Abstract
Survival motor neuron (SMN) complex is essential for the biogenesis of the small nuclear ribonucleoprotein (snRNP) complex, although the complete role of each SMN complex component for the snRNP synthesis is largely unclear. We have identified an interaction between the two components Gemin2-Gemin7 using the mammalian two-hybrid system. In vitro stability assay revealed that the known SMN-Gemin7 interaction becomes stable in the presence of Gemin2 possibly via the identified Gemin2-Gemin7 interaction. Gemin7 knockdown revealed a decrease in snRNP assembly activity and a decrease in SmE protein, a component of snRNP, in the SMN complex, which was consistent with a previous discussion that the Gemin6-Gemin7 heterodimer may serve as a surrogate for the SmD3-SmB particle in forming a subcore, the intermediate complex for snRNP. Interestingly, we found that Unrip, but not Gemin8, can remove Gemin7 from the stable SMN-Gemin2-Gemin7 ternary complex. In an in vitro snRNP assembly assay using the Unrip knockdown and the untreated cell lysates, we revealed that there was a decrease in Gemin7 and increase in SmB/B′ in the SMN complex observed in untreated cells during the assay, suggesting that the Gemin6-Gemin7 heterodimer in the subcore is exchanged by the SmD3-SmB particle to form snRNP. Surprisingly, these changes were not observed in the assay using the Unrip knockdown cell extracts, indicating the importance of Unrip in the formation of snRNP likely via removal of the Gemin6-Gemin7 from the SMN complex. Taken together, these results indicate that snRNP is synthesized by harmonization of the SMN complex components.
Survival motor neuron (SMN)2 protein is expressed in all metazoans and in all cell types of vertebrate organisms (1, 2). SMN is found in both the cytoplasm and nucleus, where it is concentrated in distinct nuclear structures termed Gems and Cajal bodies (3, 4). SMN oligomerizes and tightly associates directly and indirectly with at least six additional proteins (Gemin2-7) to form a macromolecular complex (40 S to 70 S in sucrose density gradient sedimentation) termed the SMN complex (5, 6). Recently, the proteins Gemin8 and Unrip have been associated with the SMN complex (7, 8). The SMN complex is very important for the biogenesis of small nuclear ribonucleo-proteins (snRNPs) that are essential for pre-mRNA splicing (9). It directly identifies and binds to both the protein (SmB, D1, D2, D3, E, F, and G) and RNA (U1, U2, U4, and U5 snRNA) components of snRNPs and mediates their interaction to ensure that the Sm proteins are assembled on the snRNAs (6, 10-12). Recently, Chari et al. (13) have reported that plCln, a component of the PRMT5 complex, binds to Sm proteins and acts as an assembly chaperon for Sm proteins to form a stable subcore, the intermediate complex for snRNP, in cooperation with the SMN complex. The assembled snRNPs remain attached in the SMN complex for subsequent transfer into the nucleus (14).
SMN associates directly with Gemin2, Gemin3, Gemin5, Gemin7, and Gemin8, whereas Gemin4 associates with the SMN complex via its interaction partner Gemin3, and Gemin6 and Unrip associate with the complex via their interaction partner Gemin7 (5, 15, 16). Gemin2, -3, -4, -6, -8, and SMN are important for the U1 snRNP assembly (7, 8, 10, 17, 18). Gemin6 and -7 form a Sm core protein dimer-like structure, suggesting that they have roles in assembling snRNPs (19). Gemin6, Gemin7, and Unrip form a stable cytoplasmic complex whose association with SMN requires Gemin8 (16). Gemin2 is a 30-kDa protein with no homology to any other protein and tightly associates with the amino-terminal side of SMN (20). Jablonka et al. (21) reported that homozygous deficiency of Gemin2 in mice leads to an embryonic lethal phenotype, and double heterozygous deficiency of Smn and Gemin2 in mice leads to a defect in the biogenesis of snRNPs and enhanced motor neuron loss in the spinal cord. Moreover, sedimentation analysis has revealed that the amount of SMN and Gemin2 is far greater than that of the other components of the SMN complex, strongly suggesting that the core of the SMN complex has a simple protein composition comprised of only SMN and Gemin2 (6).
The precise function and stoichiometry of the individual components and detailed conformation of the SMN complex are still not fully understood. Recently, we have discovered that Gemin2 forms a homodimer responsible for the stability of the SMN oligomer/complex required for efficient snRNP assembly (22). Surprisingly, we also found that siRNA-mediated Gemin2 knockdown causes dissociation of several components including Gemin3 and Gemin7 from the SMN complex, suggesting that Gemin2 helps to stabilize other components in the SMN complex either directly or indirectly (22).
Here, we first systematically searched for protein interactions between Gemin2 and other components of the SMN complex, and in so doing found a Gemin2-Gemin7 interaction. This interaction correlates with the stability of Gemin7 in the SMN complex, which likely occurs through the ternary complex composed of SMN, Gemin2, and Gemin7. Next we showed the importance of Gemin7 in the snRNP assembly possibly via formation of a stable subcore complex. Furthermore, we found that Unrip can remove the Gemin6-Gemin7 heterodimer from the SMN complex. Moreover, we showed that the decrease in Gemin7 and the increase in SmB were observed in the SMN complex during the snRNP assembly assay using the untreated cell extract, whereas this change was not observed using the Unrip knockdown cell extract, suggesting that Unrip plays an important role in the final step of the snRNP assembly.
EXPERIMENTAL PROCEDURES
cDNA Clones—The full-length cDNAs encoding the SMN complex component proteins were obtained from the RIKEN mouse cDNA bank (FANTOM) (23). Gene identifiers were I730032A18 for SMN, 1810015M21 for Gemin2, F630042F15 for Gemin3, C230037I02 for Gemin4, C330013N08 for Gemin5, 2810470M17 for Gemin6, 2400008I04 for Gemin7, 2410189L24 for Gemin8, and 1810063O13 for Unrip.
Mammalian Two-hybrid Assay—Mammalian two-hybrid assays, including sample construction and transfection, were carried out as previously described (22). The linear double-strand DNA constructs to express fusion proteins with the Gal4 DNA-binding domain (BIND) or the VP16 trans-activation domain (ACT) were prepared in a two-step PCR. All the combinations of Gemin2 and other components were transfected to CHO-K1 cells using Lipofectamine 2000 (Invitrogen) together with the luciferase reporter plasmid pG5luc. After a 22-h incubation, the reporter activity was measured using the Steady-Glo® Luciferase Assay System (Promega Co., Madison, WI).
Rapid in Vitro Pull-down Assay—The in vitro pull-down assay was carried out as previously described (24). The PCR products encoding protein-coding sequences were used to construct samples for in vitro transcription/translation. Briefly, independent in vitro syntheses of biotinylated and 35S-labeled proteins were carried out from the corresponding PCR constructs by using the Transcend Biotinylated lysine-tRNA (Promega), Redivue l-[35S]methionine (GE Healthcare Bio-Sciences Corp.), and TnT T7 Quick Coupled Transcription/Translation System (Promega). Biotinylated protein and 35S-labeled protein were mixed and incubated on ice. A Dynabeads streptavidin (Dynal, Invitrogen Corp.) suspension was mixed with the reaction and incubated in a rotary shaker at 4 °C. The beads were isolated with a magnet and washed with ice-cold TBST (50 mm Tris-HCl, pH 8.0, 137 mm NaCl, 2.68 mm KCl, 0.1% (w/v) Tween 20). The radiolabeled proteins that co-precipitated with biotinylated proteins were separated by SDS-PAGE and visualized by autoradiography using FLA-7000 (FUJIFILM Corporation, Tokyo, Japan). Each band density was measured by using the MultiGauge software (FUJIFILM).
Glutathione S-Transferase Pull-down Assay—Glutathione S-transferase (GST) or GST-Gemin2 was expressed in Escherichia coli and purified. In vitro synthesis of six histidine-tagged Gemin7 (His-Gemin7) was performed with the TnT T7 Quick Coupled Transcription/Translation System (Promega) in the presence of Redivue l-[35S]methionine (GE Healthcare). Synthesized 35S-labeled His-Gemin7 was purified with the MagZ Protein Purification System (Promega). Purified GST or GST-Gemin2 was immobilized on glutathione-Sepharose 4B (Amersham Biosciences). Purified His-Gemin7 was incubated with the immobilized GST or GST-Gemin2 in a lysis buffer (10 mm Tris-HCl, pH 7.8, 1% (w/v) Nonidet P-40, 150 mm NaCl, 1 mm EDTA). After washing the resin, the bound proteins were subjected to SDS-PAGE and visualized by autoradiography using FLA-7000 (FUJIFILM).
In Vitro Stability Assay—For the in vitro stability assay, free [35S]methionine in the radioisotope-labeled reaction that was not incorporated into the synthesized proteins was removed using CENTRI-SEP spin columns (Princeton Separations, Inc., Adelphia, NJ). Equal volumes of biotin-labeled and 35S-labeled proteins were mixed with in vitro synthesized unlabeled protein or the reticulocyte lysate and incubated on ice. A Dynabeads-streptavidin suspension was mixed with the reaction and incubated at 4 °C. The beads were captured using a magnet and washed once with ice-cold TBST, followed by re-suspension in TBST. After 60 min of incubation, the beads were captured and the radioactivity remaining on the beads was measured using a liquid scintillation counter.
Western Blotting—The protein samples were separated by 10-20% gradient SDS-PAGE and transferred to polyvinylidene fluoride membranes. Primary antibodies used were as follows: anti-SMN, SmE (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), Gemin7 (Abnova Corporation, Taipei City, Taiwan), polyclonal IgG and anti-SmB/B′ (Lab Vision Corporation, Fremont, CA), and anti-Gemin2 and Unrip (BD Biosciences, San Jose, CA) monoclonal antibody. Horseradish peroxidase-conjugated anti-goat (Promega) or anti-mouse IgG antibodies (Amersham Biosciences) were used as the secondary antibody. The signal was detected by using the ECL plus Western blotting detection system (Amersham Biosciences) and visualized using LAS-3000 (FUJIFILM). Each band density was measured by using the MultiGauge software (FUJIFILM).
In Vitro snRNP Assembly Assay—HeLa cells were transfected with the siRNA for Unrip using Lipofectamine 2000 according to the manufacturer's protocol. Forty-four hours after the siRNA transfection, cells were harvested and cytoplasmic extracts were prepared using Ne-Per Nuclear and Cytoplasmic Extraction Reagents (Pierce Biotechnology, Inc.). The snRNP assembly assay was carried out as described previously with modifications (17, 18), where components in the snRNP and the SMN complex were monitored before and after the snRNP assembly. Briefly, U1 snRNA was transcribed in vitro using Riboprobe Systems (Promega) in the presence of the Ribo m7G Cap Analog (Promega) for 1 h at 37 °C, followed by removal of DNA template by digestion with RQ1 RNase-free DNase (Promega). The free NTP that was not incorporated into the synthesized U1 snRNA was removed by using MicroSpin S-200 HR columns (Amersham Biosciences). Cytoplasmic extracts were incubated with U1 snRNA for 0 and 60 min at 30 °C in RSB-100 buffer (10 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2.5 mm MgCl2). The reaction mixtures were immunoprecipitated with anti-SMN antibody. The co-precipitated Gemin2, Gemin7, SmB/B′, and SmE were detected by Western blotting.
RESULTS
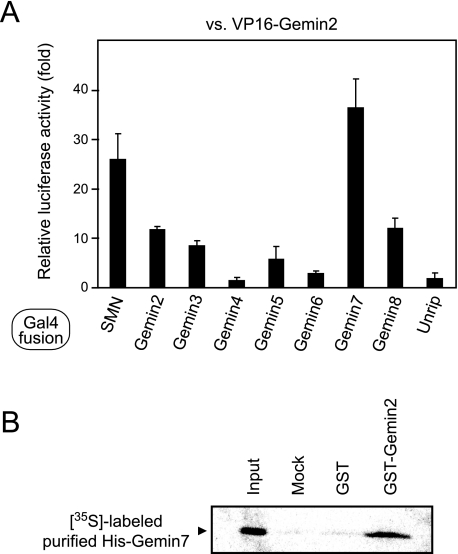
Identification of a Gemin2-Gemin7 Interaction—We recently reported that siRNA-mediated Gemin2 knockdown causes not only a decrease in oligomerization of the SMN protein but also dissociation of several components including Gemin3 and Gemin7 from the SMN complex, suggesting that Gemin2 stabilizes other components in the SMN complex (22). Therefore, we systematically explored protein interactions between Gemin2 and other components in the SMN complex by using the mammalian two-hybrid system (25). We made a fusion protein construct of the VP16 trans-activation domain with Gemin2, and fusion protein constructs of the Gal4 DNA binding domain with each member of the SMN complex (SMN and Gemin2, -3, -4, -5, -6, -7, -8, and Unrip). Together with the luciferase reporter plasmid, the VP16-Gemin2 construct was transfected into CHO-K1 cells in combination with the Gal4 fusion constructs, and then the reporter activity was measured after a 22-h incubation. In addition to the known interactions of Gemin2-SMN and Gemin2-Gemin2, we detected strong reporter activity corresponding to the interactions of Gemin2-Gemin7 and Gemin2-Gemin8 but marginal reporter activity for Gemin2-Gemin3 and Gemin2-Gemin5 (Fig. 1A). Because Gemin2-Gemin7 showed the strongest reporter activity and this interaction has been detected with at least one of three methods by another group (26), we investigated further to confirm this interaction. We used purified recombinant GST-Gemin2 and in vitro translated 35S-labeled His-Gemin7 to indicate Gemin2 association with Gemin7 (Fig. 1B). Because the purified His-Gemin7 did not contain rabbit reticulocyte lysate-derived SMN and Gemin2 (supplemental Fig. S1), it is more likely that Gemin2 associates directly with Gemin7 rather than indirect association via the rabbit SMN and/or Gemin2.
FIGURE 1.
Identification and confirmation of the interaction between Gemin2 and Gemin7. A, results of the mammalian two-hybrid assay. VP16-Gemin2 and Gal4-SMN, -Gemin2, -3, -4, -5, -6, -7, -8, and -Unrip were expressed in CHO-K1 cells. The relative luciferase activity of the reporter gene was measured. B, results of the GST pull-down assay. Purified GST or GST-Gemin2 were immobilized on glutathione-Sepharose and incubated with purified in vitro translated 35S-labeled His-Gemin7. After resolution by SDS-PAGE, the bound proteins were visualized by autoradiography. Ten percent of 35S-labeled His-Gemin7 used in a reaction was loaded as input.
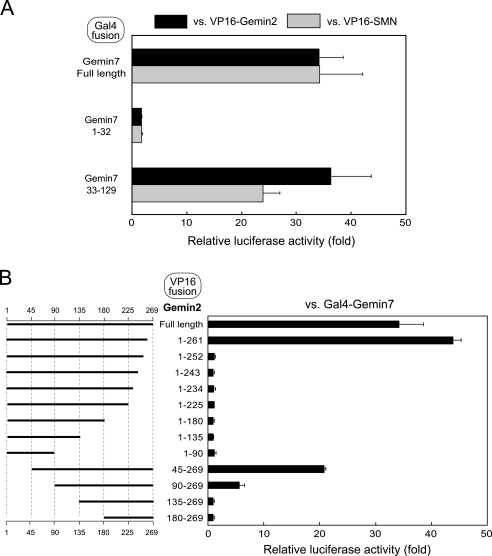
Domain Mapping of Gemin2-Gemin7 Interaction—To identify the binding domains for the Gemin2-Gemin7 interaction, we used deletion mutants for Gemin2 and Gemin7 in the mammalian two-hybrid system (Fig. 2). Ma et al. (19) reported that the amino-terminal ∼30 amino acid residues of Gemin7 may mediate the known SMN-Gemin7 interaction because the remaining region of Gemin7 is used for the association with Gemin6. However, we found that the amino-terminal region of Gemin7 has scarcely any binding availability for SMN and Gemin2, and the remaining region of Gemin7 is responsible for the association with them (Fig. 2A). Next, we examined the Gemin2 region responsible for association with Gemin7. We found that these binding domains reside almost entirely throughout the Gemin2 protein (Fig. 2B), which is similar to our previous result for Gemin2 self-association and Gemin2-SMN association. The binding availability seems to be sensitive to deletion at the carboxyl terminus as Gemin7 binding drastically decreased in the mutant Gemin21-252, a deletion mutant lacking 17 amino acid residues at the carboxyl terminus of Gemin2. In comparison, the amino-terminal deletion of Gemin2 was relatively tolerant of the binding availability; the reporter activity for Gemin2-Gemin7 association was significantly high even in Gemin245-269 and Gemin290-269 (Fig. 2B).
FIGURE 2.
Mapping of the interaction region of Gemin2 and Gemin7. A, identification of the region where Gemin7 interacts with SMN and Gemin2 by the mammalian two-hybrid assay. Gal4-fused proteins for full-length Gemin7 or Gemin7-deletion mutants were expressed in CHO-K1 cells with VP16-SMN or -Gemin2. B, identification of the region where Gemin2 interacts with Gemin7 by the mammalian two-hybrid assay. VP16-fused proteins for full-length Gemin2 or Gemin2-deletion mutants were expressed in CHO-K1 cells with Gal4-Gemin7. The relative luciferase activity of the reporter gene was measured.
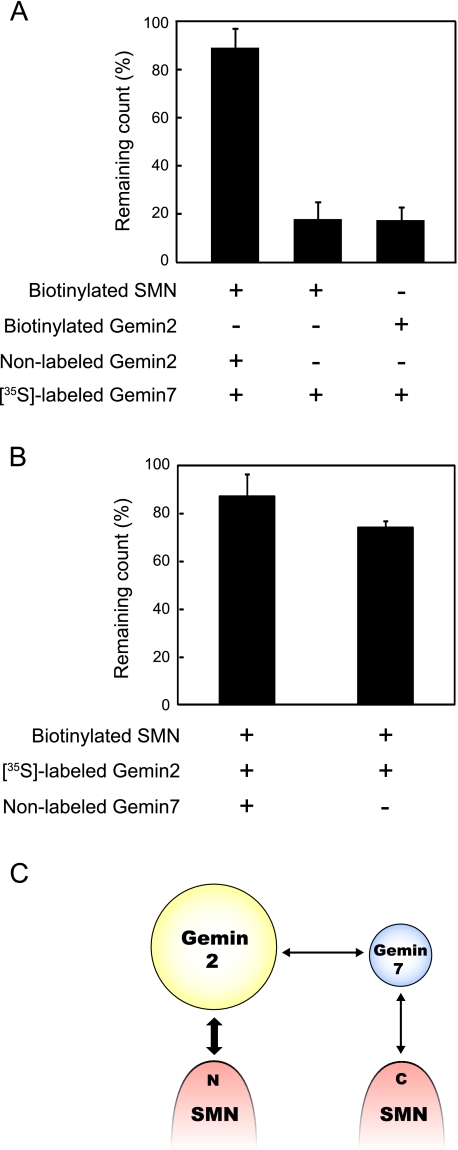
Gemin2 Stabilizes SMN-Gemin7 Association—Because Gemin7 associates with both SMN (27) and Gemin2 (this work) and Gemin2 knockdown decreased Gemin7 co-precipitation with SMN (22), these interactions together may stabilize Gemin7 in the SMN complex. To test this hypothesis, we examined the stability of SMN-Gemin7 interaction in the presence or absence of synthesized Gemin2 using the in vitro stability assay that we recently developed (22). After association with in vitro synthesized biotin-labeled protein and 35S-labeled protein, the complex was captured by streptavidin beads and suspended in assay buffer. The remaining 35S-labeled protein that was attached to the beads was measured to assay stability at 60 min. We found that the SMN-Gemin7 interaction was unstable in the absence of Gemin2; only 20% of the original 35S-labeled Gemin7 remained after 60 min of incubation (middle column in Fig. 3A). This interaction became significantly (p < 0.01) more stable in the presence of the synthesized Gemin2 protein (left column in Fig. 3A); 35S-labeled Gemin7 remained at more than 90% of the initial value even after 60 min of incubation. It is unlikely that Gemin7 was preserved in the linear complex of SMN-Gemin2-Gemin7, because Gemin2-Gemin7 association (right column in Fig. 3A) was unstable as observed in the SMN-Gemin7 association. We also performed stability experiments using biotin-SMN and 35S-labeled Gemin2 in the presence or absence of synthesized Gemin7 (Fig. 3B). Although the Gemin2-SMN interaction was relatively stable even in the absence of Gemin7 (right column in Fig. 3B), association of Gemin2 with SMN became slightly stable in the presence of Gemin7 (left column in Fig. 3B). Thus, the results indicate that SMN, Gemin2, and Gemin7 compose a stable ternary complex by association with one another (Fig. 3C).
FIGURE 3.
Stability of the interaction among Gemin2, Gemin7, and SMN. A, results from in vitro stability assays using 35S-labeled Gemin7. The sample combinations are described below the graph. Each column indicates the relative remaining count of pulled down 35S-labeled Gemin7 attached to streptavidin beads after 60 min incubation compared with the count at zero time. The errors bars represent the standard deviation. B, result of the stability assay using 35S-labeled Gemin2. C, the schematic interaction map of SMN, Gemin2, and -7 was shown based on our results and previous reports. The arrows indicate the relationship of direct interaction between each component. Thick lines show stable interaction and thin lines show unstable interactions.
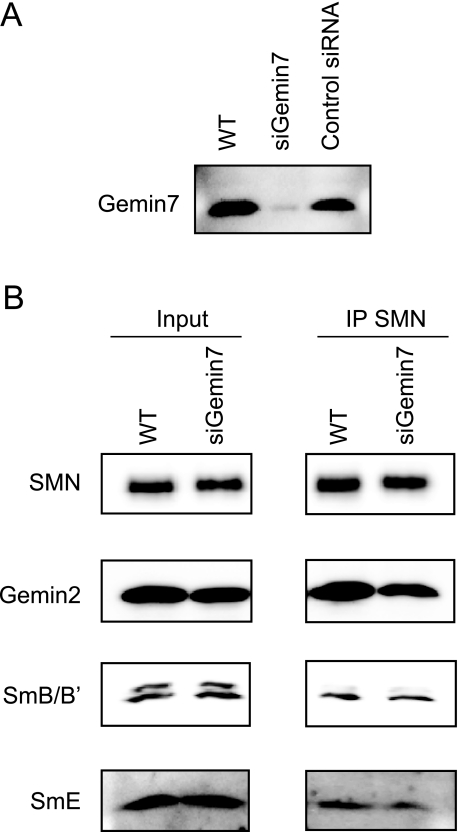
Gemin7 Knockdown Lowers in Vitro snRNP Assembly Rates—Our results, described above, indicate that SMN-Gemin7 association was stabilized by Gemin2. Because Gemin6 is involved in the SMN complex through association with Gemin7 (27) and siRNA-mediated knockdown of Gemin6 reduced snRNP assembly activity (17, 18), Gemin7 must play an important role in the biogenesis of snRNPs in the SMN complex. Shpargel and Matera (18) showed that siRNA-mediated knockdown of Gemin7 modestly inhibited the snRNP assembly, although they were unable to confirm the effect of Gemin7 siRNA on the expression of Gemin7. Therefore, we transfected siRNA against Gemin7 into HeLa cells and analyzed the knockdown effect by Western blotting. The Gemin7 expression level was reduced by Gemin7 siRNA treatment (siGemin7) compared with control cell extracts (supplemental Fig. S2A). We then performed in vitro snRNP assembly assays with 32P-labeled U1 snRNA and HeLa cell extracts with siRNA treatment to test the consequences of reduced expression levels of Gemin7 on SMN complex function in the Sm core assembly. The in vitro snRNP assembly activity was used to measure co-immunoprecipitated 32P-labeled U1 snRNA as Sm protein-U1 snRNA complexes. As shown in supplemental Fig. S2, B and C, Gemin7 siRNA treatment (siGemin7) significantly decreased snRNP assembly activity: there was 40% assembly activity compared with the untreated (WT) cells. On the other hand, in the negative control siRNA (siControl)-treated cells, the snRNP assembly activity level was comparable with untreated cells, indicating that Gemin7 is required for efficient snRNP assembly.
During snRNP synthesis, Sm protein loading on the snRNA is thought to occur on the SMN complex in a stepwise manner; SmD1, D2, E, F, and G are assembled on the snRNA and a stable subcore is formed, then the SmD3-SmB particle is added to the subcore to form the heptamer ring in snRNP (28). Recent analysis of the protein crystal structure has shown that the Gemin6-Gemin7 heterodimer is structurally very similar to the Sm-protein heterodimers, especially to SmD3-SmB/B′, and it can associate with several Sm proteins (19); thus, the Gemin6-Gemin7 heterodimer may serve as a surrogate for the SmD3-SmB particle in the formation of a subcore with other Sm proteins during snRNP assembly (19). To explore the effect of Gemin7 knockdown to subcore formation, we immunoprecipitated the SMN complex from the Gemin7 siRNA-treated and untreated cell lysates using anti-SMN antibody, then detected several SMN complex components by Western blotting. We found that SmE, a component of the subcore, was significantly decreased (average of 47% reduction in 3 experiments, p < 0.05) in the SMN complex from Gemin7 siRNA-treated cell lysates (Fig. 4B). Interestingly we also observed a slight reduction of Gemin2 in the SMN complex, which is consistent with the results in our in vitro stability assay (Fig. 3B).
FIGURE 4.
Gemin7 is important for formation of the subcore complex. A, Gemin7 gene silencing with siRNA. Cytoplasmic extracts were prepared from untreated (WT), Gemin7 siRNA transfected (siGemin7), and negative control siRNA transfected (control siRNA) HeLa cells. The extracts were analyzed by Western blotting using anti-Gemin7 antibody. B, cytoplasmic soluble fraction from the siRNA (siGemin7)-treated and untreated (WT) HeLa cells were immunoprecipitated with anti-SMN antibody. Five percent of the input (left panel) and immunoprecipitates (IP) (right panel) was analyzed by Western blotting using anti-SMN, Gemin2, SmB/B′, and SmE antibodies.
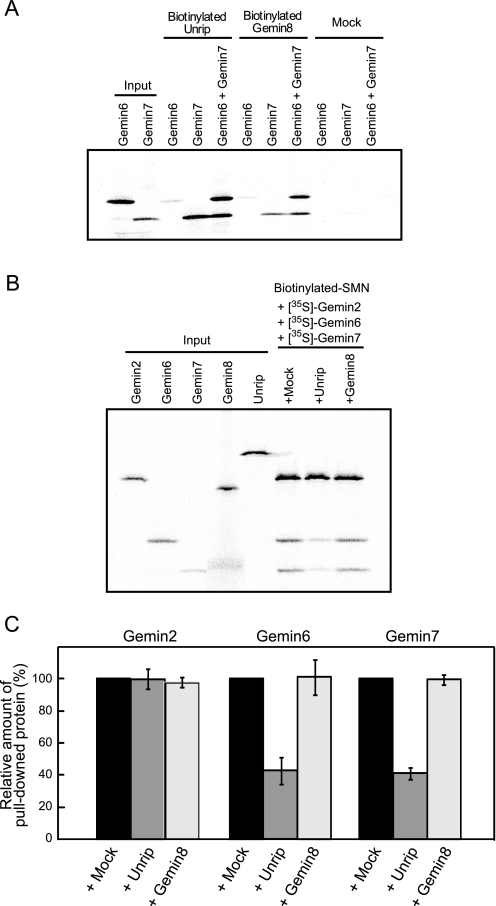
Unrip Removes Gemin7 from the Stable Complex Composed of SMN, Gemin2, and Gemin7—Recently, two novel proteins in the SMN complex, Unrip and Gemin8, have been directly associated with Gemin7; Grimmler et al. (15) reported that Unrip associates with Gemin7 or the Gemin6-Gemin7 heterodimer, and Carissimi et al. (16) reported that Gemin8 binds directly to SMN and mediates its interaction with the Gemin6-Gemin7 heterodimer. To explore the effect of Unrip and Gemin8 on the stable complex composed of SMN, Gemin2, and Gemin7, we first confirmed Gemin7 binding with both Unrip and Gemin8 using an in vitro pull-down assay (Fig. 5A). Both Unrip and Gemin8 bound with the Gemin7 and Gemin6-Gemin7 heterodimer, but binding with Gemin6 was weak. Gemin8 enhanced co-precipitation of Gemin7 in the presence of Gemin6, indicating that Gemin6-Gemin7 and Gemin8 compose a stable ternary complex. These results were similar to those of previous reports (7, 15). Next we explored the effect of Unrip and Gemin8 on the SMN-Gemin2-Gemin7 ternary complex. We incubated the biotin-SMN driver, 35S·labeled Gemin2, 35S-labeled Gemin6, and 35S-labeled Gemin7 for 60 min, then incubated them another 60 min without (mock) and with Unrip or Gemin8. After the incubation, the biotin-SMN was pulled down by streptavidin beads and analyzed by SDS-PAGE (Fig. 5B). We found that co-precipitation of the Gemin6-Gemin7 heterodimer with biotin-SMN became significantly weaker (p < 0.05) when we added Unrip (compare lanes Mock and Unrip in Fig. 5B), but we did not observe this in the addition of Gemin8 (Fig. 5C). This result shows that Unrip can remove Gemin7 from the stable SMN-Gemin2-Gemin7 ternary complex; see also Fig. 3A, which shows that the SMN-Gemin2-Gemin7 complex was stable for at least 1 h.
FIGURE 5.
Unrip removes Gemin7 from the stable SMN-Gemin2-Gemin7 ternary complex. A and B, results of rapid in vitro pull-down assay. Sham operation without biotinylated protein is shown as a negative control (Mock). Five percent of the 35S-labeled proteins used in the reaction was loaded as input. A, in vitro synthesized 35S-labeled Gemin6 and Gemin7 were pulled down using in vitro synthesized biotinylated Unrip and Gemin8 followed by SDS-PAGE. B, in vitro synthesized biotinylated SMN was incubated with 35S-labeled Gemin2 and Gemin7 for 1 h and then with in vitro synthesized 35S-labeled Gemin8 or Unrip for 1 h followed by pull-down by streptavidin beads. C, statistical analysis of B. Change in the amount of pulled down Gemin2, Gemin6, and Gemin7 in the presence of Unrip or Gemin8 is shown. Percentages show the relative amount of the pulled down proteins compared with the sham operation (+ Mock). The experiment was independently conducted three times and the errors bars represent S.D.
It was reported that Unrip is a component of the SMN complex and plays a role in snRNP assembly (8). Another report also showed that the fractionation experiment of the HeLa cell lysate using sucrose gradient centrifugation revealed that a majority of Unrip is separated from the SMN containing fractions and found in slow sedimenting fractions that contain Gemin6-Gemin7 (16). To clarify these observations, we analyzed the amount of Unrip in the SMN complex. HeLa cell lysate was immunoprecipitated using anti-SMN antibody and co-immunoprecipitated Unrip was compared with the total amount of Unrip in the lysate. We confirmed that a marginal amount of Unrip associates with the SMN complex, and most of the Unrip is free from the SMN complex, which is quite different from the observation that the majority of Gemin2 was likely to co-exist with SMN (supplemental Fig. S3). Considering the result, together with the previous ones, perhaps Unrip plays a role in snRNP assembly via a dynamic mechanism rather than as a steady component within the SMN complex.
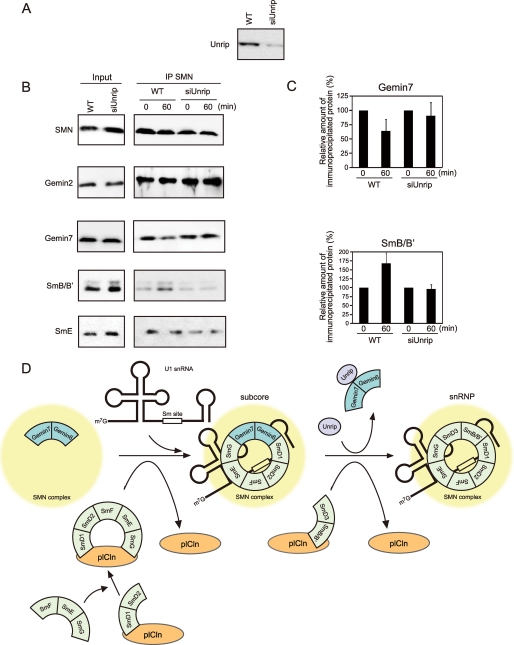
The Exchange of Gemin7 and SmB in the SMN Complex Occurs in the snRNP Assembly Assay Using the Wild-type Cell Extract—Because Unrip will remove Gemin7 from the stable SMN-Gemin2-Gemin7 ternary complex in in vitro experiments, we hypothesize that Unrip promotes the exchange between Gemin6-Gemin7 and SmD3-SmB in the SMN complex in the final step of the snRNP assembly. We therefore explored the importance of Unrip to the snRNP assembly by applying Unrip siRNA in HeLa cells. First we confirmed that Unrip siRNA affects expression of the Unrip protein. As expected, Unrip siRNA treatment decreased the Unrip protein expression (Fig. 6A), but did not affect several component proteins in the SMN complex (Input in Fig. 6B). Next, cytoplasmic extracts from the siRNA-treated and untreated cells were incubated with in vitro synthesized U1 snRNA, and the SMN complex components and the attached snRNP components were immunoprecipitated after no incubation and 60 min of incubation using an anti-SMN antibody; this was followed by Western blotting (Fig. 6, B and C). In the untreated cells (WT), Gemin7 was clearly decreased, whereas SmB/B′ increased after 60 min of incubation. The amount of co-precipitated SmE, one of the subcore components, did not change during 60 min of incubation. The results support the assumption that snRNPs are synthesized on the SMN complex by dissociation of the Gemin6-Gemin7 heterodimer from the subcore complex and incorporation of SmD3-SmB during the incubation. Surprisingly, siRNA treatment did not show a prominent change in the amount of co-precipitated Gemin7 and SmB/B′ during the incubation (siUnrip in Fig. 6, B and C), which suggests that snRNPs were not efficiently synthesized by Unrip knockdown.
FIGURE 6.
Unrip knockdown lowers exchange of Gemin7 and SmB in the SMN complex during in vitro snRNP assembly. A, Unrip gene silencing with siRNA. Cytoplasmic extracts were prepared from untreated (WT) and Unrip siRNA transfected (siUnrip) HeLa cells. The extracts were analyzed by Western blotting using anti-Unrip antibody. B, cytoplasmic extractions from the Unrip siRNA (siUnrip) treated and untreated (WT) HeLa cells were subjected to snRNP assembly assay followed by immunoprecipitation with anti-SMN antibody. Two percent of the input (left panel) and immunoprecipitates (IP) (right panel) was analyzed by Western blotting using anti-SMN, Gemin2, Gemin7, SmB/B′, and SmB antibodies. C, change in the amount of immunoprecipitated Gemin7 and SmB/B′ during snRNP assembly. Each value was calculated by band intensity from Western blotting. Percentages show the relative amount of the immunoprecipitated proteins before and after 60 min of incubation. The experiment was independently conducted three times and the errors bars represent S.D. D, a working model for snRNP assembly pathway.
DISCUSSION
In this paper, we first described identification of Gemin2-Gemin7 interaction and revealed that this discovery identifies Gemin2 as responsible for stabilizing the SMN-Gemin7 interaction. In our previous report (22), we demonstrated that Gemin2 plays important roles in stabilizing the amino-terminal SMN self-association to form the SMN oligomer mediated by SMN-Gemin2 and Gemin2-Gemin2 interactions. siRNA-mediated Gemin2 knockdown revealed the dissociation of Gemin3 and Gemin7 from the SMN complex, suggesting that Gemin2 stabilizes other components in the SMN complex. In addition to the confirmed Gemin2 interactions (SMN-Gemin2, Gemin2-Gemin2, and Gemin2-Gemin7), we showed the possible association of Gemin2 with other SMN components such as Gemin3, -5, and -8 (Fig. 1A), of which the Gemin2-Gemin5 interaction has recently been reported (26). Interestingly, the association spectrum is similar to that of SMN (see Introduction), suggesting that the unevaluated components are also stabilized in the SMN complex due to multiple associations with both SMN and Gemin2. Thus, we assume that the main function of Gemin2, a core component in the SMN complex, is to stabilize other components in the SMN complex.
We next showed the importance of Gemin7 in the snRNP assembly by the Gemin7 siRNA experiment; Gemin7 knockdown reduced snRNP assembly (supplemental Fig. S2), which was consistent with the previous report (18). Because Gemin7 knockdown reduced the amount of SmE protein that is co-immunoprecipitated with the SMN protein (Fig. 4B), the defect of subcore formation of the SMN complex by Gemin7 knockdown likely affected the reduction of snRNP assembly activity. However, because Gemin7 knockdown revealed a slight reduction in Gemin2 in the SMN complex, the snRNP assembly might be affected by the decrease in stability of the SMN complex due to the absence of Gemin7.
For further analysis of Gemin7 function, we explored Gemin7 association with other components in the SMN complex. We showed that in addition to the SMN protein and Gemin2, Unrip, and Gemin8 can also associate with Gemin7. These four Gemin7 associating proteins do not show any sequence homology with one another, suggesting that each protein possesses distinct Gemin7-association properties. Actually, we showed that SMN and Gemin2 cooperatively associate with Gemin7 (Fig. 3A) where Unrip competes for the association and takes Gemin7 away from the stable SMN-Gemin2-Gemin7 ternary complex (Fig. 5B). On the other hand, we did not observe Gemin7 removal when Gemin8 was added or Gemin8 co-precipitation together with the ternary complex (Fig. 5B). Therefore, we think the region of Gemin7 responsible for the association with SMN and Gemin2 overlaps with that for the association with Unrip and Gemin8. Interestingly, a recent study has shown that Gemin8 forms a heteromeric complex with Gemin6, Gemin7, and Unrip (7). Thus, Gemin7 associations with these proteins are very complicated, suggesting that Gemin7 plays a crucial role in the function of the SMN protein complex through these associations.
Our results, together with the previous knowledge, enable us to create a working model for the snRNP assembly (Fig. 6D). Recently, Chari et al. (13, 28) reported that the SmD1-SmD2 particle initially associates with plCln separately and then the SmE-SmF-SmG particle is recruited to the plCln-SmD1-SmD2 complex. In the next step, the transfer of the SmD1-SmD2-SmE-SmF-SmG complex onto the SMN complex coincides with displacement of plCln and a stable subcore is formed. In this step, the Gemin6-Gemin7 heterodimer, stabilized in the SMN complex (left in Fig. 6D), is considered to play an important role in the biogenesis of the subcore. Considering the recent results that the Gemin6-Gemin7 heterodimer can associate with several Sm proteins (19), the Gemin6-Gemin7 heterodimer creates stable footholds for the formation of the subcore and acts as a surrogate for the SmD3-SmB particle (middle in Fig. 6D). At the final snRNP assembly, the Gemin6-Gemin7 heterodimer, stabilized in the SMN complex, should be exchanged with the SmD3-SmB particle to form snRNP. Our results suggest that Unrip plays an important role in promoting this exchange by associating with the Gemin6-Gemin7 heterodimer and removing it from the SMN complex (right in Fig. 6D). We need to perform further analysis to construct a more convincing model. However, our present model clearly indicates that snRNP is synthesized by harmonization of the SMN complex components. We could not find the role of Gemin8 in the snRNP assembly. However, because Gemin8 is required for efficient snRNP assembly and forms the heteromeric complex with Gemin6, Gemin7, and Unrip (7), Gemin8 may function to transport the Gemin6-Gemin7 heterodimer into the SMN complex in coupling with the snRNP assembly.
Supplementary Material
Acknowledgments
We thank Toshiko Imamura for technical assistance. We also thank Dr. Mutsumi Kanamori-Katayama, Chikatoshi Kai, Makoto Aoki, and other members of the Omics Science Center.
This work was supported by a grant from CREST (Core Research for Evolutional Science and Technology) of the Japan Science and Technology Corporation (JST) (to Y. H.) and a research grant for the RIKEN Omics Science Center from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government (to Y. H.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3 and additional data.
Footnotes
The abbreviations used are: SMN, survival motor neuron; snRNPs, small nuclear ribonucleoproteins; Unrip, unr-interacting protein; GST, glutathione S-transferase; CHO, Chinese hamster ovary; siRNA, small interfering RNA; WT, wild type.
References
- 1.Zerres, K., and Rudnik-Schoneborn, S. (1995) Arch. Neurol. 52 518-523 [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre, S., Burglen, L., Reboullet, S., Clermont, O., Burlet, P., Viollet, L., Benichou, B., Cruaud, C., Millasseau, P., Zeviani, M., Le Paslier, D., Frézal, J., Cohen, D., Weissenbach, J., Munnich, A., and Melki, J. (1995) Cell 80 155-165 [DOI] [PubMed] [Google Scholar]
- 3.Liu, Q., and Dreyfuss, G. (1996) EMBO J. 15 3555-3565 [PMC free article] [PubMed] [Google Scholar]
- 4.Matera, A. G., and Frey, M. R. (1998) Am. J. Hum. Genet. 63 317-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gubitz, A. K., Feng, W., and Dreyfuss, G. (2004) Exp. Cell Res. 296 51-56 [DOI] [PubMed] [Google Scholar]
- 6.Paushkin, S., Gubitz, A. K., Massenet, S., and Dreyfuss, G. (2002) Curr. Opin. Cell Biol. 14 305-312 [DOI] [PubMed] [Google Scholar]
- 7.Carissimi, C., Saieva, L., Baccon, J., Chiarella, P., Maiolica, A., Sawyer, A., Rappsilber, J., and Pellizzoni, L. (2006) J. Biol. Chem. 281 8126-8134 [DOI] [PubMed] [Google Scholar]
- 8.Carissimi, C., Baccon, J., Straccia, M., Chiarella, P., Maiolica, A., Sawyer, A., Rappsilber, J., and Pellizzoni, L. (2005) FEBS Lett. 579 2348-2354 [DOI] [PubMed] [Google Scholar]
- 9.Will, C. L., and Luhrmann, R. (2001) Curr. Opin. Cell Biol. 13 290-301 [DOI] [PubMed] [Google Scholar]
- 10.Meister, G., Eggert, C., and Fischer, U. (2002) Trends Cell Biol. 12 472-478 [DOI] [PubMed] [Google Scholar]
- 11.Yong, J., Pellizzoni, L., and Dreyfuss, G. (2002) EMBO J. 21 1188-1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yong, J., Golembe, T. J., Battle, D. J., Pellizzoni, L., and Dreyfuss, G. (2004) Mol. Cell. Biol. 24 2747-2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chari, A., Golas, M. M., Klingenhager, M., Neuenkirchen, N., Sander, B., Englbrecht, C., Sickmann, A., Stark, H., and Fischer, U. (2008) Cell 135 497-509 [DOI] [PubMed] [Google Scholar]
- 14.Narayanan, U., Achsel, T., Luhrmann, R., and Matera, A. G. (2004) Mol. Cell 16 223-234 [DOI] [PubMed] [Google Scholar]
- 15.Grimmler, M., Otter, S., Peter, C., Muller, F., Chari, A., and Fischer, U. (2005) Hum. Mol. Genet. 14 3099-3111 [DOI] [PubMed] [Google Scholar]
- 16.Carissimi, C., Saieva, L., Gabanella, F., and Pellizzoni, L. (2006) J. Biol. Chem. 281 37009-37016 [DOI] [PubMed] [Google Scholar]
- 17.Feng, W., Gubitz, A. K., Wan, L., Battle, D. J., Dostie, J., Golembe, T. J., and Dreyfuss, G. (2005) Hum. Mol. Genet. 14 1605-1611 [DOI] [PubMed] [Google Scholar]
- 18.Shpargel, K. B., and Matera, A. G. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 17372-17377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, Y., Dostie, J., Dreyfuss, G., and Van Duyne, G. D. (2005) Structure 13 883-892 [DOI] [PubMed] [Google Scholar]
- 20.Liu, Q., Fischer, U., Wang, F., and Dreyfuss, G. (1997) Cell 90 1013-1021 [DOI] [PubMed] [Google Scholar]
- 21.Jablonka, S., Holtmann, B., Meister, G., Bandilla, M., Rossoll, W., Fischer, U., and Sendtner, M. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 10126-10131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogawa, C., Usui, K., Aoki, M., Ito, F., Itoh, M., Kai, C., Kanamori-Katayama, M., Hayashizaki, Y., and Suzuki, H. (2007) J. Biol. Chem. 282 11122-11134 [DOI] [PubMed] [Google Scholar]
- 23.Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., Oyama, R., Ravasi, T., Lenhard, B., Wells, C., Kodzius, R., Shimokawa, K., Bajic, V. B., Brenner, S. E., Batalov, S., Forrest, A. R., Zavolan, M., Davis, M. J., Wilming, L. G., Aidinis, V., Allen, J. E., Ambesi-Impiombato, A., Apweiler, R., Aturaliya, R. N., Bailey, T. L., Bansal, M., Baxter, L., Beisel, K. W., Bersano, T., Bono, H., Chalk, A. M., Chiu, K. P., Choudhary, V., Christoffels, A., Clutterbuck, D. R., Crowe, M. L., Dalla, E., Dalrymple, B. P., de Bono, B., Della Gatta, G., di Bernardo, D., Down, T., Engstrom, P., Fagiolini, M., Faulkner, G., Fletcher, C. F., Fukushima, T., Furuno, M., Futaki, S., Gariboldi, M., Georgii-Hemming, P., Gingeras, T. R., Gojobori, T., Green, R. E., Gustincich, S., Harbers, M., Hayashi, Y., Hensch, T. K., Hirokawa, N., Hill, D., Huminiecki, L., Iacono, M., Ikeo, K., Iwama, A., Ishikawa, T., Jakt, M., Kanapin, A., Katoh, M., Kawasawa, Y., Kelso, J., Kitamura, H., Kitano, H., Kollias, G., Krishnan, S. P., Kruger, A., Kummerfeld, S. K., Kurochkin, I. V., Lareau, L. F., Lazarevic, D., Lipovich, L., Liu, J., Liuni, S., McWilliam, S., Madan Babu, M., Madera, M., Marchionni, L., Matsuda, H., Matsuzawa, S., Miki, H., Mignone, F., Miyake, S., Morris, K., Mottagui-Tabar, S., Mulder, N., Nakano, N., Nakauchi, H., Ng, P., Nilsson, R., Nishiguchi, S., Nishikawa, S., Nori, F., Ohara, O., Okazaki, Y., Orlando, V., Pang, K. C., Pavan, W. J., Pavesi, G., Pesole, G., Petrovsky, N., Piazza, S., Reed, J., Reid, J. F., Ring, B. Z., Ringwald, M., Rost, B., Ruan, Y., Salzberg, S. L., Sandelin, A., Schneider, C., Schonbach, C., Sekiguchi, K., Semple, C. A., Seno, S., Sessa, L., Sheng, Y., Shibata, Y., Shimada, H., Shimada, K., Silva, D., Sinclair, B., Sperling, S., Stupka, E., Sugiura, K., Sultana, R., Takenaka, Y., Taki, K., Tammoja, K., Tan, S. L., Tang, S., Taylor, M. S., Tegner, J., Teichmann, S. A., Ueda, H. R., van Nimwegen, E., Verardo, R., Wei, C. L., Yagi, K., Yamanishi, H., Zabarovsky, E., Zhu, S., Zimmer, A., Hide, W., Bult, C., Grimmond, S. M., Teasdale, R. D., Liu, E. T., Brusic, V., Quackenbush, J., Wahlestedt, C., Mattick, J. S., Hume, D. A., Kai, C., Sasaki, D., Tomaru, Y., Fukuda, S., Kanamori-Katayama, M., Suzuki, M., Aoki, J., Arakawa, T., Iida, J., Imamura, K., Itoh, M., Kato, T., Kawaji, H., Kawagashira, N., Kawashima, T., Kojima, M., Kondo, S., Konno, H., Nakano, K., Ninomiya, N., Nishio, T., Okada, M., Plessy, C., Shibata, K., Shiraki, T., Suzuki, S., Tagami, M., Waki, K., Watahiki, A., Okamura-Oho, Y., Suzuki, H., Kawai, J., and Hayashizaki, Y. (2005) Science 309 1559-156316141072 [Google Scholar]
- 24.Suzuki, H., Ogawa, C., Usui, K., and Hayashizaki, Y. (2004) BioTechniques 37 918-920 [DOI] [PubMed] [Google Scholar]
- 25.Suzuki, H., Fukunishi, Y., Kagawa, I., Saito, R., Oda, H., Endo, T., Kondo, S., Bono, H., Okazaki, Y., and Hayashizaki, Y. (2001) Genome Res. 11 1758-1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otter, S., Grimmler, M., Neuenkirchen, N., Chari, A., Sickmann, A., and Fischer, U. (2007) J. Biol. Chem. 282 5825-5833 [DOI] [PubMed] [Google Scholar]
- 27.Baccon, J., Pellizzoni, L., Rappsilber, J., Mann, M., and Dreyfuss, G. (2002) J. Biol. Chem. 277 31957-31962 [DOI] [PubMed] [Google Scholar]
- 28.Raker, V. A., Plessel, G., and Luhrmann, R. (1996) EMBO J. 15 2256-2269 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.