Abstract
The sexual phase of the malaria parasite Plasmodium falciparum is accompanied by the coordinated expression of stage-specific adhesive proteins. Among these are six secreted proteins with multiple adhesion domains, termed P. falciparum LCCL domain-containing protein (PfCCp) proteins, which are expressed in the parasitophorous vacuole of the differentiating gametocytes and which are later associated with macrogametes. Although the majority of the PfCCp proteins are implicated in parasite development in the mosquito vector, their functions remain unknown. In the present study we investigated the molecular interactions between the PfCCp proteins during gametocyte development and emergence. Using five different gene-disruptant parasite lines, we show that the lack of one PfCCp protein leads to the loss of other PfCCp family members. Co-immunoprecipitation assays on gametocyte lysates revealed formation of complexes involving all PfCCp proteins, and affinity chromatography co-elution binding assays with recombinant PfCCp domains further indicated direct binding between distinct adhesion domains. PfCCp-coated latex beads bind to newly formed macrogametes but not to gametocytes or older macrogametes 6 or 24 h post-activation. In view of these data, we propose that the PfCCp proteins form multi-protein complexes that are exposed during gametogenesis, thereby mediating cell contacts of macrogametes.
The sexual phase of the malaria pathogen Plasmodium falciparum begins with the differentiation of predetermined blood stage parasites to gametocytes in the human host and continues with the formation of male and female gametes in the midgut of the blood-fed female mosquito vector. Subsequently, the motile male microgamete fertilizes the amotile female macrogamete, and within a day after the blood meal, the resulting zygote transforms into an infective ookinete. Gametocytogenesis and gamete formation are accompanied by the coordinated expression of numerous sexual stage proteins (1, 2), including the epidermal growth factor domain-containing proteins Pfs25 and Pfs28, as well as the cysteine motif-rich proteins Pfs230 and Pfs48/45, all of which are considered promising candidates for components of a transmission blocking vaccine (2, 3).
Among the sexual stage proteins that have been recently identified in P. falciparum are a highly conserved family of six secreted proteins that are comprised of multiple predicted adhesive domains (4–8). Five of the proteins possess a common Limulus coagulation factor C (LCCL)2 domain and are termed PfCCp1 through PfCCp5, whereas a sixth protein, PfFNPA, lacks this domain but shares architectural features that indicate its vertical relationship with PfCCp5 and warrant its inclusion as a member of the protein family. The PfCCp proteins have orthologs in other apicomplexan parasites, including Toxoplasma and Cryptosporidium, and are known as PbLAP proteins in the rodent malaria parasite Plasmodium berghei (7–9).
The six PfCCp proteins are expressed during gametocytogenesis of P. falciparum and localize within the gametocyte parasitophorous vacuole (7, 10, 11). During gamete emergence they are exposed associated to the surface of macrogametes, but expression ceases within a day (7, 11). Gene disruptions of PfCCp2 or PfCCp3 resulted in a blocked transition of sporozoites from the mosquito midgut to the salivary glands, showing that these proteins are essential for the parasite development in the vector (7). Gene disruption of PfCCp4, on the other hand, did not reveal any knock-out phenotype in the mosquito stages (11). Antibodies directed against select PfCCp adhesion domains induce a complement-mediated reduction of exflagellation, making the proteins potential candidates as subunits of transmission blocking vaccines (11).
We have recently reported that PfCCp1, PfCCp2, and PfCCp3 co-localize within the parasitophorous vacuole of mature gametocytes (10). The three proteins are co-dependently expressed at the protein but not transcript level, leading to the assumption that they might interact during expression in gametocytes and that the loss of one protein results in degradation of the other two (10). For this study, we generated novel PfCCp1 and PfFNPA gene-disruptant parasites and show that in gene-disruptant gametocytes lacking PfCCp1, PfCCp2, PfCCp3, PfCCp4, or PfFNPA, the PfCCp proteins were either absent or expressed at a decreased protein level. We further demonstrate that the six proteins undergo intermolecular interactions and are involved in cell binding of the newly emerged macrogametes. In conclusion, we propose that the PfCCp proteins form multi-protein complexes that are exposed during gamete formation and that might mediate cell contacts of macrogametes.
EXPERIMENTAL PROCEDURES
Gene Identifications—The following gene identifiers are assigned to the proteins investigated in this study: Pf39, PF11_0098; PfCCp1, PF14_0723; PfCCp2, PF14_0532; PfCCp3, PF14_0067; PfCCp4, PFI0185w; PfCCp5, PFA0445w; PfFNPA, PF14_0491; and Pfs25, PF10_0303.
Oligonucleotides—PfCCp1 5′ SpeI, 5′-ACTAGTTACATGTCTGAAGAGTCC; PfCCp1 5′ BglII, 5′-AGATCTTTACTGATTATCAGTACTTACAG; PfCCp1 3′ ClaI, 5′-ATCGATGCACAC AATTATCTGAATTG; PfCCp1 3′ NcoI, 5′-CCATGGTTAGGATGTTGTTGGGTC; PfFNPA BamHI, 5′-ATGGATCCGGGGTACATGCATTTATG; PfFNPA 5′-NotI, TAGCGGCCGCTTACGTATTGACCCAGTGATT; a, 5′-CTTCTCATCATAAGCATTGGC; b, 5′-CCCGTCCATGTTATTTCTTCATC; b′,5′-CAAAGAAGAAGCTCAGAGATTGC; c, 5′-GTAAGCAAAATGATAATTTAGCAGC; c′,5′-GTTTTGTAATTTATGGGATAGCG; d, 5′-ATGTATGTACCCATCAAACGG; e, 5′-GGAATGTTGAAAAGGAAAATGAGCAG; f, 5′-GCTGCTAAATTATCATTTTGCTTAC; g, 5′-CTGGTATGGACCATTATGTTGGG; h, 5′-CGAAATTACAGAAGAATCAACACCATG; i, 5′-TCGGATGGAGAATCCGTT; j, 5′-GTATCCCATGTCTTGTGA; k, 5′-GCCCTCATGAAACACATGATG; l, 5′-CACATCTCCTGTTGTCGC; m, 5′-ATGCCCGAACAAATCATTAAT; n, 5′-TGCTGAGGCGCATATACT; o, 5′-GGTTCGCGCGATTGGGAT; p, 5′-TTTTCCGCACACAGAATC; q, 5′-CGCTGGATTCAGTGGTAATAACC; r, 5′-CCGTATTGGTCTTTATTGGAG; s, 5′-GGTAATGTATCTACGCCT; t, 5′-AATGATGATGCCCAGGAG; u, 5′-TATTCCTAATCATGTAAATCTTAAA; v, 5′-CAATTAACCCTCACTAAAG; w, 5′-GCAACATCTCTAAGTGATACTGG; x, 5′-TGGTATCCCTGTGTCCCA; y, 5′-CTTGAACACCATGATGTA; and z, 5′-TCCACTTTCATGAGCAGG.
Parasite Culture—Mature gametocytes of the P. falciparum NF54 isolate were cultivated in vitro as described (12). Gametocyte activation and gametogenesis was induced by incubating mature gametocyte cultures in 100 μm xanthurenic acid/PBS, pH 8.5 (13, 14) or human serum for 15 min at room temperature.
Generation of Gene-disruptant Parasites—PfCCp1 gene-disruptant parasites were generated by double homologous recombination using the targeting vector pHHT-TK (ATCC, Manassas, VA) (15). Two fragments of the PfCCp1 gene corresponding to the 5′-and 3′-coding regions of 1,138 and 1,522 bp, respectively, were amplified by PCR from P. falciparum NF54 genomic DNA using the above mentioned primer pairs PfCCp1 5′ SpeI and PfCCp1 5′ BglII as well as PfCCp1 3′ ClaI and PfCCp1 3′ NcoI. These PCR products were sequentially cloned into the restriction sites SpeI/BglII and ClaI/NcoI of the pHHT-TK vector. The gene targeting disruption plasmid was introduced into the P. falciparum isolate NF54 via the erythrocyte-loading method (16), and stable transfected parasites that have integrated plasmid by double cross-over homologous recombination were selected with pyrimethamine and ganciclovir (15). A pyrimethamine-resistant polyclonal line was established, and clones were isolated by limiting dilution. Correct integration of the targeting plasmid into the genomic locus was verified by PCR and Southern blot analyses diagnostic of a disrupted locus. PCR was performed with 100 ng of genomic DNA from wild type or disruptant parasites for 35 cycles of amplification and with an annealing temperature of 50 °C, using the above mentioned primers a, b, b′,c,c′, and d (shown in supplemental Fig. S1). Southern blotting was performed with 10 μg of genomic DNA from wild type or disruptant parasites, digested with KpnI plus BamHI, separated by 0.8% agarose gel electrophoresis, and transferred to Hybond N+ (Amersham Biosciences). A 1.1-kb hybridization probe corresponding to the 5′-coding region of PfCCp1 was amplified with the primers PfCCp1 5′ SpeI and PfCCp1 5′ BglII. The probe was labeled with digoxigenin, and hybridization was performed at 48 °C using a DIG High Prime DNA labeling and detection starter kit II (Roche Applied Science), according to the manufacturer's instructions. Lack of PfCCp1 transcripts in the disruptant clone was confirmed by RT-PCR. RNA from mature wild type and disruptant gametocytes was isolated with TRIzol (Invitrogen) according to the manufacturer's instructions, treated with DNase I (Invitrogen), and purified on RNeasy MinElute Cleanup columns (Qiagen). RNA was reverse-transcribed using Superscript II that was primed with random hexanucleotides (Invitrogen). PCR was performed for 35 cycles of amplification using the oligonucleotides e and f. The absence of genomic DNA contamination was confirmed by PCR amplification on sham-treated RNA samples that lacked reverse transcriptase. Absence of protein expression was assessed by Western blot and indirect immunofluorescence assay, as described below. Furthermore, PfFNPA gene-disruptant parasites were generated by single homologous recombination using the vector pCAM-BSD (17, 18). A 568-bp fragment of the PfFNPA gene was amplified by PCR from P. falciparum NF54 genomic DNA using the primer pair PfFNPA BamHI and PfFNPA NotI, which included a stop codon. The PCR product was cloned into the restriction site BamHI/NotI of the pCAM-BSD vector. Further steps were carried out as described above, except that selection of stable transfected parasites, which had integrated plasmid by single cross-over homologous recombination, was pursued using blasticidin. PCR with genomic DNA from wild type or disruptant parasites was performed for 33 cycles of amplification and with annealing temperatures ranging from 42 to 50 °C using the primers s, t, u, and v (shown in supplemental Fig. S2). Southern blotting was carried out with 2 μg of wild type parasites or parasites of the PfFNPA-KO (knock-out) clone 1H4, digested with MfeI and NcoI. A hybridization probe of 825 bp corresponding to the 5′-coding region of PfFNPA was amplified with the primers w and x. Southern blotting was performed as mentioned above, using a hybridization temperature of 35 °C. RNA isolation and transcript level analysis was conducted as below. For RT-PCR oligonucleotides q and r were used as well as Pf39-specific oligonucleotides y and z as a control.
Antibodies—The following antibodies were used in this study: mouse antisera against PfCCp1-CC, PfCCp2-Disc, and PfCCp3-SR (7); PfCCp1rp1, PfCCp1rp5, PfCCp2rp3, PfCCp3rp3, PfCCp4rp1, PfCCp4rp3, PfCCp5rp1, PfCCp5rp2, PfFNPArp1, and PfFNPArp2 (11); mouse antisera against peptide PfCCp4pt2 (11); mouse antisera against the endoplasmic reticulum-associated protein Pf39 (19); and goat antisera against GST tag (Amersham Biosciences) or mouse antisera against His6 tag (GE Healthcare).
RNA Isolation and Transcript Level Analysis—Gametocytes were enriched by Percoll gradient purification (20), and RNA was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer's protocol. RNA preparations were treated with RNase-free DNase I (Invitrogen) to remove genomic DNA contamination, followed by phenol/chloroform extraction and ethanol precipitation. Synthesis of cDNA was accomplished using a Superscript II cDNA synthesis kit (Invitrogen). RT-PCR assays were performed with 125 ng of cDNA using gene specific primers by conventional thermocycling (25 cycles) separated by 2% agarose gel electrophoresis. The following primers were used: PfCCp1, g and h; PfCCp2, i and j; PfCCp3, k and l; PfCCp4, m and n; PfCCp5, o and p; and PfFNPA, q and r.
Western Blot Analysis—Pellets of enriched gametocytes were resuspended in PBS and SDS-PAGE loading buffer as described (11). Parasite proteins were separated by SDS-PAGE electrophoresis and transferred to Hybond ECL nitrocellulose membrane (Amersham Biosciences) according to the manufacturer's instructions. The membranes were blocked for nonspecific binding by incubation in Tris-buffered saline containing 5% skim milk and 1% bovine serum albumin fraction V, followed by immune recognition for 2 h at room temperature with mouse immune sera specific to PfCCp proteins, Pf39, or to GST- and His6/SUMO fusion partner protein control. After washing, the membranes were incubated for 1 h at room temperature with an alkaline phosphatase-conjugated secondary antibody (Sigma-Aldrich) and developed in nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate solution (Sigma-Aldrich) for 5–30 min. Scanned blots were processed using Adobe Photoshop CS software.
Indirect Immunofluorescence Assay—Activated or nonactivated gametocyte cultures were air dried on slides and fixed for 10 min in -80 °C methanol. For membrane permeabilization and blocking of nonspecific binding, the fixed cells were incubated in 0.01% saponin, 0.5% bovine serum albumin, and 1% neutral goat serum (Sigma-Aldrich) in PBS twice for 30 min each. The preparations were then incubated for 1.5 h at 37 °C with anti-PfCCp mouse immune sera. Binding of primary antibody was visualized using fluorescence-conjugated goat anti-mouse secondary antibodies (Alexa Fluor 488 and 594; Molecular Probes). Counterstaining was performed using 0.05% Evans Blue in PBS for 1 min (Sigma-Aldrich). Labeled specimens were examined by confocal laser scanning microscopy using a Zeiss LSM 510. The digital images were processed using Adobe Photoshop CS software.
Co-immunoprecipitation Assay—Enriched gametocytes were lysed in 0.5% saponin in PBS, homogenized, and sonicated for 1 min. The homogenate was centrifuged at 16,000 × g, and prepurification of the supernatant was performed by consecutive incubation with 5% (v/v) preimmune mouse sera and 20 μl of protein G-beads (Santa Cruz Biotechnology) for half an hour each at 4 °C. After centrifugation at 3,400 × g, the supernatant was then incubated for 1 h at 4 °C with 5% (v/v) polyclonal mouse antisera against PfCCp proteins or with antisera against Pf39 as a negative control. A volume of 20 μl of protein G beads was added and incubated for another hour. The beads were then centrifuged, washed five times with PBS, mixed with an equal volume of loading buffer, and loaded onto a 12% SDS gel. Precipitated proteins were then detected via Western blot analysis as described above. To investigate the ability of precipitated PfCCp proteins to bind to recombinant PfCCp adhesion domains, co-immunoprecipitation was performed as described using anti-PfCCp1 antibodies, and the precipitated complex was then incubated with purified recombinant PfCCp3rp3-GST or GST alone, both of which were prepurified with beads as described above, overnight at 4 °C (for expression and affinity purification; see below). The samples were washed five times with PBS and loaded onto 12% or 15% SDS gels. Precipitated proteins were detected via Western blot analysis as described above.
Affinity Chromatography Co-elution Binding Assay—Recombinant proteins were expressed as fusion proteins with either a GST tag using the pGEX-4T1 vector (Amersham Biosciences) or a His6/SUMO tag using the pSUMO vector as described (see Fig. 3B for domain specificity) (11). Recombinant proteins PfCCp1rp1, PfCCp1rp6, PfCCp3rp1, PfCCp3rp3, PfCCp3rp4, PfCCp4rp4, and Pf39 were tagged with GST and used as bait, whereas the recombinant proteins PfCCp1rp1, PfCCp1rp2, PfCCp1rp3, PfCCp1rp5, PfCCp2rp3, PfCCp2rp4, PfCCp3rp1, PfCCp3rp3, PfCCp5rp1, PfCCp5rp2, and PfFNPA-rp2 were expressed with a His6/SUMO tag and functioned as prey in the affinity chromatography co-elution binding experiments. The proteins were expressed in BL21 (DE3) RIL cells according to the manufacturer's protocol (Stratagene). The GST fusion protein expressing bacteria cells were lysed three times using a French press and 2 min via sonication. The lysed cells were centrifuged at 30,000 × g, and the supernatant was mixed with GST-Sepharose (Amersham Biosciences) and applied to a Poly-Prep chromatography column (Bio-Rad) at 4 °C. Supernatants containing His6-tagged potential interaction partners were loaded onto the GST fusion protein-bound column. The column was washed five times with PBS. The last washing step was investigated via Western blot analysis for the absence of GST-tagged and His6/SUMO-tagged proteins. The mutual binding abilities were investigated via co-elution, using a 50 mm Tris/HCl elution buffer, pH 8, containing 10 mm reduced glutathione. Via SDS-PAGE electrophoretic separation of eluates, the interaction partners were assayed by Western blot analysis using anti-His6 antibodies (GE Healthcare) and anti-GST antibodies (Amersham Biosciences). Co-elutions of recombinant GST tag protein with His6/SUMO tag protein or of Pf39 protein with PfCCp1rp1 protein were used as negative controls. For binding studies on endogenous PfCCp proteins, GST-tagged PfCCp3rp3 bait protein was applied to the columns. Gametocyte lysate was prepared as described for the co-immunoprecipitation assay, diluted with PBS, and loaded to the bait-bound column. Co-elution binding assays were performed as described above. The elutions were concentrated with Amicon ultra filter devices (Millipore) and analyzed for the presence of PfCCp3rp3-GST and endogenous PfCCp1 proteins by Western blotting. Blotting with antibodies against Pf39 was used as negative control.
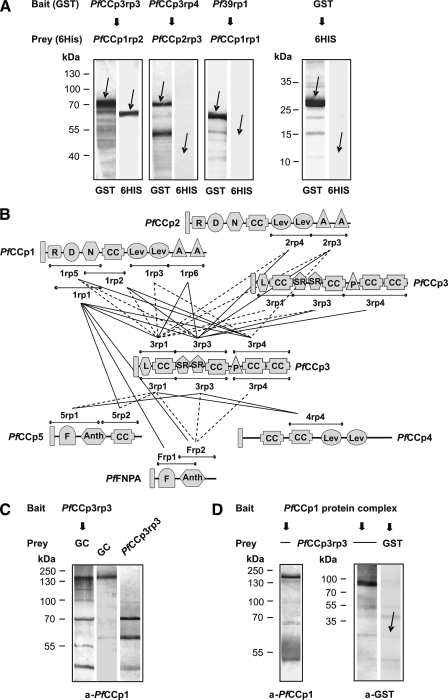
FIGURE 3.
PfCCp protein interactions through direct binding between distinct adhesion domains. A, examples of affinity chromatography co-elution binding assays using recombinant PfCCp proteins. Western blot analyses of eluted proteins, using anti-GST and anti-His6 antibodies, reveal an interaction between the two recombinant proteins PfCCp3rp3-GST and PfCCp1rp2-His6/SUMO and also show a co-elution assay with two proteins (PfCCp3rp4-GST and PfCCp2rp3-His6/SUMO), which did not interact. As negative control, bound GST-tagged Pf39 protein (termed Pf39rp1) did not interact with PfCCp1rp1-His6/SUMO protein. In another negative control, column-bound GST tag alone did not interact with His6/SUMO tag alone. The arrows either indicate the eluted recombinant protein or the approximate migration position expected for the eluted protein. B, schematic overview of all 33 implemented affinity chromatography co-elution binding assays. The lines below and above the domains represent all recombinant proteins tested. A number of 18 pairs of recombinant PfCCp proteins showed protein interaction (solid lines), whereas 15 protein pairs did not interact (dotted lines). A, apical A domain; Anth, anthrax toxin N-terminal region domain; D, discoidin domain; F, fibronectin type II domain; CC, Limulus coagulation factor C domain; Lev, levanase domain; L, lipoxygenase domain; N, neurexins and collagens domain; R, ricin domain; SR, scavenger receptor domain. C, affinity chromatography co-elution binding assay on endogenous PfCCp protein. Western blot analysis indicates binding of endogenous PfCCp1 of gametocyte lysate (GC) to the column-bound PfCCp3rp3-GST (left lane), as detected with anti-PfCCp1 antibodies. The protein band for the co-eluted PfCCp1 migrates with the same molecular mass as PfCCp1 in gametocyte lysate (center lane). Additional protein bands that are visible in lane 1 likely represent bacterial proteins of the PfCCp3rp3-expressing bacterial strain, because the anti-PfCCp1 serum would be expected to recognize contaminant bacterial proteins that were carried through the immunization regimen. Similar protein bands were visualized by the anti-PfCCp1 serum, when the bacterially expressed PfCCp3rp3 was loaded to the gel directly (right lane). D, double co-immunoprecipitation assay on recombinant PfCCp protein. The PfCCp multi-protein complex was firstly precipitated from gametocyte lysate by anti-PfCCp1 antibodies and then used for co-precipitation of PfCCp3rp3 protein. Western blot analyses show presence of endogenous PfCCp1 protein complex (left lane). Blotting with antibodies directed against the GST tag revealed that the PfCCp1 complex had bound to PfCCp3rp3 (center lane). No interaction was detected when the complex was incubated with GST alone (right lane). The arrow indicates the expected migration position of the GST protein.
Latex Bead-mediated Cell Binding Assay—For activation, 5 × 107 carboxylate-modified, yellow-green fluorescent polysterene latex beads with a diameter of 1 μm (Sigma-Aldrich) were incubated in 50 μl of 500 mm MES (2-(N-morpholino)-ethanesulfonic acid; Roth) pH 5–6, 40 μl of 100 mg/ml N-ethyl-N′-(3-dimethylaminopropyl)-carbodiimide hydrochloride (Sigma-Aldrich), and 410 μl of H2Obidest for 2 h at room temperature under rotation. After washing twice with 50 mm MES, the beads were conjugated with 5 μg of the recombinant proteins PfCCp3rp1 or PfCCp3rp3 (GST-tagged), PfCCp1rp1 or PfCCp2rp3 (His6/SUMO-tagged), or, as negative controls, with the GST-portion alone or the His6 fusion protein 5-helix (21) in 50 mm MES for 2 h at room temperature under rotation. Expression and purification of the recombinant proteins was conducted as described (see Fig. 3B for domain specificity) (11). To remove unbound proteins and block unspecific binding sites, the coated latex beads were washed twice with PBS and incubated overnight in 1% bovine serum albumin, 0.05% Tween 20 in PBS, pH 7.4, at 4 °C, followed by washing three times in PBS. The coated beads were mixed with either nonactivated gametocyte cultures or gametocyte cultures 15 min, 6 h, or 24 h post-activation or with PfCCp3-KO gametocytes 15 min post-activation, which were fixed in 4% paraformaldehyde overnight and washed three times in PBS. After incubation overnight at 4 °C, the number of macrogametes (15 min, 6 h, and 24 h post-activation), nonactivated gametocytes and erythrocytes that bound the PfCCp-coated latex beads were counted for 20 optical fields (400× magnification) using an Zeiss Axiophot microscope. This corresponds to a total number of ∼100 gametocytes or gametes and of ∼1000 erythrocytes. In total, two to four independent assays were performed, and the mean values were calculated. The respective cell types were distinguished because of their morphological appearance; mature (stage V) gametocytes were crescent-shaped, and macrogametes were round, contained multiple hemozoin particles, and were smaller and more translucent than erythrocytes. Binding of latex beads to macrogametes was subsequently verified by immunofluorescence assay using antibodies against Pfs25 in combination with Alexa Fluor 596-conjugated secondary antibodies.
RESULTS
Generation of Gene-disruptant Parasites—The PfCCp1 and PfFNPA gene loci were disrupted to complete the panel of targeted gene knock-outs of PfCCp family members. The PfCCp1 targeting disruption plasmids were introduced into the P. falciparum isolate NF54, and stable transfected parasites were selected with pyrimethamine and ganciclovir to drive plasmid integration by double cross-over homologous recombination (supplemental Fig. S1) (15). A pyrimethamine-resistant polyclonal line was established, and clonal lines were isolated by limiting dilution. Correct disruption of PfCCp1 was verified by diagnostic PCR (supplemental Fig. S1). Southern blot analysis of KpnI- and BamHI-digested genomic DNA from wild type and knock-out parasites showed correct disruption of the PfCCp1 locus and confirmed that double cross-over occurred (supplemental Fig. S1). PfCCp1 transcripts were not detected by RT-PCR in the gametocyte stages of disruptant parasites (supplemental Fig. S1), and the lack of PfCCp1 expression was confirmed by Western blot analysis of gametocyte protein extracts isolated from PfCCp1-KO parasites compared with a wild type control (Fig. 1A) and by immunofluorescence assay (supplemental Fig. S1). PfCCp1 gene-disruptant parasites were capable of propagation of asexual intraerythrocytic stages and production of gametocytes comparable with wild type, indicating that PfCCp1 is not essential for intraerythrocytic development. Moreover, gamete emergence and exflagellation of male gametes appeared normal in in vitro emergence assays.
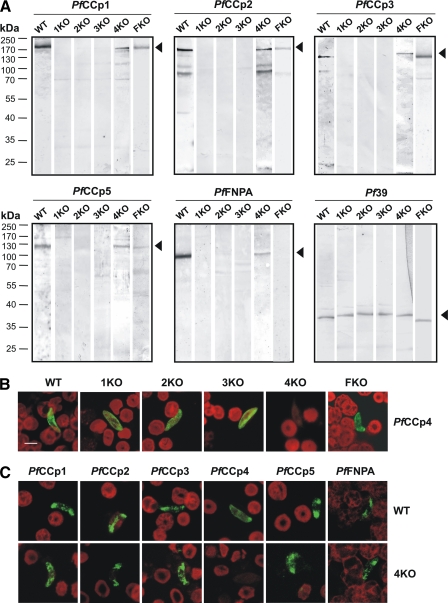
FIGURE 1.
Co-dependent expression of PfCCp proteins. A, Western blot analysis of gametocyte lysates from PfCCp1-KO, PfCCp2-KO, PfCCp3-KO, PfCCp4-KO, and PfFNPA-KO parasite lines using PfCCp-specific mouse antisera. PfCCp1, PfCCp2, PfCCp3, PfCCp5, and PfFNPA proteins are absent in the PfCCp1-KO, PfCCp2-KO, and PfCCp3-KO lines and expressed at a decreased protein level in PfCCp4-KO and PfFNPA-KO parasites. Protein bands migrate at the expected molecular masses of 185 kDa (PfCCp1, PfCCp2), 150 kDa (PfCCp3), 125 kDa (PfCCp5), and 100 kDa (PfFNPA), as indicated by arrowheads. An additional protein band at a molecular mass of 70 kDa was detectable for PfCCp2. Antibodies against the endoplasmic reticulum-associated protein Pf39 (39 kDa) were used as loading control. B, indirect immunofluorescence assay using PfCCp4 mouse antiserum demonstrated that PfCCp4 is expressed in PfCCp1-KO, PfCCp2-KO, PfCCp3-KO, and PfFNPA-KO gametocytes (green). Erythrocytes were counterstained with Evans Blue (red). C, indirect immunofluorescence assay using PfCCp-specific mouse antisera demonstrated that the proteins PfCCp1 through PfFNPA, with the exception of PfCCp4, are expressed in wild type as well as PfCCp4-KO gametocytes. PfCCp4 is not expressed in PfCCp4-KO gametocytes. 1KO, PfCCp1-KO; 2KO, PfCCp2-KO; 3KO, PfCCp3-KO; 4KO, PfCCp4-KO; FKO, PfFNPA-KO; WT, wild type. Bar, 5 μm.
Furthermore, we were able to disrupt the locus of PfFNPA by means of a pCAM-BSD targeting disruption plasmid, enabling the selection of stable transfectants with blasticidin, which lead to plasmid integration by single cross-over homologous recombination (supplemental Fig. S2). Disruption of the PfFNPA gene locus was verified for a clonal line via diagnostic PCR and Southern blot (supplemental Fig. S2). Absence of PfFNPA protein was shown by Western blot analysis of gametocyte protein extracts (Fig. 1A), as well as by immunofluorescence assay (supplemental Fig. S2), compared with protein expression in wild type gametocytes, respectively. PfFNPA gene-disruptant parasites were capable of propagation of asexual intraerythrocytic stages and production of gametocytes comparable with wild type, and gamete emergence and exflagellation appeared normal in in vitro emergence assays.
Co-dependent Expression of PfCCp Proteins—We have previously shown that protein but not transcript expression of PfCCp1 and PfCCp2 is abolished in PfCCp3 gene-disruptant lines and that PfCCp1 and PfCCp3 protein expression decreases in PfCCp2-KO clones (10). To determine whether co-dependent protein expression extends to other PfCCp proteins, we investigated gene-disruptant lines of the newly generated PfCCp1-KO and PfFNPA-KO (described above), as well as of PfCCp2-KO and PfCCp3-KO (7), and of PfCCp4-KO (11). Western blot analysis revealed that expression of PfCCp1, PfCCp2, PfCCp3, PfCCp5, and PfFNPA was abolished in the PfCCp1-KO, PfCCp2-KO, and PfCCp3-KO lines, whereas the respective PfCCp proteins were present at a decreased expression level in the PfCCp4-KO and the PfFNPA-KO (Fig. 1A). Screening with antibodies against the endoplasmic reticulum-associated protein, Pf39, was used as a control for equal loading. Because of the low efficiency of the PfCCp4 antibody in Western blot analysis, co-dependent expression for this protein was investigated by immunofluorescence assay and revealed that PfCCp4 remains homogeneously expressed associated to the surface of PfCCp1-KO, PfCCp2-KO, PfCCp3-KO, and PfFNPA-KO gametocytes (Fig. 1B). Immunofluorescence assays further revealed that all PfCCp proteins are present on the surface of PfCCp4-KO gametocytes (Fig. 1C), indicating that none of the five proteins are targeted to other compartments.
To confirm that the lack of PfCCp expression manifests at the protein but not the transcript level, mRNA expression was investigated in the respective gene-disruptant gametocytes using RT-PCR. All of the PfCCp transcripts were present in the PfCCp1-KO, PfCCp2-KO, and PfCCp3-KO parasites (supplemental Fig. S3). In the PfCCp2-KO and PfCCp3-KO, transcript was also present for the respective disrupted genes, as described (10). These gene-disruptant parasite lines were generated by single cross-over homologous recombination of the vector pDT-Tg23, and transcript expression is caused by nonspecific promoter activity of the integrated vector. In contrast, the PfCCp1-KO clone was generated using vector pHHT-TK via double cross-over homologous recombination (see above), and no transcript expression was observed for PfCCp1. Our data indicate that the six PfCCp family members are co-dependently expressed in gametocytes on the protein but not transcript level and that the lack of one of these proteins leads to either partial or to complete loss of the other PfCCp proteins.
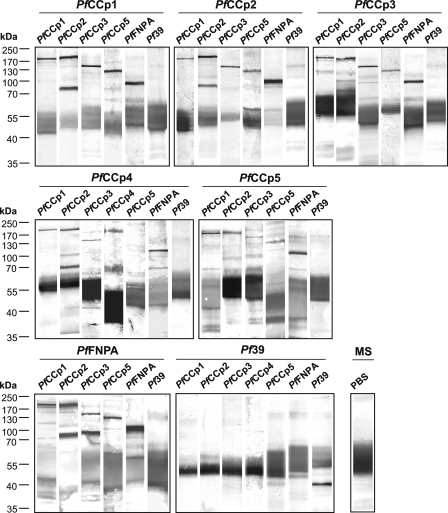
Molecular Interactions between PfCCp Proteins—Co-dependent expression of the PfCCp proteins suggested protein-protein interactions, in agreement with the previously described co-localization in the parasitophorous vacuole (7, 10, 11). To investigate possible molecular interactions, we initially performed co-immunoprecipitation assays on gametocyte lysates, as described (11). Prominent protein bands for PfCCp1, PfCCp2, PfCCp3, PfCCp5, and PfFNPA were detected by Western blot analysis of gametocyte lysates, when these were individually precipitated with antibodies against either one of these proteins (Fig. 2). Because of the above mentioned low efficiency of the PfCCp4 antibody in Western blot analysis, interactions of PfCCp4 with other PfCCp proteins were detected by precipitating with the respective PfCCp4 antibody and showed binding of the protein to PfCCp1, PfCCp2, PfCCp3, and PfFNPA (Fig. 2). The precipitated proteins migrated at the expected molecular masses of 185 kDa (PfCCp1 and PfCCp2), 150 kDa (PfCCp3), 180 kDa (PfCCp4), 125 kDa (PfCCp5), and 100 kDa (PfFNPA). For PfCCp2 and PfCCp4, second protein bands with molecular masses of ∼70 and 60 kDa were detected, respectively, as has been described (11). For PfCCp3, in some occasions a second protein band, running at a molecular mass of ∼90 kDa, was detected, which might be due to processing of the protein. No PfCCp bands were detectable when lysates were precipitated with antibodies against Pf39 (Fig. 2). An additional negative control was performed by screening with antibodies against Pf39 after precipitation with each of the six PfCCp antibodies, and no protein bands were detected (Fig. 2). Furthermore, anti-PfCCp1 mouse serum was used for precipitation in PBS instead of gametocyte lysate, and the precipitate was screened by Western blot analysis with goat anti-mouse secondary antibody only to visualize precipitating antibody. The Western blot resulted in a smeared protein band near 55 kDa, which was also observed in all other lanes (Fig. 2). The results obtained from co-immunoprecipitation assays reveal multiple interactions between the six PfCCp proteins and point to the formation of a multi-protein complex in gametocytes.
FIGURE 2.
Multi-protein complex formation of the six PfCCp proteins in gametocytes. Interactions between PfCCp proteins were determined by co-immunoprecipitation assays with wild type gametocyte lysates precipitated with mouse antibodies against PfCCp1 through PfFNPA (indicated above the horizontal line of each blot), followed by Western blot analysis using the same antibodies individually (indicated above each lane). The protein bands migrate at the expected molecular masses of 185 kDa (PfCCp1, PfCCp2), 150 kDa (PfCCp3), 180 kDa (PfCCp4), 125 kDa (PfCCp5), and 100 kDa (PfFNPA). The additional protein bands at 70, 90, and 60 kDa were detected for PfCCp2, PfCCp3, and PfCCp4, respectively. Antibodies directed against the endoplasmic reticulum-specific protein Pf39 (39 kDa) were used as a negative control and revealed no binding. Also, no PfCCp protein bands were detected in gametocyte lysates precipitated with antibodies against Pf39. As a control, mouse serum (MS) was used for precipitation, and the precipitate was blotted with PBS and the alkaline phosphatase-conjugated goat anti-mouse secondary antibody to visualize precipitating antibody, resulting in a smeared protein band at ∼55 kDa.
To investigate the molecular interactions between the PfCCp proteins in more detail, affinity chromatography co-elution binding assays were performed. Recombinant proteins corresponding to distinct adhesion domains were expressed for each of the six proteins (for domain regions, see Fig. 3B). Recombinant PfCCp3 adhesion domains, fused to a GST tag, were immobilized to Sepharose and used as baits. Other recombinant PfCCp adhesion domains, fused to a His6/SUMO tag, functioned as preys and were incubated with the Sepharose-bound PfCCp3 domains. Bound proteins were eluted after several washing steps. The eluted protein complexes were screened by Western blot analyses, in which the GST-tagged and His6/SUMO-tagged proteins were detected via antibodies directed against the respective tags. In an additional set of experiments, GST-tagged PfCCp1rp1 was used as a bait protein and investigated for its interaction with His6/SUMO-tagged PfCCp5 and PfFNPA recombinant adhesion domains. Fig. 3A shows a Western blot assay representative of a positive interaction between the recombinant proteins PfCCp3rp3-GST and PfCCp1rp2-His6/SUMO and also gives an example for a co-elution binding assay between two proteins (PfCCp3rp4-GST and PfCCp2rp3-His6/SUMO) that did not interact. During each interaction experiment, a sample of the last washing step was investigated for the presence of prey or bait proteins and shown to be negative (data not shown). In some cases, lower molecular mass protein bands of GST fusion proteins were detected in addition to the expected full size molecular mass recombinant protein. These protein bands are likely due to contamination of the recombinant protein with truncated recombinant products. In a set of negative controls, GST tag alone was immobilized to Sepharose, and the His6/SUMO tag alone was used as prey. In another negative control, GST-tagged Pf39 protein (termed Pf39rp1) was immobilized to Sepharose, and the PfCCp1rp1-His6/SUMO protein was used as prey. In both control experiments, no interactions were detectable (Fig. 3A).
In total we investigated interactions between 33 combinations of recombinant PfCCp proteins. Of these pairs, 18 showed adhesive interaction, whereas 15 recombinant protein pairs did not interact in affinity chromatography co-elution binding assays (Fig. 3B). Adhesion domains that were predominant in protein binding interactions include the LCCL domains of all PfCCp proteins and the scavenger receptor (SR) domains of PfCCp3. The two domains were involved in 32% (LCCL) and 19% (SR) of all binding events. Moreover, the apical A, neurexin, and discoidin domains of PfCCp1 and PfCCp2 might be involved in these interactions.
We additionally examined the ability of recombinant PfCCp domains to bind endogenous PfCCp. In a first set of experiments, affinity chromatography co-elution binding assays were performed using GST-tagged PfCCp3rp3 as bait protein, which was incubated with gametocyte lysate. Western blot analysis on co-eluted samples using anti-PfCCp1 sera revealed binding between recombinant PfCCp3rp3 and endogenous PfCCp1 (Fig. 3C). For control, the samples were blotted with antibodies against Pf39, and the respective blot was negative (data not shown). In a vice versa approach we examined the capability of the recombinant PfCCp3rp3 to bind to the endogenous multiprotein complex. The protein complex, which was precipitated from gametocyte lysate via co-immunoprecipitation using anti-PfCCp1 antibodies, was used as bait and incubated with purified PfCCp3rp3-GST. A prominent PfCCp3rp3 protein band was detected by Western blot analysis using anti-GST antibodies (Fig. 3D). As negative control, the multi-protein complex was incubated with GST alone, and no interactions were detected (Fig. 3D).
Taken together, we demonstrated binding between endogenous PfCCp proteins by co-immunoprecipitation assays, and by co-elution binding assays we further showed binding between recombinant PfCCp proteins and of recombinant PfCCp to endogenous PfCCp protein. In light of our interaction data we conclude that the six PfCCp proteins form multi-protein complexes in the gametocyte parasitophorous vacuole by direct binding through distinct adhesion domains.
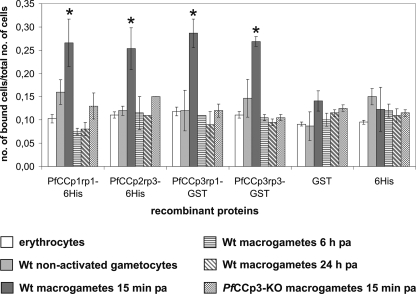
Cell Binding Properties of PfCCp Proteins—In a concluding set of experiments, we wanted to investigate whether the adhesive properties of PfCCp proteins can be observed at the cellular level. Cell binding assays were performed on exflagellating P. falciparum cultures, using PfCCp-coated fluorescent latex beads. The recombinant proteins PfCCp1rp1-His6/SUMO, PfCCp2rp3-His6/SUMO, PfCCp3rp1-GST, and PfCCp3rp3-GST (for domain regions, see Fig. 3B) were bound to latex beads, followed by incubation with paraformaldehyde-fixed activated or nonactivated gametocyte cultures (supplemental Fig. S4). The number of macrogametes, gametocytes, and erythrocytes that bound the PfCCp-coated latex beads were counted for 20 optical fields and compared with the total number of each of the investigated cell types. The cell binding assays revealed that the majority of latex beads adhered to macrogametes 15 min post-activation (supplemental Fig. S4). Approximately 25% of macrogametes bound to the beads (Fig. 4), with an average number of two beads/macrogamete. The binding of the beads to nonactivated gametocytes, macrogametes 6 or 24 h post-activation, and erythrocytes was significantly lower with only ∼10% of cells having bound to latex beads. Similarly, there was no significant binding of the PfCCp-coated latex beads to macrogametes of PfCCp3-KO parasites (Fig. 4). These gene-disruptant parasites do not express any of the PfCCp proteins except for PfCCp4 (see above). Binding of latex beads to microgametes was never observed. As negative controls, the beads were covered with GST tag protein or the His6-tagged recombinant protein 5-Helix (21) and bound to ∼10% of cells (Fig. 4). In summary, the recombinant PfCCp proteins bind to the surface of newly emerged PfCCp-expressing macrogametes, and these data provide the first evidence for the adhesive properties of macrogametes caused by PfCCp protein interactions.
FIGURE 4.
Binding of PfCCp proteins to macrogametes. The interaction of latex beads coated with the recombinant proteins PfCCp1rp1-His6/SUMO, PfCCp2rp3-His6/SUMO, PfCCp3rp1-GST, or PfCCp3rp3-GST with different cell types during exflagellation of gametocyte cultures was investigated by cell binding assay. The binding of PfCCp-coated latex beads to macrogametes 15 min post-activation is significantly increased compared with the binding of latex beads to uninfected erythrocytes, nonactivated gametocytes, and macrogametes 6 or 24 h post-activation (asterisk, Students t test, p < 0.05). Also, PfCCp-coated latex beads did not exhibit any specific binding to PfCCp3-KO macrogametes 15 min post-activation. No specific binding of latex beads coated with the control recombinant proteins GST tag alone or His6-5 helix to any of the investigated cell types was detectable. Wt, wild type.
DISCUSSION
During the last two decades a substantial number of proteins has been identified that are expressed in the sexual and mosquito stages of malaria parasites. The majority of these sexual stage proteins are initially expressed in the parasitophorous vacuole during gametocyte differentiation, and some are later exposed on the surface of emerged gametes and fertilized zygotes. Surface-associated proteins include Pfs25 and Pfs28, Pfs48/45 and Pfs47, Pfs230 and PfMR5, PfPeg3 and PfPeg4, and Pfg14.744 and Pfg14.748, as well as the six PfCCp proteins (2). Despite ongoing characterizations for most of the identified proteins, the functional basis for their expression within the parasitophorous vacuole of gametocytes, their exposure during emergence, and the role of the numerous adhesive motifs of these proteins remain unclear.
The PfCCp protein family represents prominent members of sexual stage proteins with their expression lasting for ∼10 days. The proteins are expressed in early gametocytes during differentiation and are later present associated to the surface of macrogametes (11). For PfCCp1, PfCCp2, and PfCCp3, a partial release of protein during egress from the erythrocyte was reported, and the proteins later relocate surrounding exflagellation centers (7). The genes PfCCp2–PfCCp4 have been disrupted for functional characterization via single cross-over homologous recombination (7, 11). Surprisingly, gene-disruptant parasites revealed normal exflagellation and fertilization behavior, and oocysts developed in the midgut; however, in PfCCp2-KO and PfCCp3-KO lines, midgut sporozoite transition to the salivary glands was blocked (7). In contrast, PfCCp4-KO sporozoites were later present in the glands, leading to the assumption that PfCCp4 has a nonessential function for the malaria parasite (11). The striking difference in the predominant gametocyte-specific expression of the PfCCp proteins and the knock-out phenotype of some of the proteins during sporozoite formation hitherto is not known. We hypothesize that the molecular interactions of extracellularly exposed PfCCp proteins might mediate signaling events during fertilization that ultimately lead to sporozoite formation. The phenotypes for the newly generated PfCCp1-KO and PfFNPA-KO lines during mosquito transmission are currently under investigation.
The six proteins have also been studied in the rodent P. berghei system, where they are termed LAP proteins (9, 22). Transcript analysis revealed a predominant expression in P. berghei gametocytes for the six proteins, which was confirmed by transgenic parasites expressing green fluorescent protein driven by select PbCCp/LAP promoters (23). For functional characterization, loss of function mutants were generated for PbCCp1/LAP2, PbCCp2/LAP4, PbCCp3/LAP1, and PbCCp4/LAP6 (22) as well as two double knock-outs, PbCCp1/CCp3 and PbCCp1/CCp4 (23). For all mutants, the formation of sporozoites in the midgut oocysts of parasite-infected mosquitoes was aborted (22, 23). Interestingly, sporulation of PbCCp3/LAP1 (termed PbSR in this study) appears to be normal in in vitro assays, pointing to the involvement of mosquito factors in the loss-of-function phenotype of this protein (24). Genetic cross studies with female-deficient or male-deficient parasites further indicated that the functional genes are inherited from female gametocytes (22). Green fluorescent protein fusions of PbCCp3/LAP1 (PbSR) showed protein expression in macrogametocytes and accumulation of these proteins in the ookinete crystalloid, suggesting a role in protein trafficking (24). However, no antibody-based protein expression analysis for the PbCCp/LAP proteins has so far been performed in P. berghei to support these results. In P. falciparum, PfCCp expression in the ookinete crystalloid was never observed (7, 11).3
We recently reported that PfCCp1, PfCCp2, and PfCCp3 proteins are co-dependently expressed and that the abrogation of PfCCp3 in gene-disruptant gametocytes leads to the lack of PfCCp1 and PfCCp2 (10). At that time we hypothesized that the three proteins interact to form a protein complex. In the present study we address this hypothesis and described in detail the molecular interactions between the six members of the PfCCp protein family. First, we showed that all PfCCp proteins are co-dependently expressed and that the lack of one of the proteins leads to partial or complete loss of the other family members. We subsequently revealed that the six endogenous PfCCp proteins interact by forming protein complexes that were co-precipitated using a variety of PfCCp antibodies. Protein interactions were confirmed by direct protein binding of distinct adhesion domains using bacterially expressed recombinant proteins. This is the first time that such complexes involving interactions of multiple adhesive proteins are described for the sexual stages of malaria parasites.
The PfCCp proteins are comprised of multiple adhesive domains, whose remarkable architectures are conserved throughout the apicomplexan clade (8). The complex structure of these proteins is suggestive of roles in protein, lipid, and carbohydrate interactions. Affinity chromatography co-elution binding assays performed in this study indicate that particularly the two SR domains of PfCCp3 as well as the LCCL domains are involved in protein binding. The SR domain, present in a variety of metazoan organisms such as marine sponges and mammals (25), has been reported to be involved in protein-protein binding (26). In vertebrates, the SR domain is found in receptors on the surface of immune cells and in secreted proteins that are involved in host defense (25, 27, 28), regulation of the complement cascade (29), or binding and phagocytosis of invading pathogens (30). The LCCL domain, however, was so far only thought to be involved in binding of lipopolysaccharides (31, 32). The domain was first described in Limulus factor C, the zymogene of a secreted serine protease in the horseshoe crab (33), and was later found in the human cochlear protein Coch-5b2 (34) and the late gestation lung protein Lgl1 of rats (35). An involvement of the SR and LCCL domains in immune evasion mechanisms of malaria parasites was discussed in previous studies (4, 6, 36).
Recently, another protein interaction between two sexual stage proteins, namely PfCCp4 and Pfs230, has been identified (11). Pfs230 is present on the surface of gametocytes and gametes and mediates binding of microgametes to uninfected red blood cells during exflagellation (37). The secreted Pfs230 protein is associated to the gametocyte plasma membrane by binding to the glycosylphosphatidylinositol-anchored protein Pfs48/45 within the parasitophorous vacuole (38, 39). It is possible that PfCCp4 is also involved in this binding and thereby mediates the association of all PfCCp proteins to the plasma membrane. However, surface-associated expression of the other PfCCp proteins is not dependent on PfCCp4, indicating that the PfCCp complex would be linked to the gametocyte by additional surface proteins.
In a concluding set of experiments we investigated whether the exposed PfCCp proteins are involved in cellular interactions. Using PfCCp-coated latex beads, we showed that PfCCp1, PfCCp2, and PfCCp3 specifically bind to newly formed macrogametes but not to macrogametes and zygotes 6 and 24 h post-activation. Furthermore, the binding of macrogametes is dependent on the presence of PfCCp proteins, as shown by adhesion studies on PfCCp3-KO parasites. In accord with these results, only the female macrogametes but not the male microgametes or zygotes expose PfCCp proteins on their surface, and expression decreases within a few hours after emergence (11). We therefore conclude that the recombinant PfCCp proteins bind to endogenous PfCCp proteins that are exposed on the surface of the newly emerged macrogametes. Although not all macrogametes bind the PfCCp-coated latex beads, this might be due to the fact that the bacterially expressed recombinant proteins do not exhibit all adhesion domains of the respective full-length proteins and that they might also be subject to misfolding during expression, thus not exhibiting the full adhesive functions of the endogenous proteins.
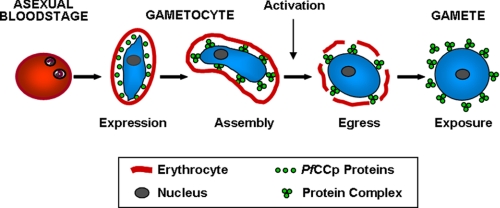
In summary, the data presented here describe molecular interactions of sexual stage adhesion proteins. Our findings support the hypothesis that the six PfCCp proteins are components of a multi-protein complex, which is involved in cell adhesion events during gametogenesis and fertilization. We propose that the proteins are expressed in the parasitophorous vacuole of the differentiating gametocyte, where the complexes are initially assembled (Fig. 5). During the blood meal of the female mosquito, the gametocytes enter the mosquito midgut and become activated because of a shift in temperature and the presence of the mosquito-derived molecule xanthurenic acid (2, 13, 14). After emergence of the activated gametocyte from the erythrocyte, the multi-protein complexes are exposed on the macrogamete surface, where they might be involved in adhesive interactions. It is tempting to speculate that the complexes mediate cell-cell contacts of macrogametes, e.g. binding of macrogametes to microgametes caused by interaction of select PfCCp proteins with Pfs230 or Pfs48/45 on the microgamete surface. In this context, a recent study described the identification of the microgamete protein HAP2, which enables fertilization in P. berghei (40). Interestingly, HAP2 is essential for gamete fusion but not for the initial binding between the two mating partners, which appears to involve other adhesion proteins. Alternatively, the adhesive properties of the PfCCp multiprotein complexes might enable the macrogamete to bind factors of the blood meal, e.g. factors of the human complement system, thus forming a protective shield that represents a barrier between the newly exposed gametes and the aggressive environment of the mosquito midgut. The possible involvement of other sexual stage adhesive proteins in the formation of the multi-protein complexes described here is currently being investigated in our laboratory.
FIGURE 5.
Formation and exposure of PfCCp multi-protein complexes. The six PfCCp proteins are expressed in the gametocyte parasitophorous vacuole associated to the plasma membrane, where they assemble to multiprotein complexes. During activation, the gametocyte egresses from the erythrocyte, and the multi-protein complexes are subsequently exposed on the macrogamete surface.
Supplementary Material
Acknowledgments
We thank Dr. Catherine Lavazec for making the PfCCp1 disruption construct, Ludmilla Sologub for technical assistance, and Andres Abreu and Roland Frank for assisting with the construction of the PfCCp expression vectors and with the affinity chromatography co-elution binding assays, respectively. We further thank David Fidock, Jean Halbert, and Christian Doerig for the opportunity to use the pCAM-BSD vector. The His6 fusion protein 5-helix was kindly provided by Marc Kirschner.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 A1054580-01A1 (to T. J. T.). This work was also supported by an Emmy Noether grant and an SFB479 grant from the Deutsche Forschungsgemeinschaft (to G. P.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1–S4.
Footnotes
The abbreviations used are: LCCL, Limulus coagulation factor C; CCp, LCCL domain-containing protein; GST, glutathione S-transferase; KO, knock-out; LAP, LCCL-lectin adhesive-like protein; PBS, phosphate-buffered saline; RT, reverse transcription; SR, scavenger receptor; Pf, P. falciparum; SUMO, small ubiquitin-like modifier; MES, 4-morpholineethanesulfonic acid.
N. Simon, S. M. Scholz, C. K. Moreira, T. J. Templeton, A. Kuehn, M.-A. Dude, and G. Pradel, unpublished observations.
References
- 1.Alano, P. (2007) Mol. Microbiol. 66 291-302 [DOI] [PubMed] [Google Scholar]
- 2.Pradel, G. (2007) Parasitology 134 1911-1929 [DOI] [PubMed] [Google Scholar]
- 3.Saul, A. (2007) Curr. Opin. Infect. Dis. 20 476-481 [DOI] [PubMed] [Google Scholar]
- 4.Delrieu, I., Waller, C. C., Mota, M. M., Grainger, M., Langhorne, J., and Holder, A. A. (2002) Mol. Biochem. Parasitol. 121 11-20 [DOI] [PubMed] [Google Scholar]
- 5.Lasonder, E., Ishihama, Y., Andersen, J. S., Vermunt, A. M., Pain, A., Sauerwein, R. W., Eling, W. M., Hall, N., Waters, A. P., Stunnenberg, H. G., and Mann, M. (2002) Nature 419 537-542 [DOI] [PubMed] [Google Scholar]
- 6.Dessens, J. T., Sinden, R. E., and Claudianos, C. (2004) Trends Parasitol. 20 102-108 [DOI] [PubMed] [Google Scholar]
- 7.Pradel, G., Hayton, K., Aravind, L., Iyer, L., Abrahamsen, M. S., Bonawitz, A., Mejia, C., and Templeton, T. J. (2004) J. Exp. Med. 199 1533-1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Templeton, T. J., Iyer, L. M., Anantharaman, V., Enomoto, S., Abrahante, J. E., Subramanian, G. M., Hoffman, S. L., Abrahamsen, M. S., and Aravind, L. (2004) Genome Res. 14 1686-1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trueman, H. E., Raine, J. D., Florens, L., Dessens, J. T., Mendoza, J., Johnson, J., Waller, C. C., Delrieu, I., Holders, A. A., Langhorne, J., Carucci, D. J., Yates, J. R. III, and Sinden, R. E. (2004) J. Parasitol. 90 1062-1071 [DOI] [PubMed] [Google Scholar]
- 10.Pradel, G., Wagner, C., Mejia, C., and Templeton, T. J. (2006) 112 263-268 [DOI] [PubMed]
- 11.Scholz, S. M., Simon, N., Lavazec, C., Dude, M. A., Templeton, T. J., and Pradel, G. (2008) Int. J. Parasitol. 38 327-340 [DOI] [PubMed] [Google Scholar]
- 12.Ifediba, T., and Vanderberg, J. P. (1981) Nature 294 364-366 [DOI] [PubMed] [Google Scholar]
- 13.Billker, O., Lindo, V., Panico, M., Etienne, A. E., Paxton, T., Dell, A., Rogers, M., Sinden, R. E., and Morris, H. R. (1998) Nature 392 289-292 [DOI] [PubMed] [Google Scholar]
- 14.Garcia, G. E., Wirtz, R. A., Barr, J. R., Woolfitt, A., and Rosenberg, R. (1998) J. Biol. Chem. 273 12003-12005 [DOI] [PubMed] [Google Scholar]
- 15.Duraisingh, M. T., Triglia, T., and Cowman, A. F. (2002) Int. J. Parasitol. 32 81-89 [DOI] [PubMed] [Google Scholar]
- 16.Deitsch, K. W., Driskill, C. L., and Wellems, T. E. (2001) Nucleic Acids Res. 29 850-853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sidhu, A. B., Valderramos, S. G., and Fidock, D. A. (2005) Mol. Microbiol. 57 913-926 [DOI] [PubMed] [Google Scholar]
- 18.Dorin-Semblat, D., Quashie, N., Halbert, J., Sicard, A., Doerig, C., Peat, E., Ranford-Cartwright, L., and Doerig, C. (2007) Mol. Microbiol. 65 1170-1180 [DOI] [PubMed] [Google Scholar]
- 19.Templeton, T. J., Fujioka, H., Aikawa, M., Parker, K. C., and Kaslow, D. C. (1997) Mol. Biochem. Parasitol. 90 359-365 [DOI] [PubMed] [Google Scholar]
- 20.Kariuki, M. M., Kiaira, J. K., Mulaa, F. K., Mwangi, J. K., Wasunna, M. K., and Martin, S. K. (1998) Am. J. Trop. Med. Hyg. 59 505-508 [DOI] [PubMed] [Google Scholar]
- 21.Koshiba, T., and Chan, D. C. (2003) J. Biol. Chem. 278 7573-7579 [DOI] [PubMed] [Google Scholar]
- 22.Raine, J. D., Ecker, A., Mendoza, J., Tewari, R., Stanway, R. R., and Sinden, R. E. (2007) PLoS Pathog. 3 e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavazec, C., Moreira, C. K., Mair, G. R., Waters, A. P., Janse, C. J., and Templeton, T. J. (2009) Mol. Biochem. Parasitol. 163 1-7 [DOI] [PubMed] [Google Scholar]
- 24.Carter, V., Shimizu, S., Arai, M., and Dessens, J. T. (2008) Mol. Microbiol. 68 1560-1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aruffo, A., Bowen, M. A., Patel, D. D., Haynes, B. F., Starling, G. C., Gebe, J. A., and Bajorath, J. (1997) Immunol. Today 18 498-504 [DOI] [PubMed] [Google Scholar]
- 26.Whitney, G. S., Starling, G. C., Bowen, M. A., Modrell, B., Siadak, A. W., and Aruffo, A. (1995) J. Biol. Chem. 270 18187-18190 [DOI] [PubMed] [Google Scholar]
- 27.Resnick, D., Pearson, A., and Krieger, M. (1994) Trends Biochem. Sci. 19 5-8 [DOI] [PubMed] [Google Scholar]
- 28.Yu, Q., Reichert, M., Brousseau, T., Cleuter, Y., Burny, A., and Kettmann, R. (1990) Nucleic Acids Res. 18 5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldberger, G., Bruns, G. A., Rits, M., Edge, M. D., and Kwiatkowski, D. J. (1987) J. Biol. Chem. 262 10065-10071 [PubMed] [Google Scholar]
- 30.Elomaa, O., Sankala, M., Pikkarainen, T., Bergmann, U., Tuuttila, A., Raatikainen-Ahokas, A., Sariola, H., and Tryggvason, K. (1998) J. Biol. Chem. 273 4530-4538 [DOI] [PubMed] [Google Scholar]
- 31.Trexler, M., Banyai, L., and Patthy, L. (2000) Eur. J. Biochem. 267 5751-5757 [DOI] [PubMed] [Google Scholar]
- 32.Liepinsh, E., Trexler, M., Kaikkonen, A., Weigelt, J., Banyai, L., Patthy, L., and Otting, G. (2001) EMBO J. 20 5347-5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muta, T., Miyata, T., Misumi, Y., Tokunaga, F., Nakamura, T., Toh, Y., Ikehara, Y., and Iwanaga, S. (1991) 266 6554-65561 [PubMed]
- 34.Robertson, N. G., Skvorak, A. B., Yin, Y., Weremowicz, S., Johnson, K. R., Kovatch, K. A., Battey, J. F., Bieber, F. R., and Morton, C. C. (1997) Genomics 46 345-354 [DOI] [PubMed] [Google Scholar]
- 35.Kaplan, F., Ledoux, P., Kassamali, F. Q., Gagnon, S., Post, M., Koehler, D., Deimling, J., and Sweezey, N. B. (1999) Am. J. Physiol. 276 1027-1036 [DOI] [PubMed] [Google Scholar]
- 36.Claudianos, C., Dessens, J. T., Trueman, H. E., Arai, M., Mendoza, J., Butcher, G. A., Crompton, T., and Sinden, R. E. (2002) Mol. Microbiol. 45 1473-1484 [DOI] [PubMed] [Google Scholar]
- 37.Eksi, S., Czesny, B., van Gemert, G. J., Sauerwein, R. W., Eling, W., and Williamson, K. C. (2006) Mol. Microbiol. 61 991-998 [DOI] [PubMed] [Google Scholar]
- 38.Kumar, N. (1987) Parasite Immunol. 9 321-335 [DOI] [PubMed] [Google Scholar]
- 39.Kumar, N., and Wizel, B. (1992) Mol Biochem. Parasitol. 53 113-120 [DOI] [PubMed] [Google Scholar]
- 40.Liu, Y., Tewari, R., Ning, J., Blagborough, A. M., Garbom, S., Pei, J., Grishin, N. V., Steele, R. E., Sinden, R. E., Snell, W. J., and Billker, O. (2008) Genes Dev. 22 1051-1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.