Abstract
The G protein-coupled lysophosphatidic acid 2 (LPA2) receptor elicits prosurvival responses to prevent and rescue cells from apoptosis. However, G protein-coupled signals are not sufficient for the full protective effect of LPA2. LPA2 differs from other LPA receptor subtypes in the C-terminal tail, where it contains a zinc finger-binding motif for the interactions with LIM domain-containing TRIP6 and proapoptotic Siva-1, and a PDZ-binding motif through which it complexes with the NHERF2 scaffold protein. In this report, we identify a unique CXXC motif of LPA2 responsible for the binding to TRIP6 and Siva-1, and demonstrate that disruption of these macromolecular complexes or knockdown of TRIP6 or NHERF2 expression attenuates LPA2-mediated protection from chemotherapeutic agent-induced apoptosis. In contrast, knockdown of Siva-1 expression enhances this effect. Furthermore, a PDZ-mediated direct interaction between TRIP6 and NHERF2 facilitates their interaction with LPA2. Together, these results suggest that in addition to G protein-activated signals, the cooperation embedded in the LPA2-TRIP6-NHERF2 ternary complex provides a novel ligand-dependent signal amplification mechanism that is required for LPA2-mediated full activation of antiapoptotic signaling.
Lysophosphatidic acid (LPA)4 is a growth factor-like lysophospholipid abundantly present in biological fluids. It mediates diverse cellular responses important for cell survival, growth, differentiation, migration, inflammation, angiogenesis, and platelet aggregation (1, 2). At least eight G protein-coupled LPA receptors have been identified: LPA1, LPA2, and LPA3 of the endothelial differentiation gene family and the structurally distinct LPA4/P2Y9, LPA5/GPR92, LPA6/GPR87, LPA7/P2Y5, and LPA8/P2Y10 of the purinergic receptor cluster (3-5). These receptors couple to Gi/o, Gq/11, Gs, and/or G12/13 proteins to activate various signaling pathways. However, the molecular mechanisms underlying the specificity and diversity with which different LPA receptors regulate these wide ranging cellular responses are not yet fully understood.
Substantial evidence has demonstrated that LPA is a survival factor that protects non-transformed cells from different stress-induced apoptosis (6) and renders cancer cells resistance to apoptosis-inducing treatments (1, 2). Among the various G protein-coupled LPA receptors, LPA2 has been shown to mediate the antiapoptotic effect of LPA in vivo. LPA -/-2 mice show significantly increased rates of radiation-induced apoptosis and less crypt survival (7). LPA protects gut epithelial cells from radiation-induced apoptosis in WT and LPA -/-1 mice but not in LPA -/-2 mice (7). The classical paradigm for G protein-coupled receptor (GPCR)-mediated prosurvival signaling involves the coupling of ligand-bound receptors to heterotrimeric G proteins that sequentially activate the downstream effectors involved in Ras/ERK, phosphatidylinositol 3-kinase (PI3K)/AKT, and NF-κB signaling pathways (6). However, recent evidence suggests that non-G protein-coupled signals mediated via the unique C-terminal binding motifs of LPA2 may be required for its antiapoptotic function. LPA2 is the only LPA receptor subtype known to interact with various molecules via the unique binding domains present in its C terminus (8). The last four amino acids, DSTL, of LPA2 bind to several PDZ proteins, including NHERF2 (Na+/H+ exchanger regulatory factor), PDZ-RhoGEF, LARG (leukemia-associated RhoGEF), and MAGI-3 (membrane-associated guanylate kinase with an inverted domain structure-3) (9-13). These scaffold proteins modulate LPA-induced activation of ERK and/or RhoA. NHERF2 can also regulate phospholipase C-β3 signaling pathways and bridge LPA2 to cystic fibrosis transmembrane regulator Cl- channel (9, 10). Through the PDZ-mediated interactions, membrane-localized NHERF2 and MAGI-3 can recruit the phosphatase and tensin homolog in close proximity to the cell surface to restrict PI3K/AKT activity (14, 15). However, NHERF2 can also serve as a scaffold protein for PDK1 (3-phosphoinositide-dependent protein kinase 1), which plays a central role in the activation of AGC family kinases, including AKT (16). It has been reported that knockdown of NHERF2 attenuates LPA-induced AKT activation in colon cancer cells (12). Thus, the function of NHERF2 in restricting or promoting PI3K/AKT signaling may depend on the relative cellular expression levels of phosphatase and tensin homolog versus PDK1.
Two zinc finger proteins, including the LIM domain-containing TRIP6 and Siva-1 proapoptotic protein, have been found to bind to the C-terminal tail of LPA2 (17, 18). The LIM domain is comprised of two zinc finger motifs, which are critical for protein-protein interactions (19). The association of TRIP6 with LPA2 enhances LPA-induced ERK activation and cell migration in a c-Src-dependent manner (20). However, it is unknown whether TRIP6 plays any role in the LPA2-mediated antiapoptotic effect. In contrast, Siva-1, a transcriptional target of p53 and E2F1, functions as a proapoptotic protein during DNA damage response (21). The binding of LPA2 to Siva-1 promotes LPA-dependent ubiquitination and down-regulation of Siva-1 expression (18).
As different LPA receptors show overlapping patterns of G protein coupling, LPA2-mediated protein-protein interactions may be particularly important in the signal amplification and diversification of this receptor subtype. Here we demonstrate that an LPA-induced ternary complex of TRIP6 NHERF2 and LPA2 regulates antiapoptotic function. We have mapped the CXXC zinc finger-binding motif of LPA2 and found the LPA2-mediated antiapoptotic effect was abolished only when both CXXC- and PDZ-binding motifs were disrupted, indicating that the supramolecular complexes formed via the C-terminal tail of LPA2 are required for the full antiapoptotic function of LPA2, and thus play a critical role in the chemoprotective action of LPA in cancer cells.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Site-directed Mutagenesis—The cDNA sequences encoding one of the deletion mutants or point mutants of FLAG-LPA2 were amplified by overlapping extension PCR and inserted into pcDNA3.1 (Invitrogen). These cDNA fragments were subsequently subcloned into pcFUW-puro lentiviral vector. The cDNA sequences encoding full-length NEHRF2, NHERF-PDZ1 (residues 8-99), or NHERF2-PDZ2 (residues 149-228) were inserted in-frame into pEGFP (Clontech) or pGEX-6P3 (Amersham Biosciences). The full-length TRIP6 cDNA sequences were inserted in-frame into pEBFP-C1. The expression vectors of GST-LPA2-CT mutants in which Cys-311 and/or Cys-314 were mutated to Ala were constructed by QuikChange site-directed mutagenesis (Stratagene) using pGEX-6P1-LPA2-CT (17) as the template. The pLVTHM (Addgene) lentiviral vector was used to direct the expression of a short hairpin RNA that specifically targets the 19-nucleotide sequences of mouse TRIP6 (siTRIP6-1) (20), human TRIP6 (siTRIP6-2, siTRIP6-3) (17), mouse Siva-1 (18), or human NHERF2 cDNA (siNHERF2-4 (10) and siNHERF2-5, 5′-CCTGCATAGTGACAAGTCC-3′), respectively. The pGIPZ lentiviral expression vector (Openbiosystems) was used to direct the expression of a miR-30-based mouse NHERF2 short hairpin RNA (siNHERF2-3) or a luciferase control short hairpin RNA, which specifically targets the 22-nucleotide sequences of mouse NHERF2 cDNA (5′-ATCAGAGAAGGACAATGAGGAT-3′) and luciferase cDNA (5′-CCACTTACGCTGAGTACTTCGA-3′), respectively. pCMV-FLAG-ΔTTDC was constructed by deleting the cDNAs encoding the distal four amino acids, TTDC, of TRIP6 using QuikChange site-directed mutagenesis (Stratagene). All of these cDNA constructs have been verified by DNA sequencing.
Stable Transfection—Primary mouse embryonic fibroblasts (MEF) were isolated from E13.5 LPA½ DKO embryos and continuously cultured to obtain the spontaneously immortalized MEF cell lines. These MEFs were transduced with an empty lentivirus or the lentivirus harboring WT or one of the FLAG-LPA2 mutants and selected with puromycin to establish the stable cell lines.
Cellular Co-immunoprecipitation, In Vitro Binding, Immunoblotting, Immunostaining, and LPA-induced Calcium Activation Assay—Experiments and purification of the recombinant Siva-1 and TRIP6 were performed as described previously (7, 17, 18, 20). To detect the interactions of LPA2, NHERF2, and TRIP6 at physiological levels, SKOV-3 cells expressing a scrambled siRNA, a TRIP6 siRNA, or a NHERF2 siRNA were starved in 0.1% fatty acid-free bovine serum albumin (BSA)-containing DMEM overnight, followed by addition of 2 μm LPA for 10 min and harvested. Endogenous LPA2 was immunoprecipitated with an anti-LPA2 rat antibody (a gift from Dr. Junken Aoki) or a control rat IgG. TRIP6 was immunoprecipitated with an anti-TRIP6 mouse monoclonal antibody (BD Biosciences) or a control mouse IgG. Proteins were resolved by SDS-PAGE and subjected to immunoblotting using an antibody specific to NHERF2, LPA2 (gifts from Dr. A. P. Naren), or TRIP6 (Bethyl Laboratories), respectively.
Palmitoylation of LPA2—HEK 293T cells expressing WT or one of the point mutants of FLAG-LPA2 were incubated with [3H]palmitic acid (60 Ci/mmol, Amersham Biosciences) at 37 °C for 2 h. FLAG-LPA2 in the whole cell lysates was immunoprecipitated with the anti-FLAG M2 monoclonal antibody-conjugated agarose beads, resolved by SDS-PAGE, and detected by autoradiography. The blot was then probed with an anti-FLAG antibody to detect FLAG-LPA2. The effect of 2-bromopalmitate (2-BP, Sigma) on the inhibition of LPA2 palmitoylation was determined by pretreating the transfected HEK 293T cells with 100 μm 2-bromopalmitate for 30 min followed by labeling with [3H]palmitic acid.
To determine whether inhibition of palmitoylation affects LPA2 binding to Siva-1, TRIP6, or NHERF2, HEK 293T cells transiently expressing GFP-Siva-1, GFP-TRIP6, or GFP-NHERF2 with or without FLAG-LPA2 were pretreated with 100 μm 2-BP in 0.1% fatty acid-free BSA-containing DMEM for 4 h, followed by addition of 2 μm LPA for 10 min. After immunoprecipitation of FLAG-LPA2, half the precipitates were resolved by SDS-PAGE. Immunoblotting was performed to detect the co-immunoprecipitated GFP-Siva-1, GFP-TRIP6, or GFP-NHERF2. The rest of the samples were subjected to the acyl-biotinyl exchange procedure (22) to determine the levels of palmitoylated LPA2. Precipitated LPA2 was pretreated with 50 mm N-ethylmaleimide (Pierce) for 1 h, followed by the acyl-biotinyl exchange with 1 m hydroxylamine and 0.2 mm EZ-link biotin-N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (HPDP) (Pierce) for another hour at room temperature. Biotinylated FLAG-LPA2 was eluted from the anti-FLAG beads using 100 μg/ml FLAG peptides (Sigma) for competition and subsequently pulled down with avidin beads (Amersham Biosciences). After SDS-PAGE, immunoblotting was performed using an anti-FLAG rabbit polyclonal antibody (Sigma).
Apoptosis Assays—Stable MEFs were starved in 0.1% fatty acid-free BSA-containing DMEM with or without 10 μm LPA for 1 h followed by the addition of 1.5 to 2 μm adriamycin for 7-9 h. Caspase-3/7 activity was determined by cleavage of the luminogenic substrate containing the DEVD sequence (Promega) and was normalized by protein concentrations. To determine the effect of pertussis toxin (PTX) on apoptosis, DKO-LPA2 MEFs were pretreated with 100 ng/ml PTX overnight before the apoptosis assay. Alternatively, apoptosis was determined by annexin V-fluorescein isothiocyanate staining (BD Biosciences) and analyzed by flow cytometry following a 14-h treatment. SKOV-3 cells transduced with the lentivirus harboring a scrambled siRNA, TRIP6 siRNA, or a NHERF2 siRNA were starved and pretreated with LPA followed by the addition of 50 μm cisplatin for 20 h. Apoptosis was determined by caspase-3/7 activity assay and immunoblotting using an antibody specific to PARP-1 (BD Biosciences).
To determine apoptosis by DNA fragmentation assay, stable MEFs were seeded on plates coated with 0.1 mg/ml poly-l-lysine overnight followed by the addition of 3 μm adriamycin in 0.5% fetal bovine serum-containing DMEM. Two μm LPA were added 1 h later. After a 6-h treatment, DNA fragmentation was measured by enzyme-linked immunosorbent assay following the procedure of the Cell Death Detection Kit (Roche).
Statistic Analysis—Statistic significance (p < 0.05) was determined using Student's t test.
RESULTS
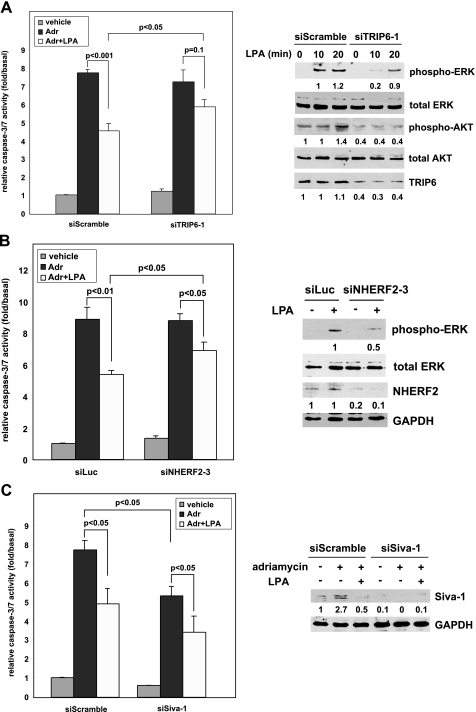
LPA2-mediated Protection from Chemotherapeutic Agent-induced Apoptosis Is Regulated by Proteins Interacting with the C terminus of LPA2—To elucidate the unique properties of LPA2 leading to the attenuation of DNA damage-induced apoptosis, we tested the hypothesis that LPA2-elicited antiapoptotic signaling could be regulated through the interactions with its C terminus-binding partners. We knocked down the expression of TRIP6, NHERF2, or Siva-1 in LPA½ double knock-out (DKO) MEFs stably transduced with a human LPA2 (designated DKO-LPA2 MEFs). These LPA½ DKO MEFs were chosen because LPA fails to induce ERK/AKT activation in these MEFs required for the antiapoptotic effect (Fig. 5C), and cannot protect them from adriamycin-induced apoptosis (Fig. 5D), although they endogenously express LPA4 and LPA7 receptor transcripts (data not shown). We found that 60% knockdown of TRIP6 expression did not further enhance adriamycin-induced caspase-3/7 activation but significantly attenuated LPA-mediated protection and the activation of ERK and AKT (Fig. 1A). Knockdown of NHERF2 expression by 80% also reduced LPA-induced chemoprotection and ERK activation (Fig. 1B). In contrast, knockdown of Siva-1 expression by more than 90% reduced adriamycin-induced caspase-3/7 activation and enhanced the LPA-mediated protective effect (Fig. 1C). These results suggest that in mouse embryonic fibroblasts, the antiapoptotic effect of LPA2 involves NHERF2, TRIP6, and Siva-1.
FIGURE 5.
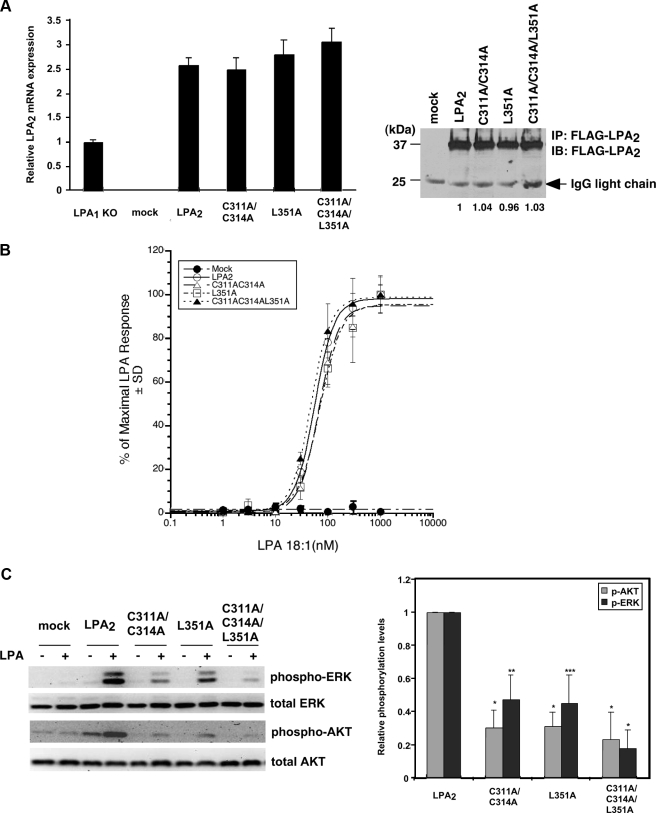
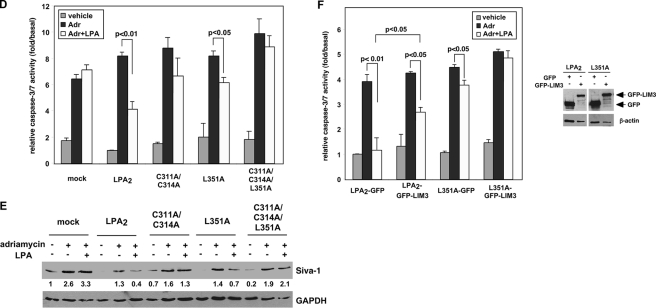
Protein-protein interactions via the CXXC- and/or PDZ-binding motifs do not affect LPA2-mediated Ca2+ response but regulate its chemoprotective effect. A, stable expression of LPA2 or one of the point mutants deficient in binding to the zinc finger proteins and/or PDZ proteins in LPA½ DKO MEFs. Total mRNAs were isolated from LPA1 KO MEFs or one of the LPA½ DKO MEF cell lines (mock, WT LPA2, C311A/C314A, L351A, or C3131A/C314A/L351A). Quantitative real-time reverse transcriptase-PCR was performed to determine the relative expression levels of LPA2 mRNA compared with that expressed in LPA1 KO MEFs and was normalized by the expression of β-actin mRNA. Results show the mean ± S.E. done in triplicates and are representative of two separate experiments. To determine total protein levels of each receptor, FLAG-LPA2 in the whole cell lysates was immunoprecipitated using anti-FLAG M2 monoclonal antibody-conjugated agarose beads and detected with an anti-FLAG rabbit polyclonal antibody. The result shown is representative of five independent experiments. B, mutation of the CXXC motif and/or PDZ-binding motif does not affect LPA2-mediated Ca2+ response. Stable LPA½ DKO MEFs were stimulated with different concentrations of LPA as indicated for 5 min. LPA-induced calcium response was determined as described previously (7). The curves for WT LPA2 and mutants were generated by normalizing the Ca2+ peak at various dilutions to the response elicited by the highest concentration of LPA (1 μm) applied. The values of the mock-transduced MEFs were normalized to that of WT LPA2-transduced MEFs. Data shown are the mean ± S.D. done in triplicates and are representative of two separate experiments. C, mutation of the CXXC motif and/or PDZ-binding motif attenuates LPA2-mediated activation of ERK and AKT. Stable LPA½ DKO MEFs were stimulated with LPA for 10 min. Immunoblotting was performed to detect the levels of phosphorylated and total ERK and AKT (left panel). The intensity of each protein was quantified to determine the relative activation fold of phospho-ERK and AKT by LPA stimulation (right panel). Data shown are the mean ± S.E. of five independent experiments. *, p < 0.001; **, p < 0.01; ***, p < 0.05 versus LPA-stimulated DKO-LPA2 MEFs (Student's t test). D, LPA-mediated protection from adriamycin-induced caspase-3/7 activation is inhibited by mutation of both CXXC- and PDZ-binding motifs. Different LPA½ DKO MEFs transfectants were pretreated with 10 μm LPA in 0.1% fatty acid-free BSA-containing medium for 1 h, followed by addition of 1.7 μm adriamycin for 8 h. Apoptosis was determined by caspase-3/7 activity assay. Data show the mean ± S.E. of four independent experiments. E, mutation of the CXXC motif abrogates LPA2-mediated protection from adriamycin-induced Siva-1 induction. Different LPA½ DKO MEFs were pretreated with 10 μm LPA for 2 h, followed by the addition of 1.5 μm adriamycin for 14 h. Immunoblotting (IB) was performed to detect the expression of Siva-1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in the whole cell lysates. The relative expression of Siva-1 was compared with that expressed in the mock cells without any treatment and was normalized by the levels of GAPDH. The result shown is representative of three independent experiments. F, inhibition of the CXXC motif-mediated interaction with TRIP6-LIM3 attenuates LPA-mediated protection from adriamycin-induced apoptosis. pEGFP or pEGFP-TRIP6-LIM3 was transiently transfected by electroporation into LPA½ DKO MEFs that expressed WT LPA2 or L351A. Cells were treated with 10 μm LPA for 1 h followed by addition of 1.5 μm adriamycin for 9 h. Caspase-3/7 activity was determined and normalized by protein concentrations in each sample. Data show the mean ± S.E. of three independent experiments. The immunoblot shows the expression of GFP-TRIP6-LIM3, GFP, and β-actin in the whole cell lysates.
FIGURE 1.
LPA2-mediated protection from adriamycin-induced apoptosis is regulated by proteins interacting with its C-terminal tail. A, knockdown of TRIP6 expression reduces LPA2-mediated ERK and AKT activation and protection from adriamycin-induced caspase-3/7 activation. LPA½ DKO MEFs stably expressing FLAG-LPA2 (DKO-LPA2) were transduced with the lentivirus harboring a mouse TRIP6 siRNA (siTRIP6-1) or a scrambled control siRNA (siScramble). After starvation for 4 h, cells were treated with 2 μm LPA for 10 or 20 min, and immunoblotting was performed to determine the levels of activated phospho-ERK or phospho-AKT, respectively (right panel). The same blot was reprobed with an antibody specific to ERK, AKT, or TRIP6, respectively. The intensity of each protein was quantified by NIH IMAGE J software and the relative expression of phospho-ERK or phospho-AKT was normalized by the levels of total ERK or AKT. The same cells were starved in 0.1% fatty acid-free BSA-containing DMEM with or without 10 μm LPA for 1 h followed by the addition of 1.7 μm adriamcyin for 9 h, and caspase-3/7 activity was determined by cleavage of the luminogenic substrate containing the DEVD sequence and was normalized by protein concentrations (left panel). Data shown are the mean ± S.E. of five independent experiments. Statistic significance (p < 0.05) was determined by Student's t test. B, knockdown of NHERF2 expression attenuates LPA2-mediated ERK activation and protection from adriamycin-induced apoptosis. DKO-LPA2 MEFs were transduced with the lentivirus harboring a mouse NHERF2 siRNA (siNHERF2-3) or a luciferase control siRNA (siLuc). The starved cells were treated with LPA for 10 min and the levels of activated ERK, total ERK, NHERF2, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in the whole cell lysates were determined by immunoblotting. Adriamycin-induced caspase-3/7 activation were assayed as described in A. Data shown are the mean ± S.E. of three independent experiments. C, inhibition of Siva-1 expression enhances LPA2-mediated protection from adriamycin-induced apoptosis. DKO-LPA2 MEFs were transduced with the lentivirus harboring a Siva-1 siRNA or a scrambled siRNA. The effect of LPA on adriamycin-induced caspase-3/7 activation was determined as described in A. Data shown are the mean ± S.E. of three independent experiments. Immunoblotting was performed to detect the expression of Siva-1 and control GAPDH in the whole cell lysates.
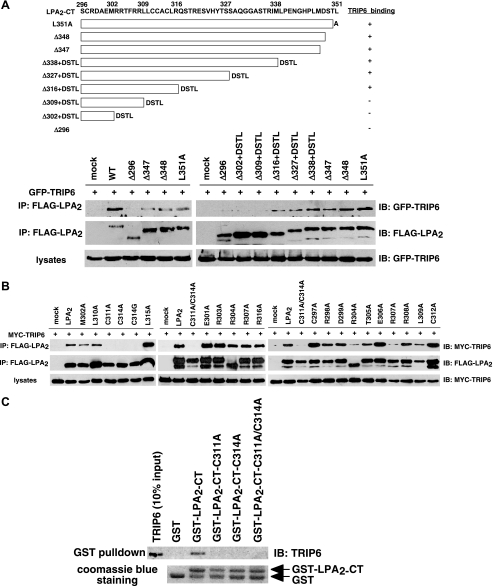
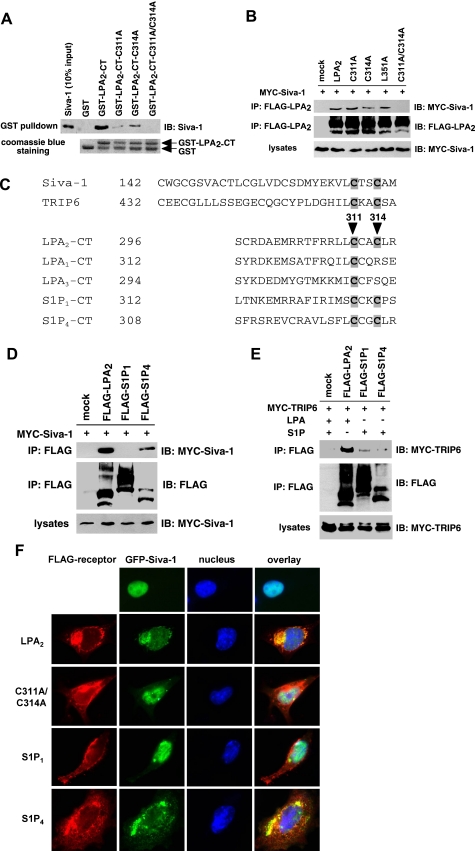
A CXXC Motif Unique to the C-terminal Tail of LPA2 Mediates the Interactions with TRIP6 and Siva-1—To investigate if the effects of these LPA2-interacting partners on LPA-elicited chemoprotection are regulated through direct interactions with LPA2, next we delineated the position of the TRIP6-interacting motif in the C terminus of LPA2. We applied deletion mutagenesis while keeping the DSTL motif intact in the mutants to permit interactions with the PDZ partners. Cellular co-immunoprecipitation demonstrated that except for Δ309+DSTL, Δ302+DSTL, and Δ296 mutants, other deletion mutants of LPA2, including Δ316+DSTL, Δ327+DSTL, Δ338+DSTL, Δ347, and Δ348, and the L351A point mutant that lacks PDZ binding were able to bind to TRIP6, although to a lesser degree than the WT LPA2 (Fig. 2A). This result indicates that the minimal sequences required for TRIP6 binding contain residues 297-316. Alanine scanning mutational analysis of the 297-316 region identified Cys-311 and Cys-314 as the residues required for the interaction with TRIP6 (Fig. 2B). In vitro binding assays confirmed that mutation of Cys-311 and/or Cys-314 to Ala abolished the direct binding of LPA2-CT to TRIP6 (Fig. 2C).
FIGURE 2.
The 311CXXC314 motif of LPA2 is required for the interaction with TRIP6. A, the TRIP6-binding motif of LPA2 is located in the region proximal to Arg-316 of the C-terminal tail. GFP-TRIP6 was expressed in HEK 293T cells with FLAG-LPA2 or one of the LPA2 deletion mutants with or without the DSTL sequences as indicated in the top panel. After stimulation of the cells with 2 μm LPA for 10 min, LPA2 was immunoprecipitated (IP) with anti-FLAG M2 antibody-conjugated agarose beads and resolved by SDS-PAGE. GFP-TRIP6 co-immunoprecipitated with LPA2 was detected by immunoblotting (IB) using an anti-GFP antibody. B, the ability of LPA2 to bind to TRIP6 is eliminated by mutation of Cys-311 and/or Cys-314 to Ala. MYC-TRIP6 was co-expressed with WT or one of the point mutants of LPA2 in HEK 293T cells. Co-immunoprecipitation was performed as described above. MYC-TRIP6 co-immunoprecipitated with LPA2 was detected with an anti-MYC polyclonal antibody. C, Cys-311 or Cys-314 of LPA2 mediates the binding to TRIP6 in vitro. Purified recombinant TRIP6 was incubated with glutathione S-transferase (GST), GST-LPA2-CT, or one of the GST-LPA2-CT mutants at 4 °C for 3 h. TRIP6 pulled down by GST-LPA2-CT was resolved by SDS-PAGE and detected by immunoblotting using an anti-TRIP6 antibody. The bottom panel shows Coomassie Blue staining of GST and GST-LPA2-CT proteins. Data shown in each figure are representative of three independent experiments.
Next we examined the interaction of Siva-1 with WT or cysteine mutants of LPA2. We found that only when both Cys-311 and Cys-314 were mutated was the interaction with Siva-1 completely abolished in vitro (Fig. 3A) and in cells (Fig. 3B). However, this binding was not affected by L351A mutation (Fig. 3B).
FIGURE 3.
Both Cys-311 and Cys-314 residues are required for LPA2 binding to Siva-1. A, in vitro binding of LPA2 to Siva-1 is partially impaired by mutation of Cys-311 or Cys-314 to Ala and is completely abolished when both cysteine residues are mutated. Purified recombinant Siva-1 was incubated with glutathione S-transferase (GST), GST-LPA2-CT, or one of the GST-LPA2-CT mutants. Siva-1 pulled down by GST-LPA2-CT was detected by immunoblotting using an anti-Siva-1 antibody. B, association of LPA2 with Siva-1 is eliminated only when both Cys-311 and Cys-314 residues are mutated to Ala. MYC-Siva-1 was co-expressed with WT or one of the point mutants of FLAG-LPA2 in HEK 293T cells. Cells were starved overnight and then harvested for co-immunoprecipitation (IP) and immunoblotting (IB) as described above. MYC-Siva-1 was detected with an anti-MYC polyclonal antibody. C, amino acid sequence alignment of the C-terminal zinc finger of Siva-1 (residues 142-172) and TRIP6-LIM3 (residues 432-462), and the proximal region of the C-terminal tail of LPA2 (residues 296-316), LPA1 (residues 312-332), LPA3 (residues 294-314), S1P1 (residues 312-322), and S1P4 (residues 308-318). D, Siva-1 interacts with S1P4 but not S1P1. HEK 293T cells expressing MYC-Siva-1 with FLAG-tagged LPA2, S1P1, or S1P4 were subjected to a co-immunoprecipitation experiment as described in B. E, TRIP6 binds weakly to S1P1 and barely associates with S1P4. HEK 293T cells expressing MYC-TRIP6 with FLAG-tagged LPA2, S1P1, or S1P4 were starved for 8 h followed by the addition of 2 μm LPA or S1P for 10 min. Co-immunoprecipitation was performed as described above. Data shown in A, B, D, and E are representative of three independent experiments. F, Siva-1 colocalizes with LPA2 and S1P4 but not S1P1 or the C311A/C314A mutant of LPA2 in the cytosol. GFP-Siva-1 was transiently co-expressed with FLAG-tagged LPA2, C311A/C314A of LPA2, S1P1, or S1P4 in LPA½ DKO MEFs. Cells were fixed, permeabilized, and then incubated with the anti-FLAG M2 monoclonal antibody followed by the Texas Red X-conjugated mouse secondary antibody to detect the FLAG-tagged receptors. GFP-Siva-1 was directly visualized by fluorescence microscopy.
Among the endothelial differentiation gene family lysophospholipid receptors, the CXXC motif is unique only to the LPA-specific LPA2 and sphingosine 1-phosphate (S1P)-specific S1P1 and S1P4 receptors (Fig. 3C). In contrast to LPA2, LPA1 and LPA3 lack the CXXC motif in the C terminus (Fig. 3C) and fail to interact with TRIP6 and Siva-1 (17, 18). S1P4 formed a complex with Siva-1 but barely bound to TRIP6; and S1P1, albeit weakly, associated with TRIP6 but not Siva-1 (Fig. 3, D and E). Thus, the CXXC motifs of distinct endothelial differentiation gene family lysophospholipid receptors are capable of mediating interactions with the C-terminal zinc finger of Siva-1 or TRIP6-LIM3 (Fig. 3C), which have been shown as the LPA2-interacting domains (17, 18); however, additional residues are required for strengthening the interactions.
Previously we have demonstrated in NIH3T3 fibroblasts that Siva-1 colocalizes with LPA2 in the cytosol and the plasma membrane, and this association prevents nuclear translocation of Siva-1 (18). Likewise, Siva-1 colocalized with LPA2 and S1P4 but not S1P1 in the cytosol in LPA½ DKO MEFs (Fig. 3F). When both Cys-311 and Cys-314 of LPA2 were mutated to Ala, colocalization was completely abolished and Siva-1 was found in the nucleus.
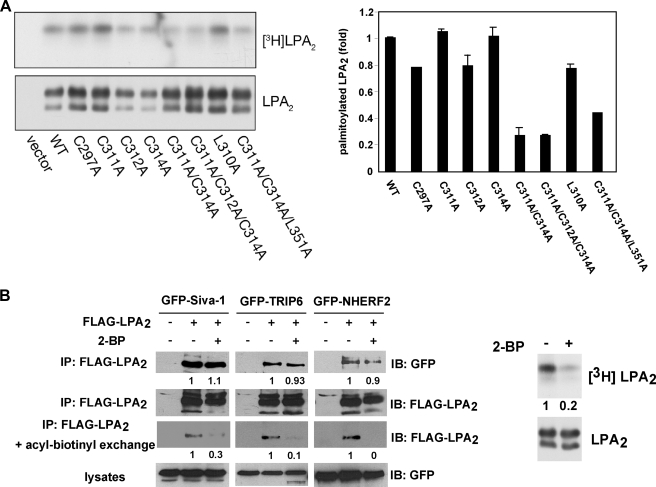
Palmitoylation of LPA2 Does Not Affect Its Ability to Interact with TRIP6, Siva-1, or NHERF2—Many GPCRs undergo cysteine palmitoylation that may affect their coupling to G proteins, cell surface receptor expression, receptor desensitization, or trafficking (23). However, the role of palmitoylation varies among different GPCRs. Among the several cysteine residues located in the C-terminal tail of LPA2, including Cys-297, Cys-311, Cys-312, and Cys-314, mutation of a single cysteine residue either did not or slightly reduced palmitoylation of LPA2 (Fig. 4A). Mutation of two or three cysteine residues partially reduced receptor palmitoylation but did not completely abolish this modification (Fig. 4A).
FIGURE 4.
Palmitoylation modification does not affect LPA2 binding to TRIP6, Siva-1, or NHERF2. A, palmitoylation of LPA2 is partially impaired by mutation of the cysteine residues in the proximal end of its C-terminal tail. WT or one of the point mutants of FLAG-LPA2 were transiently expressed in HEK 293T cells and labeled with [3H]palmitic acid at 37 °C for 2 h. FLAG-LPA2 was immunoprecipitated with anti-FLAG M2 monoclonal antibody-conjugated agarose beads, resolved by SDS-PAGE, and detected by autoradiography (left top panel), followed by immunoblotting (IB) with an anti-FLAG antibody (left bottom panel). The right panel shows the relative levels of palmitoylation of each mutant compared with WT LPA2 after normalization by total receptor expression. Data shown are the mean of two to three independent experiments. B, association of LPA2 with Siva-1, TRIP6, or NHERF2 is not affected by blocking palmitoylation of LPA2. GFP-Siva-1, GFP-TRIP6, or GFP-NHERF2 was co-expressed with FLAG-LPA2 in HEK 293T cells. After treatment with 100 μm 2-BP in 0.1% fatty acid-free BSA-containing DMEM for 4 h, cells were stimulated with LPA for 10 min and FLAG-LPA2 was immunoprecipitated with anti-FLAG M2 monoclonal antibody-conjugated agarose beads. Half the precipitates were resolved by SDS-PAGE and the immunoblot (IB) was probed with an anti-GFP polyclonal antibody to detect co-immunoprecipitated GFP-Siva-1, GFP-TRIP6, or GFP-NHERF2. The rest of the samples were subjected to the acyl-biotinyl exchange procedure to determine levels of palmitoylated LPA2 as described under “Experimental Procedures.” A separate experiment was performed to determine the effect of 2-BP on blocking palmitoylation of LPA2 by pretreating the FLAG-LPA2-expressing HEK 293T cells with 100 μm 2-BP for 30 min followed by labeling with [3H]palmitic acid for 2 h (right panel). The relative levels of Siva-1, TRIP6, or NHERF2 co-immunoprecipitated (IP) with FLAG-LPA2 were quantified and normalized to the levels of FLAG-LPA2 immunoprecipitates. Data shown are representative of three separate experiments.
To evaluate whether palmitoylation affects LPA2 binding to TRIP6, Siva-1, or NHERF2, we used 2-bromopalmitate to block palmitoylation and examined these interactions. Although 2-bromopalmitate inhibits palmitoylation of LPA2 as assayed by determining the levels of palmitoylated LPA2 with the acyl-biotinyl exchange procedure (22) and metabolic labeling of the receptor with [3H]palmitic acid, it did not significantly affect interactions of LPA2 with TRIP6, Siva-1, or NHERF2 (Fig. 4B).
G Protein-mediated LPA2 Signaling to Calcium Mobilization Is Not Affected by Mutation of the CXXC- and/or PDZ-binding Motifs—To investigate the impact of mutations of the C-terminal binding motifs on LPA2 functions, lentiviral constructs harboring WT LPA2, the C311A/C314A mutant that is unable to interact with TRIP6 and Siva-1, the L351A mutant defective in binding to PDZ proteins, or the C311A/C314A/L351A mutant that cannot bind any of the interacting partners were stably transduced into the LPA½ DKO MEFs. Compared with the endogenous LPA2 mRNA expressed in the LPA -/-1 MEFs, the WT and point mutants of LPA2 mRNA were expressed at 2.5- to 3-fold higher but comparable levels in the stable LPA½ DKO MEF cell lines (Fig. 5A). They were also expressed at similar protein levels in the whole cell lysates (Fig. 5A) and on the cell surface demonstrated by flow cytometry analysis (data not shown). LPA-induced Ca2+ mobilization showed indistinguishable dose-response curves in the MEFs expressing WT or in any of the LPA2 mutants (Fig. 5B), indicating that these binding motifs do not affect the Gq/11 signaling branch. These findings also imply that Gq/11-mediated signaling events are not altered by disruption of the palmitoylation modification of Cys-311 and Cys-314.
LPA2-mediated Chemoprotection Is Attenuated by Mutation of the CXXC- and PDZ-binding Motifs—We reasoned that if LPA2-mediated chemoprotection is regulated by these interacting partners, disruption of the interactions in itself would eliminate the function of TRIP6 and NHERF2 in promoting LPA2-mediated prosurvival signaling and allow the stabilization of Siva-1, which can enhance chemotherapeutic agent-induced apoptosis. Indeed, the efficacy of LPA2 in mediating LPA-induced ERK and AKT activation was significantly attenuated by C311A/C314A or L351A mutation and was completely abolished by the C311A/C314A/L351A mutation (Fig. 5C). These results suggest that the CXXC- and PDZ-binding motifs cooperatively regulate LPA2-mediated prosurvival signaling.
Next we examined the effect of LPA on protecting adriamycin-induced apoptosis in these MEFs. The DNA fragmentation assay showed that following a 6-h adriamycin treatment, LPA protected cells from apoptosis in the LPA½ DKO MEFs that expressed WT LPA2, C311A/C314A, or L351A but not in the mock-transfected LPA½ DKO MEFs or those expressing the C311A/C314A/L351A mutant (supplemental Fig. S1A). After an 8-h treatment, the antiapoptotic efficacy of LPA2 measured by the caspase-3/7 activity assay (Fig. 5D) was reduced by the C311A/C314A or L351A mutations and was completely abolished by the C311A/C314A/L351A mutation. A similar effect was also observed using annexin V staining after a 14-h treatment (supplemental Fig. S1B).
Previously we have shown that LPA inhibits adriamycin-induced Siva-1 expression and its proapoptotic functions. Analysis of these LPA2 mutants showed that the inhibitory effect of LPA on adriamycin-induced Siva-1 expression was eliminated by mutation of the CXXC motif but not the PDZ-binding motif, confirming that this regulation is mediated through the direct interaction of LPA2 with Siva-1 (Fig. 5E). Together, these results suggest that the CXXC- and PDZ-binding motifs differentially and cooperatively regulate LPA2 function in protecting cells from adriamycin-induced apoptosis.
The role of Siva-1 in apoptosis is well established (18); however, TRIP6 has not yet been implicated in antiapoptotic signaling and for this reason we focused our investigation on this adapter protein. The LIM3 domain of TRIP6 binds to LPA2, and has been shown to serve as a dominant-negative probe to attenuate LPA2-mediated cell migration previously (17). Overexpression of the GFP-TRIP6-LIM3 mutant reduced the chemoprotective effect of LPA in the DKO-LPA2 MEFs, and this inhibitory effect was augmented in the DKO-L351A MEFs (Fig. 5F), supporting the notion that cooperative protein-protein interactions via both CXXC- and PDZ-binding motifs are required for the maximal protective effect of LPA2. NHERF2 binds to LPA2 through its PDZ2 domain (10). However, we did not succeed in overexpressing NHERF2-PDZ2 to attenuate the protective effect of LPA (data not shown).
The results also showed that when Gi/o signaling was inhibited with PTX, LPA-mediated ERK activation and protection from adriamycin-induced apoptosis were partially attenuated in DKO-LPA2 MEFs (supplemental Fig. S2), suggesting that Gi/o signaling to some extent is involved in the LPA2-mediated antiapoptotic signaling. Nonetheless, LPA-induced recruitment of Siva-1, TRIP6, or NHERF2 was not significantly altered by treatment with PTX or the U73122 phospholipase C inhibitor (supplemental Fig. S3, A and B), suggesting that the macromolecular complex formation via the C terminus of LPA2 is independent on Gi/o or Gq/11 signaling.
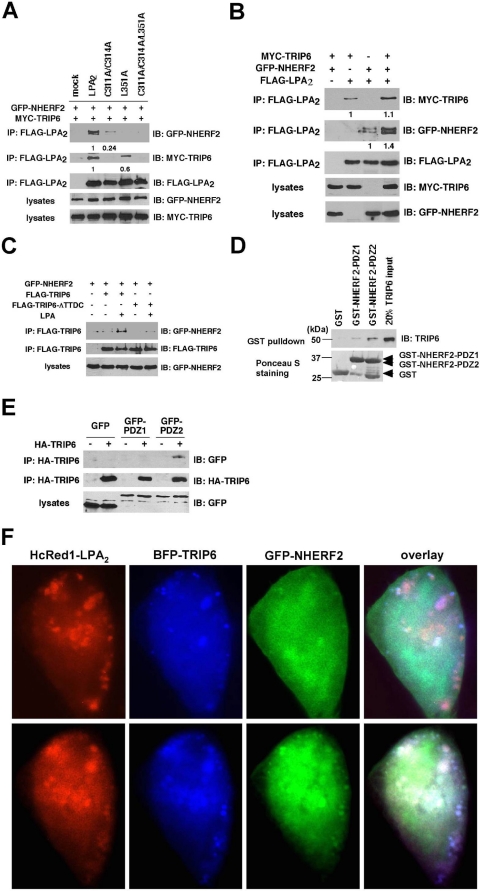
The PDZ-mediated Association of TRIP6 with NHERF2 Facilitates Their Interaction with LPA2—When we examined the interaction of different LPA2 mutants with TRIP6 or NHERF2, we noticed that the C311A/C314A mutant, which does not bind to TRIP6, also showed reduced association with NHERF2 compared with WT LPA2 (Fig. 6A). Likewise, the L351A mutant defective in binding to NHERF2 showed reduced association with TRIP6 (Fig. 6A) but not Siva-1 (Fig. 3B). Conversely, the interaction of NHERF2 or TRIP6 with LPA2 was further enhanced when all three proteins were overexpressed (Fig. 6B). TRIP6 contains a C-terminal TTDC PDZ-binding motif, potentially allowing it to interact with PDZ proteins, raising the possibility that cooperativity might exist between TRIP6 and NHERF2 in interacting with LPA2. Indeed, we found LPA induced the association of NHERF2 with TRIP6 but not the TRIP6-ΔTTDC mutant in HEK 293T cells (Fig. 6C). Domain mapping confirmed that TRIP6 preferentially binds to PDZ2 but not PDZ1 of NHERF2 in vitro (Fig. 6D) and also in HEK 293T cells (Fig. 6E). Because both LPA2 and TRIP6 bind to the PDZ2 domain of NHERF2, NHERF2 must be present in dimer form to bridge LPA2 and TRIP6. In support of this notion, it is known that NHERF2 forms oligomers through PDZ1- and/or PDZ2-mediated self-association (24).
FIGURE 6.
A PDZ-mediated association of TRIP6 and NHERF2 facilitates their interactions with LPA2. A, mutation of the CXXC (PDZ-binding) motif of LPA2 abolishes the binding to TRIP6 (NHERF2) and also reduces the association with NHERF2 (TRIP6). GFP-NHERF2 and MYC-TRIP6 were coexpressed with WT or one of the mutants of FLAG-LPA2 in HEK 293T cells. Co-immunoprecipitation (IP) was performed as described above. The relative levels of co-immunoprecipitated TRIP6 or NHERF2 were quantified and normalized to the immunoprecipitated WT or mutant of LPA2. B, overexpression of TRIP6 enhances association of NHERF2 with LPA2, and vice versa. FLAG-LPA2 was co-expressed with MYC-TRIP6 and/or GFP-NHERF2 in HEK 293T cells as indicated. After stimulation of the cells with 2 μm LPA for 10 min, co-immunoprecipitation was performed. GFP-NHERF2 and MYC-TRIP6 co-immunoprecipitated with FLAG-LPA2 were detected with an anti-GFP antibody and an anti-MYC antibody, respectively. The blot was reprobed with an anti-FLAG antibody to detect the immunoprecipitated LPA2. The relative levels of co-immunoprecipitated TRIP6 or NHERF2 were quantified and normalized to the immunoprecipitated FLAG-LPA2. C, C-terminal TTDC sequences of TRIP6 mediate the binding to NEHRF2. FLAG-TRIP6 or FLAG-TRIP6-ΔTTDC mutant lacking the C-terminal PDZ-binding motif was co-expressed with GFP-NHERF2 in HEK 293T cells. After stimulation of the cells with LPA for 10 min, WT or the ΔTTDC mutant of TRIP6 was immunoprecipitated with anti-FLAG M2 mouse monoclonal antibody-conjugated agarose beads and resolved by SDS-PAGE. Immunoblotting (IB) was performed using the antibodies specific to GFP and the FLAG epitope to detect GFP-NHERF2 and FLAG-TRIP6, respectively. D, TRIP6 binds to the PDZ domain of NHERF2 directly in vitro. Purified recombinant TRIP6 was incubated with GST, GST-NEHRF2-PDZ1, or GST-NHERF2-PDZ2 at 4 °C for 3 h. TRIP6 pulled down by glutathione S-transferase (GST) fusion proteins was detected by immunoblotting using an anti-TRIP6 antibody. The bottom panel shows expression of GST fusion proteins by Ponceau S staining. E, TRIP6 interacts with the PDZ2 but not PDZ1 domain of NHERF2 in cells. HA-TRIP6 was co-expressed with GFP, GFP-NHERF2-PDZ1, or GFP-NHERF2-PDZ2 in HEK 293T cells. TRIP6 in the whole cell lysates was immunoprecipitated with anti-HA mouse monoclonal antibody-conjugated agarose beads and resolved by SDS-PAGE. The immunoblot was probed with an anti-GFP antibody to detect GFP-NHERF2-PDZ2. The blot was reprobed with an anti-HA rabbit antibody to detect the immunoprecipitated HA-TRIP6. Data shown in each figure are representative of two to four independent experiments. HA, hemagglutinin. F, LPA2, TRIP6, and NHERF2 colocalize in cells. Hc-Red1-LPA2 was transiently co-expressed with BFP-TRIP6 and GFP-NHERF2 in LPA½ DKO MEFs. Cells were starved for 1 h, followed by addition of 2 μm LPA for 10 min. Subcellular distribution of these molecules was visualized by fluorescence microscopy.
To address whether LPA2, TRIP6, and NHERF2 are present in the same macromolecular complex, fluorescence microscopy was performed to examine subcellular distribution of these molecules. We found that HcRed1-LPA2, BFP-TRIP6, and GFP-NHERF2 formed clusters and colocalized in close proximity to the plasma membrane or inside the cytosol after LPA treatment for 10 min (Fig. 6F). Together, these results suggest that LPA2 forms a ternary macromolecular complex with TRIP6 and NHERF2 by LPA stimulation.
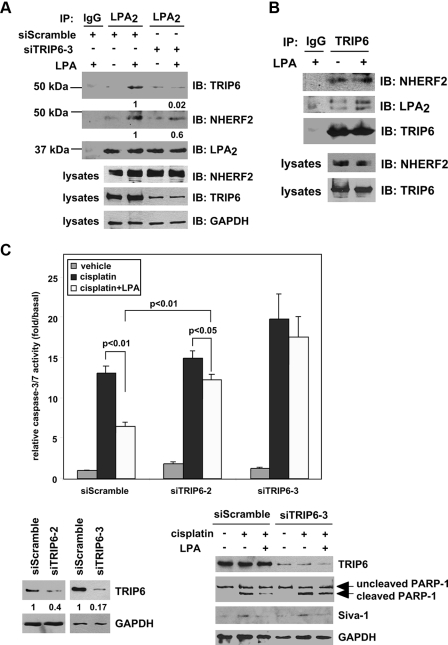
TRIP6 and NHERF2 Regulate LPA-mediated Chemoprotection in Ovarian Cancer Cells—To understand the physiological relevance of NHERF2-TRIP6-LPA2 ternary complex formation, co-immunoprecipitation was performed in SKOV-3 cells that express high levels of these three proteins. LPA induced the association of LPA2 with both TRIP6 and NHERF2 at physiological levels (Fig. 7A). When TRIP6 expression was knocked down, the interaction of NHERF2 with LPA2 was significantly attenuated, suggesting that TRIP6 facilitates this association. However, the association of TRIP6 with LPA2 was not significantly altered by knockdown of NHERF2 expression (supplemental Fig. S4), perhaps because either TRIP6 binds to LPA2 with a higher affinity or other LPA2-interacting PDZ proteins can also facilitate the association of TRIP6 with LPA2. In SKOV-3 cells, TRIP6 associated with NHERF2 constitutively. Nonetheless, formation of the ternary complex required LPA stimulation (Fig. 7B).
FIGURE 7.
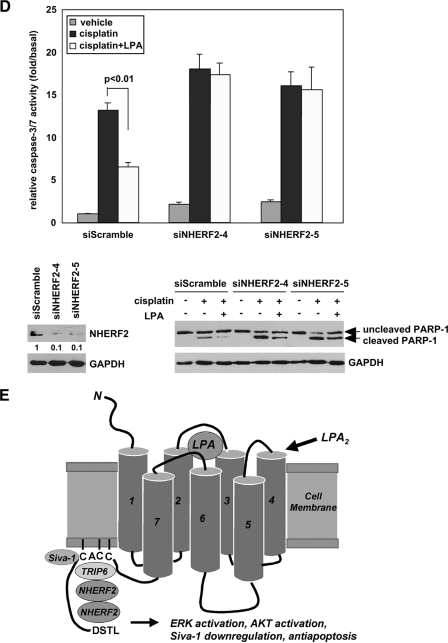
Endogenous TRIP6 and NHERF2 form a ternary complex with LPA2 by LPA stimulation, and regulate LPA-mediated chemoprotection in SKOV-3 cells. A, inhibition of TRIP6 expression reduces LPA-induced association of NHERF2 with LPA2. SKOV-3 cells expressing a scrambled siRNA or a TRIP6 siRNA (siTRIP6-3) were starved overnight followed by treatment with 2 μm LPA for 10 min. LPA2 in the whole cell lysates was immunoprecipitated (IP) with an anti-LPA2 rat antibody or a control IgG and resolved by SDS-PAGE. Immunoblotting (IB) was performed using the antibodies specific to NHERF2, TRIP6, and LPA2, respectively. The bottom three panels show expression of NHERF2, TRIP6, and control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in the whole cell lysates. The relative levels of co-immunoprecipitated TRIP6 or NHERF2 were quantified and normalized to the immunoprecipitated FLAG-LPA2. B, TRIP6 interacts with NHERF2 constitutively in SKOV-3 cells. SKOV-3 cells were treated with LPA as described in A. TRIP6 in the whole cell lysates was immunoprecipitated with an anti-TRIP6 mouse monoclonal antibody or a control IgG. After SDS-PAGE, the immunoblot was probed with an anti-NHERF2 antibody, an anti-LPA2 antibody followed by an anti-TRIP6 antibody. Results shown in A and B are representative of three independent experiments. C and D, LPA-mediated protection of SKOV-3 cells from cisplatin-induced apoptosis is eliminated by knockdown of TRIP6 or NHERF2 expression. SKOV-3 cells expressing a scrambled siRNA or one of the siRNAs that specifically target human TRIP6 (siTRIP6-2 and siTRIP6-3) (C) or NHERF2 (siNHERF2-4, siNHERF2-5) (D) as indicated were pretreated with 10 μm LPA for 1 h followed by the addition of 50 μm cisplatin for 20 h. Caspase-3/7 activity was determined. Data shown are the mean ± S.E. of three independent experiments. The knockdown effect of each TRIP6 siRNA or NHERF2 siRNA was determined by immunoblotting using an antibody specific to TRIP6 or NHERF2, respectively. Half of the lysates as indicated were subjected to immunoblotting using the antibodies specific to PARP-1, Siva-1, NHERF2, and GAPDH, respectively. E, a model for the regulation of LPA2-mediated antiapoptotic signaling through the CXXC-mediated interaction with Siva-1, and the CXXC- and PDZ-mediated LPA2-TRIP6-NHERF2 ternary complex formation.
LPA also protected cells from cisplatin-induced caspase-3/7 activation and PARP-1 cleavage in SKOV-3 ovarian cancer cells (Fig. 7C). We found 60% knockdown of TRIP6 expression attenuated LPA-mediated chemoprotection in SKOV-3 cells (Fig. 7C) as that shown in DKO-LPA2 MEFs (Fig. 1A). When TRIP6 expression was knocked down by 80-90%, the protective effect of LPA on cisplatin-induced caspase-3/7 activation and PARP-1 cleavage was almost completely eliminated (Fig. 7C). A similar effect was found by 90% knockdown of NHERF2 expression (Fig. 7D). Together, these results suggest that both TRIP6 and NHERF2 play a significant role in the LPA-mediated antiapoptotic effect in SKOV-3 cells. Lacking functional p53, cisplatin only induced modest Siva-1 expression in SKOV-3 cells (Fig. 7C). Although both TRIP6 and Siva-1 bind to the CXXC motif of LPA2, knockdown of TRIP6 expression did not alter the effect of LPA on reducing Siva-1 expression.
In summary, these data suggest that LPA induces the formation of a ternary complex containing LPA2, TRIP6, and NHERF2. Our results favor the model that in addition to binding to Siva-1 and down-regulating its activity, LPA2 forms a supramolecular complex with TRIP6 and NHERF2. Together, they coordinately regulate the antiapoptotic signaling of LPA2 (Fig. 7E).
DISCUSSION
GPCRs are increasingly viewed as a nidus for generating ligand-activated intracellular signals via interactions with G proteins and non-G protein signaling molecules (25, 26). We hypothesized that the macromolecular complex formed via the unique C-terminal binding motifs of LPA2 could be responsible for its antiapoptotic function. This hypothesis was based in part on the high degree of sequence diversity in the C termini of the endothelial differentiation gene family LPA receptors, which show only seven of 55 residues are conserved in the C terminus, in contrast to the 85% homology in their transmembrane domains. In support of this hypothesis, it has been shown that PDZ proteins, including NHERF2, interact with the C-terminal DSTL motif of LPA2 but not with other LPA receptor subtypes (10, 12, 13). Moreover, LPA2 is the only LPA receptor subtype that interacts with Siva-1 and TRIP6 (17, 18).
Using the LPA½ DKO MEFs that stably express a human LPA2 as the model system, we present evidence that siRNA-mediated knockdown of TRIP6 or NHERF2 expression attenuates LPA2-mediated chemoprotection; in contrast, knockdown of Siva-1 enhances this effect. We have mapped the 311CXXC314 motif of LPA2 required for interactions with TRIP6 and Siva-1, and demonstrated that disruption of either the CXXC motif or PDZ-binding motif attenuated LPA2-mediated chemoprotection, and only when both binding motifs were disrupted, it completely abolished this effect. Together, these data indicate that LPA2-mediated chemoprotection is regulated through these supramolecular complexes.
Palmitoylation of the four C-terminal cysteine residues of LPA2 showed that those in the CXXC motif can be lipid-modified, however, inhibition of this modification did not disrupt interaction with TRIP6 or Siva-1, indicating that palmitoylation of LPA2 is neither required nor preclusive for the interaction with TRIP6 or Siva-1. We also found that blocking Gi/o protein activation with PTX or inhibiting Gq/11 signaling with the U73122 phospholipase C inhibitor did not affect the interaction of LPA2 with Siva-1, TRIP6, or NHERF2, suggesting that G protein signaling is also not required for these protein interactions. On the other hand, mutation of the CXXC- or PDZ-binding motif of LPA2 did not affect Gq/11-mediated Ca2+ transients upon LPA stimulation, indicating that these binding motifs do not affect the Gq/11 signaling branch.
Unexpectedly, we found that truncation or point mutations that abolish the interactions of LPA2 with PDZ proteins also attenuated the binding of TRIP6 to LPA2. These observations led us to hypothesize that cooperativity might exist between TRIP6 and NHERF2 in interacting with LPA2. Indeed, we found evidence that TRIP6 interacts with the PDZ2 domain of NHERF2 via its C-terminal TTDC PDZ-binding motif. Overexpression of TRIP6 augments the complex formation between LPA2 and NHERF2, whereas knockdown of TRIP6 diminishes it. Moreover, upon ligand activation LPA2 colocalizes with both TRIP6 and NHERF2. Thus, it is likely that LPA2, TRIP6, and NHERF2 form a ternary complex in the microdomain of the plasma membrane to coordinately regulate LPA2-elicited chemoprotective effect. This complex appears to be assembled upon LPA stimulation in SKOV-3 cells at endogenous levels of the participating proteins. Our new data and previous reports (10, 12, 20, 27) show that both TRIP6 and NHERF2 are involved in LPA-induced ERK and AKT activation. SKOV-3 ovarian cancer cells show very low levels of phosphatase and tensin homolog but high activity of AKT (28, 29). When TRIP6 or NHERF2 was knocked down to a great extent, it almost completely eliminated LPA-mediated chemoprotection in SKOV-3 cells. Similarly, it has been reported that knockdown of NHERF2 expression abrogates the chemoprotective effect of LPA in colon cancer cells (27).
We also note that the CXXC motif is required for the antiapoptotic effect through the inhibition of Siva-1 signaling. However, Siva-1 interaction with LPA2 does not appear to be affected by disruption of the LPA2 interaction with PDZ proteins. Our data showed that PTX slightly attenuated LPA-induced chemoprotection, suggesting that Gi/o-mediated signals contribute to but are not sufficient for the full antiapoptotic effect of LPA2. Assembly of the LPA2-TRIP6-NHERF2 ternary complex appears to play a fundamental role in the ability of LPA to render cancer cells resistant to chemotherapeutic agents as we have demonstrated for the case of adriamycin and cisplatin.
Taken together, these data point to a novel signal amplification/diversification mechanism originating from the GPCR signal transduction hub. We favor the hypothesis that signals from protein-protein interactions via the C-terminal CXXC- and PDZ-binding motifs are integrated with G protein-activated signals to cooperatively regulate the antiapoptotic function of LPA2.
Supplementary Material
Acknowledgments
We thank Dr. Junken Aoki for providing the anti-LPA2 antibody, Dr. A. P. Naren for the anti-NHERF2 antibody and pGEX-NHERF2, and Dr. Tim Towne for sharing the flow cytometry equipment.
This work was supported, in whole or in part, by National Institutes of Health Grants CA92160 (to G. T.), AI 080405 (to G. T.), and CA100848 (to F. L.). This work was also supported by a grant from the Marsha Rivkin Center for Ovarian Cancer Research (to F. L.). G. T. is a founder of RxBio Inc. and a member of the scientific advisory board.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S4.
Footnotes
The abbreviations used are: LPA, lysophosphatidic acid; GPCR, G protein-coupled receptor; 2-BP, 2-bromopalmitate; ERK, extracellular signal-regulated kinase; PI3K, phosphatidylinositol 3-kinase; WT, wild type; siRNA, small interfering RNA; BSA, bovine serum albumin; DMEM, Dulbecco's modified Eagle's medium; PTX, pertussis toxin; MEF, mouse embryonic fibroblast; DKO, double knock-out; S1P, sphingosine 1-phosphate; GFP, green fluorescent protein.
References
- 1.Mills, G. B., and Moolenaar, W. H. (2003) Nat. Rev. Cancer 3 582-591 [DOI] [PubMed] [Google Scholar]
- 2.Moolenaar, W. H., van Meeteren, L. A., and Giepmans, B. N. (2004) Bioessays 26 870-881 [DOI] [PubMed] [Google Scholar]
- 3.Parrill, A. L. (2008) Biochim. Biophys. Acta 1781 540-546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasternack, S. M., von Kugelgen, I., Aboud, K. A., Lee, Y. A., Ruschendorf, F., Voss, K., Hillmer, A. M., Molderings, G. J., Franz, T., Ramirez, A., Nurnberg, P., Nothen, M. M., and Betz, R. C. (2008) Nat. Genet. 40 329-334 [DOI] [PubMed] [Google Scholar]
- 5.Tabata, K., Baba, K., Shiraishi, A., Ito, M., and Fujita, N. (2007) Biochem. Biophys. Res. Commun. 363 861-866 [DOI] [PubMed] [Google Scholar]
- 6.Radeff-Huang, J., Seasholtz, T. M., Matteo, R. G., and Brown, J. H. (2004) J. Cell Biochem. 92 949-966 [DOI] [PubMed] [Google Scholar]
- 7.Deng, W., E, S., Tsukahara, R., Valentine, W. J., Durgam, G., Gududuru, V., Balazs, L., Manickam, V., Arsura, M., VanMiddlesworth, L., Johnson, L. R., Parrill, A. L., Miller, D. D., and Tigyi, G. (2007) Gastroenterology 132 1834-1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin, F. T., and Lai, Y. J. (2008) Biochim. Biophys. Acta 1781 558-562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, C., Dandridge, K. S., Di, A., Marrs, K. L., Harris, E. L., Roy, K., Jackson, J. S., Makarova, N. V., Fujiwara, Y., Farrar, P. L., Nelson, D. J., Tigyi, G. J., and Naren, A. P. (2005) J. Exp. Med. 202 975-986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oh, Y. S., Jo, N. W., Choi, J. W., Kim, H. S., Seo, S. W., Kang, K. O., Hwang, J. I., Heo, K., Kim, S. H., Kim, Y. H., Kim, I. H., Kim, J. H., Banno, Y., Ryu, S. H., and Suh, P. G. (2004) Mol. Cell Biol. 24 5069-5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada, T., Ohoka, Y., Kogo, M., and Inagaki, S. (2005) J. Biol. Chem. 280 19358-19363 [DOI] [PubMed] [Google Scholar]
- 12.Yun, C. C., Sun, H., Wang, D., Rusovici, R., Castleberry, A., Hall, R. A., and Shim, H. (2005) Am. J. Physiol. 289 C2-C11 [DOI] [PubMed] [Google Scholar]
- 13.Zhang, H., Wang, D., Sun, H., Hall, R. A., and Yun, C. C. (2007) Cell. Signal. 19 261-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi, Y., Morales, F. C., Kreimann, E. L., and Georgescu, M. M. (2006) EMBO J. 25 910-920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu, Y., Dowbenko, D., Spencer, S., Laura, R., Lee, J., Gu, Q., and Lasky, L. A. (2000) J. Biol. Chem. 275 21477-21485 [DOI] [PubMed] [Google Scholar]
- 16.Chun, J., Kwon, T., Lee, E., Suh, P. G., Choi, E. J., and Sun Kang, S. (2002) Biochem. Biophys. Res. Commun. 298 207-215 [DOI] [PubMed] [Google Scholar]
- 17.Xu, J., Lai, Y. J., Lin, W. C., and Lin, F. T. (2004) J. Biol. Chem. 279 10459-10468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, F. T., Lai, Y. J., Makarova, N., Tigyi, G., and Lin, W. C. (2007) J. Biol. Chem. 282 37759-37769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bach, I. (2000) Mech. Dev. 91 5-17 [DOI] [PubMed] [Google Scholar]
- 20.Lai, Y. J., Chen, C. S., Lin, W. C., and Lin, F. T. (2005) Mol. Cell. Biol. 25 5859-5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fortin, A., MacLaurin, J. G., Arbour, N., Cregan, S. P., Kushwaha, N., Callaghan, S. M., Park, D. S., Albert, P. R., and Slack, R. S. (2004) J. Biol. Chem. 279 28706-28714 [DOI] [PubMed] [Google Scholar]
- 22.Wan, J., Roth, A. F., Bailey, A. O., and Davis, N. G. (2007) Nat. Protoc. 2 1573-1584 [DOI] [PubMed] [Google Scholar]
- 23.Qanbar, R., and Bouvier, M. (2003) Pharmacol. Ther. 97 1-33 [DOI] [PubMed] [Google Scholar]
- 24.Lau, A. G., and Hall, R. A. (2001) Biochemistry 40 8572-8580 [DOI] [PubMed] [Google Scholar]
- 25.Hur, E. M., and Kim, K. T. (2002) Cell. Signal. 14 397-405 [DOI] [PubMed] [Google Scholar]
- 26.Maudsley, S., Martin, B., and Luttrell, L. M. (2005) J. Pharmacol. Exp. Ther. 314 485-494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rusovici, R., Ghaleb, A., Shim, H., Yang, V. W., and Yun, C. C. (2007) Biochim. Biophys. Acta 1770 1194-1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longva, K. E., Pedersen, N. M., Haslekas, C., Stang, E., and Madshus, I. H. (2005) Int. J. Cancer 116 359-367 [DOI] [PubMed] [Google Scholar]
- 29.Wang, H. Q., Altomare, D. A., Skele, K. L., Poulikakos, P. I., Kuhajda, F. P., Di Cristofano, A., and Testa, J. R. (2005) Oncogene 24 3574-3582 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.