Abstract
Integrin α8β1 interacts with a variety of Arg-Gly-Asp (RGD)-containing ligands in the extracellular matrix. Here, we examined the binding activities of α8β1 integrin toward a panel of RGD-containing ligands. Integrin α8β1 bound specifically to nephronectin with an apparent dissociation constant of 0.28 ± 0.01 nm, but showed only marginal affinities for fibronectin and other RGD-containing ligands. The high-affinity binding to α8β1 integrin was fully reproduced with a recombinant nephronectin fragment derived from the RGD-containing central “linker” segment. A series of deletion mutants of the recombinant fragment identified the LFEIFEIER sequence on the C-terminal side of the RGD motif as an auxiliary site required for high-affinity binding to α8β1 integrin. Alanine scanning mutagenesis within the LFEIFEIER sequence defined the EIE sequence as a critical motif ensuring the high-affinity integrin-ligand interaction. Although a synthetic LFEIFEIER peptide failed to inhibit the binding of α8β1 integrin to nephronectin, a longer peptide containing both the RGD motif and the LFEIFEIER sequence was strongly inhibitory, and was ∼2,000-fold more potent than a peptide containing only the RGD motif. Furthermore, trans-complementation assays using recombinant fragments containing either the RGD motif or LFEIFEIER sequence revealed a clear synergism in the binding to α8β1 integrin. Taken together, these results indicate that the specific high-affinity binding of nephronectin to α8β1 integrin is achieved by bipartite interaction of the integrin with the RGD motif and LFEIFEIER sequence, with the latter serving as a synergy site that greatly potentiates the RGD-driven integrin-ligand interaction but has only marginal activity to secure the interaction by itself.
Integrins are a family of adhesion receptors that interact with a variety of extracellular ligands, typically cell-adhesive proteins in the extracellular matrix (ECM).2 They play mandatory roles in embryonic development and the maintenance of tissue architectures by providing essential links between cells and the ECM (1). Integrins are composed of two non-covalently associated subunits, termed α and β. In mammals, 18 α and 8 β subunits have been identified, and combinations of these subunits give rise to at least 24 distinct integrin heterodimers. Based on their ligand-binding specificities, ECM-binding integrins are classified into three groups, namely laminin-, collagen- and RGD-binding integrins (2, 3), of which the RGD-binding integrins have been most extensively investigated. The RGD-binding integrins include α5β1, α8β1, αIIbβ3, and αV-containing integrins, and have been shown to interact with a variety of ECM ligands, such as fibronectin and vitronectin, with distinct binding specificities.
The α8 integrin subunit was originally identified in chick nerves (4). Integrin α8β1 is expressed in the metanephric mesenchyme and plays a crucial role in epithelial-mesenchymal interactions during the early stages of kidney morphogenesis. Disruption of the α8 gene in mice was found to be associated with severe defects in kidney morphogenesis (5) and stereocilia development (6). To date, α8β1 integrin has been shown to bind to fibronectin, vitronectin, osteopontin, latency-associated peptide of transforming growth factor-β1, tenascin-W, and nephronectin (also named POEM) (7–13), among which nephronectin is believed to be an α8β1 integrin ligand involved in kidney development (10).
Nephronectin is one of the basement membrane proteins whose expression and localization patterns are restricted in a tissue-specific and developmentally regulated manner (10, 11). Nephronectin consists of five epidermal growth factor-like repeats, a linker segment containing the RGD cell-adhesive motif (designated RGD-linker) and a meprin-A5 protein-receptor protein-tyrosine phosphatase μ (MAM) domain (see Fig. 3A). Although the physiological functions of nephronectin remain only poorly understood, it is thought to play a role in epithelial-mesenchymal interactions through binding to α8β1 integrin, thereby transmitting signals from the epithelium to the mesenchyme across the basement membrane (10). Recently, mice deficient in nephronectin expression were produced by homologous recombination (14). These nephronectin-deficient mice frequently displayed kidney agenesis, a phenotype reminiscent of α8 integrin knock-out mice (14), despite the fact that other RGD-containing ligands, including fibronectin and osteopontin, were expressed in the embryonic kidneys (9, 15). The failure of the other RGD-containing ligands to compensate for the deficiency of nephronectin in the developing kidneys suggests that nephronectin is an indispensable α8β1 ligand that plays a mandatory role in epithelial-mesenchymal interactions during kidney development.
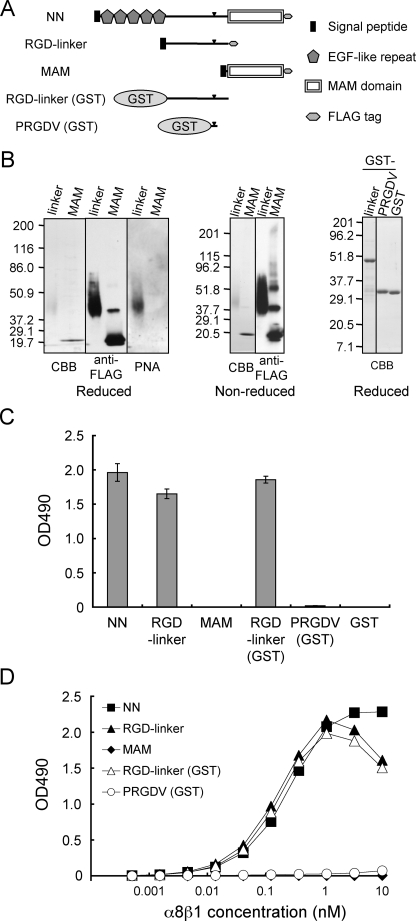
FIGURE 3.
Binding activities of α8β1 integrin to nephronectin and its fragments. A, schematic diagrams of full-length nephronectin (NN) and its fragments. RGD-linker and RGD-linker (GST), the central RGD-containing linker segments expressed in mammalian and bacterial expression systems, respectively; PRGDV, a short RGD-containing peptide modeled after nephronectin and expressed as a GST fusion protein (see Fig. 4A for the peptide sequence). The arrowheads indicate the positions of the RGD motif. B, purified recombinant proteins were analyzed by SDS-PAGE in 7–15% gradient (left and center panels) and 12% (right panels) gels, followed by Coomassie Brilliant Blue (CBB) staining, immunoblotting with an anti-FLAG mAb, or lectin blotting with PNA. The quantities of proteins loaded were: 0.5 μg (for Coomassie Brilliant Blue staining) and 0.1 μg (for blotting with anti-FLAG and PNA) in the left and center panels;1 μg in the right panel. C, recombinant proteins (10 nm) were coated on microtiter plates and assessed for their binding activities toward α8β1 integrin (10 nm) in the presence of 1 mm Mn2+. The backgrounds were subtracted as described in the legend to Fig. 2. The results represent the mean ± S.D. of triplicate determinations. D, titration curves of α8β1 integrin bound to full-length nephronectin (NN, closed squares), the RGD-linker segments expressed in 293F cells (RGD-linker, closed triangles) and E. coli (RGD-linker (GST), open triangles), the MAM domain (MAM, closed diamonds), and the PRGDV peptide expressed as a GST fusion protein in E. coli (PRGDV (GST), open circles). The assays were performed as described in the legend to Fig. 2B. The results represent the means of duplicate determinations.
Although ligand recognition by RGD-binding integrins is primarily determined by the RGD motif in the ligands, it is the residues outside the RGD motif that define the binding specificities and affinities toward individual integrins (16, 17). For example, α5β1 integrin specifically binds to fibronectin among the many RGD-containing ligands, and requires not only the RGD motif in the 10th type III repeat but also the so-called “synergy site” within the preceding 9th type III repeat for fibronectin recognition (18). Recently, DiCara et al. (19) demonstrated that the high-affinity binding of αVβ6 integrin to its natural ligands, e.g. foot-and-mouth disease virus, requires the RGD motif immediately followed by a Leu-Xaa-Xaa-Leu/Ile sequence, which forms a helix to align the two conserved hydrophobic residues along the length of the helix. Given the presence of many naturally occurring RGD-containing ligands, it is conceivable that the specificities of the RGD-binding integrins are dictated by the sequences flanking the RGD motif or those in neighboring domains that come into close proximity with the RGD motif in the intact ligand proteins. However, the preferences of α8β1 integrin for RGD-containing ligands and how it secures its high-affinity binding toward its preferred ligands remain unknown.
In the present study, we investigated the binding specificities of α8β1 integrin toward a panel of RGD-containing cell-adhesive proteins. Our data reveal that nephronectin is a preferred ligand for α8β1 integrin, and that a LFEIFEIER sequence on the C-terminal side of its RGD motif serves as a synergy site to ensure the specific high-affinity binding of nephronectin to α8β1 integrin.
EXPERIMENTAL PROCEDURES
Cells and Reagents—HT1080 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. HT1080 cells were transfected with an expression vector for human full-length α8 integrin (described below). The cells were passaged at 24 h after transfection and maintained in medium containing 0.8 mg/ml G418 to select stable transfectants. The transfectants were subjected to selection by cell adhesion to a nephronectin-coated substratum as follows. The cells were seeded onto 6-well plates (Falcon) that had been coated with 10 nm nephronectin and blocked with phosphate-buffered saline (PBS) containing 10 mg/ml bovine serum albumin (BSA), and allowed to adhere for 30 min at 37 °C. After removal of non-adherent cells using serum-free medium, the remaining cells were detached with PBS containing 0.025% trypsin and 1 mm EDTA, plated on 6-well plates and grown to confluence. The resulting cells (designated HT1080-A8 cells) were cloned by limiting dilution and used for cell adhesion assays. K562 human erythroleukemic cells stably transfected with a chicken α8 integrin cDNA (KA8 cells) were kindly provided by Dr. Louis F. Reichardt (University of California, San Francisco, CA) and maintained as described previously (7).
Human plasma fibronectin and vitronectin were purified from outdated human plasma by gelatin and heparin affinity chromatography, respectively, as described previously (20, 21). Human fibrinogen was purchased from Enzyme Research Laboratories Inc. (South Bend, IN). A polyclonal antibody against actin and a horseradish peroxidase-conjugated monoclonal antibody (mAb) against FLAG tag were obtained from Sigma. An horseradish peroxidase-conjugated polyclonal antibody against glutathione S-transferase (GST) was purchased from GE Healthcare. Horseradish peroxidase-conjugated peanut agglutinin (PNA) was purchased from Seikagaku Co. (Tokyo, Japan). mAbs against the human integrin α8 subunit were produced by fusion of Sp2 mouse myeloma cells with spleen cells from mice immunized with recombinant soluble integrin α8β1, and selected by both positive reactivity for recombinant integrin α8β1 and negative reactivity for recombinant integrin α5β1. mAb 10A8 was capable of binding to the denatured integrin α8 chain on immunoblots under reducing and non-reducing conditions. mAb 7A5 was capable of binding to the nondenatured integrin α8 chain, and was used for flow cytometry. Synthetic peptides were purchased from Biologica Co. (Nagoya, Japan) and dissolved into 100% dimethyl sulfoxide.
Expression Vectors—A mouse nephronectin cDNA was obtained from Dr. Yoshihide Hayashizaki (Riken, Yokohama, Japan). The cDNA was subcloned into the pFLAG-CMV vector (Invitrogen) in-frame with a FLAG tag at the 3′ end. For expression of a panel of nephronectin deletion mutants in mammalian cells, a cDNA fragment encoding a mouse nephronectin 5′-non-translated region and the signal peptide (nucleotides -61 to 69) was amplified by PCR with the primer set 5′-GAATTCGAGATCCCGGGACGC-3′ (forward) and 5′-GGGCCCGTCGAAGTCGGCAGC-3′ (reverse) using the full-length nephronectin cDNA as a template. This fragment was digested with EcoRI and ApaI, and fused in-frame with cDNAs encoding the RGD-linker segment or the MAM domain. The cDNAs encoding the RGD-linker and the MAM domain were amplified by PCR with primer sets 5′-GGGCCCAAAGTCATGATTGAAC-3′ (forward) and 5′-GTCGACGTCGTCCTTTACTTCCTC-3′ (reverse), and 5′-GGGCCCGGTATTCTCATACACAGC-3′ (forward) and 5′-GTCGACGCAGCGACCTCTTTTCAAG-3′ (reverse), respectively. After verification by DNA sequencing, each PCR-amplified cDNA was digested with ApaI and SalI, and inserted into the EcoRI and SalI sites of the pFLAG-CMV vector together with the cDNA encoding the 5′-non-translated region and signal peptide.
An expression vector for the RGD-linker as a GST fusion protein was prepared as follows. A cDNA encoding the RGD-linker was amplified by PCR with the primer set 5′-GAATTCCCCAAAGTCATGATTGAACCT-3′ (forward) and 5′-GTCGACTCAGTCGTCCTTTACTTCCTC-3′ (reverse) using an E13.5 mouse embryo cDNA as a template. The PCR product was subcloned into the pBluescriptII SK+ vector (Stratagene, La Jolla, CA), digested with EcoRI and SalI, and inserted into the corresponding restriction sites of the pGEX 4T-1 expression vector (GE Healthcare) (designated pGEX-RGD-linker). cDNAs encoding a series of deletion mutants and substitution mutants of the RGD-linker region were amplified by PCR using pGEX-RGD-linker encoding the GST-RGD-linker fusion protein as a template. A list of the primer sequences used for PCR is available upon request. The PCR products were digested with EcoRI and SalI and inserted into the corresponding restriction sites of pGEX 4T-1.
A cDNA encoding the human integrin α8 subunit was amplified by PCR with the primer set 5′-AAGGAAGCTTCCACCATGTCGCCCGGGGCCAGCCGCGG-3′ (forward) and 5′-AATCACTCGAGTGCCTCAGGGGTCTTGTCATTTG-3′ (reverse) using human fetal cDNA (Clontech) as a template. The PCR-amplified cDNA was subcloned into pBluescript II SK+ and verified by DNA sequencing. A cDNA encoding the intact α8 integrin subunit was prepared by addition of a stop codon and insertion into the pcDNA 3.1 vector (Invitrogen). An expression vector for an integrin α8 subunit that was truncated before the transmembrane domain and fused to the “ACID” α-helical coiled-coil peptide with a FLAG tag was prepared as described previously (22, 23). Expression vectors for recombinant soluble αV, β1, and β3 integrin subunits were previously described (24, 25).
Expression and Purification of Recombinant Proteins—For purification of recombinant nephronectin, its mutants and integrins, Freestyle™ 293-F cells were transiently transfected with individual plasmids using a Freestyle 293 Expression system (Invitrogen) according to the manufacturer's instructions. Conditioned media were collected at 48–72 h after transfection and centrifuged to remove cells and debris. For purification of nephronectin and its mutants, EDTA (5 mm), phenylmethylsulfonyl fluoride (1 mm), and sodium azide (0.02%) were added to the conditioned media. The media were then incubated with anti-FLAG M2 affinity beads (Sigma) overnight with gentle agitation at 4 °C. The beads were transferred into an empty column and washed with PBS. Bound proteins were eluted with 100 μg/ml FLAG peptide dissolved in PBS. The purified proteins were dialyzed against 2 mm CAPS buffer (pH 11.4) containing 0.5 mm EDTA, and used in the following assays. For purification of recombinant integrins, phenylmethylsulfonyl fluoride (1 mm) and sodium azide (0.02%) were added to the conditioned media. The media were then incubated with nickel-nitrilotriacetic acid-agarose beads (Qiagen, Valencia, CA), followed by washing with Tris-buffered saline (TBS). Bound proteins were eluted with TBS containing 200 mm imidazole. The eluted proteins were applied to anti-FLAG M2 affinity beads, and the bound proteins were eluted with 100 μg/ml FLAG peptide and dialyzed against TBS.
GST fusion proteins were induced in Escherichia coli by overnight incubation with 0.1 mm isopropyl β-d-thiogalactopyranoside at 25 °C. The cells were then lysed by sonication, and the supernatants were passed over a glutathione-Sepharose 4B column (GE Healthcare). Bound proteins were eluted with 50 mm Tris-HCl (pH 8.0) containing 10 mm glutathione, and then dialyzed against 2 mm CAPS buffer containing 0.5 mm EDTA. Protein concentrations were determined by the Bradford method, except for those of the nephronectin deletion mutants expressed in 293-F cells, which were determined by immunoblotting with an anti-FLAG antibody because of the very low reactivity of the highly glycosylated RGD-linker with the Bradford reagent.
SDS-PAGE and Western Blotting—SDS-PAGE was carried out according to Laemmli (26) using 8, 12, or 7–15% gradient gels. Separated proteins were visualized by Coomassie Brilliant Blue staining or transferred onto polyvinylidene difluoride (for immunoblotting) or nitrocellulose (for lectin blotting) membranes. The membranes were probed with antibodies (for immunoblotting) or PNA (for lectin blotting), followed by visualization with an ECL detection kit (GE Healthcare).
Gel Filtration—Purified nephronectin (500 μl) was loaded on a Superose 6 gel filtration column (10 × 300 mm; GE Healthcare) equilibrated in PBS. Fractions were collected at a flow rate of 0.2 ml/min and monitored for their absorbance at 280 nm. The column was calibrated with the following molecular mass standards: chymotrypsinogen, 25 kDa; ovalbumin, 43 kDa; catalase, 232 kDa; ferritin, 440 kDa; thyroglobulin, 669 kDa.
Integrin Binding Assay—Integrin binding assays were performed as described previously (23). Briefly, microtiter plates were coated with various substrate proteins (10 nm) overnight at 4 °C, and then blocked with 10 mg/ml BSA. In trans-complementation assays, the plates were coated with the first substrate protein, washed with PBS, coated with the second substrate proteins overnight at 4 °C, and then blocked with BSA. The plates were incubated with integrins in the presence of 1 mm MnCl2 with or without 10 mm EDTA. In some experiments, integrins were incubated on the plates in the presence of 1 mm CaCl2 and 1 mm MgCl2 to see the effects of divalent cations or in the presence of synthetic peptides at various concentrations to evaluate their inhibitory activities. The plates were washed with TBS containing 1 mm MnCl2, 0.1% BSA, and 0.02% Tween 20 with or without 10 mm EDTA, followed by quantification of bound integrins by an enzyme-linked immunosorbent assay using a biotinylated rabbit anti-Velcro (ACID/BASE coiled-coil) antibody and horseradish peroxidase-conjugated streptavidin. Dissociation constants were calculated by saturation binding assays as described previously (27).
Flow Cytometry—Flow cytometric analyses were performed as described previously (28). Suspended cells were incubated with anti-integrin mAbs for 30 min at 4 °C. After washing with PBS, the cells were incubated with an Alexa Fluor® 488-labeled secondary antibody (Molecular Probes, Eugene, OR) for 30 min at 4 °C and subjected to flow cytometric analysis using a FACScan flow cytometer (BD Biosciences).
Cell Adhesion Assays—Microtiter plates were coated overnight at 4 °C with substrate proteins diluted to various concentrations in PBS in a volume of 50 μl/well. The coating efficiencies of GST fusion proteins were determined by their reactivities with an anti-GST polyclonal antibody. The plates were blocked with blocking medium (serum-free Dulbecco's modified Eagle's medium containing 10 mg/ml BSA and 10 mm HEPES, pH 7.5) for 1 h at 37 °C. HT1080-A8 cells resuspended in blocking medium were plated at 4.0 × 104 cells/well and incubated for 30 min at 37 °C in a humidified atmosphere of 5% CO2 in air. For cell adhesion inhibition assays, cells were preincubated with synthetic peptides in blocking medium at room temperature for 10 min prior to plating. After removal of non-adherent cells by washing with blocking medium, the attached cells were fixed with 3.7% formaldehyde, washed three times with PBS, and stained for 10 min with 0.1% toluidine blue in PBS. After lysis in 1% SDS, the attached cells were quantified by their absorption at 595 nm.
RESULTS
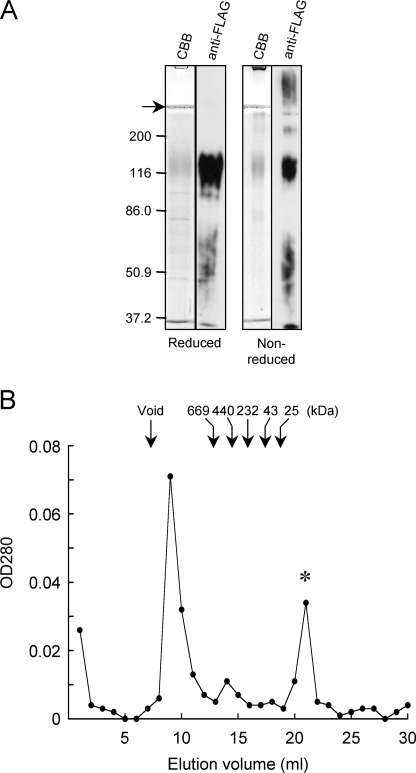
α8β1 Integrin Preferentially Binds to Nephronectin—To examine the binding specificities of α8β1 integrin toward nephronectin and other RGD-containing ECM ligands, we expressed and purified recombinant nephronectin using a mammalian expression system. A FLAG tag was added to the C terminus of nephronectin to facilitate affinity purification of the recombinant protein. The authenticity of the purified nephronectin was verified by SDS-PAGE and immunoblotting against the FLAG tag (Fig. 1A). Purified nephronectin gave a broad band at ∼120 kDa under reducing conditions, which was significantly larger than the predicted protein size (61 kDa). The apparent discrepancy was probably caused by modification with a large number of O-linked sugar chains, which has been predicted to occur at the mucin-like region within the central linker segment (10, 11). The recombinant nephronectin gave a less prominent band at ∼120 kDa upon immunoblotting with an anti-FLAG mAb under non-reducing conditions (Fig. 1A), possibly due to the occurrence of disulfide-bonded multimers that did not enter the resolving gel. Multimer formation by nephronectin was further confirmed by gel filtration chromatography using a Superose 6 column. Nephronectin eluted at a position slightly after the void volume (Fig. 1B). The second major peak that eluted at 21 ml was identified by N-terminal amino acid sequencing as the FLAG peptide used for elution of recombinant nephronectin from the immunoaffinity column (data not shown). Multimer formation by recombinant nephronectin was further confirmed by native PAGE (data not shown).
FIGURE 1.
Expression and purification of recombinant nephronectin. A, 0.5 μg (for Coomassie Brilliant blue (CBB) staining) or 0.1 μg (for immunoblotting with a mAb against FLAG tag) of purified nephronectin was subjected to SDS-PAGE in 8% gels under reducing (left) and non-reducing (right) conditions. The positions of molecular size markers are shown in the left margin. The arrow points to the interface between the stacking and resolving gels. B, 500 μl of purified nephronectin was applied to a Superose 6 column immediately after elution from an anti-FLAG affinity column. Fractions were collected at 1 ml/tube and monitored for their absorbance at 280 nm. The elution positions of molecular size markers are indicated by arrows. The asterisk points to the peak arising from the FLAG peptide used for elution of nephronectin from the anti-FLAG affinity column.
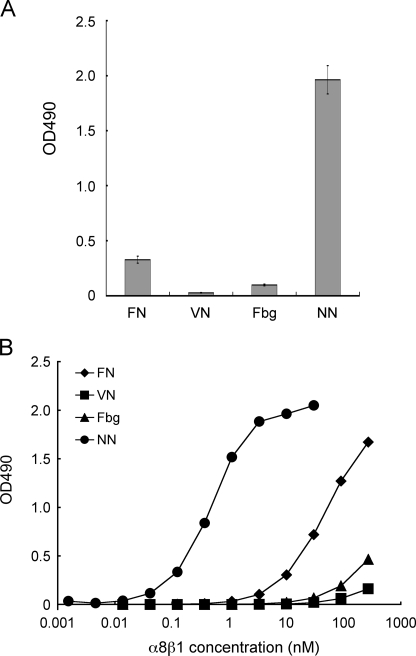
Purified recombinant nephronectin was subjected to α8β1 integrin binding assays along with other RGD-containing proteins including fibronectin, vitronectin, and fibrinogen. The assays were performed in the presence of 1 mm Mn2+ to fully activate integrins. Although α8β1 integrin was previously reported to bind to fibronectin and vitronectin (7, 8), the binding activity of α8β1 integrin toward nephronectin far exceeded those toward fibronectin, vitronectin, and fibrinogen (Fig. 2A). Saturation binding assays revealed that α8β1 integrin bound to nephronectin with a dissociation constant of 0.28 ± 0.01 nm, which was approximately 2 orders of magnitude lower than that for fibronectin (Fig. 2B). Vitronectin and fibrinogen exhibited only marginal binding activities, even at the highest α8β1 integrin concentration examined. The low binding affinities of α8β1 integrin toward fibronectin, vitronectin, and fibrinogen were not due to inactivation of these adhesive proteins because they retained their abilities to bind to their specific integrin receptors, namely α5β1, αVβ3, and αIIbβ3, respectively (data not shown). Taken together, these results demonstrate that nephronectin is a preferred ligand for α8β1 integrin among RGD-containing proteins known to bind to α8β1 integrin.
FIGURE 2.
Binding activities of α8β1 integrin toward nephronectin and other RGD-containing proteins. A, 96-well microtiter plates were coated with various RGD-containing proteins (10 nm) and then incubated with α8β1 integrin (10 nm) in the presence of 1 mm Mn2+. FN, fibronectin; VN, vitronectin; Fbg, fibrinogen; NN, nephronectin. Bound integrins were quantified as described under “Experimental Procedures.” The amounts of integrin bound in the presence of 10 mm EDTA were taken as negative controls, and subtracted as backgrounds. The results represent the mean ± S.D. of triplicate determinations. B, titration curves of recombinant α8β1 integrin bound to fibronectin (diamonds), vitronectin (squares), fibrinogen (triangles), and nephronectin (circles). Microtiter plates coated with individual proteins (10 nm) were incubated with increasing concentrations of α8β1 in the presence of 1 mm Mn2+, followed by quantification of the bound integrins under “Experimental Procedures.” The results represent the means of duplicate determinations.
The RGD-linker Segment of Nephronectin Harbors the α8β1 Binding Activity—Because the RGD motif is situated within the linker segment connecting the N-terminal epidermal growth factor-like repeats and the C-terminal MAM domain, we examined whether the RGD-linker segment could recapitulate the high-affinity binding to α8β1 integrin. To this end, we expressed the RGD-linker in mammalian cells or as a GST fusion protein in bacteria (Fig. 3A). We also expressed the C-terminal MAM domain in mammalian cells and a GST fusion protein with a C-terminal extension of the PRGDV sequence of nephronectin in bacteria as controls. The RGD-linker expressed in 293-F cells gave a broad band migrating at 35–60 kDa upon SDS-PAGE, whereas the RGD-linker expressed as a GST fusion protein in bacteria gave a sharp band at 50 kDa (Fig. 3B). The apparent heterogeneity of the RGD-linker expressed in mammalian cells was probably caused by extensive O-linked glycosylation because the RGD-linker was strongly reactive with PNA, a lectin that recognizes mucin-type O-linked sugar chains (Fig. 3B). The recombinant MAM domain, with a predicted mass of 17 kDa, gave a major band migrating at ∼20 kDa under both reducing and non-reducing conditions (Fig. 3B). Additional bands were detected at 40 and 60 kDa by immunoblotting with an anti-FLAG mAb under non-reducing conditions, indicating that the MAM domain tends to form dimers and trimers that are resistant to dissociation under the denaturing conditions used, i.e. boiling in the presence of SDS.
The purified RGD-linkers were assayed for their binding activities toward α8β1 integrin (Fig. 3C). Not only the full-length nephronectin but also the RGD-linkers, irrespective of the cells used for recombinant protein expression, bound strongly to α8β1 integrin, whereas the MAM domain did not show any binding activity. Furthermore, the PRGDV peptide expressed at the C terminus of GST had only marginal, if any, binding activity toward α8β1 integrin, even though it contained the RGD motif. Because the GST-fused PRGDV peptide retained the ability to bind to αVβ3 integrin (see Fig. 4D), these results indicate that the RGD motif is necessary but not sufficient for binding of nephronectin to α8β1 integrin. Saturation binding assays revealed that the titration curves of the RGD-linkers were essentially the same as that of full-length nephronectin (Fig. 3D), yielding dissociation constants of 0.2–0.3 nm (Table 1). We also performed saturation binding assays in the presence of 1 mm Ca2+ and Mg2+, instead of 1 mm Mn2+, and confirmed that the activities of the RGD-linkers to bind to α8β1 integrin were the same as that of full-length nephronectin (data not shown). These results indicate that the linker segment is sufficient for the high-affinity recognition of nephronectin by α8β1 integrin. However, the PRGDV peptide modeled after nephronectin was barely active in binding to α8β1 integrin, even at the highest α8β1 integrin concentration tested, suggesting that in addition to the RGD motif, the linker segment contains a region that potentiates the binding activity of nephronectin to α8β1 integrin.
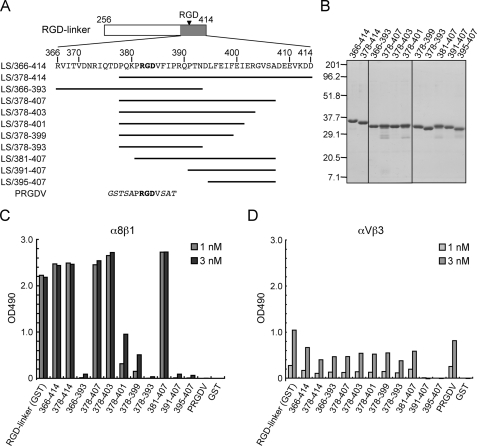
FIGURE 4.
Binding activities of α8β1 integrin toward deletion mutants of the RGD-linker segment. A, amino acid sequences of deletion mutants of the RGD-linker segment. The mutant proteins were expressed in bacteria as GST fusion proteins. The RGD motif is indicated in bold. The vector-derived sequences are shown in italics. B, SDS-PAGE profiles of the mutant proteins (1 μg/lane) stained with Coomassie Brilliant Blue. The positions of molecular size markers are shown in the left margin. C and D, binding activities of α8β1 (C) and αVβ3 (D) integrins to the mutant proteins. Microtiter plates were coated with the individual mutant proteins (10 nm) and incubated with the integrins at the indicated concentrations. Bound integrins were quantified as described under “Experimental Procedures.” The results represent the means of duplicate determinations.
TABLE 1.
Dissociation constants of α8β1 integrin toward nephronectin and its deletion mutants
| Ligands | Kda |
|---|---|
| nm | |
| NN | 0.28 ± 0.01 |
| RGD-linker | 0.23 ± 0.05 |
| MAM | ND (–)b |
| RGD-linker (GST) | 0.22 ± 0.04 |
| PRGDV (GST) | NDc |
| LS/366–414 | 0.32 ± 0.03 |
| LS/378–414 | 0.33 ± 0.02 |
| LS/366–393 | ND |
| LS/378–407 | 0.33 ± 0.01 |
| LS/378–403 | 0.44 ± 0.04 |
| LS/378–401 | 7.1 ± 0.6 |
| LS/378–399 | 13 ± 1 |
| LS/378–393 | ND |
| LS/381–407 | 0.42 ± 0.08 |
| LS/391–407 | ND |
| LS/395–407 | ND |
Values represent the mean ± S.D. of three independent experiments
ND, not determined. The dissociation constant was not determined owing to the absence of significant binding
The dissociation constants were not determined owing to only partial saturation at the highest integrin concentration examined
Identification of the Region within the Linker Segment That Warrants High Affinity Binding to α8β1 Integrin—To explore the region within the linker segment that potentiates the RGD-dependent binding of nephronectin to α8β1 integrin, we produced a series of RGD-linker deletion mutants as GST fusion proteins (Fig. 4A). Because the linker segment expressed as a GST fusion protein was fully competent in high-affinity binding to α8β1 integrin and devoid of the mucin-like sugar chains that have been predicted to attach to the N-terminal two-thirds of the linker segment (based on Net-O-Glyc (29)), we first produced a deletion mutant comprising the C-terminal one-third of the linker segment, designated linker segment (LS)/366–414. LS/366–414 bound avidly to α8β1 integrin with a dissociation constant of 0.32 ± 0.03 nm, comparable with that of the full-length RGD-linker (Fig. 4C and Table 1). Given the retention of the full α8β1 binding activity within LS/366–414, we employed this fragment as a template for the production of a series of deletion mutants (Fig. 4A). The authenticities of the resulting mutant proteins were verified by SDS-PAGE (Fig. 4B).
The purified mutant proteins were assayed for their binding activities toward α8β1 integrin. LS/378–414, a mutant protein lacking the N-terminal 12 amino acid residues of LS/366–414, was capable of binding to α8β1 integrin with a comparable potency to that of LS/366–414 (Fig. 4C and Table 1). On the other hand, LS/366–393, a deletion mutant lacking the C-terminal 21 amino acid residues, was barely active in binding to α8β1 integrin, although the RGD motif remained intact in this mutant. These results indicate that the C-terminal 21 residues (394–414) are required for the high-affinity binding of nephronectin to α8β1 integrin. To further narrow down the residues required for the high-affinity binding to α8β1 integrin, we produced a series of C-terminal deletion mutants of LS/378–414, i.e. LS/378–407, LS/378–403, LS/378–401, LS/378–399, and LS/378–393 (Fig. 4A). Among these C-terminal deletion mutants, LS/378–407 and LS/378–403 retained comparable integrin binding activities to that of LS/378–414, whereas LS/378–401, LS/378–399, and LS/378–393 exhibited stepwise reductions in their integrin binding activities, resulting in an almost complete loss of the activity after deletion of residues 394–414 (Fig. 4C and Table 1). We also assessed the binding activities of α8β1 integrin to the C-terminal deletion mutants of the RGD-linker in the presence of 1 mm Ca2+ and Mg2+, instead of 1 mm Mn2+. The stepwise reductions of the binding activities of α8β1 integrin to LS/378–401, LS/378–399, and LS/378–393 were reproduced in the assays performed in the presence of Ca2+ and Mg2+, although the binding activities of the mutants were less pronounced than those attained in the presence of Mn2+ (data not shown). Because LS/378–403 retained almost full activity, these results indicate that residues 394–403 (DLFEIFEIER) are involved in the high-affinity binding to α8β1 integrin.
The RGD motif has been shown to be a prerequisite for the integrin binding activity of nephronectin (10, 11). To confirm the critical role of the RGD motif, we produced a series of N-terminal deletion mutants of LS/378–407, i.e. LS/381–407, LS/391–407, and LS/395–407. Among these mutants, LS/391–407 and LS/395–407, both lacking the RGD motif but retaining the (D)LFEIFEIER sequence, were barely active in binding to α8β1 integrin, whereas LS/381–407, an N-terminal deletion mutant possessing both the RGD motif and the DLFEIFEIER sequence, was fully active in binding to α8β1 integrin (Fig. 4C). Taken together, these results indicate that both the RGD motif and the DLFEIFEIER sequence at 10 residues to the C-terminal side of the RGD motif are required for the high-affinity binding of nephronectin to α8β1 integrin.
In contrast to the clear dependence of α8β1 integrin on both the RGD motif and the DLFEIFEIER sequence for binding to nephronectin, the binding activities of the mutant fragments to αVβ3 integrin, another integrin known to bind to nephronectin (10), remained almost unchanged regardless of the presence or absence of the DLFEIFEIER sequence, as long as the fragments contained the RGD motif (Fig. 4D). Therefore, the DLFEIFEIER sequence is involved in nephronectin recognition by α8β1 integrin but not αVβ3 integrin.
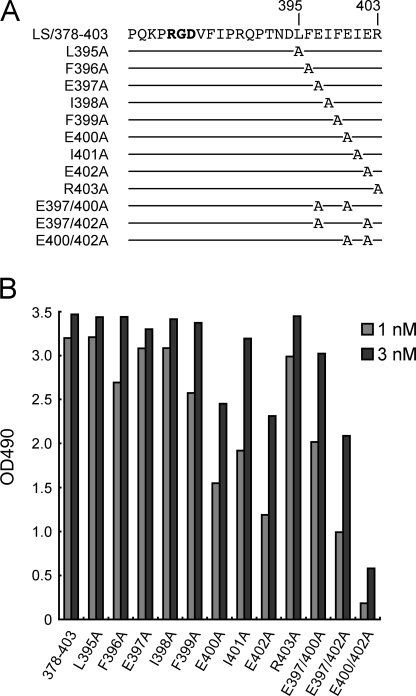
Identification of Critical Amino Acid Residues in the DLFEIFEIER Sequence—To further define the amino acid residues in the DLFEIFEIER sequence that are critical for high-affinity binding to α8β1 integrin, we produced a series of alanine substitution mutants of LS/378–403, in which individual amino acid residues of the DLFEIFEIER sequence were substituted with alanine (Fig. 5A), except for Asp-394, which is not conserved in nephronectin among different species (supplemental Fig. S1). Although the L395A, E397A, I398A, and R403A substitutions did not cause any detectable decreases in the integrin binding activity of LS/378–403, the E400A, I401A, and E402A mutants exhibited moderate reductions (Fig. 5B), resulting in 2-, 3-, and 5-fold decreases in their integrin binding affinities, respectively, compared with that of LS/378–403 (Table 2). Small, yet reproducible, decreases were also observed for the F396A and F399A mutants.
FIGURE 5.
Effects of alanine substitutions within the LFEIFEIER sequence on the integrin binding activity. A, amino acid sequences of LS/378–403 mutants with alanine substitutions. The mutant proteins are designated by the substituted residues. B, binding activities of LS/378–403 and its alanine-substituted mutants toward α8β1 integrin. Microtiter plates were coated with the alanine-substituted mutants and incubated with α8β1 integrin (1 or 3 nm) in the presence of 1 mm Mn2+. Bound integrins were quantified as described under “Experimental Procedures.” The results represent the means of duplicate determinations.
TABLE 2.
Dissociation constants of α8β1 integrin toward LS/378 – 403 after alanine scanning mutagenesis
| Ligands | Kda |
|---|---|
| nm | |
| LS/378–403 | 0.44 ± 0.04 |
| L395A | 0.44 ± 0.08 |
| F396A | 0.84 ± 0.22 |
| E397A | 0.42 ± 0.06 |
| I398A | 0.38 ± 0.04 |
| F399A | 0.72 ± 0.12 |
| E400A | 0.95 ± 0.12 |
| I401A | 1.3 ± 0.2 |
| E402A | 2.5 ± 0.4 |
| R403A | 0.51 ± 0.08 |
| E397/400A | 0.81 ± 0.23 |
| E397/402A | 4.4 ± 0.6 |
| E400/402A | NDb |
Values represent the mean ± S.D. of three independent experiments
ND, not determined. The dissociation constant was not determined due to only partial saturation at the highest integrin concentration examined
Given the involvement of two glutamic acid residues (Glu-400 and Glu-402) in nephronectin recognition by α8β1 integrin, we substituted both of these Glu residues with alanine. The resulting double-substitution mutant showed a significant decrease in α8β1 binding activity, although no such additive effects were found when E397A substitution was combined with E400A or E402A substitutions. Therefore, the marked loss of α8β1 binding activity upon the E400A/E402A double substitutions was not simply due to an extra loss of negative charges but due to the involvement of both residues in binding to α8β1 integrin. Despite the substantial impact of the E400A/E402A double mutations on binding to α8β1 integrin, these mutations did not compromise the binding affinity of LS/378–403 toward the αVβ3 integrin (supplemental Fig. S2), underscoring a role of Glu-400/Glu-402 in the high-affinity binding of nephronectin to α8β1 integrin. Given that I401A substitution also caused a moderate decrease in the α8β1 integrin binding activity, these results raised the possibility that the EIE sequence serves as an auxiliary recognition site within nephronectin that directly interacts with α8β1 integrin in concert with the RGD motif.
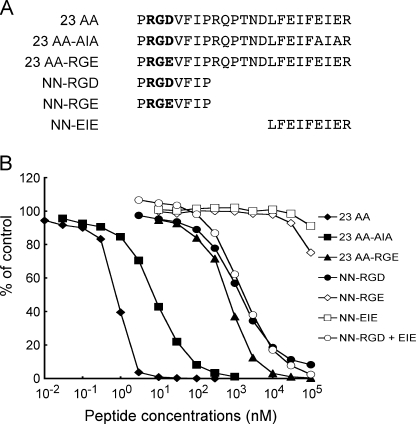
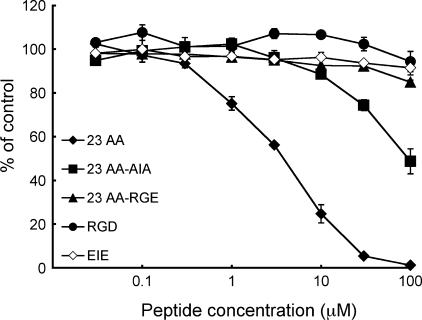
Effects of the LFEIFEIER Peptide on the Interaction of α8β1 Integrin with Nephronectin—To address the role of the LFEIFEIER sequence as one of the bipartite α8β1 integrin recognition sites in nephronectin, we examined whether a synthetic peptide modeled after the sequence was able to inhibit the interaction of nephronectin with α8β1 integrin. We synthesized a 23-mer peptide encompassing the region from the RGD motif to the LFEIFEIER sequence and its mutant forms with RGD → RGE and EIE → AIA substitutions (Fig. 6A), and examined their effects on the binding of nephronectin to α8β1 integrin (Fig. 6B). The 23-mer peptide strongly inhibited the binding of α8β1 integrin to nephronectin with an IC50 of ∼0.6 nm (Fig. 6B and Table 3). Because this IC50 value was comparable with the dissociation constant of α8β1 integrin for nephronectin, the 23-mer peptide appeared to be fully competent in high-affinity binding to α8β1 integrin. Substitution of the EIE motif with AIA resulted in an ∼10-fold decrease in the potency of the peptide to inhibit the integrin-nephronectin interaction, whereas substitution of the RGD motif with RGE resulted in an ∼1000-fold decrease. These results indicate that, although both RGD and EIE motifs are involved in the binding of α8β1 integrin to nephronectin, the contribution of the RGD motif is much greater than that of the EIE motif in securing the association of the synthetic peptide with α8β1 integrin. We also synthesized two shorter peptides harboring the RGD or EIE motifs (Fig. 6A), and assessed their potencies to inhibit the binding of α8β1 integrin to nephronectin. The PRGDVFIP peptide containing only the RGD motif was moderately inhibitory toward binding with an IC50 of 1.2 μm, ∼2000-fold less potent than the 23-mer peptide. On the other hand, the LFEIFEIER peptide containing only the EIE motif was barely inhibitory, in good agreement with the dominant contribution of the RGD motif over the EIE motif in the association of the 23-mer peptide with α8β1 integrin. The LFEIFEIER peptide did not potentiate the inhibitory activity of the PRGDVFIP peptide when mixed with an equimolar amount of the RGD-containing peptide, suggesting that the RGD and EIE motifs need to be aligned in tandem within the same polypeptide to exert their potency to competitively inhibit the binding of α8β1 integrin to nephronectin.
FIGURE 6.
Inhibition of α8β1 integrin binding to nephronectin by synthetic peptides. A, amino acid sequences of the synthetic peptides tested. The RGD motifs are shown in bold. B, α8β1 integrin (1 nm) was incubated on microtiter plates coated with full-length nephronectin (10 nm) in the presence of increasing concentrations of synthetic peptides. To prevent precipitation of the peptides, the integrin binding assays were performed in the presence of 10% dimethyl sulfoxide. The amounts of bound α8β1 integrin are shown as percentages relative to the control, in which α8β1 integrin was incubated on nephronectin-coated plates in the presence of 10% dimethyl sulfoxide. The results represent the means of duplicate determinations. Closed diamonds, 23 AA (23-mer containing both the RGD motif); and the LFEIFEIER sequence; closed squares, 23 AA-AIA (23-mer with the E400A/E402A double mutation); closed triangles, 23 AA-RGE (23-mer with the RGD → RGE mutation); closed circles, NN-RGD; open diamonds, NN-RGE; open squares, NN-EIE; open circles, NN-RGD plus an equal amount of NN-EIE.
TABLE 3.
Inhibition of α8β1-nephronectin interaction by synthetic peptides
| Peptides | IC50a |
|---|---|
| nm | |
| 23 AA | 0.63 ± 0.14 |
| 23 AA-AIA | 7.0 ± 0.9 |
| 23 AA-RGE | 650 ± 50 |
| NN-RGD | 1200 ± 200 |
| NN-RGE | NDb |
| NN-EIE | NDb |
| NN-RGD + NN-EIE | 1300 ± 500 |
Determined based on the data in Fig. 5B. The values represent the mean ± S.D. of three independent determinations
ND, not determined
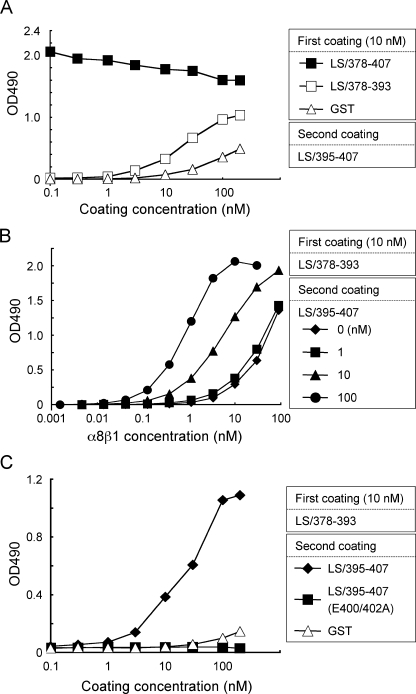
Trans-complementation of Activity between the RGD and EIE Motifs—To further explore the role of the EIE motif in the high-affinity binding of nephronectin to α8β1 integrin, we employed trans-complementation assays (30) using GST fusion proteins containing only the RGD (LS/378–393) or EIE (LS/395–407) motifs. Plates were coated with LS/378–393, followed by a second coating with increasing concentrations of LS/395–407, and then subjected to integrin binding assays. Plates coated with LS/378–407 or GST were also subjected to a second coating with LS/395–407 and subsequent integrin binding assays as controls. Although LS/378–393 alone was inactive in binding to α8β1 integrin, it became active upon the second coating with increasing concentrations of LS/395–407 (Fig. 7A). A second coating with LS/395–407 at >10 nm also endowed GST with integrin binding activity. However, the activity restored to LS/378–393 was significantly greater than the activity restored to the GST-coated plates, indicating that close juxtaposition of LS/395–407 harboring the EIE motif and LS/378–393 harboring the RGD motif synergistically potentiated their integrin binding activities. Saturation integrin binding assays demonstrated that the affinity of LS/378–393 for α8β1 integrin was significantly enhanced upon a second coating with increasing concentrations of LS/395–407 (Fig. 7B) and reached a dissociation constant of 1.2 nm, which was comparable with that of LS/378–407 containing both the RGD and EIE motifs within the same polypeptide chain. These results support the conclusion that nephronectin has a bipartite α8β1 integrin binding site comprising the RGD and EIE motifs, of which the EIE motif plays an auxiliary role in stabilizing the complex of nephronectin and α8β1 integrin. To further confirm the importance of the EIE motif in high-affinity binding of α8β1 integrin to nephronectin, we performed trans-complementation assays using an LS/395–407 mutant in which the two glutamic acid residues in the EIE motif were replaced with alanine, designated LS/395–407(E400A/E402A). The LS/395–407 mutant was incapable of potentiating the integrin binding activity of LS/378–393, even at the highest concentrations used (Fig. 7C). No such synergism was observed in trans-complementation assays with αVβ3 integrin (supplemental Fig. S3), consistent with the specific role of the EIE motif in selective recognition of nephronectin by α8β1 integrin.
FIGURE 7.
Trans-complementation assays of recombinant nephronectin fragments. A, 96-well microtiter plates were coated with LS/378–407 (closed squares), LS/378–393 (open squares), and GST (open triangles), washed with PBS, and then coated a second time with increasing concentrations of LS/395–407 lacking the RGD motif. The plates were subjected to integrin binding assays using 1 nm α8β1 integrin as described under “Experimental Procedures.” B, titration curves of α8β1 integrin bound to substrates coated with LS/378–393, followed by a second coating with LS/395–407 at 0 (diamonds), 1 (squares), 10 (triangles), and 100 nm (circles). Note that the affinities ofα8β1 integrin for LS/378–393 are synergistically enhanced by the presence of LS/395–407. C, microtiter plates were coated with 10 nm LS/378–393, washed with PBS, and then coated a second time with increasing concentrations of LS/395–407 (closed diamonds), LS/395–407 (E400A/E402A) (closed squares), and GST (open triangles). The plates were subjected to integrin binding assays using 1 nm α8β1 integrin as described under “Experimental Procedures.” The results represent the means of duplicate determinations.
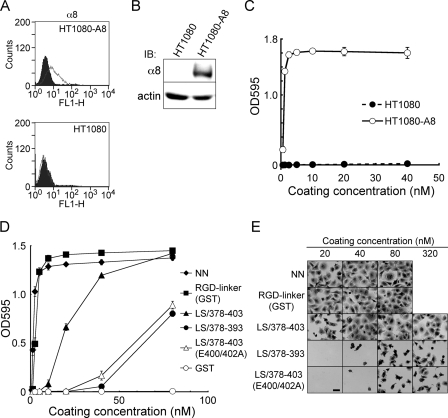
Involvement of the EIE Motif in α8β1 Integrin-dependent Cell Adhesion—The role of the EIE motif in nephronectin recognition by α8β1 integrin was further assessed by cell adhesion assays. To this end, we transfected HT1080 human fibrosarcoma cells with an α8 integrin cDNA and isolated stable transfectants expressing α8 integrin. Expression of the exogenous α8 integrin in the transfectants (designated HT1080-A8 cells) was verified by flow cytometry (Fig. 8A) and immunoblotting (Fig. 8B). HT1080-A8 cells were readily adherent to nephronectin upon coating on substrates at >1 nm, whereas untransfected HT1080 cells did not adhere to nephronectin even after coating on substrates at 40 nm (Fig. 8C), confirming that the adhesion of HT1080-A8 cells to nephronectin was dependent on α8β1 integrin.
FIGURE 8.
Cell-adhesive activities of recombinant nephronectin and its fragments. A, flow cytometric analyses of α8 integrin on HT1080 cells. Control and α8 integrin-transfected HT1080 cells (the latter designated HT1080-A8 cells) were incubated with an anti-humanα8 mAb (7A5) and stained with an Alexa Fluor 488-conjugated secondary antibody. The expressions of α8 integrin are shown as gray lines, whereas the black areas indicate the negative controls incubated with mouse IgG instead of an anti-α8 mAb. B, cell lysates were subjected to SDS-PAGE, followed by immunoblotting (IB) with an anti-human α8 integrin mAb (10A8) (upper panel) or anti-actin polyclonal antibody (lower panel). C, HT1080 (closed circles) and HT1080-A8 (open circles) cells were incubated at 37 °C for 30 min on 96-well microtiter plates coated with increasing concentrations of nephronectin. Adherent cells were fixed and stained as described under “Experimental Procedures.” The results represent the mean ± S.D. of triplicate assays. D, HT1080-A8 cells were incubated at 37 °C for 30 min on 96-well microtiter plates coated with increasing concentrations of nephronectin (NN, closed diamonds), the RGD-linker segment expressed as a GST fusion protein (RGD-linker (GST), closed squares), LS/378–403 (closed triangles), LS/378–393 (closed circles), LS/378–403 (E400/402A) (open triangles), and GST (open circles). Adherent cells were fixed and stained as described under “Experimental Procedures.” The results represent the mean ± S.D. of triplicate assays. E, representative images of HT1080-A8 cells adhering to substrates coated with the indicated proteins. Bar = 50 μm.
To assess the α8β1 integrin binding activities of the RGD-linker segment and its mutant proteins on the basis of cell adhesion assays, HT1080-A8 cells were allowed to adhere to substrates coated with increasing concentrations of the linker segment and its mutants with deletions and/or amino acid substitutions. The RGD-linker was equally active to intact nephronectin in promoting the adhesion of HT1080-A8 cells, whereas LS/378–403, which harbors both the RGD and EIE motifs and retains comparable α8β1 integrin binding activity to that of intact nephronectin, was significantly less active than intact nephronectin and the control RGD-linker in promoting the adhesion of HT1080-A8 cells (Fig. 8D). The difference between the cell-adhesive activities of the full-length RGD-linker and LS/378–403 could be due to the involvement of non-integrin-type adhesion receptors that possibly recognize the N-terminal region of the linker segment absent from LS/378–403. LS/378–393, which contains the RGD motif but not the EIE motif, was significantly less active than LS/378–403. A similar reduction in cell-adhesive activity was also observed with LS/378–393 in cell adhesion assays performed in the presence of Mn2+ (data not shown), together corroborating the importance of the EIE motif in the potent cell-adhesive activity of nephronectin. In support of the critical role of the EIE motif, alanine substitution of the two Glu residues in the EIE motif of LS/378–403 resulted in a marked reduction in the cell-adhesive activity, leading to comparable activity to that of LS/378–393.
Integrin-mediated adhesion has been shown to transduce signals that induce reorganization of the actin cytoskeleton, thereby leading to cell spreading on substrates. Cells adhering to LS/378–403 exhibited well spread polygonal shapes at coating concentrations of >40 nm, whereas cells were poorly spread on LS/378–393 and LS/378–403(E400/402A) even after coating at 320 nm (Fig. 8E). These results indicate that LS/378–403 is competent in stimulating actin cytoskeleton reorganization through binding to α8β1 integrin, and that the EIE motif within the fragment is indispensable for stimulation of integrin-mediated signaling events. Similar results were obtained in cell adhesion assays using K562 erythroleukemic cells stably transfected with chick α8 integrin (data not shown).
The importance of the EIE motif for α8β1 integrin-dependent cell adhesion to nephronectin was further addressed by cell adhesion inhibition assays using synthetic peptides. Adhesion of HT1080-A8 cells to nephronectin was strongly inhibited by the 23-mer peptide with an apparent IC50 of 3.8 μm (Fig. 9). Substitution of the EIE motif with AIA resulted in an ∼30-fold decrease in the potency of the 23-mer peptide to inhibit the cell adhesion on nephronectin, whereas substitution of the RGD motif to RGE resulted in an almost complete loss of the inhibitory activity. These results were in line with those obtained in the peptide inhibition assays of direct binding of α8β1 integrin to nephronectin (Fig. 6), underscoring the greater contribution of the RGD motif over the EIE motif in stabilizing the association of nephronectin with α8β1 integrin. Shorter peptides containing either the RGD or EIE motifs did not exert inhibitory effects, even at the highest concentrations examined, consistent with the requirement for both the RGD and EIE motifs within the same peptide for efficient inhibition of the interaction between nephronectin and α8β1 integrin.
FIGURE 9.
Inhibition of α8β1 integrin-mediated cell adhesion by synthetic peptides. HT1080-A8 cells were incubated on plates coated with 3 nm nephronectin for 30 min in the presence of increasing concentrations of the following synthetic peptides: 23-mer containing both the RGD motif and LFEIFEIER sequence (23 AA, closed diamonds); 23-mer with the E400A/E402A double mutation (23 AA-AIA, closed squares); 23-mer with the RGD → RGE mutation (23 AA-RGE, closed triangles); PRGDVFIP (RGD, closed circles); and LFEIFEIER (EIE, open diamonds). Adherent cells were fixed and stained as described under “Experimental Procedures.” Adhesion of cells in the presence of 0.5% dimethyl sulfoxide was taken as 100%. The results represent the mean ± S.D. of triplicate determinations.
DISCUSSION
In the present study, we investigated the interactions of α8β1 integrin with a panel of RGD-containing proteins and found that α8β1 integrin binds strongly and preferentially to nephronectin with an ∼100-fold higher affinity than those to other RGD-containing proteins including fibronectin and vitronectin. In addition to the RGD-containing proteins examined, α8β1 integrin has been shown to bind to osteopontin, the latency-associated peptide of transforming growth factor-β, tenascin-W, MAEG, and QBRICK/Frem1 in RGD-dependent manners (9, 12, 31–33). Although the binding affinities for these ligand proteins remain to be determined, the following observations, together with our data, indicate that nephronectin is the most preferred ligand for α8β1 integrin with the highest binding affinity. Recombinant α8β1 integrin bound to nephronectin, but not to vitronectin or osteopontin, in far-Western blotting assays with tissue extracts (10). Mice deficient in nephronectin expression displayed kidney agenesis or hypoplasia and were associated with a marked reduction in glial cell line-derived neurotrophic factor expression in the metanephric mesenchyme, resembling the phenotypes observed in mice deficient in α8 integrin expression (5, 14). Consistent with the similarities in phenotypes between the nephronectin-deficient and α8 integrin-deficient mice, the distribution patterns of nephronectin in embryonic lungs and kidneys overlapped with those of α8β1 integrin (5, 10, 34, 35). Recently, we examined the distributions of α8β1 ligands by integrin overlay assays using frozen mouse tissues and found that the signals for α8β1 ligands significantly overlapped with those for nephronectin visualized by immunohistochemistry,3 further supporting the possibility that nephronectin is the physiological ligand for α8β1 integrin with the highest binding affinity among the RGD-containing proteins identified to date.
Our results showed that the RGD-linker segment connecting the N-terminal five epidermal growth factor-like repeats and the C-terminal MAM domain harbors full activity to bind to α8β1 integrin. Although the RGD motif in the linker segment is a prerequisite for the integrin binding activity, several lines of evidence indicate that the LFEIFEIER sequence at ∼10 amino acid residues downstream of the RGD motif is required for the high-affinity binding of the linker segment to α8β1 integrin. First, deletion of the LFEIFEIER sequence from the linker segment resulted in a dramatic loss of the integrin binding activity, even though the RGD motif remained unperturbed. Second, trans-complementation assays demonstrated that almost full binding activity toward α8β1 integrin was restored to LS/378–393, a deletion mutant containing only the RGD motif, upon a second coating with LS/395–407, another deletion mutant containing only the LFEIFEIER sequence. Third, a synthetic 23-mer peptide covering the region from the RGD motif to the LFEIFEIER sequence strongly inhibited the binding of α8β1 integrin to nephronectin, whereas shorter peptides containing either the RGD motif or the LFEIFEIER sequence were only poorly inhibitory. Taken together, these results point to the conclusion that the LFEIFEIER sequence synergizes with the RGD motif to ensure the high-affinity binding of nephronectin to α8β1 integrin.
Alanine scanning mutagenesis of the LFEIFEIER sequence revealed that two Glu residues within the sequence, i.e. Glu-400 and Glu-402, play critical roles in the synergy of this sequence with the RGD motif. Alanine substitution of these individual Glu residues resulted in moderate decreases in the integrin binding activity of the linker segment, whereas double substitution of both Glu residues severely impaired the activity. In support of the critical roles of these two Glu residues, E400A/E402A double mutation abrogated the activity of LS/395–407, a mutant fragment containing the LFEIFEIER sequence but not the RGD motif, to potentiate the integrin binding activity of LS/378–393, a fragment containing only the RGD motif, in trans-complementation assays. In addition to these two Glu residues, several hydrophobic residues in the LFEIFEIER sequence may also be involved in nephronectin recognition by α8β1 integrin, because alanine substitution of Phe-396, Phe-399, and Ile-401 caused moderate reductions in the integrin binding activity of the RGD-linker segment. Recently, DiCara et al. (19) reported that hydrophobic residues downstream of the RGD motif are involved in potentiation of the binding affinity of αVβ6 integrin toward RGD-containing oligopeptides modeled after its natural ligands, i.e. the coat protein of foot-and-mouth disease virus and the latency-associated peptides of transforming growth factor-β proteins. They demonstrated that oligopeptides capable of binding to αVβ6 integrin with high affinities had an RGDLXXL or RGDLXXI motif, of which LXX(L/I) forms a stable helix with two hydrophobic residues (i.e. Leu and Leu/Ile) exposed almost in apposition on one face of the helix, suggesting that these hydrophobic residues, together with the RGD motif, directly interact with the ligand binding pocket of αVβ6 integrin (19). Given the similarity between nephronectin and the natural ligands for αVβ6 integrin in the requirement for a post-RGD sequence for high-affinity integrin binding, the hydrophobic residues within the LFEIFEIER sequence may also be directly involved in ligand recognition by α8β1 integrin. Consistent with this possibility, the LFEIFEIER sequence was predicted to form a helix by PAPIA, an algorithm for protein secondary structure prediction (36).
The requirement for an auxiliary site for selective high-affinity recognition of RGD-containing ligands by integrins has been documented for other integrin-ligand pairs. Fibronectin has been the prototype for such ligands harboring a so-called synergy site, requiring not only the 10th type III module containing the RGD motif but also the preceding type III module (FNIII9) for its high-affinity binding to α5β1 integrin (18, 37). The PHSRN sequence in the FNIII9 module was shown to be critical for the synergistic activity (18). The natural ligands for αVβ6 integrin also possess such an auxiliary site immediately on the C-terminal side of the RGD motif as described above (19). Despite the available evidence, including the data in the present study, for the requirement of synergy sites for high-affinity binding of RGD-containing ligands to integrins, it remains to be elucidated how the synergy sites potentiate the binding affinities between integrins and their RGD-containing ligands. This is in striking contrast to the interactions of the RGD motif with integrins, because determination of the crystal structures of integrins complexed with cyclic RGD-like peptides revealed that the arginine side chain of the RGD motif fits into a cleft in the β-propeller domain of integrin α subunits, whereas the aspartate side chain coordinates the divalent cation at the metal ion-dependent adhesion site (designated MIDAS) of β subunits (38, 39). It should be noted, however, that in addition to the residues defined by the crystal structures to directly interact with the RGD motif, a group of residues located on the upper or side faces of the β-propeller domain of the α subunits have also been implicated in ligand binding by integrins. Specifically, epitope mapping of function-blocking anti-integrin antibodies as well as mutational analyses of the integrin α subunits indicated that the ligand-binding specificities of α5β1 and αVβ1 integrins were dependent on residues in blades 2 and 3 of the β-propeller domain of their α subunits (40–42), of which Tyr-208 and Ile-210 in blade 3 of the integrin α5 subunit were shown to recognize the synergy site of fibronectin (43). Furthermore, mutations that disrupted ligand binding by αIIbβ3 integrin were found to be clustered on the top or side of the β-propeller domain of the αIIb subunit, and mostly located within blades 2 and 3 (44). Similarly, the acidic clusters located on blade 3 of the β-propeller domain of the α7 subunit were shown to determine the ligand-binding specificity of α7β1 integrin (45). Given the involvement of the upper or side faces of the β-propeller domain, particularly those involving blades 2 and 3, high-affinity ligand recognition by integrins, the same regions of the β-propeller domain of the α8 subunit may be involved in recognition of the LFEIFEIER sequence, thereby ensuring the high-affinity binding of nephronectin to α8β1 integrin. It is interesting to note that the residues comprising the upper or side loops of the β-propeller domain differ significantly among RGD-recognizing integrins. The α8 integrin subunit is unique in that it possesses evolutionarily conserved basic amino acid clusters at the loops connecting blades 1 and 2 and within blade 3 of its β-propeller domain (supplemental Fig. S4), the latter being equivalent to the loops involved in the ligand-binding specificities of α5β1 and α7β1 integrins (43, 45). Because two Glu residues within the LFEIFEIER sequence are required for high-affinity binding of nephronectin to α8β1 integrin and evolutionarily conserved among vertebrates (supplemental Fig. S1), it is tempting to speculate that the cluster of basic amino acid residues in the loop either between blades 1 and 2 or within blade 3 of the α8 β-propeller domain form salt bridges with the acidic residues of the EIE motif, thereby sustaining the high-affinity recognition of nephronectin by α8β1 integrin.
Synthetic RGD-containing peptides have been widely used as probes that specifically block the interactions of RGD-binding integrins with their ligands in a variety of biochemical and cell biological assays (46–48). However, the RGD peptides are equally inhibitory toward all RGD-binding integrins, and therefore cannot be used as specific probes for the biological consequences of interactions between defined RGD-binding integrins and their physiological ligands. Furthermore, the absence of the synergy sequences that potentiate the integrin binding affinities of RGD-containing ligands leads to a requirement for high concentrations of RGD peptides to block the integrin-ligand interactions. For instance, RGD peptide concentrations of >1 mm are required to effectively block α5β1-mediated cell adhesion onto fibronectin-coated substrates (46–48). In this regard, the 23-mer peptide containing both the RGD motif and the LFEIFEIER sequence fully mimics the interaction of nephronectin with α8β1 integrin and completely blocks the interaction at a concentration of 10 nm, but has no inhibitory effects on the interaction between fibronectin and α5β1 integrin.4 Given the remarkable potency of the 23-mer peptide in specifically blocking the nephronectin-α8β1 integrin interaction at nanomolar concentrations, the 23-mer peptide represents a promising probe for elucidating the biological functions of this integrin-ligand interaction in both physiological and pathological processes.
In summary, we have shown that nephronectin is the most preferred high-affinity ligand for α8β1 integrin and that the specificity and high-affinity binding toward the integrin are fully recapitulated by a 23-amino acid residue fragment harboring both the RGD motif and the LFEIFEIER sequence, the latter serving as an auxiliary site that ensures the specific high-affinity binding of nephronectin to α8β1 integrin in concert with the RGD motif. Although the mechanism by which the synergy site potentiates the integrin binding affinity remains to be validated by determination of the three-dimensional structure of the fully active RGD-linker segment at the atomic level, these results provide, for the first time, the molecular basis of the specific interaction between nephronectin and α8β1 integrin, and highlight the bipartite nature of the integrin recognition site of nephronectin.
Supplementary Material
Acknowledgments
We thank Dr. Yoshihide Hayashizaki for cDNA encoding mouse nephronectin, Dr. Aki Osada for cDNA encoding human α8 integrin, and Dr. Louis F. Reichardt for K562 cells transfected with chick α8 integrin. We are grateful to Yumi Yoshimura for peptide sequence analyses and Noriko Sanzen and Tasuku Tsukamoto for assistance in the production of anti-human α8 integrin mAbs.
This work was supported in part by Grants-in-aid for Scientific Research 17082005 and 20370046 (to K. S.) and Research Contract 06001294-0 with the New Energy and Industrial Technology Development Organization of Japan.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: ECM, extracellular matrix; BSA, bovine serum albumin; GST, glutathione S-transferase; mAb, monoclonal antibody; MAM, meprin-A5 protein-receptor protein-tyrosine phosphatase μ; PBS, phosphate-buffered saline; PNA, peanut agglutinin; RGD-linker, RGD-containing linker segment; TBS, Tris-buffered saline; CAPS, N-cyclohexyl-3-aminopropane sulfonic acid; LS, linker segment.
M. Takeichi and D. Kiyozumi, unpublished observations.
Y. Sato, unpublished observations.
References
- 1.Hynes, R. O. (1992) Cell 69 11-25 [DOI] [PubMed] [Google Scholar]
- 2.Hynes, R. O. (2002) Cell 110 673-687 [DOI] [PubMed] [Google Scholar]
- 3.Takagi, J. (2007) Curr. Opin. Cell Biol. 19 557-564 [DOI] [PubMed] [Google Scholar]
- 4.Bossy, B., Bossy-Wetzel, E., and Reichardt, L. F. (1991) EMBO J. 10 2375-2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller, U., Wang, D., Denda, S., Meneses, J. J., Pedersen, R. A., and Reichardt, L. F. (1997) Cell 88 603-613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Littlewood Evans, A., and Muller, U. (2000) Nat. Genet. 24 424-428 [DOI] [PubMed] [Google Scholar]
- 7.Muller, U., Bossy, B., Venstrom, K., and Reichardt, L. F. (1995) Mol. Biol. Cell 6 433-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnapp, L. M., Hatch, N., Ramos, D. M., Klimanskaya, I. V., Sheppard, D., and Pytela, R. (1995) J. Biol. Chem. 270 23196-23202 [DOI] [PubMed] [Google Scholar]
- 9.Denda, S., Reichardt, L. F., and Muller, U. (1998) Mol. Biol. Cell 9 1425-1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandenberger, R., Schmidt, A., Linton, J., Wang, D., Backus, C., Denda, S., Muller, U., and Reichardt, L. F. (2001) J. Cell Biol. 154 447-458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morimura, N., Tezuka, Y., Watanabe, N., Yasuda, M., Miyatani, S., Hozumi, N., and Tezuka, K. (2001) J. Biol. Chem. 276 42172-42181 [DOI] [PubMed] [Google Scholar]
- 12.Lu, M., Munger, J. S., Steadele, M., Busald, C., Tellier, M., and Schnapp, L. M. (2002) J. Cell Sci. 115 4641-4648 [DOI] [PubMed] [Google Scholar]
- 13.Scherberich, A., Tucker, R. P., Samandari, E., Brown-Luedi, M., Martin, D., and Chiquet-Ehrismann, R. (2004) J. Cell Sci. 117 571-581 [DOI] [PubMed] [Google Scholar]
- 14.Linton, J. M., Martin, G. R., and Reichardt, L. F. (2007) Development 134 2501-2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekblom, P. (1981) J. Cell Biol. 91 1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takagi, J. (2004) Biochem. Soc. Trans. 32 403-406 [DOI] [PubMed] [Google Scholar]
- 17.Ruoslahti, E. (1996) Annu. Rev. Cell Dev. Biol. 12 697-715 [DOI] [PubMed] [Google Scholar]
- 18.Aota, S., Nomizu, M., and Yamada, K. M. (1994) J. Biol. Chem. 269 24756-24761 [PubMed] [Google Scholar]
- 19.DiCara, D., Rapisarda, C., Sutcliffe, J. L., Violette, S. M., Weinreb, P. H., Hart, I. R., Howard, M. J., and Marshall, J. F. (2007) J. Biol. Chem. 282 9657-9665 [DOI] [PubMed] [Google Scholar]
- 20.Sekiguchi, K., Hakomori, S., Funahashi, M., Matsumoto, I., and Seno, N. (1983) J. Biol. Chem. 258 14359-14365 [PubMed] [Google Scholar]
- 21.Yatohgo, T., Izumi, M., Kashiwagi, H., and Hayashi, M. (1988) Cell Struct. Funct. 13 281-292 [DOI] [PubMed] [Google Scholar]
- 22.Ido, H., Nakamura, A., Kobayashi, R., Ito, S., Li, S., Futaki, S., and Sekiguchi, K. (2007) J. Biol. Chem. 282 11144-11154 [DOI] [PubMed] [Google Scholar]
- 23.Nishiuchi, R., Takagi, J., Hayashi, M., Ido, H., Yagi, Y., Sanzen, N., Tsuji, T., Yamada, M., and Sekiguchi, K. (2006) Matrix Biol. 25 189-197 [DOI] [PubMed] [Google Scholar]
- 24.Takagi, J., Erickson, H. P., and Springer, T. A. (2001) Nat. Struct. Biol. 8 412-416 [DOI] [PubMed] [Google Scholar]
- 25.Takagi, J., Petre, B. M., Walz, T., and Springer, T. A. (2002) Cell 110 599-611 [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 27.Nishiuchi, R., Murayama, O., Fujiwara, H., Gu, J., Kawakami, T., Aimoto, S., Wada, Y., and Sekiguchi, K. (2003) J. Biochem. (Tokyo) 134 497-504 [DOI] [PubMed] [Google Scholar]
- 28.Gu, J., Sumida, Y., Sanzen, N., and Sekiguchi, K. (2001) J. Biol. Chem. 276 27090-27097 [DOI] [PubMed] [Google Scholar]
- 29.Hansen, J. E., Lund, O., Tolstrup, N., Gooley, A. A., Williams, K. L., and Brunak, S. (1998) Glycoconj. J. 15 115-130 [DOI] [PubMed] [Google Scholar]
- 30.Obara, M., Kang, M. S., and Yamada, K. M. (1988) Cell 53 649-657 [DOI] [PubMed] [Google Scholar]
- 31.Scherberich, A., Tucker, R. P., Degen, M., Brown-Luedi, M., Andres, A. C., and Chiquet-Ehrismann, R. (2005) Oncogene 24 1525-1532 [DOI] [PubMed] [Google Scholar]
- 32.Kiyozumi, D., Osada, A., Sugimoto, N., Weber, C. N., Ono, Y., Imai, T., Okada, A., and Sekiguchi, K. (2005) Exp. Cell Res. 306 9-23 [DOI] [PubMed] [Google Scholar]
- 33.Osada, A., Kiyozumi, D., Tsutsui, K., Ono, Y., Weber, C. N., Sugimoto, N., Imai, T., Okada, A., and Sekiguchi, K. (2005) Exp. Cell Res. 303 148-159 [DOI] [PubMed] [Google Scholar]
- 34.Wagner, T. E., Frevert, C. W., Herzog, E. L., and Schnapp, L. M. (2003) J. Histochem. Cytochem. 51 1307-1315 [DOI] [PubMed] [Google Scholar]
- 35.Manabe, R., Tsutsui, K., Yamada, T., Kimura, M., Nakano, I., Shimono, C., Sanzen, N., Furutani, Y., Fukuda, T., Oguri, Y., Shimamoto, K., Kiyozumi, D., Sato, Y., Sado, Y., Senoo, H., Yamashina, S., Fukuda, S., Kawai, J., Sugiura, N., Kimata, K., Hayashizaki, Y., and Sekiguchi, K. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 12849-12854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akiyama, Y., Onizuka, K., Noguchi, T., and Ando, M. (1998) Genome Inform. Ser. Workshop Genome Inform. 9 131-140 [PubMed] [Google Scholar]
- 37.Redick, S. D., Settles, D. L., Briscoe, G., and Erickson, H. P. (2000) J. Cell Biol. 149 521-527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong, J. P., Stehle, T., Zhang, R., Joachimiak, A., Frech, M., Goodman, S. L., and Arnaout, M. A. (2002) Science 296 151-155 [DOI] [PubMed] [Google Scholar]
- 39.Xiao, T., Takagi, J., Coller, B. S., Wang, J. H., and Springer, T. A. (2004) Nature 432 59-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mould, A. P., Askari, J. A., and Humphries, M. J. (2000) J. Biol. Chem. 275 20324-20336 [DOI] [PubMed] [Google Scholar]
- 41.Humphries, J. D., Askari, J. A., Zhang, X. P., Takada, Y., Humphries, M. J., and Mould, A. P. (2000) J. Biol. Chem. 275 20337-20345 [DOI] [PubMed] [Google Scholar]
- 42.Humphries, M. J., Symonds, E. J., and Mould, A. P. (2003) Curr. Opin. Struct. Biol. 13 236-243 [DOI] [PubMed] [Google Scholar]
- 43.Mould, A. P., Symonds, E. J., Buckley, P. A., Grossmann, J. G., McEwan, P. A., Barton, S. J., Askari, J. A., Craig, S. E., Bella, J., and Humphries, M. J. (2003) J. Biol. Chem. 278 39993-39999 [DOI] [PubMed] [Google Scholar]
- 44.Kamata, T., Tieu, K. K., Irie, A., Springer, T. A., and Takada, Y. (2001) J. Biol. Chem. 276 44275-44283 [DOI] [PubMed] [Google Scholar]
- 45.von der Mark, H., Poschl, E., Lanig, H., Sasaki, T., Deutzman, R., and von der Mark, K. (2007) J. Mol. Biol. 371 1188-1203 [DOI] [PubMed] [Google Scholar]
- 46.Pierschbacher, M. D., and Ruoslahti, E. (1984) Nature 309 30-33 [DOI] [PubMed] [Google Scholar]
- 47.Akiyama, S. K., and Yamada, K. M. (1985) J. Biol. Chem. 260 10402-10405 [PubMed] [Google Scholar]
- 48.Manabe, R., Ohe, N., Maeda, T., Fukuda, T., and Sekiguchi, K. (1997) J. Cell Biol. 139 295-307 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.