Abstract
Proteins evolve in a fitness landscape encompassing a complex network of biological constraints. Because of the interrelation of folding, function, and regulation, the ground-state structure of a protein may be inactive. A model is provided by insulin, a vertebrate hormone central to the control of metabolism. Whereas native assembly mediates storage within pancreatic β-cells, the active conformation of insulin and its mode of receptor binding remain elusive. Here, functional surfaces of insulin were probed by photocross-linking of an extensive set of azido derivatives constructed by chemical synthesis. Contacts are circumferential, suggesting that insulin is encaged within its receptor. Mapping of photoproducts to the hormone-binding domains of the insulin receptor demonstrated alternating contacts by the B-chain β-strand (residues B24-B28). Whereas even-numbered probes (at positions B24 and B26) contact the N-terminal L1 domain of the α-subunit, odd-numbered probes (at positions B25 and B27) contact its C-terminal insert domain. This alternation corresponds to the canonical structure of aβ-strand (wherein successive residues project in opposite directions) and so suggests that the B-chain inserts between receptor domains. Detachment of a receptor-binding arm enables photo engagement of surfaces otherwise hidden in the free hormone. The arm and associated surfaces contain sites also required for nascent folding and self-assembly of storage hexamers. The marked compression of structural information within a short polypeptide sequence rationalizes the diversity of diabetes-associated mutations in the insulin gene. Our studies demonstrate that photoscanning mutagenesis can decode the active conformation of a protein and so illuminate cryptic constraints underlying its evolution.
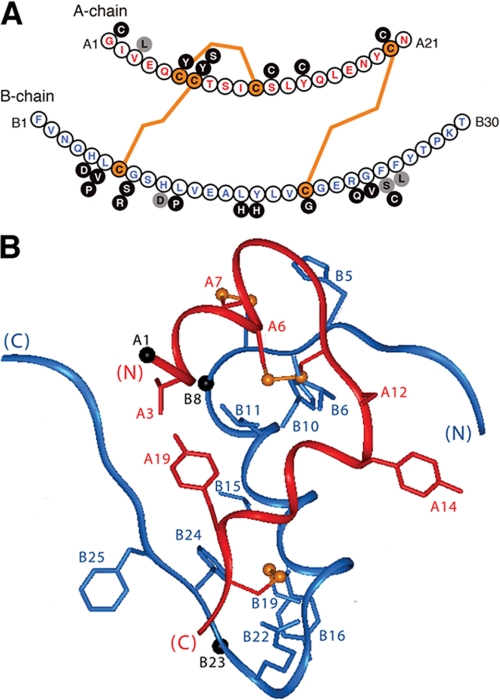
Insulin is a small globular protein containing two chains, A (21 residues) and B (30 residues). Although its unbound structure is well characterized (1-3), how the hormone binds to the insulin receptor (IR)2 is not well understood. In this study, we investigated the receptor-binding surface of insulin by synthetic photoscanning. An extensive set of photoactivable insulin analogs was prepared, each containing a single para-azidoPhe (Pap) substitution (Fig. 1A) (4). The modular structure of the IR (5) permits assignment of contact points to the major hormone-binding elements of the receptor α-subunit (6). Analysis of photoproducts provides evidence for the structural reorganization of insulin within the crux of the receptor ectodomain (7). An alternating pattern of contacts by the B-chain β-strand provides a signature of its insertion between receptor domains.
FIGURE 1.
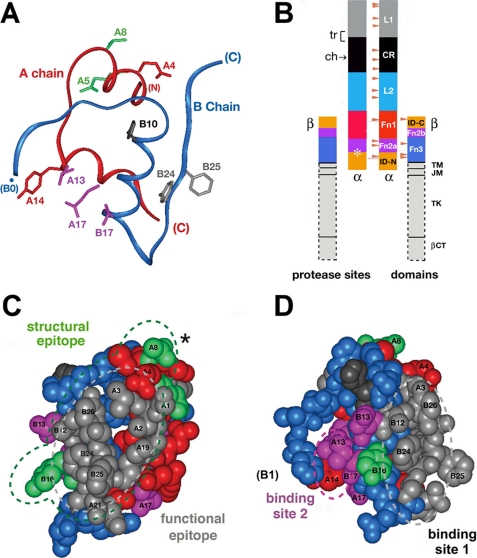
Structure of insulin and domain organization of the insulin receptor. A, ribbon model of insulin (as T-state protomer; Protein Data Bank code 4INS) with the A-chain in red and the B-chain in blue. Selected side chains are shown: GluA4 and TyrA14 (red); GlnA5 and ThrA8 (green); LeuA13, LeuA17, and ValB17 (magenta); HisB10 (black); and PheB24 and PheB25 (gray). B, domain organization of the IR as an (αβ)2-dimer. Color-coded segments indicate structural domains; at left are selected sites of limited proteolysis of photocross-linked complexes: trypsin (tr, bracket at the L1-CR domain junction) and chymotrypsin (ch, arrow within the CR domain), and tryptic and chymotryptic sites at the Fn2a-ID-N junction (asterisk). Beige arrowheads indicate sites of N-linked glycosylation in the extracellular portion of the IR. This figure was adapted from Ref. 5 with the permission of the authors. Dashed lines outline domains (light gray) not present in the crystal structure of the IR ectodomain (Fig. 7); these span the transmembrane α-helix (TM), the juxtamembrane segment (JM), tyrosine kinase (TK), and the C-terminal tail of the β-subunit (βCT). C, space-filling model of the insulin protomer (with residues B27-B30 removed) showing the functional epitope (gray) and its extended structural epitope (green). D, view of the insulin protomer rotated by 90° about the vertical axis with classification of binding sites 1 (gray) and 2 (magenta) as proposed by De Meyts (18, 73). Residues B27-B30 were deleted from the coordinate file to enable better visualization of IleA2 and ValA3 in accord with the detachment model (preceding article (31)).
The IR is a disulfide-linked dimer (designated (αβ)2) in which extracellular α-subunits bind insulin and transmembrane β-subunits contain the cytoplasmic tyrosine kinase domain (Fig. 1B) (5). Mutagenesis has established that the major hormone-binding regions are the N-terminal L1 β-helix and insert domain-derived tail (ID-N) of the α-subunit (8). Whereas the cysteine-rich (CR) domain and L2 β-helix contribute primarily to the architecture of the ectodomain, insulin may also contact the fibronectin homology domains of the α-subunit (Fn1 and part of Fn2) (9, 10). The homologous type I insulin-like growth factor (IGF) receptor exhibits a similar domain organization. Studies of chimeric receptors suggest that the IR L1 domain (large β-helical domain 1 (residues 1-158)) confers specificity for insulin (relative to IGF-I) (11-13). Although co-crystals of the insulin-IR complex have not been obtained to date, evidence for selected domain-specific contacts has been provided by photocross-linking (4, 14-17). The modular structure of the α-subunit enables convenient assignment of photoadducts to L1 or ID-N by limited proteolysis (6). Key tryptic and chymotryptic sites (Fig. 1B, left) are exposed at domain boundaries (supplemental Figs. S1 and S2), leading to photoadducts of characteristic molecular mass (see Table 1).
TABLE 1.
Key receptor fragments in photomapping strategy
|
Protease
|
Photoadduct
massa
|
Assignment
|
Ref.
|
||
|---|---|---|---|---|---|
| Glycosylated | Deglycosylated | ||||
| kDa | |||||
| Chymotrypsin | 50 (47) | 31 | L1/part of CR domainb | 6 | |
| Chymotrypsin | 34 | 23 | C-terminal fragmentc | 34 | |
| Chymotrypsin | 20 | 17 | ID-Nd | 34 | |
| Trypsin | 31 (28) | 20 (17) | L1e | 6 | |
Molecular masses in parentheses are exclusive of the respective A- or B-chain adduct.
The fragment contains ∼250-260 residues (supplemental Fig. S2).
The fragment contains (from the C to N terminus) ID-N, Fn2a, and part of Fn1.
The fragment is likely to begin within Fn2a.
The fragment is likely to contain initial residues of the CR domain (supplemental Fig. S1).
Insight into the receptor-binding surface of insulin has been obtained from analysis of species variants, chemical modification, and mutagenesis (Fig. 1, C and D) (5, 18). The classical binding surface spans the N-terminal A-chain α-helix (residues A1-A8), the central B-chain α-helix (residues B9-B19), and the C-terminal B-chain β-strand (residues B24-B28) (1, 19-21). Mutagenesis defines a functional epitope (by analogy to growth hormone) (22) containing GlyA1, IleA2, ValA3, GlnA5, TyrA19, AsnA21, ValB12, GlyB23, PheB24, PheB25, and TyrB26 (light gray in Fig. 1, C and D) (18, 23); an extended structural epitope (i.e. contacts at which Ala substitutions have minor effect) (24) comprises the contiguous side chains of ThrA8 and TyrB16 (green in Fig. 1, C and D) (25-27). In addition to the classical surface (site 1 in Fig. 1D), kinetic analysis of insulin analogs has revealed a second and distinct functional surface (SerA12, IleA13, GluA17, GluB13, and LeuB17) (site 2 in Fig. 1D) (18). By analogy to growth hormone (28), binding sites 1 and 2 are proposed to function in trans within the (αβ)2-receptor dimer, creating a single high affinity complex (20). HisB10 (dark gray in Fig. 1, C and D) may also contribute to receptor binding, as to a limited extent (<4-fold), substitutions can augment (29) or impair (23) receptor binding. Mutagenesis has been precluded at some sites by impaired foldability (23), and its interpretation at other sites can be confounded by structural perturbations (30).
In the preceding article (31), we demonstrated that stereospecific detachment of the C-terminal β-strand of the B-chain enhances the activity of insulin. In this study, we provide evidence that the detached segment inserts between receptor domains. Our approach is based on photomapping. To this end, a set of >20 photoactivable insulin derivatives (each containing residue-specific Pap substitutions at single sites3) was prepared (see Fig. 2A). The photoactivable insulin derivatives were found to exhibit a range of cross-linking efficiencies. Sites of photocross-linking within the IR were mapped to L1, ID-N, or intervening domains. Such mapping yielded a striking pattern of alternating N- and C-terminal contacts by the conserved B-chain β-strand (residues B24-B28) accompanied by C-terminal engagement of the underling A-chain α-helix (residues A1-A8). These results illustrate the potential of synthetic photomapping to decode active but cryptic protein conformations.
FIGURE 2.
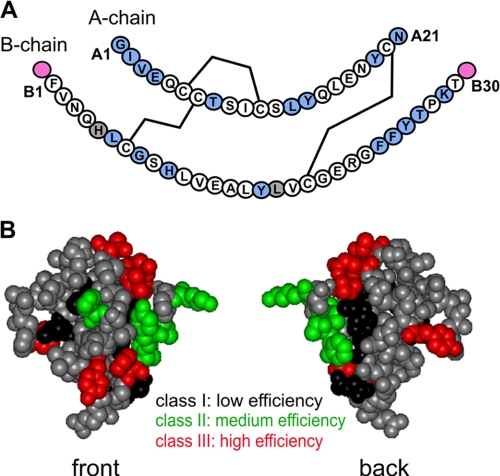
Sites of photoprobes in insulin and summary of cross-linking efficiencies. A, sequence of human insulin showing the A-chain (upper) and B-chain (lower); in each case, the N-terminal residue is at left, and C-terminal residue is at right. The three disulfide bridges (cysteines A6-A11, A7-B7, and A20-B19) are shown as black lines. Sites of Pmp derivatives are highlighted in light blue (within A- or B-chains) or magenta (B-chain extensions B0 and B31). Gray shading (residues B5 and B17) indicates failed syntheses due to impaired chain combination. The PmpB8 analog contained the d-isomer; both l- and d-Pmp derivatives were individually prepared at positions A1 and B24. Not shown are DKP substitutions in the B-chain to yield a monomeric template (AspB10, LysB28, and ProB29) and biotin tags at possible attachment sites (B0, B1, A1, or via the ε-NH2 moiety of a d-LysA1 substituent; see “Experimental Procedures” and supplemental Table S1). B, front and back surfaces of insulin color-coded by efficiency of photocross-linking. The front surface is predominantly composed of B-chain residues, and the back surface of A-chain residues. Sites of high, medium, and low photocross-linking efficiency are shown in red, green, and black, respectively; sites not tested are shown in gray. The structure shown is based on a T6 crystallographic protomer (2Zn molecule 1; Protein Data Bank code 4INS).
Insulin provides a classical paradigm for studies of protein structure and evolution (1, 19), yet ground-state structures provide an incomplete account of functional relationships. Compressed within the short sequences of the A- and B-chains lies information encoding a complex conformational life cycle, extending from nascent folding to storage and induced fit (1, 2, 31). This life cycle both reflects and imposes an interlocking set of evolutionary constraints. Diabetes-associated mutations in the insulin gene highlight the interrelation of foldability, structure, activity, and the overarching threat of toxic misfolding.
EXPERIMENTAL PROCEDURES
Preparation of Insulin Analogs—A summary of insulin analogs is provided in supplemental Table S1. Chemical synthesis of variant A- and B-chains containing photostable precursor para-amino-Phe (Pmp) was performed by manual solid-phase synthesis (4). B-chains each contained three “DKP” substitutions to prevent self-association of insulin (HisB10 → Asp, ProB28 → Lys, and LysB29 → Pro) (2, 32). In selected cases, human and porcine B-chains, obtained by sulfitolysis from human insulin and porcine insulin (kindly provided by Lilly), were employed in combination with synthetic A-chain analogs. Pmp substitutions were introduced in the A-chain at positions A1-A4, A8, A13, A14, A19, and A21 and in the B-chain at positions B0 (an N-terminal extension), B5, B6, B8, B10, B16, B17, B24-B27, B29, and B31 (a C-terminal extension). Individual d- and l-Pmp stereoisomers were introduced at positions A1 and B24. At position B8 (occupied in native insulin by an invariant Gly with a positive ϕ angle) (33), only the d-isomer was prepared. The synthesis and characterization of protoprobes at positions A1, A3, A21, B0, B16, B24, and B25 have been described previously (6, 17, 25, 26, 34, 35).
Biotin Labeling—To permit detection of photoproducts by NeutrAvidin™ (Pierce), an N-terminal biotin tag was included in each analog (supplemental Table S1). Tags were generally introduced by means of a caproyl linker either through Nα of PheB1 (in the case of B-chain Pmp derivatives) (25) or through Nε of d-LysA1 (in the case of A-chain Pmp derivatives) (34); A1 and B0 analogs were biotinylated through Nα of the N-terminal Pmp residue.
Chain Combination—Insulin chain combination and protein purification were performed as described (25, 34). Fidelity of synthesis was verified by matrix-assisted laser desorption time-of-flight mass spectrometry.
Receptor Binding Assays—The activities of insulin analogs were evaluated in a competitive displacement assay using a human placental membrane preparation as described (36). The percentage of tracer bound in the absence of competing ligand was <15% to avoid ligand depletion artifacts. A summary of receptor-binding affinities of the Pmp-insulin analogs is provided in supplemental Table S1. Assays were performed in duplicate (positions B29 and B31) or triplicate (the remaining 29 analogs).
Photocross-linking Studies—Conversion of Pmp-substituted insulin analogs to the corresponding photoactivable Pap derivatives was performed as described (4) and verified by mass spectrometry. Photocross-linking of biotin-labeled Pap analogs to the purified receptor ectodomain (17) or IR (isoform B) (6, 25) was induced by UV irradiation (4); photoproducts were characterized by SDS-PAGE and Western blotting (6, 17, 25). Gels were probed with NeutrAvidin to detect biotin-labeled photoproducts and with polyclonal antiserum recognizing the N-terminal segment of the IR α-subunit (designated IRα-N; Santa Cruz Biotechnology). Hormone-receptor complexes were formed at sufficiently high protein concentrations (∼200 nm receptor and photoreactive insulin analog; >100-fold greater than the highest (weakest) dissociation constant) to ensure >95% receptor occupancy in each case. Assays were performed in duplicate.
Specificity of Photocross-linking—Control experiments verifying specificity were performed to demonstrate competition between binding of the Pap analogs and native ligands (human insulin and IGF-I). Additional control experiments demonstrated that no photocross-linking reactions occurred between Pap derivatives and heterologous proteins (lysozyme and immunoglobulin G) or between the photostable Pmp precursors and the ectodomain under the same conditions (17).
Mapping of Photo Contacts—Domain mapping in holoreceptor complexes was established by limited proteolysis with trypsin and chymotrypsin (6, 25). The apparent molecular masses of proteolytic fragments (with and without N-linked glycosylation) (Table 1) were inferred by SDS-PAGE in relation to standards. Insert domain-specific photocross-linking by the complementary analog PapB25-DKP-insulin (6) was mapped in parallel as a control. Key sites of proteolysis by trypsin and chymotrypsin are indicated in Fig. 1B (bracket and arrow on the left and asterisk at Fn2a/ID-N junction); structural rationales are provided in supplemental Figs. S1 and S2.
RESULTS
Photoprobes were introduced through chemical synthesis of A- or B-chain analogs. Insulin chain combination was generally robust to substitution by the photostable precursor Pmp (yields of >25% relative to wild-type chain combination). Lower yields were obtained at A3, A21, B6, and B24; Pmp substitutions at B5 and B17 blocked disulfide pairing (gray circles in Fig. 2A). Substitution of ValA3 by Pmp yielded native and non-native disulfide isomers, resulting in a relative yield of 10-15%; products were distinguished by receptor-binding activity (35, 37). Substitution of GlyB8 by d-Pmp enhanced the efficiency of disulfide pairing; substitution by l-Pmp was not attempted due to low l-specific yields (33). Pmp substitutions were likewise not attempted at sites (B12 and B15) previously associated with impaired chain combination or attenuated cellular expression of single-chain insulin analogs (23, 38, 39). Each photoactivable analog contained a biotin tag to permit identification of photoadducts by an avidin-based reagent.
The receptor-binding affinities of Pmp analogs were highly variable, impaired in some cases and enhanced in others (range of 0.2-240% relative to human insulin) (supplemental Table S1). 20 of 27 analogs exhibited relative affinities of >25%. The highest affinities were conferred by PmpA8 and d-PmpB24 (>100% relative to the parent analogs), and the lowest by PmpA2 and d-PmpB8 (<1%). Such site-specific modulation of binding is in accord with trends in studies of mutant insulins (1, 23, 33, 40), including stereospecific effects of d- and l-Pmp substitutions at B24 (see accompanying article (31)). The impaired binding of GluA4 → Pmp and TyrA14 → Pmp analogs was unanticipated by Ala scanning (23). The activities of the corresponding Pap analogs were not determined. Because wild-type insulin binds with a dissociation constant of <0.1 nm, however, relative affinities of >1% would correspond to dissociation constants of <10 nm, sufficient at the protein concentrations employed (200 nm) to permit predominant formation of a photoreactive hormone-receptor complex. UV exposure for 20 s led in each case to essentially complete photolysis or cross-linking. No correlation was observed between the relative affinities of Pmp analogs and the extent of photocross-linking by the corresponding Pap derivatives. Control experiments demonstrated that site-specific photocross-linking was competed by unmodified human insulin or (at higher protein concentration) by IGF-I.
Photocross-linking Efficiencies
Relative photocross-linking efficiencies are mapped in Fig. 2B onto the front and back surfaces of insulin in its classical conformation. High efficiency (>20%) was achieved by photoprobes at positions A1 (d-isomer), A3, A4, A8, A14, B16, B24 (d- > l-isomer), B27, and B31 (red surface in Fig. 2B; B31 extension not shown). Of these derivatives, the highest efficiencies were observed at positions A3 (with a biotin tag at d-LysA1 Nε), A8, and B24 (d-isomer), at which sites 30-40% of UV-irradiated complexes gave rise to a covalent photoproduct (31, 34, 35). Inefficient photocross-linking (<5%) was observed at positions A1 (l-isomer), A13, A19, A21, B0, B6, and B8 (d-isomer) (black surface in Fig. 2B). Of these, the lowest efficiencies were exhibited by probes at A21 and B8 (d-isomer). Intermediate photocross-linking efficiencies were obtained in studies of Pap derivatives at positions A2, B10, B25, B26, and B29 (green surface in Fig. 2B). With the exception of the poorly binding PapA2 analog (see below), the extent of photocross-linking could not be appreciably increased by raising ligand concentrations, suggesting that relative efficiencies are intrinsic to the structure of the hormone-receptor complex rather than determined by initial binding affinities. Photocross-linking efficiency can in principle be influenced by both proximity of an interface and accessibility of nitrogenous groups (which favor covalent free radical insertion) (41).
Mapping of Photoproducts
The modular IR structure enables mapping of photocross-links (i) to L1 by limited tryptic digestion and (ii) to ID-N by limited chymotryptic digestion (supplemental Figs. S1 and S2) (6). Following reduction by dithiothreitol (DTT), proteolytic fragments were resolved by SDS-PAGE. Bands containing a photoadduct were identified using a biotin-specific reagent (NeutrAvidin). N-terminal fragments of the receptor α-subunit (and hence containing L1) were detected by Western blotting using anti-peptide antiserum (IRα-N); C-terminal fragments (containing ID-N) were characterized by B25 photocross-linking, previously shown to contact the C-terminal 15-residue peptide (4). The latter proteolytic signature has been confirmed using engineered midi-receptors (35).
L1 Contact Sites—Tryptic mapping yielded a unique 31-kDa cross-linked fragment in L1-cross-linked B-chain complexes. The molecular mass of this fragment (excluding glycosylation and the tethered B-chain) indicates that it contains <170 amino acids and so consists of the L1 domain (residue 1-158) and flanking tryptic site. The structure of the ectodomain exhibits exposed basic side chains at the outer L1-CR domain junction (supplemental Fig. S1), as illustrated in the preceding article (31). The following photoprobes cross-linked to L1: B16, B24-L, B24-D, and B26. L1 photoadducts were not observed in studies of PapB25 or A-chain derivatives.
Limited chymotryptic digestion yielded distinct N- and C-terminal signatures. N-terminal contacts are characterized by a fragment spanning L1 and part of the CR domain (apparent glycosylated molecular mass of 50 kDa and deglycosylated mass of 31 kDa, including the insulin chain adduct). Observation of this fragment corroborated tryptic assignment of the above L1 contacts; the outer surface contains solvent-exposed Phe side chains at appropriate positions (CR domain residues 256 and 258) (supplemental Fig. S2). Further digestion of the B24-cross-linked complex yielded a light 19-kDa band recognized by IRα-N; its estimated mass (<11 kDa, exclusive of the B-chain adduct and two N-linked carbohydrates) suggests that the site of cross-linking is likely to occur within the first 100 residues of the L1 domain, in accord with sites of Ala substitution in L1 that markedly impair insulin binding (8, 42).
ID-N Contacts—Photocross-linking of PapB25 has been shown previously to map to the extreme C-terminal peptide of the IR α-subunit (4) and so provides a standard. Unlike L1-specific contacts, PapB25 photocross-linking is characterized by (a) a C-terminal fragment with apparent molecular mass of 34 kDa (glycosylated) and 23 kDa (deglycosylated), which, upon further digestion, yields (b) a C-terminal fragment of 20 kDa (17 kDa upon deglycosylation) (6). The larger fragment encompasses Fn1-N and ID-N (residues 590-731 in the B-receptor isoform). Its difference in apparent mass upon deglycosylation (11 kDa) represents three N-linked carbohydrates (positions 606, 624, and 671) (43). The structure of the ectodomain suggests possible cleavage at exposed Trp side chains at positions 551 and 559 (supplemental Fig. S2). The smaller fragment contains a single N-linked carbohydrate and so begins after residue 624. Its mass is consistent with the sum of the B-chain adduct (3.7 kDa) and a fragment containing ID-N (residues 638-731; glycosylated mass of 16 kDa) and at most a small number of residues from the Fn2a junction.
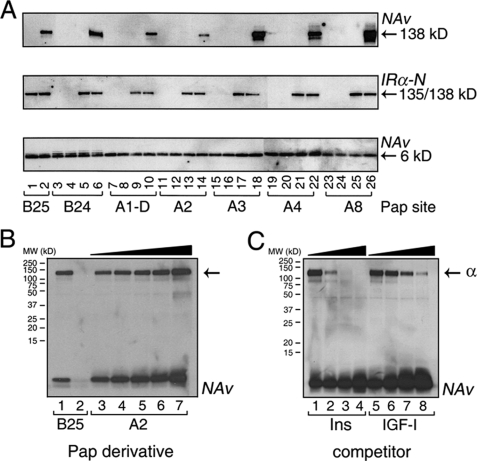
Mapping studies using limited chymotryptic digestion demonstrated the following ID-N contacts: A1-D, A2, A3, A4, A8, A14, B25, and B27. Probes at B29 and B31 were observed to photocross-link to both L1 and ID-N (and possibly to intervening domains), presumably due to flexibility in the hormone-receptor complex. The B10 probe exhibited neither L1-CR domain nor Fn2a-ID-N signatures and so must have intervening contact sites. Adducts A1-L, A13, A19, A21, B0, B6, and B8 were not mapped due to their low photocross-linking efficiencies.
Induced Fit of Insulin
To investigate the active conformation of insulin, we focused in turn on the C-terminal β-strand of the B-chain (residues B24-B28) and the N-terminal α-helix of the A-chain (residues A1-A8).
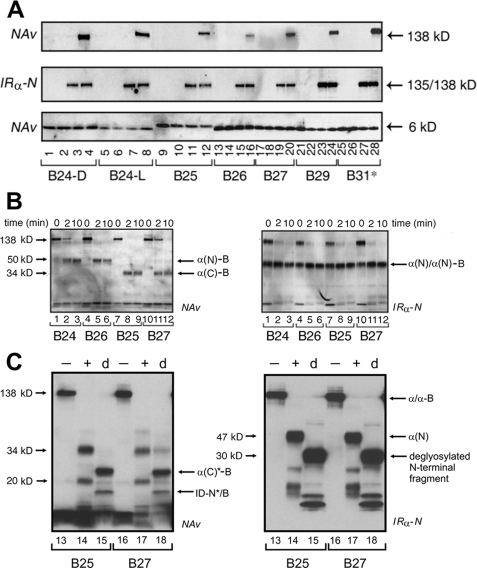
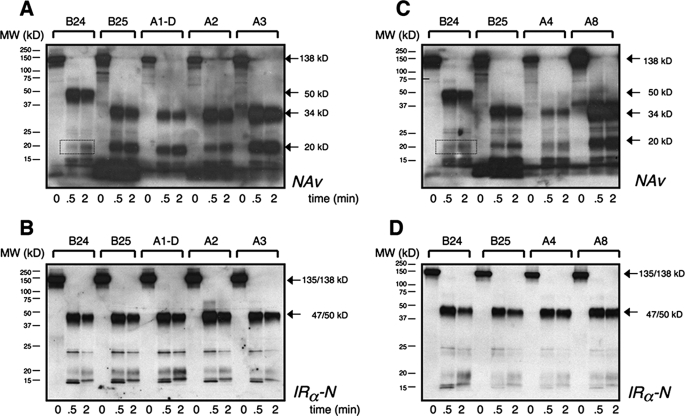
B-chain β-Strand—Photocross-linking by photoprobes B24-B30 (extended to B31) exhibited moderate to high efficiencies (Fig. 3A, upper panel); loading controls as probed by IRα-N and NeutrAvidin are shown (middle and lower panels, respectively). In Fig. 3A, each site is represented by four lanes; the first three provide negative controls (containing the indicated Pap derivative unirradiated in the absence of receptor, irradiated in the absence of receptor, or unirradiated in the presence of receptor, respectively), and the fourth lane contains the products of the photocross-linking reaction. Covalent hormone-receptor complexes are thus observed only in lanes 4, 8, 12, 16, 20, 24, and 28 (Fig. 3A, upper panel).
FIGURE 3.
Photocross-linking studies of B-chain derivatives. A, Western blots showing subunits of the holoreceptor (α2β2, 460 kDa; α, 135 kDa; β, 95 kDa) cross-linked with biotin-labeled insulin derivatives containing Pap at positions B24-D (lanes 1-4), B24-L (lanes 5-8), B25 (lanes 9-12), B26 (lanes 13-16), B27 (lanes 17-20), and B29 (lanes 21-24) or at extended site B31 (lanes 25-28). Samples were treated without (odd-numbered lanes) or with (even-numbered lanes) UV irradiation. After cross-linking and reduction with DTT, samples were resolved by SDS-PAGE and detected either with alkaline phosphatase-conjugated NeutrAvidin (NAv; upper panel) or with anti-receptor antibody (IRα-N; middle panel); the latter demonstrates equal amounts of IR. Control blots probed with NeutrAvidin (lower panel; without DTT reduction) demonstrate equal amounts of insulin analog. In the control lanes, in each case, photocross-linking was not observed in the absence of the IR and UV irradiation (lanes 1, 5, 9, 13, 17, 21, and 25), in the absence of the IR and presence of UV irradiation (lanes 2, 6, 10, 14, 18, 22, and 26), or in the presence of the IR and absence of UV irradiation (lanes 3, 7, 11, 15, 19, 23, and 27). B, limited chymotryptic digestion of PapB24-, PapB25-, PapB26-, and PapB27-photocross-linked hormone-receptor complexes (6). At the indicated time points, aliquots were mixed with an equal volume of Laemmli sample buffer containing 100 mm DTT, heated at 95 °C for 5 min, and resolved by SDS-PAGE. Photoproducts and their fragments were blotted onto nitrocellulose membrane and probed with NeutrAvidin (left panel) or IRα-N (right panel). The apparent molecular masses were as follows: α(N)-B, 47 kDa; α(C)-B, 34 kDa; and α(N)/α(N)-B, 47/50 kDa (Table 1). C, deglycosylation of B25 and B27 chymotryptic fragments as probed with NeutrAvidin (left panel) or IRα-N (right panel). The IR was cross-linked to PapB25 or PapB27 derivatives (lanes 13 and 16; - indicates the absence of protease), digested with chymotrypsin (lanes 14, 15, 17, and 18; +), and then subjected to enzymatic deglycosylation (lanes 15 and 18; labeled d) (25). The apparent molecular masses were as follows: α(C)*-B, 23 kDa; and ID-N*/B, 17 kDa (Table 1).
Limited chymotryptic digestion of the photocross-linked B24-B27 products (Fig. 3B, left panel) yielded distinct NeutrAvidin-detected N- and C-terminal patterns, whereas detection by IRα-N yielded similar N-terminal patterns as a loading control and verification of digestion (Fig. 3B, right panel). PapB24 and PapB26 derivatives gave rise to an L1-CR* domain pattern (Fig. 3B, band α(N)-B in lanes 2 and 3 and lanes 5 and 6, respectively). By contrast, B25 and B27 photocross-links gave rise to similar C-terminal patterns (Fig. 3B, band α(C)-B in lanes 8 and 9 and lanes 11 and 12, respectively). Thus, the odd-numbered probes did not photocross-link to the 50-kDa N-terminal chymotryptic fragment characteristic of even-numbered probes. Further chymotryptic digestion of the B25 and B27 photoadducts in each case yielded glycosylated 20-kDa signature fragments (Fig. 3C, lanes 14 and 17) and deglycosylated 17-kDa fragments (band ID-N*/B in lanes 15 and 18). In these reactions, distinct glycosylated and deglycosylated N-terminal fragments were detected by antiserum IRα-N (Fig. 3D). The tryptic signature of PapB26-derived photoadducts was similar to that of B24 (data not shown).
A-chain α-Helix—Photoprobes at residues A1 (d-chirality), A2, A3, A4, and A8 cross-linked to the IR with varying efficiencies (Fig. 4A, upper panel); negative and loading controls are as in Fig. 3A. Sites of photocross-linking thus include both the polar (GluA4 and ThrA8) and nonpolar (IleA2 and ValA3) faces of the amphipathic A1-A8 α-helix. Chymotryptic mapping of covalent complexes A1 (d-isomer), A2, A3, A4, and A8 each yielded 34- and 20-kDa glycosylated fragments similar to those characteristic of the B25-cross-linked complex (Fig. 5). (Their slightly increased SDS-PAGE mobilities are consistent with the smaller size of the A-chain (21 residues) relative to the B-chain (30 residues).) None of the cross-linked A-chain complexes gave rise to the 50-kDa N-terminal adduct characteristic of the PapB24-mediated photoproduct between the B-chain and an L1-CR* domain fragment (first three lanes in Fig. 5, A-D). Fine mapping of PapA3-mediated photo contacts to the same C-terminal peptide (the C-terminal peptide of the IR α-subunit comprising residues 704-718 (referred to as αCT)) as contacted by PapB25 has been demonstrated in engineered midi-receptors (35).
FIGURE 4.
Photocross-linking studies of A-chain derivatives. A, IR cross-linking of insulin derivatives containing Pap at positions A1-D (lanes 7-10), A2 (lanes 11-14), A3 (lanes 15-18), A4 (lanes 19-22), and A8 (lanes 23-26) in relation to B-chain derivatives B25 (lanes 1 and 2) and B24 (lanes 3-6). Samples were resolved by SDS-PAGE prior to (odd-numbered lanes) or following (even-numbered lanes) UV irradiation. A-chain Pap derivatives (except A1) had a biotin tag attached to the ε-NH2 moiety of a d-LysA1 substituent; A1 d- and l-Pap derivatives were labeled with biotin at Nα. The format is as described for Fig. 3A. Cross-linked adducts were detected after DTT reduction by NeutrAvidin (NAv; upper panel). Control blots probed by IRα-N (middle panel) demonstrate equal amounts of IR. Control blots probed with NeutrAvidin (lower panel; without DTT) demonstrate equal amounts of insulin derivative. In the control lanes, in each case, the photocross-linking band was not detected in the absence of the IR and UV irradiation (lanes 3, 7, 11, 15, 19, and 23), in the absence of the IR and presence of UV irradiation (lanes 4, 8, 12, 16, 20, and 24), or in the presence of the IR and absence of UV irradiation (lanes 1, 5, 9, 13, 17, 21, and 25). B, Western blots showing A2-specific IR photocross-linking at successive concentrations of PapA2 derivative (lanes 3-7, corresponding to ligand concentrations of 200, 240, 320, 400, and 600 nm). PapB25-specific photocross-linking is also shown (lane 1; lane 2 is empty). C, competition experiment. A2-specific photocross-linking could be competed by unmodified insulin (Ins) or IGF-I, added prior to UV irradiation. Successive concentrations of insulin (lanes 1-4) or IGF-I (lanes 5-8) were 0, 3, 30, and 300 times that of PapA2 (600 nm).
FIGURE 5.
Chymotryptic mapping of A-chain-specific photocross-linked complexes. Photoadducts were obtained at the indicated positions and digested with chymotrypsin as described (6). At successive time points, aliquots were reduced by treatment with DTT, heat-denatured, and resolved by SDS-PAGE. Fragments were blotted onto nitrocellulose membrane and detected by NeutrAvidin (NAv; A and C) or antiserum IRα-N (B and D). Dotted boxes in A and C indicate the 19-kDa N-terminal L1-derived tryptic fragment estimated to contain ∼100 residues (and three N-linked carbohydrates). Molecular masses and domain assignments are otherwise as indicated.
Photocross-linking by PapA2 is of particular interest in relation to classical packing of IleA2 within the hydrophobic core (supplemental Fig. S3). Increasing the concentration of the PapA2 derivative from 200 to 600 nm (at a constant receptor concentration of 200 nm) enhanced the extent of photocross-linking (Fig. 4B), suggesting that its intrinsic photocross-linking efficiency is high but limited by partial receptor occupancy. The specificity of such photocross-linking was demonstrated by competition with unmodified insulin or IGF-I (Fig. 4C). Structural interpretation of A2 photocross-linking is uncertain. Because of the small size of insulin, molecular modeling suggests that analogous packing of PapA2 with the hydrophobic core could allow protrusion of the azido group from the protein surface. Direct engagement of IleA2 is nonetheless supported by mutagenesis (34, 40, 44).
DISCUSSION
Classical models of insulin binding (19, 21, 45) were based on crystal structures of zinc insulin hexamers (46, 47). Such hexamers represent the storage form of the hormone within the glucose-regulated secretory vesicles of pancreatic β-cells (48). Because insulin functions in the bloodstream as a monomer, however, its active conformation has long been the subject of speculation. Although the conformation of an engineered insulin monomer in solution closely resembles that of a crystallographic protomer (2, 3), its flexibility has left open the possibility of induced fit (49). In the preceding article (31), we have shown that the activity of insulin can be enhanced by chiral destabilization of the B-chain. Here, we have exploited chemical protein synthesis to obtain a photomap of the receptor-binding surface. The pattern of photocross-linking provides a signature of the hormone's active conformation.
Induced Fit Extends the Receptor-binding Surface—Evidence for induced fit within the B-chain has been provided by the very low activities of native-like analogs containing tethers between the C terminus of the B-chain and the N terminus of the A-chain (50-53). Such impairment suggests that the tethered sites separate upon receptor binding (Fig. 6A) (54). The detachment model thus posits that the C-terminal β-strand of the B-chain detaches from the α-helical core of the hormone (54-56). This strand is anchored to the core by an invariant aromatic side chain (PheB24). Strikingly, as shown in the accompanying article (31), d-amino acid substitutions at this site enhance activity (55, 57) by segmental unfolding. Together, these observations suggest that the C-terminal β-strand adopts an inhibitory conformation within classical structures of insulin. A conformational switch at B24 leading to detachment of the C-terminal segment would also rationalize the high activities of foreshortened analogs (58-61).
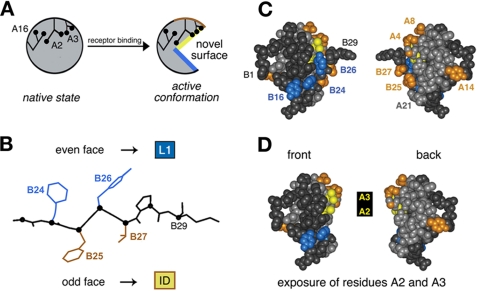
FIGURE 6.
Mapping of photo contacts and relationship to induced fit. A, schematic model of insulin fit. The insulin monomer is proposed to undergo a change in conformation from its closed unbound state (left) to a more open state (right) in which detachment of the C-terminal B-chain β-strand (residues B24-B28) exposes conserved aliphatic side chains IleA2 and ValA3. B, alternating pattern of photo contacts by the C-terminal B-chain β-strand in which its even-numbered face (PheB24 and TyrB26) contacts the L1 domain of the IR (α-subunit residues 1-158), whereas the odd-numbered face (PheB25 and ThrB27) contacts the C-terminal insert domain (ID)-derived tail of the α-subunit. C, space-filling model of insulin depicting its front and back surfaces (left and right). Putative L1 contact residues TyrB16, PheB24, and TyrB26 are shown in blue, and insert domain contact residues (GlyA1, IleA2, ValA3, GluA4, ThrA8, TyrA14, PheB25, and ThrB27) are shown in yellow (A2 and A3) or gold. The A-chain is otherwise shown in light gray, and the B-chain in dark gray. D, corresponding molecular surfaces following removal of residues B26-B30 to simulate exposure of IleA2 and ValA3 upon detachment of the C-terminal B-chain β-strand in the hormone-receptor complex. The color code is the same as described for C.
Ala scanning mutagenesis of the IR has demonstrated that the N- and C-terminal domains of the α-subunit (L1 and ID-N) contain the major hormone-binding sites (8, 42, 62). Remarkably, photoprobes at positions B24-B27 exhibit an alternating pattern of L1 and ID-N contacts. Because odd- and even-numbered side chains in a β-strand project in opposite directions (Fig. 6B), these observations provide evidence that this segment inserts as a β-strand between the L1 and ID-N domains. Detachment of the β-strand from the hydrophobic core of insulin would in turn be expected to expose the conserved inner surface of the A1-A8 α-helix (IleA2 and ValA3) (Fig. 6, C and D). Although largely inaccessible in the unbound hormone, the side chains of IleA2 and ValA3 are critical to biological activity (40, 63). Direct contacts between these aliphatic side chains and the receptor (Fig. 7) would rationalize the marked effects of even subtle substitutions. An example is provided by inversion of Cβ chirality at A2 (yielding allo-IleA2 analogs): this modification is readily accommodated within a native-like core but impairs receptor binding by 50-fold (34, 44). Similarly, substitution of ValA3 by Leu is structurally conservative (34) but associated with a 500-fold decrease in receptor binding (40, 64). In this study, we have provided evidence that the A1-A8 α-helix contacts IN-D at multiple points, including A2 and A3.
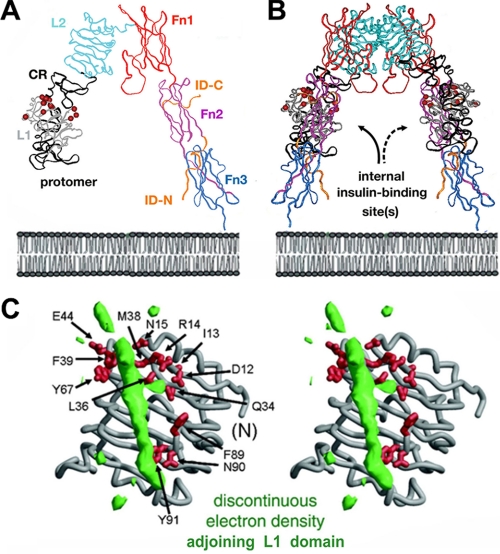
FIGURE 7.
Structure of the IR ectodomain and possible proximity of L1 and ID-N. A, ribbon model of the component protomer (Protein Data Bank code 2DTG). Individual domains L1, CR, L2, Fn1, Fn2, and Fn3 are shown in gray, black, light blue, red, purple, and dark blue, respectively. The insert domain (ID) exhibits missing or discontinuous electron density (residues 655-755; IR isoform A); only respective N- and C-terminal subsegments of ID-N and ID-C are well defined (orange). The color code is in accord with the schematic model in Fig. 1B. B, inverted V-shaped dimer. The position of the plasma membrane is indicated at the bottom in a schematic lipid bilayer. C, stereo model showing the L1 domain (gray ribbon) in relation to unassigned and discontinuous electron density (green), potentially from αCT; density may represent ∼20 residues of the insert domain. αCT- and ID-N-related density may reorganize upon insulin binding. Residues in L1 critical to hormone binding (as inferred from Ala scanning mutagenesis) (62) are shown as red sticks. C was kindly provided by C. Ward.
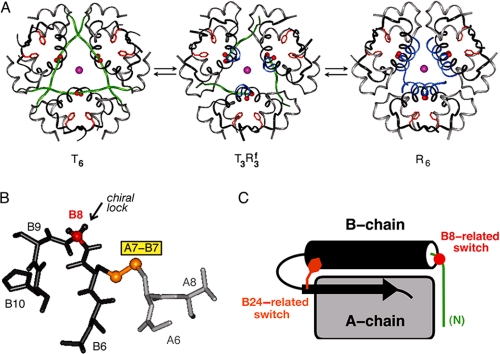
A classical model for induced fit has been provided by the allosteric transition among T6, T3 Rf3, and R6 hexamers (Fig. 8A) (65). In this conformational equilibrium, the N-terminal segment (residues B1-B8) undergoes a dramatic change in secondary structure (66, 67): extended in the T-state (green) and α-helical in the R-state (blue). The relationship between the TR transition and the mechanism of receptor binding is unclear, as activity and allostery may be uncoupled by mutagenesis (68). In addition, the N-terminal five residues of the B-chain may be deleted without significant change in activity (69). Evidence for an R-state-related change in conformation at GlyB8 has nonetheless been provided by chiral mutagenesis. The glycine (red in Fig. 8B) exhibits a positive ϕ dihedral angle (like a d-amino acid) in T-states (including T-like monomers in solution) but a negative ϕ angle (like an l-amino acid) in R-states. d-Amino acid substitutions at B8 (replacing the pro-d-Hα atom) (arrow in Fig. 8B) stabilize the T-state but impair receptor binding (33, 70). The low efficiency of photocross-linking by d-Pap at this site suggests that such loss of activity is due to chiral impairment of a B8-specific conformational change upon receptor binding. By contrast, a frustrated l-SerB8 analog retains high affinity despite marked thermodynamic instability (70). We imagine that the B8-related conformational switch (red in Fig. 8C) alters the topography of cystine A7-B7 (gold in Fig. 8B) and, like the B24-related switch (tawny in Fig. 8C), extends the nonpolar receptor-binding surface of insulin. Molecular modeling suggests that displacement of the N-terminal arm of the B-chain from its T-state conformation would expose LeuB6 and part of the A-chain (IleA10 and LeuA13).
FIGURE 8.
Classical insulin allostery and induced fit. A, conformational equilibria among three families of zinc insulin hexamers: T6 (left), T3Rf3 (middle), and R6 (right). In each structure, the A-chains are shown in light gray; residues B9-B30 in dark gray; the side chain of PheB24 in tawny; and axial zinc ions (overlaid) in magenta. The adjustable conformations of residues B1-B8 in the T- and R-states are highlighted in green and blue, respectively. Coordinates were obtained from Protein Data Bank codes 4INS, 1TRZ, and 1TNJ. B, T-state-specific B7-B10 β-turn and adjoining B7-A7 disulfide bridge (gold). Substitution of the invariant GlyB8 (Cα in red) by d-amino acids stabilizes the T-state but markedly impairs receptor binding (33, 70). The corresponding l-amino acid substitutions destabilize the T-state but can be highly active. The arrow indicates pro-d-Hα of GlyB8. A- and B-chain residues (B6-B10 and A6-A8) are otherwise shown in light and dark gray, respectively; selected residue numbers are provided. C, schematic model of insulin T-state showing proposed sites of conformational change upon receptor binding: an R-state-related B8-related switch (red, right) within the N-terminal segment of the B-chain (green) and a B24-related switch (tawny, right) as revealed by stereospecific unfolding of the C-terminal segment of the B-chain (see preceding article (31)). The A-chain is represented as a gray rectangle, and the central B-chain α-helix (cylinder) and C-terminal β-strand (arrow) are shown in black.
Limitations of Photoscanning—The above interpretations assume that photocross-linking provides a valid probe for native contacts within the hormone-receptor complex. A general limitation is nonetheless posed by possible structural perturbations associated with the photoprobe. Although Pap is among the smallest of available azido reagents, its substitution at sites containing smaller amino acids (such as Gly, Leu, Val, and Thr) or at sites containing formal charges (Glu and Lys) can in principle lead to local or non-local conformational changes in the hormone. Fortuitously, the sequence and structure of insulin facilitate photoscanning. Key sites contain either aromatic side chains similar in shape and size to Pap (TyrA14, TyrA19, PheB1, TyrB16, PheB24, PheB25, and TyrB26) or non-aromatic side chains positioned such that a Pap substituent would be expected to project into solvent (d-PapA1, PapA4, PapA8, PapB0, d-PapB8, PapB28, and PapB29). The conclusions of this study are thus likely to be robust to probe-induced distortions. Structural perturbations are nonetheless likely at A2 and A3. The side chain of IleA2 packs against the aromatic ring of TyrA19 within the core, whereas ValA3 projects within an interchain crevice adjoining TyrB26. Crystal structures of Pmp or related photostable analogs have not been determined.4 Despite the uncertain structural effects of Pap at these sites, prior studies of insulin analogs support the direct engagement of IleA2 and ValA3 at the receptor interface (30, 35, 44, 63).
Relationship to the Structure of the Receptor—The photoactive surface of insulin spans both its front and back surfaces. Such circumferential binding is consistent with its proposed mode of binding within the crux of the inverted V-shaped dimeric ectodomain (Fig. 7, A and B) (7). How insulin binds within the interior of the ectodomain is unclear. Modeling is limited by the absence of continuous electron density spanning the insert domain, including the C-terminal ID-N-derived peptide contacted by PapB25 and PapA3 (αCT; residues 716-731 in isoform B). An intriguing but presently uninterpretable feature of the crystal structure is a low resolution tube of electron density adjoining the hormone-binding face of L1 (Fig. 7C). Alternation of B24-B27 photo contacts suggests that this anomalous feature represents αCT. It is possible that, upon binding of insulin, this region of the receptor becomes better organized with changes in the relative orientation of L1 and αCT.
The overall features of the ectodomain structure, supported by complementation analysis between mutations in L1 and ID-N (71), suggest that the tail of one subunit is near the head (L1) of the other (7). Such head-to-tail dimerization would imply that the B-chain β-strand inserts in trans between α-subunits. The proposed bivalency of insulin may regulate the orientation of subunits in the α2β2-holoreceptor and in turn initiate transmembrane signaling (20, 72). Bridging L1/αCT contacts are unrelated to binding site 2 as defined in kinetic studies of insulin analogs by De Meyts (18).
Contacts between insulin and the IR are likely to extend beyond L1 and ID-N. Although the contribution of such contacts to binding affinity may be smaller than that of classical contacts (as defined by Ala scanning mutagenesis) (23, 24), they may be integral to the mechanism of signal transduction. The present mapping studies indicate that the B10 photoprobe contacts neither L1-CR* nor Fn2-ID-N. In addition, a previous study of a B1 photoprobe by Brandenburg and co-workers (16) identified a contact in a fragment spanning parts of L2 and Fn1 (residues 390-488). Evidence for potential contacts in Fn1 is provided by a recent mutagenesis study (10); these contacts may relate to site 2 in insulin (residues A12, A13, B10, B14, and B17), which influences the kinetic properties of the insulin-receptor complex disproportionately to effects on affinity (18, 73). The head-to-tail architecture of the ectodomain implies that any such contacts (like those to ID-N) would be in trans with L1.
Evolution of Insulin and Human Genetics—Most of the photocross-linking sites identified in this study are broadly conserved among vertebrate insulins and IGFs. Such conservation is likely to reflect independent evolutionary constraints simultaneously imposed by structural requirements of (a) protein folding in the endoplasmic reticulum (ER), (b) subcellular trafficking and prohormone processing (leading to self-assembly in the secretor granule), and (c) receptor binding at target tissues (74). A given residue (as illustrated by PheB24 in the previous article (31)) may play distinct roles at each stage of biosynthesis and signaling. The presence of interlocking constraints implies that the fitness landscape of insulin is sharply peaked.
Advances in human genetics have identified several monogenic forms of diabetes mellitus (DM) due to mutations in the insulin gene. Disease-associated substitutions are predicted in each region of the hormone and its precursors (the signal peptide, A- and B-chains, dibasic processing sites, and connecting domain) (75-81). The classical insulinopathies (LeuA3, SerB24, and LeuB25) (gray circles in Fig. 9A) occur at sites of efficient receptor photocross-linking (6, 17); these substitutions lead to mutant hyperinsulinemia and adult-onset DM of variable penetrance (75). Substitution of HisB10 by Asp in contrast enhances in vitro activity but, within the β-cell, leads to mistrafficking of the mutant proinsulin to a constitutive (i.e. non-glucose-regulated) granule, causing adult-onset DM with mutant hyperproinsulinemia. The majority of mutations mapping within the A- and B-domains are associated with permanent-onset neonatal DM, presumably due to misfolding of the variant proinsulin (78-81). Of these, most are due to the addition or removal of a cysteine, unbalancing disulfide pairing. One human mutation (CysA7 → Tyr) corresponds to a rodent model of DM associated with endoreticular stress (the Akita mouse) (82).
FIGURE 9.
Sites of diabetes-associated mutations in human insulin. A, sequence of human insulin with the A-chain (upper) in red and the B-chain (lower) in blue. Disulfide bridges are shown in orange. Black circles represent mutations associated with neonatal DM due to presumed folding defects in proinsulin (78-81); gray circles represent mutations that permit disulfide pairing in the ER with subsequent circulation of mutant proteins. Whereas substitutions at positions A3, B24, and B25 lead to variant hyperinsulinemias, the B10 substitution interferes with protein trafficking, leading to mutant hyperproinsulinemia (77). B, positions of mutation sites in the T-state of insulin (crystallographic protomer 1 of 2Zn insulin; Protein Data Bank code 4INS) (1). Black spheres indicate Cα atoms of Gly at A1, B8, and B23.
Clinical mutations associated with neonatal-onset DM (and hence with a block to disulfide pairing) also occur at or near sites of induced fit: within either the N-terminal segment of the B-chain (residues B5, B6, and B8) or its C-terminal segment (residues B22 and B23). We speculate that at these sites kinetic determinants of foldability in the ER are at odds with conformational requirements of receptor binding, a conflict resolved by induced fit (31). Evidence for this conflict and its resolution has been provided by comparative analysis of d- and l-amino acid substitutions at conserved “hot spot-associated” glycines flanking both N- and C-terminal segments (GlyB8 and GlyB23) (black spheres in the B-chain in Fig. 9B). Chiral mutagenesis has demonstrated that native-like positive ϕ dihedral angles facilitate pairing of neighboring cysteines (B7-A7 and B20-A19, respectively) but are dispensable for receptor binding (33, 70, 83). Whereas d-substitutions at B8 enhance the efficiency of disulfide pairing in peptide models (33, 70, 83), the inactivity of such analogs (see above) suggests that nascent structural relationships essential for foldability in the ER and prominent features of storage hexamers may be reorganized upon receptor binding.
Impairment of disulfide pairing in nascent proinsulin can in principle be severe or mild, depending on the site of mutation and properties of the substituted side chain. Indeed, whereas neonatal-onset DM represents a block to folding, analogous mutations have been identified in patients presenting with auto-antibody-negative type I DM in the second decade of life (maturity-onset diabetes of the young) (78-81). One such mutation is ArgB22 → Gln, which alters a solvent-exposed site in the B21-B24 β-turn (81). Although this mutation highlights the general importance of the C-terminal β-turn, ArgB22 is not needed for receptor binding (23), and its specific contribution to folding is not apparent. The wild-type side chain is not well ordered in crystals or in solution (1-3); its guanidinium group does not form hydrogen bonds or salt bridges within the protomer.
We speculate that globular proteins may contain cryptic determinants of foldability whose transient roles in folding are not apparent in the native state. It is possible, for example, that the conserved positive charge of ArgB22 participates in stabilization of a thiolate intermediate in the mechanism of disulfide pairing and is dispensable once folding is achieved. Another example of a mild folding defect may be provided by the classical variant SerB24-insulin, which is expressed, processed, and secreted (75). As a seeming paradox, this mutant insulin (unlike LeuA3- and LeuB25-insulins) exhibits reduced but non-negligible receptor-binding activity, sufficient to avoid a diabetic phenotype (77). Our findings that B24 substitutions partially impair insulin chain combination and predispose to aggregation-coupled misfolding (preceding article (31)) suggest that mild chronic ER stress may contribute to this associated adult-onset syndrome: its variable genetic penetrance may be regarded as the hallmark of a gene that modifies the survival of β-cells as a multigenic trait.
Concluding Remarks—The present photomap of insulin provides evidence that its receptor-binding surface is extended by conformational changes in the B-chain. Photocross-linking sites span the nonpolar surfaces of both A- and B-chains. The C-terminal β-strand of the B-chain exhibits a striking alternation of photo contacts between the C-terminal L1 domain of the receptor α-subunit (contacted by probes at B24 and B26) and its N-terminal ID-N domains (contacted by probes at B25 and B27). This even-odd alternation corresponds to the canonical structure of a β-strand in which successive side chains project in opposite directions. We propose that insertion of the C-terminal segment of the B-chain between these receptor domains (likely to occur in trans) represents the first step in a complex choreography of conformational changes in the holo-receptor leading to transmembrane activation of the tyrosine kinase.
The classical structure of insulin represents an inactive conformation that mediates native self-assembly in the β-cell. Whereas its structural relationships predict nascent interactions pertinent to the mechanism of disulfide pairing in biosynthesis, reorganization of the B-chain upon receptor binding is likely to involve both its N- and C-terminal segments. We envisage that conserved side chains play distinct roles in folding, assembly, and receptor binding. It would thus be of future interest to investigate the evolution of the insulin gene in relation to these interlocking structural constraints. Such investigation will require structural elucidation of the conformational life cycle of insulin from folding to receptor binding.
Supplementary Material
Acknowledgments
We thank Q.-X. Hua for collaborative interactions; W. Jia for assistance with the figures; B. Li for assistance with receptor purification; N. F. Phillips for advice; and P. Arvan, S. Chan, P. De Meyts, G. G. Dodson, M. Lawrence, D. F. Steiner, and C. Ward for discussion. We are grateful to S. Chan and D. F. Steiner for communication of results prior to publication.
This work was supported, in whole or in part, by National Institutes of Health Grants DK065980 (to J. W.), DK56673 (to P. G. K.), and DK040949 and DK069764 (to M. A. W.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3 and Table S1.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: IR, insulin receptor; Pap, para-azido-Phe; CR, cysteine-rich; IGF, insulin-like growth factor; Pmp, para-amino-Phe; DTT, dithiothreitol; ER, endoplasmic reticulum; DM, diabetes mellitus.
Pap is small, rigid, and readily accommodated at diverse sites. Its systematic incorporation by total chemical synthesis thus circumvents two key limitations of conventional azido modification: (a) nonspecific cross-linking by flexible swinging arms and (b) restricted potential sites of derivatization.
Native-like crystal structures of l-TrpA1- and d-TrpA1-insulin analogs (84) suggest that l- and d-PapA1 derivatives can be accommodated without transmitted perturbations. That PapA3 may likewise be accommodated is suggested by the ready crystallization of TyrA3-insulin as a canonical zinc hexamer under conditions similar to those used in crystallization of wild-type insulin (Z.-l. Wan and M. A. Weiss, unpublished data).
References
- 1.Baker, E. N., Blundell, T. L., Cutfield, J. F., Cutfield, S. M., Dodson, E. J., Dodson, G. G., Hodgkin, D. M., Hubbard, R. E., Isaacs, N. W., and Reynolds, C. D. (1988) Philos. Trans. R. Soc. Lond. B Biol. Sci. 319 369-456 [DOI] [PubMed] [Google Scholar]
- 2.Hua, Q.-x., Hu, S.-Q., Frank, B. H., Jia, W., Chu, Y.-C., Wang, S., Burke, G. T., Katsoyannis, P. G., and Weiss, M. A. (1996) J. Mol. Biol. 264 390-403 [DOI] [PubMed] [Google Scholar]
- 3.Olsen, H. B., Ludvigsen, S., and Kaarsholm, N. C. (1996) Biochemistry 35 8836-8845 [DOI] [PubMed] [Google Scholar]
- 4.Kurose, T., Pashmforoush, M., Yoshimasa, Y., Carroll, R., Schwartz, G. P., Burke, G. T., Katsoyannis, P. G., and Steiner, D. F. (1994) J. Biol. Chem. 269 29190-29197 [PubMed] [Google Scholar]
- 5.De Meyts, P., and Whittaker, J. (2002) Nat. Rev. Drug Discov. 1 769-783 [DOI] [PubMed] [Google Scholar]
- 6.Xu, B., Hu, S.-Q., Chu, Y.-C., Huang, K., Nakagawa, S. H., Whittaker, J., Katsoyannis, P. G., and Weiss, M. A. (2004) Biochemistry 43 8356-8372 [DOI] [PubMed] [Google Scholar]
- 7.McKern, N. M., Lawrence, M. C., Streltsov, V. A., Lou, M. Z., Adams, T. E., Lovrecz, G. O., Elleman, T. C., Richards, K. M., Bentley, J. D., Pilling, P. A., Hoyne, P. A., Cartledge, K. A., Pham, T. M., Lewis, J. L., Sankovich, S. E., Stoichevska, V., Da Silva, E., Robinson, C. P., Frenkel, M. J., Sparrow, L. G., Fernley, R. T., Epa, V. C., and Ward, C. W. (2006) Nature 443 218-221 [DOI] [PubMed] [Google Scholar]
- 8.Whittaker, J., Sorensen, H., Gadsboll, V. L., and Hinrichsen, J. (2002) J. Biol. Chem. 277 47380-47384 [DOI] [PubMed] [Google Scholar]
- 9.Ward, C., Lawrence, M. C., Streltsov, V., Garrett, T., McKern, N., Lou, M. Z., Lovrecz, G., and Adams, T. (2008) Acta Physiol. (Oxf.) 192 3-9 [DOI] [PubMed] [Google Scholar]
- 10.Whittaker, L. J., Hao, C., Fu, W., and Whittaker, J. (2008) Biochemistry 47 12900-12909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen, A. S., Kjeldsen, T., Wiberg, F. C., Vissing, H., Schaffer, L., Rasmussen, J. S., De Meyts, P., and Moller, N. P. (1992) J. Biol. Chem. 267 13681-13686 [PubMed] [Google Scholar]
- 12.Kjeldsen, T., Andersen, A. S., Wiberg, F. C., Rasmussen, J. S., Schaffer, L., Balschmidt, P., Moller, K. B., and Moller, N. P. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 4404-4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumacher, R., Soos, M. A., Schlessinger, J., Brandenburg, D., Siddle, K., and Ullrich, A. (1993) J. Biol. Chem. 268 1087-1094 [PubMed] [Google Scholar]
- 14.Yip, C. C., Hsu, H., Patel, R. G., Hawley, D. M., Maddux, B. A., and Goldfine, I. D. (1988) Biochem. Biophys. Res. Commun. 157 321-329 [DOI] [PubMed] [Google Scholar]
- 15.Wedekind, F., Baer-Pontzen, K., Bala-Mohan, S., Choli, D., Zahn, H., and Brandenburg, D. (1989) Biol. Chem. 370 251-258 [DOI] [PubMed] [Google Scholar]
- 16.Fabry, M., Schaefer, E., Ellis, L., Kojro, E., Fahrenholz, F., and Brandenburg, D. (1992) J. Biol. Chem. 267 8950-8956 [PubMed] [Google Scholar]
- 17.Xu, B., Hu, S.-Q., Chu, Y.-C., Wang, S., Wang, R.-Y., Nakagawa, S. H., Katsoyannis, P. G., and Weiss, M. A. (2004) Diabetes 53 1599-1602 [DOI] [PubMed] [Google Scholar]
- 18.De Meyts, P. (2008) Trends Biochem. Sci. 33 376-384 [DOI] [PubMed] [Google Scholar]
- 19.Pullen, R. A., Lindsay, D. G., Wood, S. P., Tickle, I. J., Blundell, T. L., Wollmer, A., Krail, G., Brandenburg, D., Zahn, H., Gliemann, J., and Gammeltoft, S. (1976) Nature 259 369-373 [DOI] [PubMed] [Google Scholar]
- 20.De Meyts, P. (1994) Diabetologia 37 S135-S148 [DOI] [PubMed] [Google Scholar]
- 21.Liang, D. C., Chang, W. R., and Wan, Z.-l. (1994) Biophys. Chem. 50 63-71 [DOI] [PubMed] [Google Scholar]
- 22.Cunningham, B. C., and Wells, J. A. (1993) J. Mol. Biol. 234 554-563 [DOI] [PubMed] [Google Scholar]
- 23.Kristensen, C., Kjeldsen, T., Wiberg, F. C., Schaffer, L., Hach, M., Havelund, S., Bass, J., Steiner, D. F., and Andersen, A. S. (1997) J. Biol. Chem. 272 12978-12983 [DOI] [PubMed] [Google Scholar]
- 24.Clackson, T., and Wells, J. A. (1995) Science 267 383-386 [DOI] [PubMed] [Google Scholar]
- 25.Huang, K., Xu, B., Hu, S.-Q., Chu, Y.-C., Hua, Q.-x., Qu, Y., Li, B., Wang, S., Wang, R.-Y., Nakagawa, S. H., Theede, A. M., Whittaker, J., De Meyts, P., Katsoyannis, P. G., and Weiss, M. A. (2004) J. Mol. Biol. 341 529-550 [DOI] [PubMed] [Google Scholar]
- 26.Wan, Z., Xu, B., Huang, K., Chu, Y.-C., Li, B., Nakagawa, S. H., Qu, Y., Hu, S.-Q., Katsoyannis, P. G., and Weiss, M. A. (2004) Biochemistry 43 16119-16133 [DOI] [PubMed] [Google Scholar]
- 27.Glendorf, T., Sorensen, A. R., Nishimura, E., Pettersson, I., and Kjeldsen, T. (2008) Biochemistry 47 4743-4751 [DOI] [PubMed] [Google Scholar]
- 28.Cunningham, B. C., Ultsch, M., De Vos, A. M., Mulkerrin, M. G., Clauser, K. R., and Wells, J. A. (1991) Science 254 821-825 [DOI] [PubMed] [Google Scholar]
- 29.Schwartz, G. P., Burke, G. T., and Katsoyannis, P. G. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 6408-6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakagawa, S. H., and Tager, H. S. (1991) J. Biol. Chem. 266 11502-11509 [PubMed] [Google Scholar]
- 31.Hua, Q.-x., Xu, B., Huang, K., Hu, S.-Q., Nakagawa, S., Jia, W., Wang, S., Whittaker, J., Katsoyannis, P. G., and Weiss, M. A. (2009) J. Biol. Chem. 284 14586-14596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss, M. A., Hua, Q.-x., Lynch, C. S., Frank, B. H., and Shoelson, S. E. (1991) Biochemistry 30 7373-7389 [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa, S. H., Zhao, M., Hua, Q.-x., Hu, S.-Q., Wan, Z.-l., Jia, W., and Weiss, M. A. (2005) Biochemistry 44 4984-4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan, Z.-l., Huang, K., Xu, B., Hu, S.-Q., Wang, S., Chu, Y.-C., Katsoyannis, P. G., and Weiss, M. A. (2005) Biochemistry 44 5000-5016 [DOI] [PubMed] [Google Scholar]
- 35.Huang, K., Chan, S. J., Hua, Q.-x., Chu, Y.-C., Wang, R.-Y., Klaproth, B., Jia, W., Whittaker, J., De Meyts, P., Nakagawa, S. H., Steiner, D. F., Katsoyannis, P. G., and Weiss, M. A. (2007) J. Biol. Chem. 282 35337-35349 [DOI] [PubMed] [Google Scholar]
- 36.Weiss, M. A., Hua, Q.-x., Jia, W., Nakagawa, S. H., Chu, Y.-C., Hu, S.-Q., and Katsoyannis, P. G. (2001) J. Biol. Chem. 276 40018-40024 [DOI] [PubMed] [Google Scholar]
- 37.Hua, Q.-x., Gozani, S. N., Chance, R. E., Hoffmann, J. A., Frank, B. H., and Weiss, M. A. (1995) Nat. Struct. Biol. 2 129-138 [DOI] [PubMed] [Google Scholar]
- 38.Hu, S.-Q., Burke, G. T., Schwartz, G. P., Ferderigos, N., Ross, J. B., and Katsoyannis, P. G. (1993) Biochemistry 32 2631-2635 [DOI] [PubMed] [Google Scholar]
- 39.Hua, Q.-x., Liu, M., Hu, S.-Q., Jia, W., Arvan, P., and Weiss, M. A. (2006) J. Biol. Chem. 281 24889-24899 [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa, S. H., and Tager, H. S. (1992) Biochemistry 31 3204-3214 [DOI] [PubMed] [Google Scholar]
- 41.Eberle, A. N., and de Graan, P. N. E. (1985) Methods Enzymol. 109 129-157 [Google Scholar]
- 42.Mynarcik, D. C., Yu, G. Q., and Whittaker, J. (1996) J. Biol. Chem. 271 2439-2442 [DOI] [PubMed] [Google Scholar]
- 43.Sparrow, L. G., Lawrence, M. C., Gorman, J. J., Strike, P. M., Robinson, C. P., McKern, N., and Ward, C. (2008) Proteins 71 426-439 [DOI] [PubMed] [Google Scholar]
- 44.Xu, B., Hua, Q.-x., Nakagawa, S. H., Jia, W., Chu, Y.-C., Katsoyannis, P. G., and Weiss, M. A. (2002) J. Mol. Biol. 316 435-441 [DOI] [PubMed] [Google Scholar]
- 45.Blundell, T. L., Cutfield, J. F., Cutfield, S. M., Dodson, E. J., Dodson, G. G., Hodgkin, D. C., and Mercola, D. A. (1972) Diabetes 21 492-505 [DOI] [PubMed] [Google Scholar]
- 46.Blundell, T. L., Cutfield, J. F., Cutfield, S. M., Dodson, E. J., Dodson, G. G., Hodgkin, D. C., Mercola, D. A., and Vijayan, M. (1971) Nature 231 506-511 [DOI] [PubMed] [Google Scholar]
- 47.The Peking Insulin Structure Group (1971) Peking Rev. 40 11-16 [Google Scholar]
- 48.Dodson, G., and Steiner, D. (1998) Curr. Opin. Struct. Biol. 8 189-194 [DOI] [PubMed] [Google Scholar]
- 49.Hua, Q.-x., Ladbury, J. E., and Weiss, M. A. (1993) Biochemistry 32 1433-1442 [DOI] [PubMed] [Google Scholar]
- 50.Markussen, J., Jorgensen, K. H., Sorensen, A. R., and Thim, L. (1985) Int. J. Pept. Protein Res. 26 70-77 [PubMed] [Google Scholar]
- 51.Nakagawa, S. H., and Tager, H. S. (1989) J. Biol. Chem. 264 272-279 [PubMed] [Google Scholar]
- 52.Derewenda, U., Derewenda, Z., Dodson, E. J., Dodson, G. G., Bing, X., and Markussen, J. (1991) J. Mol. Biol. 220 425-433 [DOI] [PubMed] [Google Scholar]
- 53.Huang, Y., Liang, Z., and Feng, Y. (2001) Sci. China Ser. C Life Sci. 44 593-600 [DOI] [PubMed] [Google Scholar]
- 54.Hua, Q.-x., Shoelson, S. E., Kochoyan, M., and Weiss, M. A. (1991) Nature 354 238-241 [DOI] [PubMed] [Google Scholar]
- 55.Kobayashi, M., Ohgaku, S., Iwasaki, M., Maegawa, H., Shigeta, Y., and Inouye, K. (1982) Biochem. Biophys. Res. Commun. 107 329-336 [DOI] [PubMed] [Google Scholar]
- 56.Mirmira, R. G., Nakagawa, S. H., and Tager, H. S. (1991) J. Biol. Chem. 266 1428-1436 [PubMed] [Google Scholar]
- 57.Mirmira, R. G., and Tager, H. S. (1989) J. Biol. Chem. 264 6349-6354 [PubMed] [Google Scholar]
- 58.Schwartz, G. P., Burke, G. T., and Katsoyannis, P. G. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 458-461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lenz, V., Gattner, H. G., Sievert, D., Wollmer, A., Engels, M., and Hocker, H. (1991) Biol. Chem. Hoppe-Seyler 372 495-504 [DOI] [PubMed] [Google Scholar]
- 60.Kurapkat, G., Siedentop, M., Gattner, H. G., Hagelstein, M., Brandenburg, D., Grotzinger, J., and Wollmer, A. (1999) Protein Sci. 8 499-508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zakova, L., Barth, T., Jiracek, J., Barthova, J., and Zorad, S. (2004) Biochemistry 43 2323-2331 [DOI] [PubMed] [Google Scholar]
- 62.Williams, P. F., Mynarcik, D. C., Yu, G. Q., and Whittaker, J. (1995) J. Biol. Chem. 270 3012-3016 [DOI] [PubMed] [Google Scholar]
- 63.Kitagawa, K., Ogawa, H., Burke, G. T., Chanley, J. D., and Katsoyannis, P. G. (1984) Biochemistry 23 1405-1413 [DOI] [PubMed] [Google Scholar]
- 64.Kobayashi, M., Takata, Y., Ishibashi, O., Sasaoka, T., Iwasaki, T. M., Shigeta, Y., and Inouye, K. (1986) Biochem. Biophys. Res. Commun. 137 250-257 [DOI] [PubMed] [Google Scholar]
- 65.Brader, M. L., and Dunn, M. F. (1991) Trends Biochem. Sci. 16 341-345 [DOI] [PubMed] [Google Scholar]
- 66.Bentley, G., Dodson, E., Dodson, G., Hodgkin, D., and Mercola, D. (1976) Nature 261 166-168 [DOI] [PubMed] [Google Scholar]
- 67.Derewenda, U., Derewenda, Z., Dodson, E. J., Dodson, G. G., Reynolds, C. D., Smith, G. D., Sparks, C., and Swenson, D. (1989) Nature 338 594-596 [DOI] [PubMed] [Google Scholar]
- 68.Wan, Z.-l., Huang, K., Whittaker, J., and Weiss, M. A. (2008) J. Biol. Chem. 283 21198-21210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwartz, G., and Katsoyannis, P. G. (1978) Biochemistry 17 4550-4556 [DOI] [PubMed] [Google Scholar]
- 70.Hua, Q.-x., Nakagawa, S. H., Hu, S.-Q., Jia, W., Wang, S., and Weiss, M. A. (2006) J. Biol. Chem. 281 24900-24909 [DOI] [PubMed] [Google Scholar]
- 71.Chan, S. J., Nakagawa, S., and Steiner, D. F. (2007) J. Biol. Chem. 282 13754-13758 [DOI] [PubMed] [Google Scholar]
- 72.Schäffer, L. (1994) Eur. J. Biochem. 221 1127-1132 [DOI] [PubMed] [Google Scholar]
- 73.De Meyts, P. (2004) BioEssays 26 1351-1362 [DOI] [PubMed] [Google Scholar]
- 74.Steiner, D. F., and Chan, S. J. (1988) Horm. Metab. Res. 20 443-444 [DOI] [PubMed] [Google Scholar]
- 75.Shoelson, S., Haneda, M., Blix, P., Nanjo, A., Sanke, T., Inouye, K., Steiner, D., Rubenstein, A., and Tager, H. (1983) Nature 302 540-543 [DOI] [PubMed] [Google Scholar]
- 76.Carroll, R. J., Hammer, R. E., Chan, S. J., Swift, H. H., Rubenstein, A. H., and Steiner, D. F. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 8943-8947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Steiner, D. F., Tager, H. S., Chan, S. J., Nanjo, K., Sanke, T., and Rubenstein, A. H. (1990) Diabetes Care 13 600-609 [DOI] [PubMed] [Google Scholar]
- 78.Stoy, J., Edghill, E. L., Flanagan, S. E., Ye, H., Paz, V. P., Pluzhnikov, A., Below, J. E., Hayes, M. G., Cox, N. J., Lipkind, G. M., Lipton, R. B., Greeley, S. A., Patch, A. M., Ellard, S., Steiner, D. F., Hattersley, A. T., Philipson, L. H., and Bell, G. I. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 15040-15044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Colombo, C., Porzio, O., Liu, M., Massa, O., Vasta, M., Salardi, S., Beccaria, L., Monciotti, C., Toni, S., Pedersen, O., Hansen, T., Federici, L., Pesavento, R., Cadario, F., Federici, G., Ghirri, P., Arvan, P., Iafusco, D., Barbetti, F., and Early Onset Diabetes Study Group of the Italian Society of Pediatric Endocrinology and Diabetes. (2008) J. Clin. Investig. 118 2148-2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Edghill, E. L., Flanagan, S. E., Patch, A. M., Boustred, C., Parrish, A., Shields, B., Shepherd, M. H., Hussain, K., Kapoor, R. R., Malecki, M., MacDonald, M. J., Stoy, J., Steiner, D. F., Philipson, L. H., Bell, G. I., Neonatal Diabetes International Collaborative Group, Hattersley, A. T., and Ellard, S. (2008) Diabetes 57 1034-104218162506 [Google Scholar]
- 81.Molven, A., Ringdal, M., Nordbo, A. M., Raeder, H., Stoy, J., Lipkind, G. M., Steiner, D. F., Philipson, L. H., Bergmann, I., Aarskog, D., Undlien, D. E., Joner, G., Sovik, O., Norwegian Childhood Diabetes Study Group, Bell, G. I., and Njolstad, P. R. (2008) Diabetes 57 1131-113518192540 [Google Scholar]
- 82.Oyadomari, S., Koizumi, A., Takeda, K., Gotoh, T., Akira, S., Araki, E., and Mori, M. (2002) J. Clin. Investig. 109 525-532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakagawa, S. H., Hua, Q.-x., Hu, S.-Q., Jia, W., Wang, S., Katsoyannis, P. G., and Weiss, M. A. (2006) J. Biol. Chem. 281 22386-22396 [DOI] [PubMed] [Google Scholar]
- 84.Wan, Z.-L., and Liang, D. (1988) Sci. China Ser. B Chem. Life Sci. Earth Sci. 31 1426-1438 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.