Abstract
The production of erythrocytes requires the massive synthesis of red cell-specific proteins including hemoglobin, cytoskeletal proteins, as well as membrane glycoproteins glycophorin A (GPA) and anion exchanger 1 (AE1). We found that during the terminal differentiation of human CD34+ erythroid progenitor cells in culture, key components of the endoplasmic reticulum (ER) protein translocation (Sec61α), glycosylation (OST48), and protein folding machinery, chaperones BiP, calreticulin (CRT), and Hsp90 were maintained to allow efficient red cell glycoprotein biosynthesis. Unexpected was the loss of calnexin (CNX), an ER glycoprotein chaperone, and ERp57, a protein-disulfide isomerase, as well as a major decrease of the cytosolic chaperones, Hsc70 and Hsp70, components normally involved in membrane glycoprotein folding and quality control. AE1 can traffic to the cell surface in mouse embryonic fibroblasts completely deficient in CNX or CRT, whereas disruption of the CNX/CRT-glycoprotein interactions in human K562 cells using castanospermine did not affect the cell-surface levels of endogenous GPA or expressed AE1. These results demonstrate that CNX and ERp57 are not required for major glycoprotein biosynthesis during red cell development, in contrast to their role in glycoprotein folding and quality control in other cells.
The production of red blood cells involves the terminal differentiation of hematopoietic stem cells in the bone marrow followed by release into the peripheral blood (1, 2). Red blood cells remain in circulation for ∼120 days and require the prior production of abundant red cell-specific proteins including hemoglobin, cytoskeletal proteins, and membrane glycoproteins such as anion exchanger 1 (AE1)3 and glycophorin A (GPA). During differentiation, erythroid progenitor cells undergo extensive remodeling of their cytoskeleton and loss of nuclei and other organelles like the endoplasmic reticulum (ER). AE1 and GPA are known to be synthesized late in differentiation when these key cellular components are lost (3). The efficient biosynthesis of these red cell membrane glycoproteins, however, is expected to require robust ER assembly machinery involving protein translocation, N-glycosylation, and protein folding chaperones.
The proper folding of membrane glycoproteins engages the quality control function of cytosolic and ER chaperone proteins (4, 5). Newly synthesized proteins undergo cycles of binding and release with chaperones, minimizing aggregation and facilitating folding. Chaperones also play a role in the retention and degradation of misfolded proteins and in apoptosis (6-8). The membrane-bound ER chaperone calnexin (CNX) and its luminal paralog calreticulin (CRT) interact with folding intermediates via their lectin and protein binding domains, thereby preventing aggregation (9). A wide variety of glycoprotein substrates have been identified, with some binding to one or both chaperones, and both have been shown to be vital in the prevention of aggregation and proper maturation of membrane glycoproteins (9, 10). Disruption of interactions with CNX and CRT can allow misfolded membrane glycoproteins to escape the ER and traffic to the plasma membrane (9).
In the present study, we examined the integrity of the ER protein translocation, N-glycosylation, and quality control machinery during the differentiation of human CD34+ erythroid cells in culture. We found that specific components of the protein quality control system were completely lost (CNX and ERp57) or diminished (Hsc70 and Hsp70) before the production of the major glycoproteins, AE1 and GPA, was completed. Components of the protein translocation (Sec61α) and N-glycosylation machinery (OST48) were, however, maintained. Chaperones that play other roles in erythrocyte maturation and survival (CRT, BiP, and Hsp90) were also retained (11). AE1 was found to traffic efficiently to the plasma membrane in mouse embryonic fibroblasts completely lacking the ER chaperone CNX or CRT. Furthermore, disruption of CNX/CRT-glycoprotein interactions in human K562 cells did not affect the cell-surface expression of GPA or AE1. These results demonstrate that CNX and ERp57 are not required for the efficient synthesis and folding of red cell membrane glycoproteins during terminal erythropoiesis. The lack of engagement with the quality control and disulfide folding machinery may allow the more rapid production of red cell glycoproteins late in differentiation, sacrificing quality for quantity.
EXPERIMENTAL PROCEDURES
Antibodies—The primary antibodies used include mouse monoclonal anti-AE1 (IVF12) directed against the membrane domain of AE1, a kind gift from Dr. Michael Jennings, a polyclonal rabbit anti-CNX directed against the luminal domain (12), and the mouse monoclonals AE1/CD233/BRIC6 (9439), GPA/CD235a/BRIC163 (9410), and GPA/BRIC256 (9415) from the International Blood Group Reference Laboratory. Commercial antibodies include CNX (SPA-865), CRT (SPA-600), Hsc70 (SPA-815), Hsp70 (SPA-810), Hsp90 (SPA-830), and ER protein-disulfide isomerase (PDI; SPA-891) from Stressgen Laboratories; ERp57 (sc-23886), Hsp27 (sc-13132), Hsc70 (sc-7298), OST48 (sc-255558) from Santa Cruz Biotechnology; actin (MAB1501), glyceraldehyde-3-phosphate dehydrogenase (MAB374), carbonic anhydrase II (MAB1828), and calpain (MAB3083) from Chemicon; BiP/Grp78 (610978) from BD Transduction; hemagglutinin (HA) (MMS-101R) from Covance, Inc.; and Sec61α (07-204) from Upstate. Conjugated antibodies include CD71-fluorescein isothiocyanate (555536) and GPA/CD235a-phycoerythrin (555570) from Pharmingen. Secondary antibodies include SYTO16 (S-7578), goat anti-mouse Alexa 488 (A-11001), and TO-PRO-3 iodide (T3605) from Molecular Probes; donkey anti-rabbit Cy3 (code 711-165-152) and mouse anti-rat Cy5 (code 212-176-168) from Jackson ImmunoResearch Laboratories; and horseradish peroxidase-linked anti-mouse IgG antibody (7076) and horseradish peroxidase-linked anti-rabbit IgG antibody (7074) from Cell Signaling.
Primary Human Erythroid Cultures—CD34+ early hematopoietic progenitors were either purchased from Stem Cell Technologies (mPB015F) or initially isolated from growth factor-mobilized peripheral blood (purchased from AllCells, Berkeley, CA) using an Isolex 300i cell selection device. Cells were cultured in media containing 15% fetal bovine serum, 15% human serum, Iscove's modified Dulbecco's medium, 10 ng/ml interleukin-3, 2 units/ml erythropoietin, and 50 ng/ml stem cell factor (13). No new interleukin-3 was added after day 0, and the amount of stem cell factor was gradually decreased at each feeding (day 3, 25 ng/ml; day 6, 10 ng/ml; and day 8, 2 ng/ml).
Flow Cytometry of CD34+ Cells—Cell-surface expression of GPA, transferrin receptor (TfR; CD71), and AE1 (BRIC6) was monitored by flow cytometry using FACSCalibur to confirm differentiation. SYTO16 (Invitrogen), a DNA stain, was used to monitor enucleation. Unfixed cells were sampled at various days following induction with 2 units/ml erythropoietin, and cells were probed for 0.5 h with antibodies in Hanks' balanced salt solution and 1% bovine serum albumin.
Immunofluorescence and Microscopy—Differentiating CD34+ cells were cytospun onto glass slides and fixed in 3.7% formaldehyde for 10 min. Permeabilized cells were blocked in 3% bovine serum albumin and incubated with antibodies in 1× phosphate-buffered saline and 1% bovine serum albumin for 1 h. Cells were then incubated with secondary antibodies and/or TO-PRO-3 for nuclear staining for 1 h. Cells were mounted with DakoCytomation fluorescent mounting medium before observation at magnification ×100 using a laser scanning confocal Zeiss LSM 510 microscope. Imaging equipment includes Zeiss AxioCam, AxioVision, and LSM image browser. Reagents used for counterstaining microscopy include Gill's hematoxylin (2×) (23-245654) from Fisher and O-dianisidine/benzidine (D9143) from Sigma. Cells were observed using phase contrast at magnification ×100 on a Zeiss deconvolution microscope.
Immunoblot Analysis of CD34+ Cell Lysates—Cell pellets isolated at various days were lysed in Pierce mammalian protein extraction reagent (78503) supplemented with 100 μm phenylmethylsulfonyl fluoride (Sigma), 1 μm aprotinin, 1 μm leupeptin, and 1 μm pepstatin (Roche Applied Science) for 30 min on ice. Lysates were centrifuged (16,000 × g for 10 min), and the protein concentration was determined using the Bio-Rad protein assay (500-0006). Total protein lysate (40 μg) was solubilized in 2× SDS-PAGE sample buffer (130 mm Tris, 2% SDS, 20% glycerol, 2% 2-mercaptoethanol, and 0.002% bromphenol blue, pH 6.8), resolved on 8% SDS-polyacrylamide gels, and analyzed by immunoblotting. The BM chemiluminescence blotting substrate (POD) from Roche Applied Science was used for visualization.
Statistical Analysis—Statistical analyses were performed on band densities within the linear range from separate immunoblots and quantified using NIH ImageJ Version 1.32j. Percent expression was calculated relative to the highest level of protein expression in either earlier stages of differentiation (days 6-7) for chaperones and cellular proteins and at later stages of differentiation (days 13-15) for red cell-specific membrane proteins. Data are from three or more separate experiments and are expressed as mean ± S.D. p values were calculated with paired, two-tailed Student's t test.
mRNA Detection and PCR Amplification—Total RNA isolated from CD34+ cells on day 6 and from GPA+ sorted cells at later time points during the 14-day culture period was used to synthesize cDNA. PCR amplification was performed with specific forward and reverse primers for CNX (forward, 5′-GAGGCTAGAGATCATGGAAG-3′; reverse, 5′-GCTCTTCAGCTGCCTCGATC-3′) (14) and AE1(SLC4A1) (forward, 5′-CCAGACTCCAGCTTCTACAAGG-3′; reverse, 5′-GGAAGGAGAAGATCTCCTGG-3′) (15), with the 18 S rRNA gene as a control (forward, 5′-CGCCGCTAGAGGTGAAATTCT-3′; reverse, 5′-CAATCTCGGGTGGCTGAAC-3′) (13). The PCR conditions were as follows: 30 cycles of 1-min annealing at 58 °C and a 2-min extension at 72 °C.
Expression of AE1 in K562 Cells—HEK-293 cells were cotransfected with the pVPack vectors (Stratagene) and pFBNeo constructs of HA-tagged AE1 using Lipofectamine 2000 (Invitrogen). 48 h later, virus-containing supernatant was collected and added to K562 cells in the presence of 8 μg/ml polybrene (Sigma). The infected cells were selected with 2 mg/ml geneticin (G418; Sigma) over a 3-week period. K562 cells were monitored by flow cytometry for cell-surface expression of AE1 using the engineered HA tag on the third extracellular loop. Flow cytometry and immunofluorescence confirmed the intracellular localization of the hereditary spherocytosis (HS) R760Q mutant.
Interaction of AE1 with Chaperones in K562 Cells—5 × 106 K562 cells expressing AE1 were lysed in 1% digitonin supplemented with proteasome inhibitors as above for 30 min on ice. Cell lysates were centrifuged (16,000 × g for 10 min), and the supernatant was collected. Co-immunoprecipitation was then performed, and immunoblotting identified co-immunoprecipitated AE1. Cell-surface expression of GPA, TfR, and HA-tagged AE1 were used to monitor the effect of 1 mm castanospermine (Calbiochem) treatment (18 h) on membrane protein trafficking in K562 cells. Flow cytometry was performed as described above.
Expression of AE1 in Mouse Embryonic Fibroblasts (MEFs)—CNX-deficient (9KO; CNX-/-), wild type (K41), and CRT-deficient (K42) MEFs were provided by Dr. Marek Michalak (University of Alberta) (16, 17). 9KO MEFs were infected with the virus generated as above using the pQCXIH vector containing either wild type CNX cDNA or an empty vector control. Infected cells were selected for using 100 μg/ml hygromycin B (Invitrogen). Small interfering RNA (siRNA) corresponding to the mouse CNX DNA sequence, AATGTGGTGGTGCCTATGTGA (Qiagen), was also transfected into K41 and K42 cells using Oligofectamine (Invitrogen). AE1 tagged with HA in the third extracellular loop was transiently expressed in 9KO ± CNX, K41, and K42 MEFs using Lipofectamine 2000. 48 h post-transfection, protein expression was monitored by flow cytometry and immunoblotting. To confirm the glycosylation status of AE1, cell extracts were treated with 1,000 units of endoglycosidase H, which cleaves high mannose oligosaccharides, or N-glycosidase F, which cleaves all N-linked oligosaccharides (New England Biolabs).
RESULTS
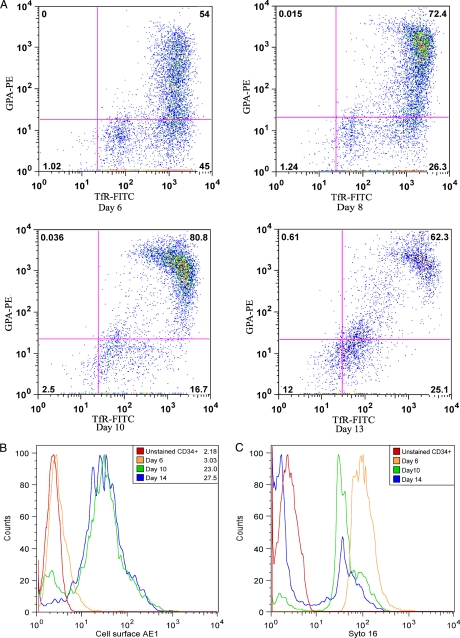
Differentiation of CD34+ Cells in Culture—The purpose of this study was to examine the integrity of the ER protein translocation, N-glycosylation, and quality control machinery during the terminal differentiation of human CD34+ erythroid progenitor cells grown in culture for over 12-18 days. The CD34 antigen is specific for early pluripotent hematopoietic stem cells at the colony-forming unit-erythroid stage of differentiation. As CD34+ cells differentiate to reticulocytes, the complement of cell-surface proteins change, and flow cytometry can be used to monitor differentiation. Flow cytometry was performed using antibodies that recognize the external epitopes of TfR and GPA (Fig. 1A) (18). TfR is present in these progenitor cells, whereas the proportion of GPA-positive cells increases during differentiation (Fig. 1A). The expression of AE1 was detected at the cell surface late in differentiation at days 10 and 14 with no expression at day 6 (Fig. 1B). Enucleation was monitored using SYTO16, a fluorescent DNA stain (Fig. 1C). An initial downshift in fluorescence intensity was seen from days 6 to 10, likely due to DNA condensation, and a loss of fluorescence by day 14 indicated enucleation of the majority of the cells.
FIGURE 1.
Flow cytometry analysis and AE1 expression in differentiating CD34+ erythroid progenitors. A, representative flow cytometry analysis used to monitor differentiation of CD34+ cells using cell-surface markers TfR and GPA. Numbers indicate the percent of positive cells in each quadrant. B, histogram of the increase in AE1 expression in differentiating CD34+ cells as measured by flow cytometry. Geometric mean fluorescence intensities are shown. C, histogram of the loss of DNA by SYTO16 dye binding in differentiating CD34+ cells as measured by flow cytometry.
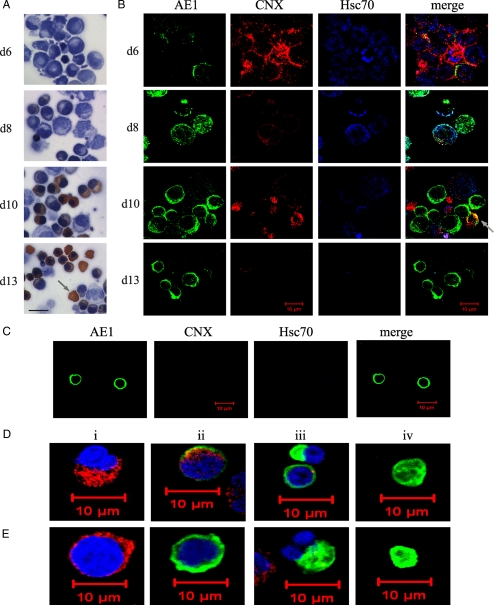
Differentiation of CD34+ cells in culture produced a heterogeneous cell population, and the timing of expression of red cell-specific proteins varied from experiment to experiment. However, late in differentiation (day 13), over 80% of the cells were hemoglobin-positive by orange benzidine staining, and evidence of enucleation was visible by loss of hematoxylin counter-staining (Fig. 2A, arrow). By immunofluorescence and confocal microscopy, few AE1-positive cells were seen at day 6, whereas most early progenitor cells stained positive for the ER chaperone CNX and cytosolic chaperone Hsc70 (Fig. 2B). During differentiation, the number and intensity of CNX- and Hsc70-positive cells decreased, whereas the number of smaller AE1-positive cells increased. An occasional cell stained positive for both CNX and AE1 (Fig. 2B, d10 Merge, arrow); however, most AE1-positive cells were CNX-negative. AE1-positive cells isolated by fluorescence-activated cell sorting on day 14 were negative for both Hsc70 and CNX when examined by confocal microscopy (Fig. 2C).
FIGURE 2.
Calnexin and Hsc 70 expression decreases as AE1 is expressed in differentiating CD34+ cells. A, shown is a cell morphology of differentiating CD34+ cells (days (d) 6, 8, 10, and 13). Cells were counterstained with hematoxylin to detectloss of nuclei (blue) and stained with benzidine to detect expression of hemoglobin (orange). The arrow highlights a hemoglobin-positive, enucleated reticulocyte. Scale bar, 10 μm. B, shown are the immunofluorescence images of differentiating CD34+ cells to detect expression of AE1 (green), calnexin (red), and Hsc70 (blue). C, immunofluorescence staining of permeabilized AE1+ cells no longer contain calnexin and Hsc70. Fluorescence-activated cell sorting was used to sort AE1 (BRIC6)-positive CD34+ differentiating cells. Green, AE1; red, calnexin; blue, Hsc70. D and E, shown are the single cell immunofluorescence images of differentiating CD34+ cells at various late stages (days13-15) used to monitor ER integrity. i-iv, shown are the images of individual cells at subsequent stages of enucleation. D, AE1 expression is shown in green, and ERp57 is shown in red. Loss of nuclei (blue) by TO-PRO-3 staining is shown in panels iii and iv. E, shown are AE1 expression (green) and calnexin (red) staining in differentiating CD34+ cells. In panels iii and iv, the loss of nuclei (blue) by TO-PRO-3 staining is shown.
ER Is Maintained during Terminal Differentiation of CD34+ Cells—The integrity of the ER during terminal differentiation was determined in individual cells by immunofluorescence staining of AE1 and ER proteins as observed by confocal microscopy. The ER-localized disulfide isomerase, ERp57, was detected (Fig. 2D, panel ii), as was the luminal ER chaperone CRT (data not shown), in nucleated and AE1-positive cells, indicating that the ER was still present in these cells. CNX, on the other hand, was not present in nucleated cells expressing AE1 (Fig. 2E, panel ii). In cells that were enucleating (Fig. 2, D-E, panel iii, blue TO-PRO-3-stained nucleus), none of the ER marker proteins, CRT, ERp57, or CNX, were present, suggesting that the ER was removed during enucleation. In fully enucleated cells (Fig. 2, D-E, panel iv) neither ERp57 nor CNX were detected, as expected, as the ER is not present in human reticulocytes or mature red cells.
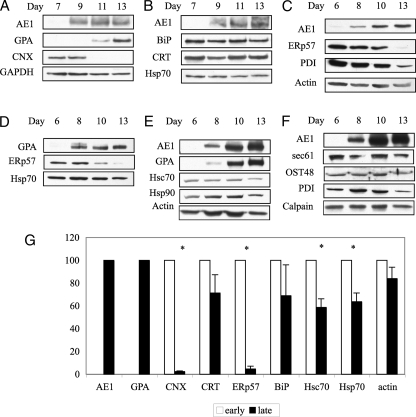
Immunoblot Analysis Shows Loss of Specific Chaperones during CD34+ Differentiation—To provide a quantitative measure of the ER components during differentiation, immunoblot analyses of whole cell extracts from differentiating CD34+ cells were carried out. Several independent differentiation experiments were performed to quantify the levels of AE1, GPA, and various components of the protein translocation and quality control machinery during red cell production (Fig. 3, panels A-F). The blots were scanned, and the expression levels were averaged (Fig. 3G). Data are divided into early (days 6-7; no AE1 or GPA detected) and late (days 13-15; maximal AE1 and GPA) stages. As expected, AE1 was not detectable at the early stages of differentiation but increased dramatically at the late stages (days 8-13) (Fig. 3, A-C, E, and F). A similar pattern was found for GPA (Fig. 3, A, D, and E). The levels of actin (Fig. 3, C and E) and glyceraldehyde-3-phosphate dehydrogenase (Fig. 3A), proteins found in the mature red cell, only decreased slightly at later stages relative to total protein (mostly hemoglobin). The ER chaperone CNX decreased dramatically during terminal differentiation and disappeared during maximal AE1 expression (Fig. 3A). The cytosolic chaperones Hsc70 and Hsp70, decreased by 40% between the early and late stages of differentiation (p = 0.001) (Fig. 3, B, D, E, and G). In contrast, the levels of cytosolic Hsp90 and the ER chaperones, BiP and CRT, were maintained during the 2-week period (Fig. 3, B and E).
FIGURE 3.
Specific molecular chaperones are lost during differentiation. A-F, immunoblot analysis of six differentiation experiments monitoring the expression of membrane proteins, AE1 and GPA, various molecular chaperones, and protein synthesis machinery. Equal amounts of total protein (40 μg of whole cell extracts) were loaded from various time points throughout differentiation. Individual blots were probed with a specific antibody, stripped, and then sequentially reprobed with different antibodies. G, percent relative expression as determined by densitometric scanning of multiple immunoblots (n = 3, not all shown in A-F). Open bars, early stage of differentiation (days 6-7); filled bars, late stage of differentiation (days 13-15), relative to maximum protein expression, either at the early or late stages. The asterisk represents significant protein loss. p < 0.001. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
The synthesis of high levels of AE1 and GPA requires a robust ER translocation and N-glycosylation machinery. The key components needed to assemble membrane glycoproteins including the translocon (Sec61α) and oligosaccharyltransferase (OST48) were maintained during the terminal stages of red cell differentiation (Fig. 3F). The levels of ER protein disulfide isomerases, PDI and ERp57, however, diminished dramatically at the later stages of differentiation (Fig. 3, C, D, and F, Day 13). ERp57 was almost completely lost, with PDI levels dropping to 20%. Because ERp57 interacts with CNX in a complex (19), the loss of CNX may result in the reduction of ERp57 levels. Calpain, a red cell calcium-activated protease involved in membrane remodeling, was maintained during the differentiation period.
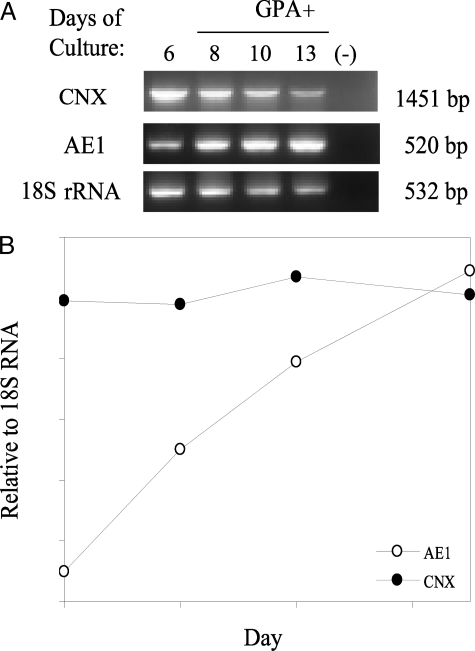
CNX Is Lost Post-translationally in Differentiating CD34+ Cells—We used reverse transcription-PCR to measure CNX mRNA levels in CD34+ cells over the 2-week differentiation period (Fig. 4A). The AE1 mRNA levels increased relative to 18S rRNA levels, whereas the ratio of CNX mRNA to 18S rRNA remained constant through differentiation (Fig. 4B). The loss in CNX is therefore not due to a decrease in gene transcription and mRNA production. CNX is a very stable and long lived protein (20), and its loss during terminal differentiation may be due to translational control or a specific mechanism of targeted protein degradation.
FIGURE 4.
Calnexin mRNA is detectable during late stages of CD34+ differentiation. A, reverse transcription-PCR analysis of the expression pattern of calnexin and AE1 mRNA in differentiating primary erythroid progenitors. As an RNA control, the 18S rRNA gene was also amplified by reverse transcription-PCR. B, relative RNA expression of calnexin and AE1 to 18S rRNA during erythroid differentiation. WT, wild type; IP, immunoprecipitation.
AE1 and Mutants Can Interact with CNX in Vivo—We have shown previously using cell-free translation and transfected HEK-293 cells that AE1 can interact with CNX in a glycosylation-dependent manner (21). To examine the role of chaperones in the retention of AE1 mutants, we chose two mutations in human AE1 that cause two dominantly inherited red blood cell diseases, Southeast Asian ovalocytosis (SAO) and HS. The non-functional SAO AE1 protein that arises from a nine-amino acid deletion (Δ400-408), however, is present in nearly equal amounts to the normal protein in the plasma membrane of red blood cells of heterozygotes (22). HS point mutations in the membrane domain of AE1 lead to a protein that is retained in the ER of transfected HEK-293 cells (23). The HS mutant, AE1 R760Q (Prague II), is completely absent in mature red blood cells (24), indicating defective trafficking to the plasma membrane during erythropoiesis.
Using cell-free translation and co-immunoprecipitation, we confirmed that AE1 and AE1 SAO and the HS mutant R760Q can interact with CNX in a glycosylation-dependent manner, but interestingly, not with CRT (supplemental Fig. S1). Wild type and mutant AE1 were also expressed in HEK-293 cells to study their interaction with CNX. The oligosaccharide on AE1 in HEK-293 cells remains in the high mannose form and is sensitive to digestion with endoglycosidase H. At steady state, about 30% of wild type AE1 in the high mannose form is present at the cell surface with the remainder in the ER (25). The expression of SAO and HS R760Q in HEK-293 cells results in their intracellular retention in the ER (23, 26); however, no enhanced interaction of chaperones with the mutants relative to the wild type protein was detected by co-immunoprecipitation (supplemental Fig. S2). Furthermore, pulse-chase experiments in HEK-293 cells using [35S]methionine showed that AE1, AE1 SAO, and AE1 HS interact with CNX in a similar transient manner (supplemental Fig. S3). These results indicate that the AE1 glycoprotein is a potential CNX substrate.
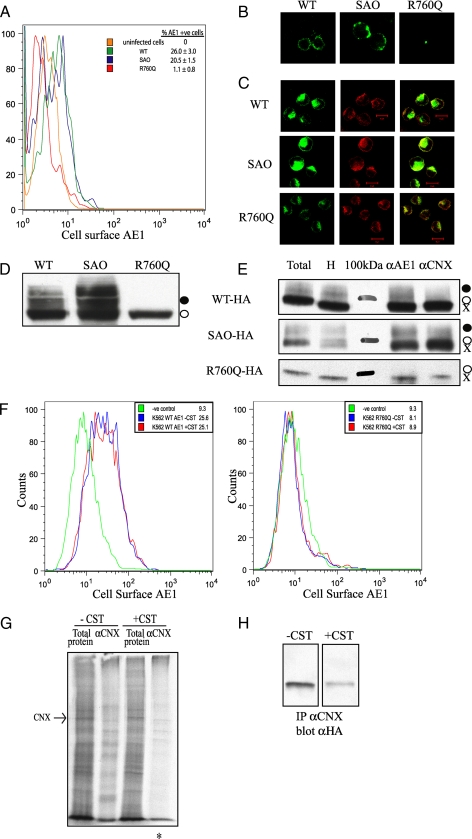
We next examined the role of CNX in the folding and trafficking of membrane glycoproteins in K562 cells, an erythroleukemia cell line that expresses the endogenous red cell proteins GPA and transferrin receptor TfR. GPA has been shown to enhance the cell-surface expression of wild type AE1 and SAO AE1 when expressed in Xenopus oocytes (27-29). The HS mutant, AE1 R760Q (Prague II) is not rescued by GPA, as it is not present in the plasma membrane of HS red cells (24). Wild type, SAO AE1, and the HS R760Q mutant AE1 with an external HA tag on the third extracellular loop were expressed in K562 cells. Flow cytometry and immunofluorescence confirmed that both the wild type and SAO AE1 were able to traffic to the plasma membrane (Fig. 5, A and B), whereas HS R760Q was seen to co-localize with CNX in the ER, indicating a trafficking defect in these cells (Fig. 5C). A portion of the wild type and SAO proteins adopts a complex oligosaccharide as they traffic through the Golgi to the plasma membrane, whereas the R760Q mutant maintains a high mannose oligosaccharide consistent with ER localization (Fig. 5D). Both wild type and mutant AE1 interact with CNX as shown by co-immunoprecipitation; however, no enhanced interaction of the mutants relative to the wild type protein was detected (Fig. 5E). This lack of difference may be due to the inefficient exit of wild type AE1 from the ER in these cells.
FIGURE 5.
Calnexin is not required for cell-surface expression of membrane glycoproteins in K562 cells. A, flow cytometry shows the cell-surface expression of AE1 and SAO but not R760Q in K562 cells. B, shown is the immunofluorescence of non-permeabilized K562 cells demonstrating cell-surface expression of the wild type (WT) and SAO AE1 and not R760Q. C, shown is the immunofluorescence of permeabilized K562 cells demonstrating intracellular co-localization with calnexin. D, shown is an immunoblot of total protein lysate from K562 cells expressing AE1 constructs containing an HA tag in the third extracellular loop. Expressed AE1 contains primarily the high mannose oligosaccharide (○), with wild type and SAO but not R760Q possessing complex oligosaccharide (•). E, K562 cells expressing AE1 were lysed in 1% digitonin for co-immunoprecipitation with antibodies against AE1 (αAE1) and calnexin (αCNX). Total, total cell extract; H, endoglycosidase H-treated cell extract; 100kDa, 100-kDa protein molecular masss marker. Immunoprecipitated (IP) proteins were run on 8% SDS-polyacrylamide gels and immunoblotted using an anti-HA antibody to detect AE1. F, the inhibition of calnexin binding does not promote cell-surface expression of membrane glycoproteins. Flow cytometry was used to monitor the cell-surface expression of HA-tagged AE1 (left panel, wild type; right panel, HS R760Q), in the presence and absence of 1 mm castanospermine. Geometric mean fluorescence intensities are shown. G, shown is the co-immunoprecipitation of radiolabeled proteins in K562 cells with calnexin in the presence and absence of 1 mm castanospermine. An asterisk indicates less radiolabeled proteins can be associated with calnexin in the presence of castanospermine. H, shown is a co-immunoprecipitation (IP) of HS R760Q AE1 with calnexin in the absence and presence of 1 mm castanospermine.
To determine whether or not CNX is required for membrane glycoprotein biosynthesis, castanospermine was used to inhibit glucose trimming by glucosidase I, thereby preventing the lectin interaction of glycoprotein substrates by CNX and CRT (9). The level of wild type AE1 expression at the cell surface was unaltered (Fig. 5F, left panel), and no R760Q mutant was detected at the cell surface following castanospermine treatment (Fig. 5F, right panel). Co-immunoprecipitation studies confirmed less radiolabeled proteins associated with CNX in the presence of castanospermine (Fig. 5G), and co-immunoprecipitation followed by immunoblotting for R760Q interacting with CNX showed a decrease of 75% in CNX-AE1 interactions by castanospermine (Fig. 5H). Overnight treatment with 1 mm castanospermine also had little or no effect on the cell-surface expression of TfR and GPA, as determined by flow cytometry (data not shown). CNX can therefore interact with normal and mutant AE1 during biosynthesis; however, it does not play a significant role in the trafficking of these membrane glycoproteins to the cell surface of K562 cells.
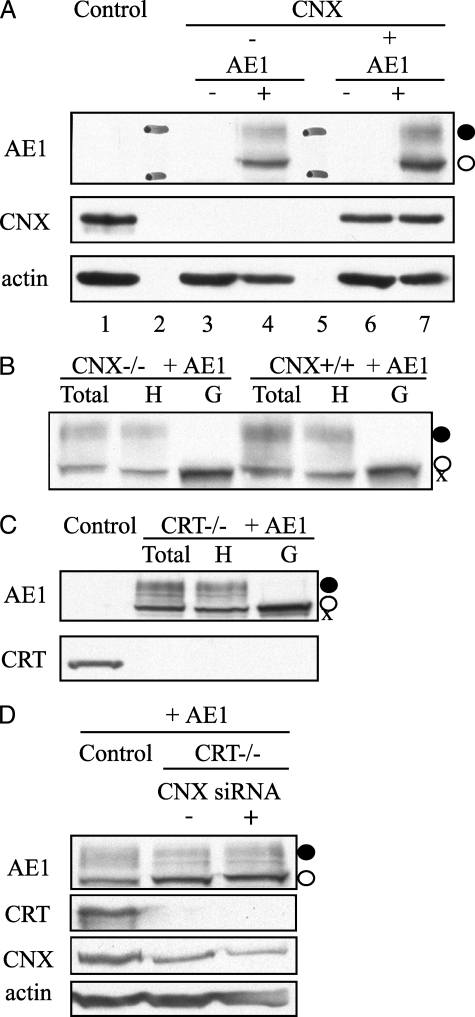
CNX and CRT Are Not Required for Cell-surface Expression of AE1—Wild type AE1 was also expressed in MEFs completely lacking CNX or CRT. The overall expression levels of AE1 were slightly lower (75%) in the CNX-deficient cells, compared with MEFs with CNX reintroduced (Fig. 6A, lane 4 versus 7). Digestion of the oligosaccharide with N-glycosidase F and endoglycosidase H revealed that AE1 can adopt a complex oligosaccharide in both cell lines (Fig. 6B), indicative of trafficking to the Golgi. The complex oligosaccharide was also seen on AE1 in MEF cells completely devoid of CRT (Fig. 6C). ImageJ quantification of the percent of oligosaccharide in the complex form on AE1 revealed similar levels in the absence of CNX, 37 versus 43% in the CNX+/+ cells, and 37% of the complex oligosaccharide on AE1 was expressed in CRT-/- cells. The lower high mannose band likely represents AE1 that is located in the ER of these transfected cells.
FIGURE 6.
AE1 adopts a complex oligosaccharide when expressed in calnexin- and calreticulin-deficient MEFs. A, expression of AE1 in calnexin-deficient (CNX-/-) and cells with CNX reintroduced. Lane 1, wild type MEFs; lane 2, 130-kDa (upper) and 95-kDa (lower) molecular mass markers in AE1 panel; lane 3, CNX-/- cells; lane 4, AE1 transfected CNX-/- cells; lane 5, molecular mass markers; lane 6, CNX+/+ cells; lane 7, AE1-transfected CNX+/+ cells. AE1 adopts a complex oligosaccharide both in the absence (lane 4) and the presence (lane 7) of calnexin. B, deglycosylation of AE1 expressed in MEFs in the absence and presence of calnexin. Total lysate was treated with either endoglycosidase H (H) to remove high mannose oligosaccharides (○) or N-glycosidase F (G) to remove both complex (•) and high mannose oligosaccharides to produce non-glycosylated AE1 (X). C, AE1 also adopts complex oligosaccharide in MEFs completely devoid of calreticulin (CRT-/-). D, siRNA of CNX in CRT-/- cells. 3 days following knockdown using either a control or CNX siRNA, AE1 was transfected and detected by immunoblotting 48 h later. When normalized to actin levels, the level of CNX was 30% of that seen using a control siRNA. AE1 trafficking to the Golgi was unaffected as it adopts a complex oligosaccharide (•) when CNX levels are reduced and CRT is completely absent.
siRNA was used to knock down CNX levels in CRT-deficient cells to deplete cells of both chaperones (Fig. 6D). Three days following knockdown, AE1 was transiently expressed in cells transfected with either a control siRNA or siRNA directed at CNX. Knockdown levels of CNX were 30% of that seen in control siRNA-treated CRT-/- cells. The level of complex oligosaccharide on AE1 was not affected by the knockdown of CNX. By reducing levels of CNX in CRT-/- cells, these results indicate that CNX and CRT do not compensate for each other and do not affect AE1 oligosaccharide processing and trafficking from the ER.
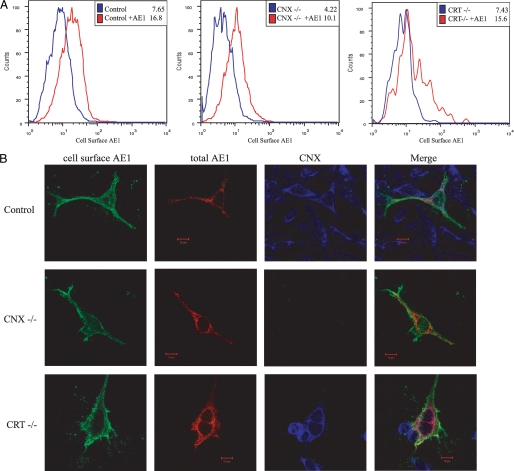
When compared with transfection of an empty cDNA vector, wild type AE1 can be detected at the cell surface in both control MEF cells and CNX- and CRT-deficient cells by flow cytometry (Fig. 7A, higher mean fluorescence intensity of red histograms). Overton subtraction (30) revealed similar levels of AE1 at the cell surface in both CNX- and CRT-deficient cell lines versus control cells, similar to the ∼40% of AE1-adopting complex oligosaccharide by immunoblotting.
FIGURE 7.
AE1 traffics to the cell surface in cells devoid of CNX or CRT. A, flow cytometry of MEFs expressing AE1 (red) at the cell surface in control cells (left panel), calnexin-deficient cells (CNX-/-; middle panel), and calreticulin-deficient cells (CRT-/-; right panel). Mean fluorescence intensities for cell-surface expression of HA-tagged AE1 are shown. B, confocal microscopy of MEFs expressing AE1 in control cells or CNX-/- or CRT-/- cells. Cells were stained for cell-surface AE1 (green) followed by fixation, permeabilization, and staining for intracellular AE1 (red) and calnexin (blue).
AE1 staining using an HA tag on the third extracellular loop in control fibroblast cells demonstrated that AE1 is present at the cell surface in these transfected cells (Fig. 7B, first Control panel) and also co-localizes with CNX in the ER (Fig. 7B, last Merge panel). Immunofluorescence staining of non-permeabilized cells also confirmed that AE1 is able to traffic to the cell surface in CNX- and CRT-deficient fibroblasts (Fig. 7B, column 1). Following fixation and permeabilization, AE1 was also detected intracellularly in CNX-/- cells and CRT-/- cells, as determined by immunofluorescence (Fig. 7B, second column). These results demonstrate that CNX and CRT are not essential for the trafficking of AE1 to the cell surface in these cell lines and that a fraction of AE1 localizes in the ER, even in the absence of these chaperones.
DISCUSSION
In this study, we examined the fate of chaperones and other ER components involved in glycoprotein biosynthesis during hematopoiesis. Surprisingly, we discovered that an ER chaperone, CNX, and protein disulfide isomerases, ERp57 and PDI, were lost during the terminal stages of differentiation of human CD34+ cells when maximal membrane glycoprotein synthesis occurs. Other chaperones and components of the ER protein translocation and N-glycosylation machinery were, however, maintained, as expected due to their requirement in membrane protein biosynthesis. This is in contrast to B lymphocytes that up-regulate cytosolic and ER chaperones to deal with protein synthesis and assist in IgM maturation during their differentiation to Ig-secreting plasma cells (31).
The absence of CNX during the production of AE1 and GPA indicates that this ER chaperone is not essential for glycoprotein synthesis and folding during terminal red cell production. The differentiation of CD34+ cells appears to be a novel system in which CNX has been effectively knocked down at the time of maximal glycoprotein biosynthesis. Even though CNX is a very stable protein (20), it is specifically targeted for removal, which may be a post-transcriptional event, perhaps involving translational inhibition or targeted proteolytic degradation. The mechanism of down-regulation of CNX in CD34+ cells is a subject of further study.
As CD34+ cells are primary pluripotent and not transformed cells, transfection and siRNA knockdown experiments are not feasible. To confirm that CNX is not essential in the biosynthesis of AE1, we have used murine fibroblasts completely devoid of CNX to show that the cell-surface expression of transfected AE1 was unaffected. This experiment is superior to siRNA knockdown experiments, which often result in incomplete depletion of the target protein. Our studies confirm that even though CNX can interact with AE1, the lack of CNX in MEF cells and the inhibition of N-linked oligosaccharide trimming with castanospermine in K562 cells had no effect on the level of cell-surface expression of AE1 or GPA.
The loss of CNX during differentiation may provide a protective role against apoptosis in erythrocytes. CNX interacts with Bap31, an ER integral membrane protein that is cleaved by caspase-8 following apoptosis activation (32). CNX-deficient leukemic cells, however, are resistant to Bap31 cleavage following thapsigargin-induced apoptosis, indicating that CNX may have an effect on later apoptotic events (32). Therefore, a loss of CNX expression from erythrocyte precursors would render them resistant to apoptosis induced by ER stress or protein overload. This may be important during the terminal differentiation of erythroid progenitors, as these cells must avoid apoptosis as they produce large amounts of membrane glycoproteins that could induce ER stress.
CRT, on the other hand, was maintained during differentiation. It has a role in mature erythrocytes by preventing the perforin-mediated lysis of mature red blood cells by cytotoxic lymphocytes (11). We examined whether CRT is able to partially compensate for the loss of CNX and aid in the folding of red cell glycoproteins. Even though CRT can interact with AE1 in HEK-293 cells, cell-free translation studies demonstrated that CNX interacts with AE1 in a glycosylation-dependent manner (21), yet no such interaction could be detected with CRT (supplemental Fig. S1), suggesting that AE1 may not be a lectin-dependent substrate for CRT. Indeed, it has been shown that CNX and CRT have different interactions with glycoprotein substrates, with CRT typically binding to more distal sites from the membrane (33). The oligosaccharide on AE1 is attached to Asn642 in a relatively small extracellular loop, which may not be accessible to CRT. Finally, the level of cell-surface expression of AE1 was not impaired in fibroblasts completely devoid of CRT or when CNX was reduced by siRNA in these cells, showing that these chaperones are not required for the trafficking of AE1 to the cell surface.
Castanospermine treatment of K562 cells, which disrupts both CNX and CRT interactions with glycoprotein substrates (9), had no effect on the cell-surface expression levels of TfR, GPA, or AE1. Non-glycosylated AE1 can traffic to the plasma membrane in transfected HEK cells (34) and when expressed in Xenopus oocytes (35), suggesting that an oligosaccharide-dependent interaction with CNX or CRT is not essential for cell-surface expression. Our results indicate that although CRT is maintained during the terminal differentiation of red cells, it likely does not substitute for the loss of CNX. This loss of protein quality control may allow misfolded glycoproteins such as AE1 SAO to escape the ER in red cell precursors. This possibility could be studied if CD34+ cells obtained from patients with this red cell defect become available.
The loss of CNX and ERp57 during red cell differentiation may compromise protein quality control but may have some benefit. Interestingly, neither AE1 nor GPA contain disulfide bonds and therefore do not require the disulfide protein folding pathway maintained by the CNX, ERp57, and PDI complex. Disulfide formation is often a rate-limiting step in protein folding, and the lack of this pathway could allow AE1 and GPA to bypass the cycles of binding and release and be expedited from the ER to the cell surface.
TfR is lost during reticulocyte maturation via exosomes, and this glycoprotein contains a single disulfide linkage between the homodimer (36). The loss of TfR during the terminal stages of red cell development limits iron uptake as hemoglobin synthesis is completed. A loss of disulfide formation due to loss of disulfide isomerases could therefore cause TfR to misfold and be targeted for degradation, terminating differentiation.
Even though the levels of the cytosolic chaperones, Hsc70 and Hsp70, decreased, 60% of the protein still remains during the late stages of CD34+ differentiation, consistent with a protective role in red cells against complement-mediated lysis (37). Proteomic analysis has confirmed the presence of Hsp70 and Hsp90 in mature human erythrocytes (38). The decrease in Hsc70 levels may be a result of its interaction with the transferrin receptor by which both proteins are secreted in exosomes during the final stages of erythrocyte maturation (39, 40). Hsp70 expression is up-regulated in membranopathies and in hemin-induced K562 cells and prevents the caspase-mediated proteolysis of the transcription factor, GATA-1, in erythroid precursors (41-43).
The level of calpain was maintained during terminal red cell development, allowing for this calcium-dependent proteolytic degradation pathway to be present in mature red cells (44). Calpastatin, an inhibitor that modulates the calpain-mediated proteolytic degradation pathway active in aging red blood cells, has also been found in erythrocytes by proteomic analysis (38). Red blood cells remain in circulation for ∼120 days and are removed by antibody recognition of protein aggregation in the form of Heinz bodies (45). The protein components of the red cell, including the cell-surface glycoproteins GPA and AE1, are very stable and do not turnover. Red cells that contain mutant or deficient membrane components, however, have a shorter lifespan and often result in hemolysis (46). Thus, quality control of erythrocytes occurs by removing damaged or aged cells from circulation rather than degrading individual components.
In conclusion, we have shown that as major membrane glycoproteins in red cell precursors are expressed, specific chaperones (CNX and ERp57) are eliminated, whereas other essential chaperones (BiP and CRT) are maintained. The lack of disulfide bonds in AE1 and GPA may facilitate rapid folding and trafficking by not requiring interactions with the disulfide isomerases mediated by CNX. Even though CNX is not required for the folding and trafficking of AE1, GPA interacts with AE1 in a “chaperone-like” manner, forming a complex that may promote AE1 trafficking to the cell surface. Our results indicate that red blood cells may be a unique cellular system in which specific chaperones are selectively removed to enable the massive synthesis of large amounts of stable membrane glycoproteins, thereby sacrificing quality for quantity.
Supplementary Material
Acknowledgments
We thank our colleagues Dr. Ulf Brockmeier for generating the reconstituted CNX MEF cell line and Ling Li and Dr. Walter Kahr for reagents and equipment. The idea to examine chaperones during the terminal differentiation of erythrocytes originated in discussions with Dr. Lesley Bruce (Bristol) and others at the Red Cell Club Meeting in Chicago, 2005.
This work was supported in part by Canadian Institutes of Health Research Grant FRN15266 (to R. A. F. R.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1-S3, and Refs. 1 and 2.
Footnotes
The abbreviations used are: AE1, anion exchanger 1; CNX, calnexin; CRT, calreticulin; ER, endoplasmic reticulum; GPA, glycophorin A; HA, hemagglutinin; HEK, human embryonic kidney; HS, hereditary spherocytosis; MEF, mouse embryonic fibroblast; PDI, ER protein-disulfide isomerase; SAO, Southeast Asian ovalocytosis; TfR, transferrin receptor.
References
- 1.Geminard, C., de Gassart, A., and Vidal, M. (2002) Biocell. 26 205-215 [PubMed] [Google Scholar]
- 2.Izak, G. (1977) Prog. Hematol. 10 1-41 [PubMed] [Google Scholar]
- 3.Wickrema, A., Koury, S. T., Dai, C. H., and Krantz, S. B. (1994) J. Cell. Physiol. 160 417-426 [DOI] [PubMed] [Google Scholar]
- 4.Trombetta, E. S., and Parodi, A. J. (2003) Annu. Rev. Cell Dev. Biol. 19 649-676 [DOI] [PubMed] [Google Scholar]
- 5.Krebs, M. P., Noorwez, S. M., Malhotra, R., and Kaushal, S. (2004) Trends Biochem. Sci. 29 648-655 [DOI] [PubMed] [Google Scholar]
- 6.Young, J. C., Barral, J. M., and Hartl, U. F. (2003) Trends Biochem. Sci. 28 541-547 [DOI] [PubMed] [Google Scholar]
- 7.Demaurex, N., Frieden, M., and Arnaudeau, S. (2003) in Calreticulin (Eggleton, P., and Michalak, M., eds) 2nd Ed., pp. 134-142, Eureka, Austin, TX
- 8.Arrigo, A. P. (2005) J. Cell. Biochem. 94 241-246 [DOI] [PubMed] [Google Scholar]
- 9.Williams, D. B. (2006) J. Cell Sci. 119 615-623 [DOI] [PubMed] [Google Scholar]
- 10.Helenius, A., Trombetta, E. S., Hebert, D. N., and Simons, J. F. (1997) Trends Cell Biol. 7 193-20017708944 [Google Scholar]
- 11.Fraser, S. A., Michalak, M., Welch, W. H., and Hudig, D. (1998) Biochem. Cell Biol. 76 881-887 [DOI] [PubMed] [Google Scholar]
- 12.Vassilakos, A., Michalak, M., Lehrman, M. A., and Williams, D. B. (1998) Biochemistry 37 3480-3490 [DOI] [PubMed] [Google Scholar]
- 13.Uddin, S., Kang, J.-A., Ulaszek, J., Mahmud, D., and Wickrema, A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 147-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeates, L. C., and Powis, G. (1997) Biochem. Biophys. Res. Commun. 238 66-70 [DOI] [PubMed] [Google Scholar]
- 15.Dudeja, P. K., Hafez, N., Tyagi, S., Gailey, C. A., Toofanfard, M., Alrefai, W. A., Nazir, T. M., Ramaswamy, K., and Al-Bazzaz, F. J. (1999) Am. J. Physiol. 276 L971-L978 [DOI] [PubMed] [Google Scholar]
- 16.Okiyoneda, T., Niibori, A., Harada, K., Kohno, T., Michalak, M., Duszyk, M., Wada, I., Ikawa, M., Shuto, T., Suico, M. A., and Kai, H. (2008) Biochim. Biophys. Acta 1783 1585-1594 [DOI] [PubMed] [Google Scholar]
- 17.Nakamura, K., Zuppini, A., Arnaudeau, S., Lynch, J., Ahsan, I., Krause, R., Papp, S., De Smedt, H., Parys, J. B., Muller-Esterl, W., Lew, D. P., Krause, K. H., Demaurex, N., Opas, M., and Michalak, M. (2001) J. Cell Biol. 154 961-972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Socolovsky, M., Nam, H., Fleming, M. D., Haase, V. H., Brugnara, C., and Lodish, H. F. (2001) Blood 98 3261-3273 [DOI] [PubMed] [Google Scholar]
- 19.Oliver, J. D., van der Wal, F. J., Bulleid, N. J., and High, S. (1997) Science 275 86-88 [DOI] [PubMed] [Google Scholar]
- 20.David, V., Hochstenbach, F., Rajagopalan, S., and Brenner, M. B. (1993) J. Biol. Chem. 268 9585-9592 [PubMed] [Google Scholar]
- 21.Popov, M., and Reithmeier, R. A. (1999) J. Biol. Chem. 274 17635-17642 [DOI] [PubMed] [Google Scholar]
- 22.Sarabia, V. E., Casey, J. R., and Reithmeier, R. A. (1993) J. Biol. Chem. 268 10676-10680 [PubMed] [Google Scholar]
- 23.Quilty, J. A., and Reithmeier, R. A. (2000) Traffic 1 987-998 [DOI] [PubMed] [Google Scholar]
- 24.Jarolim, P., Rubin, H. L., Brabec, V., Chrobak, L., Zolotarev, A. S., Alper, S. L., Brugnara, C., Wichterle, H., and Palek, J. (1995) Blood 85 634-640 [PubMed] [Google Scholar]
- 25.Quilty, J. A., Li, J., and Reithmeier, R. A. (2002) Am. J. Physiol. 282 F810-F820 [DOI] [PubMed] [Google Scholar]
- 26.Cheung, J. C., Cordat, E., and Reithmeier, R. A. (2005) Biochem. J. 392, 425-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young, M. T., Beckmann, R., Toye, A. M., and Tanner, M. J. (2000) Biochem. J. 350 53-60 [PMC free article] [PubMed] [Google Scholar]
- 28.Groves, J. D., and Tanner, M. J. (1992) J. Biol. Chem. 267 22163-22170 [PubMed] [Google Scholar]
- 29.Chernova, M. N., Jarolim, P., Palek, J., and Alper, S. L. (1995) J. Membr. Biol. 148 203-210 [DOI] [PubMed] [Google Scholar]
- 30.Overton, W. R. (1988) Cytometry 9 619-626 [DOI] [PubMed] [Google Scholar]
- 31.van Anken, E., Romijn, E. P., Maggioni, C., Mezghrani, A., Sitia, R., Braakman, I., and Heck, A. J. (2003) Immunity 18 243-253 [DOI] [PubMed] [Google Scholar]
- 32.Zuppini, A., Groenendyk, J., Cormack, L. A., Shore, G., Opas, M., Bleackley, R. C., and Michalak, M. (2002) Biochemistry 41 2850-2858 [DOI] [PubMed] [Google Scholar]
- 33.Hebert, D. N., Zhang, J. X., Chen, W., Foellmer, B., and Helenius, A. (1997) J. Cell Biol. 139 613-623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, J., Quilty, J., Popov, M., and Reithmeier, R. A. (2000) Biochem. J. 349 51-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groves, J. D., and Tanner, M. J. (1993) Mol. Membr. Biol. 11 31-38 [DOI] [PubMed] [Google Scholar]
- 36.Johnstone, R. M. (1992) Biochem. Cell Biol. 70 179-190 [DOI] [PubMed] [Google Scholar]
- 37.Fishelson, Z., Hochman, I., Greene, L. E., and Eisenberg, E. (2001) Int. Immunol. 13 983-991 [DOI] [PubMed] [Google Scholar]
- 38.Kakhniashvili, D. G., Bulla, L. A., Jr., and Goodman, S. R. (2004) Mol. Cell. Proteomics 3 501-509 [DOI] [PubMed] [Google Scholar]
- 39.Davis, J. Q., Dansereau, D., Johnstone, R. M., and Bennett, V. (1986) J. Biol. Chem. 261 15368-15371 [PubMed] [Google Scholar]
- 40.Geminard, C., Nault, F., Johnstone, R. M., and Vidal, M. (2001) J. Biol. Chem. 276 9910-9916 [DOI] [PubMed] [Google Scholar]
- 41.Singh, M. K., and Yu, J. (1984) Nature 309 631-633 [DOI] [PubMed] [Google Scholar]
- 42.Ribeil, J. A., Zermati, Y., Vandekerckhove, J., Cathelin, S., Kersual, J., Dussiot, M., Coulon, S., Moura, I. C., Zeuner, A., Kirkegaard-Sorensen, T., Varet, B., Solary, E., Garrido, C., and Hermine, O. (2007) Nature 445 102-105 [DOI] [PubMed] [Google Scholar]
- 43.Zaric, J., Lusic, M., Durkovic, A., Glisin, V., and Popovic, Z. (1998) Comp. Haematol. Int. 8 205-209 [Google Scholar]
- 44.Berg, C. P., Engels, I. H., Rothbart, A., Lauber, K., Renz, A., Schlosser, S. F., Schulze-Osthoff, K., and Wesselborg, S. (2001) Cell Death Differ. 8 1197-1206 [DOI] [PubMed] [Google Scholar]
- 45.Low, P. S., Waugh, S. M., Zinke, K., and Drenckhahn, D. (1985) Science 227 531-533 [DOI] [PubMed] [Google Scholar]
- 46.Lux, S. E. in Blood: Principles and Practice of Hematology (Handin, R. I., Lux, S. E., and Stossel, T. P., eds) pp. 1383-1397, J. B. Lippincott Co., Philadelphia
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.