE 2.
Loss of PRIP inhibits tumor formation. A, PRIP-/- tumor cells or wild type tumor cells (1 × 106 cells) were injected into nude mice. The experiment was performed in triplicate, and the average tumor sizes were calculated. The tumors from PRIP-/- tumor cells were much smaller than that from wild type PRIPLoxp/Loxp tumor cells. a, representative tumors formed by PRIP-/- tumor cells (KO) and wild type tumor cells (WT). b, the curve of tumor growth in nude mice. B, PRIP deficiency reduced anchorage-independent growth of Ras-induced tumor cells. PRIP-/- tumor cells or wild type tumor cells (5 × 103 cells) were inoculated into soft agar for a week. a, number of colonies formed by PRIP-deficient cells and wild type tumor cells. b, representative photographs of soft agar colonies. C, loss of PRIP decreased the migration and invasion capabilities. For cell migration assay, PRIP-deficient cells and wild type cells were added to Transwell migration chambers coated with collagen I. After 14 h, the cells on the bottom side of the filter were fixed and counted. Data are expressed relative to wild type (a). Cell invasion assay used Matrigel-coated chambers. Data are presented relative to wild type tumor cells (b).
Altered Gene Expression by Knock-out of PRIP—In an effort to identify potential PRIP target genes that might reveal the mechanism by which PRIP regulates tumor formation, a microarray analysis was performed with wild type and PRIP-/- Ras-induced tumor cells. We identified a total of 37 genes down-regulated with the loss of PRIP (Table 1) and a total of 35 genes up-regulated with the loss of PRIP (Table 2). The expression of Csh1 (chorionic somatomammotropin hormone 1 (placental lactogen)) (25) was increased more than 40-fold. Also increased was the expression of Xist, which is involved in X chromosome inactivation (26, 27). Among the down-regulated genes are six histone genes and trp53. Notably, the FOS and FOSB were markedly decreased with the loss of PRIP. Both FOS and FOSB genes are immediate responsive genes (20). The promoters of FOS and FOSB genes share similar regulatory elements. Although FOS and FOSB mRNA showed similar -fold decrease with PRIP deficiency, the level of FOS mRNA was about 20 times as that of FOSB. The microarray data showed that the FOS gene was one of most highly expressed transcription factors in this cell line.
TABLE 1.
Genes with significantly decreased expression in PRIP-deficient mammary tumor cells
| GenBank™ No. | Gene name | PRIP–/–/PRIP+/+ |
|---|---|---|
| -fold | ||
| NM_010917 | Nidogen 1 (nid1) | 0.0065 |
| BC028550 | H4 histone family, member A | 0.077 |
| AF177041 | Aldo-keto reductase a (akra) | 0.13 |
| NM_013778 | Aldo-keto reductase family 1, member C13 | 0.13 |
| BB119196 | Carnitine almitoyltransferase 1 | 0.15 |
| NM_011728 | xpa | 0.17 |
| BG967663 | Creatine kinase, brain | 0.17 |
| M25487 | H2B histone family, member S | 0.18 |
| BI151886 | Sialic acid synthase (sas) | 0.20 |
| NM_015786 | H1 histone family, member 2 | 0.20 |
| NM_010373 | Granzyme E | 0.22 |
| NM_008598 | mgmt | 0.23 |
| AV026617 | FBJ osteosarcoma oncogene | 0.25 |
| NM_009213 | smpd2 | 0.26 |
| BC011440 | H2B histone family, member A | 0.26 |
| AV297651 | Histone cluster 3, H2a | 0.27 |
| NM_016861 | PDZ and LIM domain 1 (elfin) | 0.27 |
| NM_007494 | Arginosuccinate synthetase 1 | 0.30 |
| NM_021883 | Tropomodulin 1 | 0.30 |
| BB828014 | trp53 | 0.31 |
| NM_007705 | Cold-inducible RNA-binding protein | 0.32 |
| NM_007646 | cd38 | 0.32 |
| AW495711 | Stress-induced protein | 0.33 |
| NM_008857 | Protein kinase Cλ | 0.33 |
| BC010564 | H2A histone family, member O | 0.33 |
| NM_008885 | Peripheral myelin protein, 22 kDa | 0.34 |
| AW539044 | pard6g | 0.35 |
| AA275072 | Integral membrane protein 2A | 0.36 |
| NM_011568 | RNA and export factor binding protein 1 | 0.36 |
| NM_018871 | ywhag | 0.37 |
| NM_008036 | FBJ osteosarcoma oncogene B | 0.37 |
| AA560093 | ramp2 | 0.38 |
| BI739353 | Phosphomannomutase 1 | 0.38 |
| BG965405 | btg2 | 0.38 |
| NM_009481 | usp9x | 0.39 |
| BF782863 | F-box and leucine-rich repeat protein 3a | 0.39 |
| NM_019671 | Neuroepithelial cell transforming gene 1 | 0.40 |
TABLE 2.
Genes with significantly increased expression in PRIP-deficient mammary tumor cells
| GenBank™ No. | Gene name | (PRIP–/–/PRIP+/+) |
|---|---|---|
| -fold | ||
| NM_008864 | csh1 | 41.8 |
| NM_007984 | Fascin homolog 1 | 37.9 |
| L04961 | Inactive X-specific transcripts | 12.6 |
| NM_007541 | γ-Carboxyglutamate protein 1 | 10.3 |
| NM_016905 | Galactokinase | 9.1 |
| NM_008908 | Peptidylprolyl isomerase C | 8.6 |
| U12889 | ly49H | 5.6 |
| NM_013473 | Annexin A8 | 5.1 |
| NM_008365 | Interleukin 18 receptor 1 | 5.0 |
| NM_007868 | Dystrophin | 4.7 |
| NM_023785 | Pro-platelet basic protein | 4.6 |
| NM_009647 | Adenylate kinase 4 | 4.6 |
| NM_138758 | tmlh | 4.0 |
| NM_019703 | Phosphofructokinase, platelet | 3.8 |
| AW049660 | Nuclear factor IX | 3.7 |
| BB233670 | Regulator of G-protein signaling 19 | 3.7 |
| NM_008216 | Hyaluronan synthase 2 | 3.6 |
| NM_009416 | Tropomyosin 2β | 3.5 |
| BB230853 | Serpin b5 | 3.4 |
| BB043407 | gja1 | 3.3 |
| NM_020332 | Progressive ankylosis (ank) | 3.3 |
| AB029929 | Caveolin-1 α-isoform | 3.3 |
| AI462635 | Aldehyde dehydrogenase 2 | 3.2 |
| M36277 | N-myc | 3.2 |
| AB021226 | Type-5 matrix metalloproteinase | 3.0 |
| NM_009801 | Carbonic anhydrase 2 | 3.0 |
| BC019379 | G protein-coupled receptor kinase 5 | 2.9 |
| AV091354 | Interleukin-1 receptor-associated kinase | 2.9 |
| BF683028 | Glycerol kinase | 2.8 |
| AV075715 | Clusterin | 2.8 |
| BC002008 | Fatty acid-binding protein 5 | 2.7 |
| BM250666 | Procollagen, type IV, α5 | 2.7 |
| NM_008827 | Placental growth factor | 2.6 |
| NM_009541 | Zinc finger protein 100 | 2.6 |
| D13695 | ST2L protein | 2.5 |
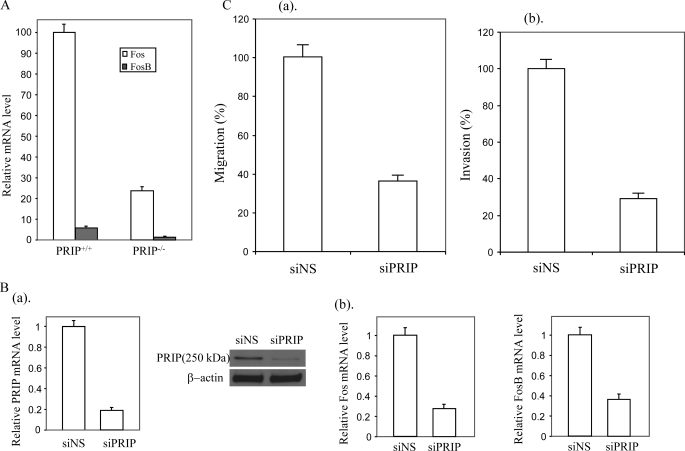
PRIP Is Required for Expression of the FOS Gene—The decreased expression of FOS and FOSB due to loss of PRIP was validated by real time RT-PCR (Fig. 3A). BT474 is breast cancer cell line with overexpression of PRIP due to the amplification of the PRIP gene. To find if PRIP is required for FOS and FOSB expression in human breast cancers, BT474 cells were transfected with siRNAs targeting PRIP, which reduced PRIP transcript at least 75% compared with controls (Fig. 3B). Real time PCR showed that FOS and FOSB expression was decreased with the suppression of PRIP expression (Fig. 3B). Similar to Ras-induced tumor cells, suppression of PRIP expression also inhibited the invasion (Fig. 3C) and migration (Fig. 3D) capability of BT474 cells.
FIGURE 3.
Loss of PRIP led to deceased expression of FOS and FOSB. A, quantitative real time RT-PCR was performed to determine the FOS and FOSB mRNA levels, which were normalized to control β-actin mRNA levels. WT, wild type tumor cells; KO, PRIP-/- tumor cells. B, suppression of PRIP inhibited FOS and FOSB expression in human breast cancer cell line BT474. BT474 cells were transfected with siRNAs targeting PRIP (siPRIP) or control siRNA (siNS). The suppression of PRIP was demonstrated by real time PCR (a, left) and confirmed by Western blot (a, right). Real time RT-PCR was performed to determine the human FOS and FOSB mRNA levels (b). C, suppression of PRIP decreased the migration and invasion capabilities of BT474 cells. BT474 cells transfected with siRNA targeting PRIP or control siRNA were inoculated into Transwell migration chambers coated with collagen (a) or Matrigel (b). The cells on the bottom side of the filter were counted 14 h later.
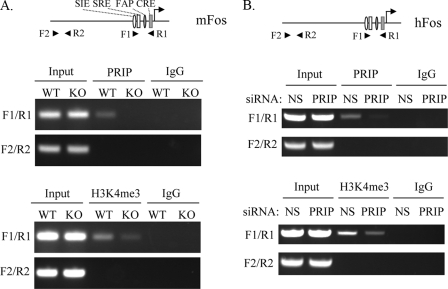
Direct Recruitment of PRIP to the FOS Promoter—ChIP assays were performed to find if PRIP is directly recruited to the FOS gene promoter in vivo. Formaldehyde-cross-linked chromatin-protein complexes from wild type tumor cells or PRIP-/- tumor cells were immunoprecipitated using antibodies against PRIP and analyzed by semiquantitative PCR. We used two different PCR primer sets, which span two different regions of the FOS promoter. Primer set F1/R1 (-348 to -50 bp) spans the region containing the SRE site, c-Sis-inducible element site, FOS activator protein 1 site, and Cre site, whereas primer set F2/R2 (-2840 to -2580 bp) is located upstream of promoter containing no identified regulatory elements. We observed occupancy of PRIP at the FOS promoter containing regulatory elements and its absence upstream of promoter in wild type tumor cells (Fig. 4A). As a negative control, no amplification of the FOS promoter was observed in PRIP-/- tumor cells. The PRIP is a component of a complex carrying histone methyltransferase activity. We performed chromatin immunoprecipitation to find if the recruitment of PRIP alters the methylated status of histone in the FOS promoter. We observed a marked increase of H3K4 trimethylation in the promoter of FOS in wild type tumor cells compared with PRIP-/- tumor cells (Fig. 4A).
FIGURE 4.
PRIP is recruited to the FOS promoter and enhances H3K4-trimethylation at the FOS promoter. A, PRIP is recruited to the FOS promoter and increases its H3K4-trimethylation in Ras-induced tumor cells. Primers located in the promoter (F1/R1) or upstream of promoter (F2/R2) were indicated with the mouse FOS promoter and its regulatory elements (top). ChIP was performed with wild type tumor cells (WT) and PRIP-/- tumor cells (KO), anti-PRIP (middle), and anti-H3K4me3 (bottom). B, association of PRIP with the FOS promoter and increased H3K4 trimethylation due to the recruitment of PRIP in breast cancer BT474 cells. One pair of primers for the human FOS promoter region (F1/R1) and another pair of primers upstream of promoter (F2/R2) were used in ChIP assays (top). ChIP was performed with BT474 cells transfected with nonspecific siRNA (siNS) or siRNA against PRIP (siPRIP), anti-PRIP (middle), and anti-H3K4me3 (bottom).
We then examined if PRIP is recruited to the FOS gene promoter in human breast cancer cell line BT474. ChIP assays were performed with anti-PRIP. We observed amplification with the primer set covering the regulatory elements of the FOS gene and no amplification with the primer set located upstream of the FOS promoter (Fig. 4B), indicating that PRIP is also recruited to the FOS gene in human breast cancers. Similarly, we found that H3K4 trimethylation in the human FOS promoter was decreased in BT474 cells with suppressed expression of PRIP (Fig. 4B).
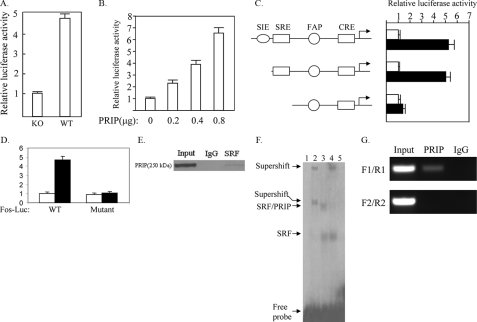
PRIP Enhances FOS Promoter Activity through Serum Response Factor—To determine the mechanism by which PRIP regulates FOS expression, we analyzed whether PRIP regulates the promoter activity of the FOS gene. We transfected PRIP-/- or wild type cells with FOS promoter-luciferase construct. As shown in Fig. 5A, the promoter activity was dramatically decreased in PRIP-/- cells compared with wild type cells. The re-expression of PRIP increased the FOS promoter activity in a dose-dependent manner, suggesting that PRIP enhances FOS promoter activity (Fig. 5B).
FIGURE 5.
PRIP regulates the FOS promoter activity through SRF. A, decreased activity of the FOS promoter in PRIP-/- tumor cells. PRIP-/- tumor cells (KO) or wild type tumor cells (WT) were transfected with the FOS-Luc reporter and the β-galactosidase expression vector. Luciferase activity was normalized to β-galactosidase activity. B, expression of PRIP increased the activity of the FOS promoter. PRIP-/- cells were transfected with the FOS-Luc reporter, different amounts of PRIP expression vector as indicated, and the β-galactosidase expression vector as the control. C, PRIP activated the FOS promoter through SRE. PRIP-/- cells were transfected with different FOS-Luc reporters, as indicated, control empty vector (white bars), or PRIP expression vector (black bars) and β-galactosidase expression vector. D, confirmation that SRE is required for activation of the FOS promoter by PRIP. Wild type FOS-Luc or FOS-Luc with a mutation in its SRE (Mutant) were transfected into PRIP-/- cells with control empty vector (white bars) or PRIP expression vector (black bars). E, the direct interaction between PRIP and SRF in vivo. The nuclear extract from wild type tumor was prepared and precipitated with anti-SRF or control IgG. The precipitate was subject to Western blot analysis using anti-PRIP. F, PRIP and SRF form a complex on SRE. Gel shift was performed with 32P-labeled SRE, nuclear extracts from Ras-induced tumor cells, and antibodies against SRF or PRIP. Lane 1, no nuclear extract; lane 2, nuclear extract plus anti-SRF; lane 3, nuclear extract; lane 4, nuclear extract plus anti-PRIP; lane 5, nuclear extract plus a 50-fold molar excess of unlabeled competitor SRE oligonucleotide. G, sequential ChIP assay demonstrating PRIP and SRF protein complexes over SRE in the FOS promoter. The first-step ChIP was performed with Ras-induced tumor cells and anti-SRF. The second step ChIP was carried out with the eluates of the initial ChIP, anti-PRIP, and IgG. PCR was performed using primer set F1/R1 and F2/R2, as described above.
The FOS promoter is under the control of several transcription factors, including signal transducers and activators of transcription, AP-1, Cre, and SRF (28). PRIP has been reported to promote the transcriptional activity of Cre, AP-1, and SRF (3, 14). To find which factor PRIP acts on to regulate FOS gene expression, we transfected the reporter gene with the FOS promoter carrying different regulatory elements and PRIP expression vector into PRIP-deficient tumor cells. PRIP enhanced the FOS promoter activity in the absence of the c-Sis-inducible element, which interacts with signal transducers and activators of transcription (Fig. 5C). When the SRE was not included in the reporter gene, PRIP had minimal effect on the FOS promoter, which still contained Cre and the FOS activator protein 1 site, indicating that PRIP acts through transcription factor SRF to promote FOS gene expression. To confirm the role of SRE in mediating activation of the FOS gene by PRIP, the SRE in the Fos-Luc reporter was mutated. As expected, the mutation of SRE abolished the enhancement of FOS promoter activity by PRIP (Fig. 5D). To explore if PRIP directly interacts with SRF, we performed immunoprecipitation with anti-SRF or control serum. The precipitates were subjected to Western blot analysis with anti-PRIP. We found that anti-SRF rather than control serum pulled down PRIP (Fig. 5E), suggesting that SRF interacts with PRIP in intact cells. The interaction of PRIP with SRF on SRE was examined by gel shift assays (Fig. 5F). Two complexes were formed with nuclear extracts from Ras-induced tumor cells. Unlabeled SRE competitor abolished both bands, indicating that both complexes resulted from the specific binding of SRF to SRE. Anti-SRF caused the supershift for both complexes, confirming that both complexes contained SRF. The inclusion of anti-PRIP led to the supershift of the band with slower mobility, indicating that this complex was formed by PRIP, SRF, and SRE. Sequential ChIP was performed to address if PRIP and SRF form a complex on the FOS promoter in mammary tumor cells. The eluated chromatin precipitated by anti-SRF was subjected to a second immunoprecipitation with anti-PRIP or control IgG. There was specific enrichment of DNA corresponding to the FOS promoter compared with IgG controls (Fig. 5G). However, we did not observe any specific enrichment of DNA 2.5 kb upstream of the FOS promoter.
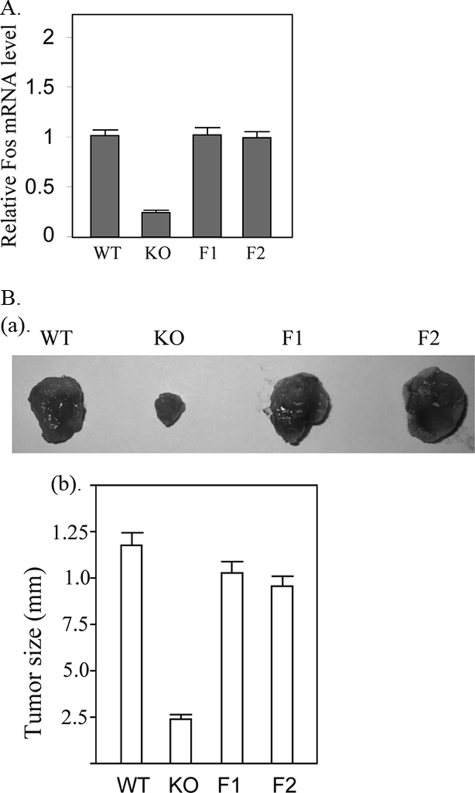
Re-expression of FOS Promotes the Tumor Formation—Given that the FOS gene is highly expressed in Ras-induced tumor cells and is substantially down-regulated with PRIP deficiency, we further tested if reduced FOS expression leads to attenuated tumor formation by PRIP-/- tumor cells. We re-expressed FOS in the PRIP-/- tumor cells (Fig. 6A). Two clones expressing FOS at levels similar to that from wild type tumor cells were inoculated into nude mice for tumor formation. The PRIP-/- cells expressing exogenous FOS showed tumor formation at a growth rate similar to that by wild type tumor cells (Fig. 6B), confirming that reduced FOS expression is responsible for the decreased tumor formation as a result of PRIP deficiency.
FIGURE 6.
Re-expression of Fos enhanced the tumor formation. A, re-expression of Fos in PRIP-/- cells. PRIP-/- cells stably re-expressing FOS were established by transfecting PRIP-/- with FOS expression vector. The FOS mRNA levels were measured by real time RT-PCR. Two clones (F1 and F2) expressed FOS mRNA at a level similar to that of wild type cells (WT). PRIP-/- cells integrated with an empty vector served as a control (KO). B, wild type tumor cells, control PRIP-/- tumor cells, and clones re-expressing FOS (F1 and F2) (1 × 106 cells) were injected into nude mice. The experiment was performed in triplicate, and tumor sizes were measured. The tumors formed by PRIP-/- tumor cells were much smaller than those from wild type tumor cells and PRIP-/- tumor cells re-expressing Fos (F1 and F2). a, representative tumors formed by injected tumor cells. b, average tumor sizes were calculated.
DISCUSSION
To understand the role of PRIP in mammary tumorigenesis, we created a mammary tumor cell line carrying a conditional flox PRIP gene. By deleting the PRIP gene, we found that PRIP is not essential for the proliferation of mammary tumor cells in vitro on plates. However, PRIP is important for in vivo invasive growth, since loss of PRIP decreases the tumor cell migration and invasion capability. Further studies found that PRIP is required for the expression of the FOS gene by interacting with the SRF factor and promoting its activity.
The PRIP gene is frequently amplified and overexpressed in breast cancers. In addition to Ras-induced tumor cells, we found that PRIP is also required for FOS expression in human breast cancer BT474 cells. BT474 cells, which contain no Ras mutation but possess an oncogenic mutation on PIK3CA, carry an amplified PRIP gene with overexpression of PRIP (3). Therefore, PRIP promotes breast tumorigenesis at least partly by enhancing the FOS gene expression in human breast tumors with PRIP overexpression.
SRF also regulates other immediate response genes (20). Among these genes, we found that JUNB, EGR1, CYR61, and NURR1 are expressed in the Ras-induced tumor cells (data not shown). However, the expression of these genes is not affected due to the loss of PRIP expression. It appears that PRIP regulates gene expression through SRF in a gene-specific fashion. The SRF activity is affected by cofactors mainly including members of the ternary complex factor family of Ets domain proteins and the myocardin-related transcription factors (29). These two families of SRF cofactors are regulated by separate signaling pathways and control the SRF targets differentially. One possibility is that one of these collaborating transcription factors is required for PRIP action.
The FOS promoter is regulated by a number of transcription factors, including AP-1, signal transducers and activators of transcription, Cre, and SRF. PRIP has been shown to increase the transcriptional activity of AP-1, Cre, and SRF using minimal enhancer-containing reporters in a transient transfection assay. However, we found that PRIP acted through SRF to regulate FOS gene expression while having no effect on a truncated FOS promoter containing binding sites for AP-1 and Cre. The functions of PRIP on AP-1 and Cre, just like SRF, seem to be affected by other regulatory elements of target genes.
Notably, microarray studies revealed that the expression of CSH1, which is normally expressed in placenta, was highly increased in PRIP-deficient tumor cells. PRIP is a component of the complex ASCOM, which includes MLL3 or MLL4. MLL3 and MLL4 are involved in epigenetic regulation of genes. It is likely that loss of PRIP disrupts the epigenetic regulation of certain genes, which leads to aberrant gene expression. Microarray analysis also showed that the expression of six histone genes was decreased with the loss of PRIP. It is consistent with the notion that histone genes are regulated coordinately (30). Despite reduced expression of six histone genes, the proliferation of the tumor cells is not affected. PRIP could serve as a coactivator for certain transcriptional factors regulating histone expression.
The FOS gene plays an important role in tumorigenesis. Depending on the cell types, FOS may promote cell proliferation, migration and invasion, and angiogenesis. In breast cancer, we found that the expression of Fos is critical for the invasive growth of Ras-induced tumor cells, and PRIP is critical for FOS gene expression. Given that increased FOS gene expression is commonly present in human cancers, including breast cancers, inhibiting PRIP activity could be a potential way to reduce FOS activity for the breast cancer treatment.
This work was supported, in whole or in part, by National Institutes of Health Grant CA 88898 (to Y. J. Z.).
Footnotes
The abbreviations used are: SRF, serum-responsive factor; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; siRNA, small interfering RNA; RT, reverse transcription; SRE, serum-responsive element; ChIP, chromatin immunoprecipitation.
References
- 1.Zhu, Y., Kan, L., Qi, C., Kanwar, Y. S., Yeldandi, A. V., Rao, M. S., and Reddy, J. K. (2000) J. Biol. Chem. 275 13510-13516 [DOI] [PubMed] [Google Scholar]
- 2.Mahajan, M. A., and Samuels, H. H. (2000) Mol. Cell. Biol. 20 5048-5063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ko, L., Cardona, G. R., and Chin, W. W. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6212-6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caira, F., Antonson, P., Pelto-Huikko, M., Treuter, E., and Gustafsson, J. A. (2000) J. Biol. Chem. 275 5308-5317 [DOI] [PubMed] [Google Scholar]
- 5.Lee, S. K., Anzick, S. L., Choi, J. E., Bubendorf, L., Guan, X. Y., Jung, Y. K., Kallioniemi, O. P., Kononen, J., Trent, J. M., Azorsa, D., Jhun, B. H., Cheong, J. H., Lee, Y. C., Meltzer, P. S., and Lee, J. W. (1999) J. Biol. Chem. 274 34283-34293 [DOI] [PubMed] [Google Scholar]
- 6.Goo, Y. H., Sohn, Y. C., Kim, D. H., Kim, S. W., Kang, M. J., Jung, D. J., Kwak, E., Barlev, N. A., Berger, S. L., Chow, V. T., Roeder, R. G., Azorsa, D. O., Meltzer, P. S., Suh, P. G., Song, E. J., Lee, K. J., Lee, Y. C., and Lee, J. W. (2003) Mol. Cell. Biol. 23 140-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Issaeva, I., Zonis, Y., Rozovskaia, T., Orlovsky, K., Croce, C. M., Nakamura, T., Mazo, A., Eisenbach, L., and Canaani, E. (2007) Mol. Cell. Biol. 27 1889-1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, Y. W., Hong, T., Hong, S., Guo, H., Yu, H., Kim, D., Guszczynski, T., Dressler, G. R., Copeland, T. D., Kalkum, M., and Ge, K. (2007) J. Biol. Chem. 282 20395-20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agger, K., Cloos, P. A., Christensen, J., Pasini, D., Rose, S., Rappsilber, J., Issaeva, I., Canaani, E., Salcini, A. E., and Helin, K. (2007) Nature 449 731-734 [DOI] [PubMed] [Google Scholar]
- 10.Hong, S., Cho, Y. W., Yu, L. R., Yu, H., Veenstra, T. D., and Ge, K. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 18439-18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu, Y., Qi, C., Cao, W. Q., Yeldandi, A. V., Rao, M. S., and Reddy, J. K. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10380-10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung, D. J., Na, S. Y., Na, D. S., and Lee, J. W. (2002) J. Biol. Chem. 277 1229-1234 [DOI] [PubMed] [Google Scholar]
- 13.Iwasaki, T., Chin, W. W., and Ko, L. (2001) J. Biol. Chem. 276 33375-33383 [DOI] [PubMed] [Google Scholar]
- 14.Lee, S. K., Na, S. Y., Jung, S. Y., Choi, J. E., Jhun, B. H., Cheong, J., Meltzer, P. S., Lee, Y. C., and Lee, J. W. (2000) Mol. Endocrinol. 14 915-925 [DOI] [PubMed] [Google Scholar]
- 15.Zhu, Y. J., Crawford, S. E., Stellmach, V., Dwivedi, R. S., Rao, M. S., Gonzalez, F. J., Qi, C., and Reddy, J. K. (2003) J. Biol. Chem. 278 1986-1990 [DOI] [PubMed] [Google Scholar]
- 16.Kuang, S. Q., Liao, L., Zhang, H., Pereira, F. A., Yuan, Y., DeMayo, F. J., Ko, L., and Xu, J. (2002) J. Biol. Chem. 277 45356-45360 [DOI] [PubMed] [Google Scholar]
- 17.Antonson, P., Schuster, G. U., Wang, L., Rozell, B., Holter, E., Flodby, P., Treuter, E., Holmgren, L., and Gustafsson, J. A. (2003) Mol. Cell. Biol. 23 1260-1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi, C., Kashireddy, P., Zhu, Y. T., Rao, S. M., and Zhu, Y. J. (2004) J. Biol. Chem. 279 33696-33701 [DOI] [PubMed] [Google Scholar]
- 19.Karin, M., Liu, Z., and Zandi, E. (1997) Curr. Opin. Cell Biol. 9 240-246 [DOI] [PubMed] [Google Scholar]
- 20.Eferl, R., and Wagner, E. F. (2003) Nat. Rev. Cancer 3 859-868 [DOI] [PubMed] [Google Scholar]
- 21.Chai, J., and Tarnawski, A. S. (2002) J. Physiol. Pharmacol. 53 147-157 [PubMed] [Google Scholar]
- 22.Miano, J. M., Long, X., and Fujiwara, K. (2007) Am. J. Physiol. Cell Physiol. 292 70-81 [DOI] [PubMed] [Google Scholar]
- 23.Yang, J., Richards, J., Guzman, R., Imagawa, W., and Nandi, S. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 2088-2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shang, Y., Hu, X., DiRenzo, J., Lazar, M. A., and Brown, M. (2000) Cell 103 843-852 [DOI] [PubMed] [Google Scholar]
- 25.Roskam, W. G., and Rougeon, F. (1979) Nucleic Acids Res. 7 305-320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chow, J. C., Yen, Z., Ziesche, S. M., and Brown, C. J. (2005) Annu. Rev. Genomics Hum. Genet. 6 69-92 [DOI] [PubMed] [Google Scholar]
- 27.Heard, E., and Disteche, C. M. (2006) Genes Dev. 20 1848-1867 [DOI] [PubMed] [Google Scholar]
- 28.Sng, J. C., Taniura, H., and Yoneda, Y. (2004) Biol. Pharm. Bull. 27 606-612 [DOI] [PubMed] [Google Scholar]
- 29.Posern, G., and Treisman, R. (2006) Trends Cell Biol. 16 588-596 [DOI] [PubMed] [Google Scholar]
- 30.Marzluff, W. F. (2005) Curr. Opin. Cell Biol. 17 274-280 [DOI] [PubMed] [Google Scholar]