Abstract
Biallelic mutations in the NBS1 gene are responsible for the Nijmegen breakage syndrome (NBS), a rare autosomal recessive disorder characterized by chromosome instability and hypersensitivity to ionising radiation (IR). Epidemiological data evidence that the NBS1 gene can be considered a susceptibility factor for cancer development, as demonstrated by the fact that almost 40% of NBS patients have developed a malignancy before the age of 21. Interestingly, also NBS1 heterozygotes, which are clinically asymptomatic, display an elevated risk to develop some types of malignant tumours, especially breast, prostate and colorectal cancers, lymphoblastic leukaemia, and non-Hodgkin’s lymphoma (NHL). So far, nine mutations in the NBS1 gene have been found, at the heterozygous state, in cancer patients. Among them, the 657del5, the I171V and the R215W mutations are the most frequently described. The pathogenicity of these mutations is presumably connected with their occurrence in the highly conserved BRCT tandem domains of the NBS1 protein, which are present in a large superfamily of proteins, and are recognized as major mediators of processes related to cell-cycle checkpoint and DNA repair.
This review will focus on the current state-of-knowledge regarding the correlation between carriers of NBS1 gene mutations and the proneness to the development of malignant tumours.
Key Words: NBS1, 657del5 mutation, R215W mutation, I171V mutation, IVS11+2insT mutation, heterozygous, cancer predisposition, lymphoma, breast cancer, prostate cancer, colorectal cancer.
INTRODUCTION
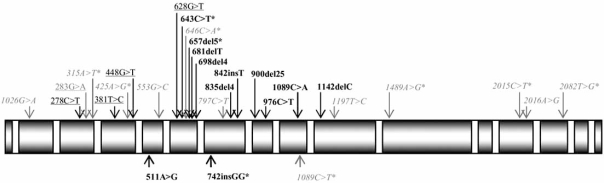
Mutations at the homozygous status in the NBS1 gene (known also as NBN) are responsible for a rare disease known as Nijmegen breakage syndrome (NBS; OMIM 251260), an autosomic recessive disorder whose signs are a distinct facial appearance, microcephaly, immunodeficiency, chromosome rearrangements and sensitivity to ionising radiation [1]. In NBS, a defective response to face DNA damage is associated with chromosomal instability, and in turn with a strong predisposition to develop malignancy, in particular lymphomas [2, 3]. The majority of them are non Hodgkin’s lymphoma (NHL) (with a high incidence of diffuse large B-cell lymphomas), lymphoblatic anaemia, and Hodgkin lymphoma (HL). Among solid tumours, medulloblastoma has been observed in four patients [4], and rhabdomiosarcoma of the perianal region in three others [5]. Since the latter is extremely uncommon among children, a strong association with NBS has been suggested [5]. All the 11 disease-causing mutations so far identified in NBS1 gene have been found within exons 6-10 (Fig. 1), and 8 of them result in premature truncation of the NBS1 protein, with the possible synthesis of NBS1 variants of lower molecular weight [6].
Fig. (1).
Coding sequence variants of NBS1 gene identified to date (modified from http://www.nijmegenbreakagesyndrome.net). Grey: high frequency polymorphisms; grey*: low frequency polymorphisms; black: mutations found in NBS patients; black*: mutations also found in cancer patients at the heterozygous state; black: mutations found in cancer patients only; grey: mutation found both in cancer patients and in healthy controls.
As a measure of cancer incidence in NBS, the Polish registry report that 40% of patients suffering for NBS developed lymphoma within the first two decades of life [3]. Though NBS is a recessive disease and one would not expect any cellular feature or clinical symptom, a growing number of papers report higher spontaneous and induced chromosome instability and an increased incidence of tumours among NBS carriers. The FISH chromosome painting analysis revealed that NBS carriers display a 3-fold higher rate of chromosome translocations compared with non-carriers [7]. Furthermore, the same authors reported that after irradiation, the response of cells from heterozygous carriers was intermediate between that of NBS homozygous and normal individuals, and could be clearly differentiated from those of the other groups in double-coded studies. Moreover, NBS heterozygosity can be distinguished from other genotypes by the number of the long-lived stable aberrations in NBS cells [8]. Radiation hypersensitivity in NBS carriers seems anyhow restricted to cells irradiated in the G1-phase, whereas the number of chromatid-aberrations scored in G2-phase-treated NBS heterozygous cells is in the range of normal cells or slightly higher [9].
Nine mutations localized in the coding sequence of the NBS1 gene have been found, at the heterozygous state, in cancer patients (Fig. 1). The 657del5 (or founder mutation), the 511A>G (I171V), the 643C>T (R215W), and the 742insGG mutations were found both in NBS patients and in cancer patients. Four mutations have been found in cancer patients only (278C>T, 381C>T, 448G>T, and 628G>T). The 283G>A (D95N) mutation, has been identified both in cancer patients and in healthy controls [10], but is not listed as known NBS1 polymorphism by the NCBI website [11].
NBS1 MUTATIONS AND CANCER RISK
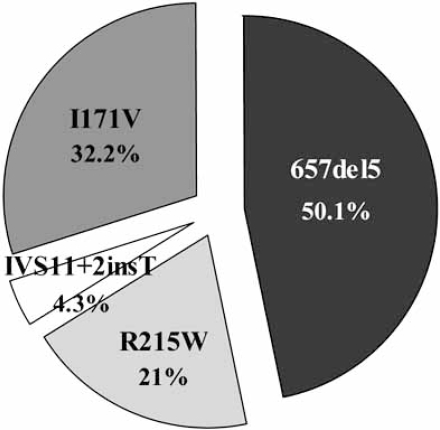
The first evidence of a possible correlation between NBS1 carriers and cancer risk, came from family data studies, indicating that blood relatives of NBS patients with the 657del5 founder mutation had a high probability to develop malignancy [12]. From 1998, several studies have examinated the frequency of the NBS1 mutations in cancer patients. In Table 1, we have collected all the existing data relative to the frequency of cancer in heterozygotes for the commonest NBS1 gene mutations, in particular the 657del5, the R125W, the I171V, and the IVS11+2insT mutations. The relative distribution of these mutations in NBS1 heterozygous cancer patients is illustrated in Fig. (2).
Table 1.
Frequency of the 657del5, R215W, I171V, and IVS11+2insT NBS1 Gene Mutations-Type Among NBS1 Heterozygotes Tumour and Control Subjects (NHL: Non-Hodgkin Lymphoma; ALL: Acute Lymphoblastic Leukaemia; HL: Hodgkin Lymphoma). (nad: Not Available Data)
| NBS1 Mutation | Patient Age | Cancer Type | N° of NBS1 Mutation Carriers Among Tumour Patients | N° of NBS1 Mutation Carriers Among Control Subjects | Statistical Analysis | Ref. |
|---|---|---|---|---|---|---|
| 657del5 | adult | colorectal | 3/234 (1.3%) | 10/1620 (0.6%) | OR: 2.091, p=0.2197 | [15] |
| breast | 4/224 (1.8%) | 10/1620 (0.6%) | OR: 2.927, p=0.0795 | [15] | ||
| 11/562 (1.96%) | nad | OR: 3.21, p = 0.0107 | [48] | |||
| 1/477 (0.2%) | 1/866 (0.1%) | OR: 1.8, p=0.76 | [49] | |||
| 7/873 (0.8%) | 2/692 (0.3%) | OR: 2.8, p=0.32 | [50] | |||
| 2/2012 (0.8%) | 18/4000 (0.5%) | OR: 1.9, p=0.09 | [51] | |||
| 2/150 (1.3%) | 3/530 (0.56%) | χ2test, p<0.0001 | [51] | |||
| 3/80 (3.75%) | 3/530 (0.56%) | χ2 test, p<0.0001 | [51] | |||
| melanoma | 4/105 (3.8%) | 10/1620 (0.6%) | OR: 6.376, p=0.0081 | [15] | ||
| 1/376 (0.26%) | 0 | nad | [23] | |||
| NHL | 2/42 (4.8%) | 10/1620 (0.6%) | OR: 8.05, p=0.0351 | [15] | ||
| 8/228 (3.5%) | 10/1620 (0.6%) | OR: 5.85, p=0.0001 | [22] | |||
| prostate | 5/56 (9%) (familial) | 9/1500 (0.6%) | OR: 16, p>0.0001 | [52] | ||
| 7/305 (2.2%)(non familial) | 9/1500 (0.6%) | OR: 3.9, p=0.01 | [52] | |||
| gastrointestinal lymphoma | 4/37 (10.8%) | 10/1620 (0.6%) | OR: 19.52, p=0.0002 | [22] | ||
| children | NHL | 1/68 (1.5%) | 11/2261 (0.49%) | χ2 test, p<0.0001 | [53] | |
| 2/212 (0.9%) | 42/6984 (0.6%) | OR: 1.57, p=0.041 | [54] | |||
| ALL | 3/270 (1.1%) | 42/6984 (0.6%) | OR: 1.85, p=0.035 | [54] | ||
| 1/68 (1.5%) | 11/2261 (0.49%) | χ2test, p<0.0001 | [53] | |||
| ALL+NHL+HL | 5/545 (0.9%) | 42/6984 (0.6%) | OR: 1.48, p=0.037 | [54] | ||
| ALL+NHL | 5/482 (1.03%) | 42/6984 (0.6%) | OR: 1.73, p=0.029 | [54] | ||
| 2/68 (2.9%) | 0 | nad | [55] | |||
| R215W | adult | colorectal | 3/234 (1.3%) | 4/1620 (0.2%) | OR: 5.247, p=0.0472 | [15] |
| breast | 9/1588 (0.6%) | 5/1014 (0.5%) | OR: 1.9, p=0.18 | [24] | ||
| 9/1076 (0.8%) | 2/1017 (0.2%) | OR: 1.9, p=0.18 | [24] | |||
| melanoma | 1/376 (0.26%) | nad | nad | [23] | ||
| NHL | 2/186 (1.07%) | 10/1620 (0.6%) | nad | [22] | ||
| prostate | 6/338 (1,78%) (sporadic) | 3/208 (1.4%) | OR: 1.24, p =0.77 | [21] | ||
| 2/139 (1,44%) (non familial) | 3/208 (1.4%) | OR: 1, p=1 | [21] | |||
| children | HL | 1/39 (2.6%) | nad | nad | [20] | |
| ALL | 1/47 (2.1%) | nad | nad | [10] | ||
| I171V | adult | larynx cancer | 4/176 (2.3%) | 1/500 (0.2%) | OR: 11.7, p=0.0175 | [26] |
| multiple primary tumors | 5/93 (5.4%) | 1/500 (0.2%) | OR: 28.35, p=0.0005 | [26] | ||
| breast | 20/1636 (1.2%) | 18/1014 (1.8%) | OR: 0.68, p=0.3 | [24] | ||
| 10/1048 (0.9%) | 7/1017 (0.7%) | OR: 1.39, p=0.7 | [24] | |||
| 5/270 (1.8%) | 1/500 (0.2%) | OR: 9.42, p=0.02 | [27] | |||
| head and neck | 5/81 (6.17%) | 1/600 (0.17%) | OR: 39.41, p=0.0001 | [28] | ||
| colorectal carcinoma | 3/131(2.29%) | 1/600 (0.17%) | OR: 14.39, p=0,0196 | [28] | ||
| IVS11+2insT | adult | lung | 2/532 (0.4%) | 2/2348 (0.08%) | OR: 4.43, p=0.16 | [29] |
| gastric cancer | 2/472 (0.4%) | 2/2348 (0.08%) | OR: 25, p<0.0001 | [29] | ||
| colorectal cancer | 3/472 (0.6%) | 2/2348 (0.08%) | OR: 9.43, p=0.02 | [29] |
Fig. (2).
Distribution of the 657del5, R215W, I171V, and IVS11+ 2insT NBS1 gene mutations-type among NBS1 heterozygotes affected with tumour.
The 657del5 Mutation
The 657del5 hypomorphic germ line mutation in exon 6 of NBS1 gene, accounts for more than 90% of all mutant alleles in NBS. The highest frequency of heterozygous carriers of the 657del5 mutation has been found in the Slavic population of Central Europe, with an average frequency of 1/177 [13]. A total number of 81 carriers of 7474 cancer patients (1.8%), have been so far identified (Table 1).
The evidence of a strong correlation between 657del5 mutation heterozygous carriers and cancer risk, has been strengthened by a large study on 344 blood relatives (first-through fourth-degree) of NBS patients, in 24 different NBS families of Czech Republic and Slovakia, from 1998 to 2003 [14]. Thirteen blood relatives developed malignancies of any type, among them eleven were carriers of the 657del5 NBS1 mutation, compared with 6 expected. In this study, the most frequently type of cancer observed was stomach and colorectal cancer. Breast cancers were also reported, though at a lower frequency [14].
The cancer risk of the 657del5 mutation carriers has been also assessed in cancer patients with no NBS cases in the family. It was found that carriers of the 657del5 mutation were about twice more frequent among cancer patients than among matched controls [15]. Most of the 657del5 carriers were found among patients with melanoma (3.8%; OR: 6.376, p=0.0081), NHL (4.8%; OR: 8.05, p=0.0351), breast cancer (1.8%; OR: 2.927, p=0.0795), and colorectal cancer (1.3%; OR: 2.091, p=0.2197) [15]. Moreover, malignant tumours among parents and siblings of 657del5 carriers were twice more frequent (14/77) than in population control [15]. Interestingly, in a study on 2.400 healthy NBS1 heterozygous Polish women, emerged a frequency of 96/10.603 (8.8%) malignant tumours among parents and siblings. This suggested that first-degree relatives of the 657del5 mutation carriers may have an elevated risk of cancer [15]. Heterozygotes for the 657del5 mutation are about three times more frequent among non-selected breast cancer patients than expected. Since an elevated risk of breast cancer has been also observed among carriers of mutations in the BRCA1, p53 and ATM genes [16, 17], and because these gene products interact with each other and with NBS1 [18], these findings suggest that NBS1 is another gene that might be associated with increased risk of breast cancer in heterozygotes [15].
The R215W Mutation
The R215W mutation has been considered for a long time a polymorphism of NBS1, and only recently its severe pathogenicity is emerged with the identification of compound heterozygous 657del5/R215W NBS patients [19].
The R215W missense mutation was first described in a case of acute lymphoblastic leukemia (ALL) and in 9 probands of Slavic origin from a population-based study [10]. Subsequently, a high frequency of heterozygous carriers of the R215W mutation was found not only among children affected by ALL (2.1%), but also by HL (2.6%) (Table 1) [20].
The R215W mutation has been also detected among several cancer patients, complexively in 34 of 4251 individuals with tumours (0.85%) (Table 1). Several studies conducted among Poland, Germany, Czech Republic and United Kingdom, report that heterozygous carriers of the R215W missense mutation have an increased risk of colorectal cancer (1.3%), prostate cancer (1.6%), NHL (1.07%), and breast cancer (0.6%) (Table 1) [15, 21-24].
The I171V Mutation
The 511A>G (I171V) germ-line mutation was identified for the first time in 5 of 47 children with ALL [10]. These children were all characterized by late prognoses due to a late relapses [10]. The same mutation, at the homozygous state, was detected in a Japanese patient with aplastic anaemia, but with no other clinical signs of NBS [25]. In a large study aimed to assess the frequency of NBS1 mutations in patients with larynx cancer and multiple primary tumours, is emerged that the frequency of the I171V mutation carriers is significantly higher than in population controls (2.3% in larynx cancer patients, p=0.0175; 5.4% in multiple primary tumours, p=0.0005). These results imply that the I171V mutation contributes significantly to the overall incidence of larynx carcinoma [26].
An investigation in the Polish population has provided evidences that the I171V mutation could be associated with an increased breast cancer risk (1.8% of I171V carriers, p=0.02) [27]. In particular, this association concerns patients with breast cancer, whose first-degree relatives also had diagnosis of those malignancies [27]. However, in an association study in two large hospital-based case-control settings from Germany and Belarus, is emerged that the I171V missense mutation does not significantly increase the breast cancer risk (0.9% of I171V carriers, p=0.7) [24].
Very recently, it has been shown that the 2.58% of studied patients with malignancies are carriers of the I171V mutation, compared to the 0.17% in the control group (p=0.0002). The percentage of the mutation carriers is particularly high among patients with neck and head tumours (6.17%, p=0.0001), thus suggesting that the I171V mutation in NBS1 gene may be susceptibility factor in solid tumours [28].
The IVS11+2insT Mutation
The IVS11+2insT mutation has been described for the first time in heterozygous Japanese subjects [29], with an increased risk of gastrointestinal cancer that originate in the stomach (2% of carriers, p<0.0001), and in the colorectum (0.8% of carriers, p=0.02) [29]. Interestingly, even if there is not a statistical significant correlation between carriers of the IVS11+2insT mutation and development of lung cancer, it is noteworthy that patients with this type of tumour and heterozygous for the IVS11+2insT mutation, are characterized by a relatively early onset [29]. Since it has been suggested the existence of a mendelian inheritance in the pathogenesis of lung cancer [30, 31], it has been proposed the possibility that the rare autosomal gene contributing to the early onset of lung cancer may be NBS1 [29].
The IVS11+2insT mutation represent the first reported example of a germ line mutation that strongly predisposes to gastric cancer development in the Japanese population. Taking into account that each year 100.000 new case of gastric cancer arises in Japan [32], it is possible to hypothesize that heterozygous carriers of the IVS11+2insT mutation are susceptible to the development of gastric and colon carcinomas [29].
BIOCHEMICAL EFFECTS OF THE NBS1 MUTATION ON THE STRUCTURE AND FUNCTION OF THE NBS1 PROTEIN
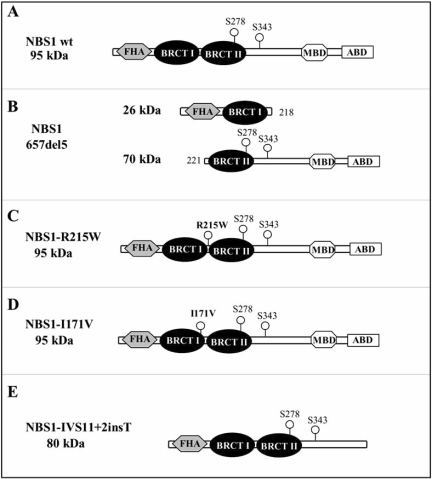
The NBS1 protein is part of a nuclear multi-protein complex composed also by MRE11 and RAD50 (MRN complex), which plays a crucial role in the response to DNA double strand breaks (DSBs), a lesion generated by both endogenous factors and environmental agents as ionising radiations [33, 34]. NBS1 consists of 754 amino acids, has a molecular weight of 95kDa, and is composed by three regions: the N-terminus, the central region, and the C-terminus (Fig. 3A). The N-terminal region contains a fork-head associated (FHA) domain (amino acids 24-109) and two breast cancer C-terminus (BRCT) domain (BRCT1: amino acids 114-183; BRCT2: amino acids 221-291) [35-37]. The central region contains several consensus sequences for phosphorylation by ATM or ATR kinases [38-40]. The C-terminal region (amino acids 665-693) contains a MRE11 binding domain [41], and an ATM recruitment motif [42].
Fig. (3).
Wild type and mutated NBS1 proteins. (A) Structure of the NBS1 wild-type protein. (B) The 657del5 mutation, which splits up the tandem BRCT domains, determines the expression of two truncated proteins of 26 and 70kDa. (C) The R215W mutation occurs in the linker region that connects the two BRCT domains, and determines the substitution at position 215 of an arginine (R) with a tryptophan (W). (D) The I171V missense mutation occurs in the first BRCT domain, and determines the substitution at position 171 of an isoleucine (I) with a valine (V). (E) The NBS1 protein arising from the IVS11+2insT mutation, is characterised by the absence of the MRE11- and ATM-binding domain at the C-terminus. (MBD: MRE11 binding domain; ABD: ATM binding domain).
Low expression of abbreviated polypeptides of both N-terminal and C-terminal NBS1 has been demonstrated in NBS lymphoblastoid cell lines with different mutations. Particularly, C-terminal peptides of lower molecular weight than 95kDa, which maintain the ability to interact with MRE11, has been detected by means of a co-immunoprecipitation assay in lymphoblastoid NBS cell lines [6, 9]. NBS cells characterised by the presence of the classical mutation 657del5 in the NBS1 gene, show two alternative forms of NBS1 with a lower molecular weight, of approximately 26 and 70 kDa. In particular, the 5bp deletion in position 657 splits the BRCT tandem domain exactly in the linker region that connects the two BRCT domains. The 26kDa protein includes the region 1-218 of the NBS1 protein, and comprises the FHA and the first BRCT domains. The 70kDa protein is produced by an alternative initiation of translation upstream the 5bp deletion: after a 18 residue extension at the N-terminus, the sequence is identical to that of the wild type NBS1, from the amino acid 221 to the end, and contains the second BRCT domain and the C-terminal half of NBS1 [37,43] (Fig. 3B).
The R215W missense mutation determines the substitution at position 215 of an arginine (R) with a tryptophan (W). The 215 residue of NBS1 protein is located at the C-terminus of the BRCT1 domain, right before the linker region which connects the two BRCT domains, and seems to be pivotal for the relative orientation of the NBS1 BRCT domains [44] (Fig. 3C). Since tryptophan is a hydrophobic and bulky residue, it could lead to a perturbation of the relative geometry of the tandem BRCT domains. It has been demonstrated, in fact, that the R215W mutation in NBS1 impairs histone γ-H2AX binding after induction of DNA damage, leading to a delay in DNA-DSB rejoining [44].
The pathogenic character of the I171V mutation is presumably connected with its occurrence in the BRCT1 domain of NBS1 (Fig. 3D). So, similarly to the R215W mutation, the I171V may perturb the proper geometry of the tandem BRCT domain, thus impairing the binding to γ-H2AX and the delocalization of the MRN complex to the vicinity of the DNA damage site [26,45].
The IVS11+2insT mutation determines the lacking of the MRE11- and ATM-binding domain at the C-terminus of the NBS1 protein [29] (Fig. 3E). This determines the synthesis of an 80kDa protein, defective in the interaction with MRE11, MDC1, BRCA1 and ATM [29].
CONCLUSIONS
Epidemiological data so far collected point to an increased risk of cancer incidence in heterozygous carriers of the 657del5, R215W, I171V, and IVS11+2insT mutations of the NBS1 gene.
The only NBS1 657del5 founder mutation frequency among newborns is 1:154 in Czech Republic, 1:182 in Ukraine (Lvov region), 1:190 in Poland, with a mean prevalence of 1:177 for the three populations tested [13], whereas the NBS1 R215W mutation frequency among Czech newborns is 1:234 [46]. A moderately elevated risk in heterozygous carriers would results in hundreds of new cancer cases in these populations every year. Because it cannot be excluded that cancer patients who carry germ-line NBS1 mutations may show a specific sensitivity to treatment with ionising radiation or cytostatic drugs, as recently shown [8], systematic studies are now under way to protocol their responses to radio- and chemotherapy.
Experimental support that NBS1 heterozygosity predispose cells to malignancy, come from a study in which the mouse homologue of the human NBS1 gene, Nbn, was disrupted in mice [47]. Nbn+/- mice showed a significantly increased occurrence of spontaneous solid tumours (epithelial tumours affecting the liver, prostate and mammary glands, and gonad malignancy) in addition to lymphoma. Moreover, ionising radiation dramatically increased cancer formation in Nbn+/- mice, especially thyroid tumours. These data provide a clear relationship between NBS1 heterozygosity, radiation sensitivity and increased cancer risk. Interestingly, examination of the tumours gave no evidence for loss or mutation of the wild-type allele, suggesting that haploinsufficiency is the presumed pathogenic mechanism. In contrast, for human heterozygotes, the possible existence of a truncated protein produced by alternative translation [6], and capable of interaction with MRE11 would be compatible with a dominant negative mechanism.
ACKNOWLEDGEMENTS
This work was partly supported by grants from ISS-NIH 526/R2.
ABBREVIATIONS
- NBS1
= Nijmegen breakage syndrome 1 mutated gene
- NBS
= Nijmegen breakage syndrome
- IR
= Ionising radiation
- BRCT
= BRCA1 C-terminal domain
- NHL
= Non Hodgkin lymphoma
- HL
= Hodgkin lymphoma
- FISH
= Fluorescence in situ hybridization
- OR
= Odds ratio
- ATM
= Ataxia telangiectasia mutated gene
- ALL
= Acute lymphoblastic leukaemia
- MRN
= MRE11/RAD50/NBS1 complex
- DSBs
= Double strand breaks
- FHA
= Forkhead associated domain
- ATR
= Ataxia telangiectasia related
- MDC1
= Mediator of DNA damage checkpoint
REFERENCES
- 1.Weemaes CM, Hustinx TW, Scheres JM, Van Munster PJ, Bakkeren JA, Taalman RD. A new chromosomal instability disorder: the Nijmegen breakage syndrome. Acta Paediatr. Scand. 1981;70:557–564. doi: 10.1111/j.1651-2227.1981.tb05740.x. [DOI] [PubMed] [Google Scholar]
- 2.The International Nijmegen Breakage Syndrome Study Group, Nijmegen breakage syndrome. The International Nijmegen Breakage Syndrome Study Group. Arch. Dis. Child. 2000;82:400–406. doi: 10.1136/adc.82.5.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Digweed M, Sperling K. Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks. DNA Repair. 2004;3:1207–1217. doi: 10.1016/j.dnarep.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Distel L, Neubauer S, Varon R, Holter W, Grabenbauer G. Fatal toxicity following radio- and chemotherapy of medulloblas-toma in a child with unrecognized Nijmegen breakage syndrome. Med. Pediatr. Oncol. 2003;41:44–48. doi: 10.1002/mpo.10275. [DOI] [PubMed] [Google Scholar]
- 5.Meyer S, Kingston H, Taylor AM, Byrd PJ, Last JI, Bren-nan BM, Trueman S, Kelsey A, Taylor GM, Eden OB. Rhabdomyosarcoma in Nijmegen breakage syndrome: strong association with perianal primary site. Cancer Genet. Cytogenet. 2004;154:169–174. doi: 10.1016/j.cancergencyto.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 6.Maser RS, Zinkel R, Petrini JH. An alternative mode of trans-lation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat. Genet. 2001;27:417–421. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- 7.Stumm M, Neubauer S, Keindorff S, Wegner RD, Wieacker P, Sauer R. High frequency of spontaneous translocations revealed by FISH in cells from patients with the cancer-prone syndromes ataxia telangiectasia and Nijmegen breakage syndrome. Cytogenet. Cell Genet. 2001;92:186–191. doi: 10.1159/000056900. [DOI] [PubMed] [Google Scholar]
- 8.Neubauer S, Arutyunyan R, Stumm M, Dork T, Bendix R, Bremer M, Varon R, Sauer R, Gebhart E. Radiosensitivity of ataxia telangiectasia and Nijmegen breakage syndrome homozygotes and heterozygotes as determined by three-color FISH chromosome painting. Radiat. Res. 2002;157:312–321. doi: 10.1667/0033-7587(2002)157[0312:roatan]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 9.Tanzarella C, Antoccia A, Spadoni E, di Masi A, Pecile V, Demori E, Varon R, Marseglia GL, Tiepolo L, Maraschio P. Chromosome instability and nibrin protein variants in NBS heterozygotes. Eur. J. Hum. Genet. 2003;11:297–303. doi: 10.1038/sj.ejhg.5200962. [DOI] [PubMed] [Google Scholar]
- 10.Varon R, Reis A, Henze G, von Einsiedel HG, Sperling K, Seeger K. Mutations in the Nijmegen Breakage Syndrome gene (NBS1) in childhood acute lymphoblastic leukemia (ALL) Cancer Res. 2001;61:3570–3572. [PubMed] [Google Scholar]
- 11.http://www.nijmegenbreakagesyndrome.net
- 12.Seemanova E. An increased risk of neoplasm in heterozygotes for a syndrome of microcephaly, normal intelligence, growth retardation, remarkable facies, immunodeficiency and chromosomal instability. Mutat. Res. 1990;238:321–324. doi: 10.1016/0165-1110(90)90024-6. [DOI] [PubMed] [Google Scholar]
- 13.Varon R, Seemanova E, Chrzanowska K, Hnateyko O, Pieku-towska-Abramczuk D, Krajewska-Walasek M, Sykut-Cegielska J, Sperling K, Reis A. Clinical ascertainment of Nijmegen breakage syndrome (NBS) and prevalence of the major mutation, 657del5, in three Slav populations. Eur. J. Hum. Genet. 2000;8:900–902. doi: 10.1038/sj.ejhg.5200554. [DOI] [PubMed] [Google Scholar]
- 14.Seemanova E, Jarolim P, Seeman P, Varon R, Digweed M, Swift M, Sperling K. Cancer risk of heterozygotes with the NBN founder mutation. J. Natl. Cancer Inst. 2007;99:1875–1880. doi: 10.1093/jnci/djm251. [DOI] [PubMed] [Google Scholar]
- 15.Steffen J, Varon R, Mosor M, Maneva G, Maurer M, Stumm M, Nowakowska D, Rubach M, Kosakowska E, Ruka W, Nowecki Z, Rutkowski P, Demkow T, Sadowska M, Bidzinski M, Gawrychowski K, Sperling K. Increased cancer risk of heterozygotes with NBS1 germline mutations in Poland. Int. J. Cancer. 2004;111:67–71. doi: 10.1002/ijc.20239. [DOI] [PubMed] [Google Scholar]
- 16.Kairouz R, Clarke RA, Marr PJ, Watters D, Lavin MF, Kearsley JH, Lee CS. ATM protein synthesis patterns in sporadic breast cancer. Mol. Pathol. 1999;52:252–256. doi: 10.1136/mp.52.5.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broeks A, Urbanus JH, Floore AN, Dahler EC, Klijn JG, Rutgers EJ, Devilee P, Russell NS, van Leeuwen FE, van't Veer LJ. ATM-heterozygous germline mutations contribute to breast cancer-susceptibility. Am. J. Hum. Genet. 2000;66:494–500. doi: 10.1086/302746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong Q, Chen CF, Li S, Chen Y, Wang CC, Xiao J, Chen PL, Sharp ZD, Lee WH. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- 19.Seemanova E, Sperling K, Neitzel H, Varon R, Hadac J, Butova O, Schrock E, Seeman P, Digweed M. Nijmegen breakage syndrome (NBS) with neurological abnormalities and without chromosomal instability. J. Med. Genet. 2006;43:218–224. doi: 10.1136/jmg.2005.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor GM, O’Brien HP, Greaves MF, Ravetto PF, Eden OB. Mutations in the Nijmegen breakage syndrome gene (NBS1) in childhood acute lymphoblastic leukaemia. Cancer Res. 2003;63:6563–6564. [PubMed] [Google Scholar]
- 21.Hebbring SJ, Fredriksson H, White KA, Maier C, Ewing C, McDonnell SK, Jacobsen SJ, Cerhan J, Schaid DJ, Ikonen T, Autio V, Tammela TL, Herkommer K, Paiss T, Vogel W, Gielzak M, Sauvageot J, Schleutker J, Cooney KA, Isaacs W, Thibodeau SM. Role of the Nijmegen Breakage Syndrome 1 gene in familial and sporadic prostate cancer. Cancer Epidemiol. Biomarkers Prev. 2006;15:935–938. doi: 10.1158/1055-9965.EPI-05-0910. [DOI] [PubMed] [Google Scholar]
- 22.Steffen J, Maneva G, Poplawska L, Varon R, Mioduszewska O, Sperling K. Increased risk of gastrointestinal lymphoma in carriers of the 657del5 NBS1 gene mutation. Int. J. Cancer. 2006;119:2970–2973. doi: 10.1002/ijc.22280. [DOI] [PubMed] [Google Scholar]
- 23.Meyer P, Stepelmann H, Frank B, Varon R, Burwinkel B, Schmitt C, Boettger MB, Klaes R, Sperling K, Hemminki K, Kammerer S. Molecular genetic analysis of NBS1 in German Melanoma patients. Melanoma Res. 2007;17:109–116. doi: 10.1097/CMR.0b013e3280dec638. [DOI] [PubMed] [Google Scholar]
- 24.Bogdanova N, Feshcenko S, Schürmann P, Waltes R, Wie-land B, Hillemanns P, Rogov YI, Dammann O, Bremer M, Karstens JH, Sohn C, Varon R, Dörk T. Nijmegen Breakage Syndrome mutations and risk of breast cancer. Int. J. Cancer. 2008;122:802–806. doi: 10.1002/ijc.23168. [DOI] [PubMed] [Google Scholar]
- 25.Shimada H, Shimizu K, Mimaki S, Sakiyama T, Mori T, Shimasaki N, Yokota J, Nakachi K, Ohta T, Ohki M. First case of aplastic anemia in a Japanese child with a homozygous missense mutation in the NBS1 gene (I171V) associated with genomic instability. Hum. Genet. 2004;115:372–376. doi: 10.1007/s00439-004-1155-1. [DOI] [PubMed] [Google Scholar]
- 26.Ziolkowska I, Mosor M, Wierzbika M, Rydzanicz M, Pernak-Schwarz M, Nowak J. Increased risk of larynx cancer in heterozygous carriers of the I171V mutation of the NBS1 gene. Cancer Sci. 2007;98:1701–1705. doi: 10.1111/j.1349-7006.2007.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roznowski K, Januszkiewicz-Lewandowska D, Mosor M, Pernak M, Litwiniuk M, Nowak J. I171V germline mutation in the NBS1 gene significantly increases risk of breast cancer. Breast Cancer Res. Treat. 2007 Sep 26; doi: 10.1007/s10549-007-9734-1. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 28.Nowak J, Mosor M, Ziolkowska I, Wierzbicka M, Pernak-Schwarz M, Prziborska M, Roznowski K, Plawski A, Slomski R, Januszkiewicz D. Heterozygous carriers of the I171V mutation of the NBS1 gene have a significantly increased risk of solid malignant tumours. Eur. J. Cancer. 2008;44:627–630. doi: 10.1016/j.ejca.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Ebi H, Matsuo K, Subito N, Suzuki M, Osada H, Tajima K, Ueda R, Takahashi T. Novel NBS1 heterozygous germline mutation causino MRE11-binding domain loss predispose sto common types of cancer. Cancer Res. 2007;67:11158–11165. doi: 10.1158/0008-5472.CAN-07-1749. [DOI] [PubMed] [Google Scholar]
- 30.Sellers TA, Bailey-Wilson JE, Elston RC, Wilson AF, Elson GZ, Ooi WL, Rothschild H. Evidence for mendelian inheritance in the pathogenesis of lung cancer. J. Natl. Cancer Inst. 1990;82:1272–1279. doi: 10.1093/jnci/82.15.1272. [DOI] [PubMed] [Google Scholar]
- 31.Yang P, Schwartz AG, McAllister AE, Swanson GM, Aston CE. Lung cancer risk in families of nonsmoking probands: heterogeneity by age at diagnosis. Genet. Epidemiol. 1999;17:253–273. doi: 10.1002/(SICI)1098-2272(199911)17:4<253::AID-GEPI2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 32.Ajiki W, Yamamoto S. Age-standardized cancer incidence rates in Japan. Jpn. J. Clin. Oncol. 1999;90:631–636. doi: 10.1093/jjco/29.1.54. [DOI] [PubMed] [Google Scholar]
- 33.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates 3rd JR, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 34.Petrini JH, Stracker TH. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 2003;13:458–462. doi: 10.1016/s0962-8924(03)00170-3. [DOI] [PubMed] [Google Scholar]
- 35.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 36.Callebaut I, Mornon JP. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 37.Becker E, Meyer V, Madaoui H, Guerois R. Detection of a tandem BRCT in Nbs1 and Xrs2 with functional implications in the DNA damage response. Bioinformatics. 2006;22:1289–1292. doi: 10.1093/bioinformatics/btl075. [DOI] [PubMed] [Google Scholar]
- 38.Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase check-point pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 39.Zhao S, Weng YC, Yuan SS, Lin YT, Hsu HC, Lin SC, Gerbino E, Song MH, Zdzienicka MZ, Gatti RA, Shay JW, Ziv Y, Shiloh Y, Lee EY. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature. 2000;405:473–477. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]
- 40.Wu X, Ranganathan V, Weisman DS, Heine WF, Ciccone DN, O'Neill TB, Crick KE, Pierce KA, Lane WS, Rathbun G, Livingston DM, Weaver DT. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature. 2000;405:477–482. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- 41.Desai-Mehta A, Cerosaletti KM, Concannon P. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol. Cell Biol. 2001;21:2184–2191. doi: 10.1128/MCB.21.6.2184-2191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 43.Williams BR, Mirzoeva OK, Morgan WF, Lin J, Dunnick W, Petrini JH. A murine model of Nijmegen breakage syndrome. Curr. Biol. 2002;12:648–653. doi: 10.1016/s0960-9822(02)00763-7. [DOI] [PubMed] [Google Scholar]
- 44.di Masi A, Viganotti M, Ponticelli F, Ascenzi P, Tanzarella C, Antoccia A. The R215W mutation in NBS1 impairs γ-H2AX binding and affects DNA repair: molecular bases for the severe phenotype of 657del5/R215W Nijmegen breakage syndrome patients. Biochem. Biophys. Res. Commun. 2008;369:835–840. doi: 10.1016/j.bbrc.2008.02.129. [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr. Biol. 2002;12:1846–1851. doi: 10.1016/s0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- 46.Seemanova E, Radvanska J, Stary J, Seeman P, Gebertova G, Varon R, Sperling K. Nositele mutaci NBS1 genu mezi pacienty detske onkologie. Csl. Pediatr. 2004;59:242–245. [Google Scholar]
- 47.Dumon-Jones V, Frappart PO, Tong WM, Sajithlal G, Hulla W, Schmid G, Herceg Z, Digweed M, Wang ZQ. Nbn heterozygosity renders mice susceptible to tumor formation and ionizing radiation-induced tumorigenesis. Cancer Res. 2003;63:7263–7269. [PubMed] [Google Scholar]
- 48.Steffen J, Nowakowska D, Niwinska A, Czapczak D, Kluska A, Piatkowska M, Wisniewska A, Paszko Z. Germline mutations 657del5 of the NBS1 gene contribute significantly to the incidence of breast cancer in Central Poland. Int. J. Cancer. 2006;119:472–475. doi: 10.1002/ijc.21853. [DOI] [PubMed] [Google Scholar]
- 49.Carlomagno F, Chang-Claude J, Dunning AM, Ponder BA. Determination of the frequency of the common 657del5 Nijmegen breakage syndrome mutation in the German population: no association with risk of breast cancer. Genes Chromosomes Cancer. 1999;25:393–395. doi: 10.1002/(sici)1098-2264(199908)25:4<393::aid-gcc12>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 50.Buslov KG, Iyevleva AG, Chekmariova EV, Suspitsin EN, Togo AV, Kuligina ESh, Sokolenko AP, Matsko DE, Turkevich EA, Lazareva YR, Chagunava OL, Bit-Sava EM, Semiglazov VF, Devilee P, Cornelisse C, Hanson KP, Imyanitov EN. NBS1 657del5 mutation may contribute only to a limited fraction of breast cancer cases in Russia. Int. J. Cancer. 2005;114:585–589. doi: 10.1002/ijc.20765. [DOI] [PubMed] [Google Scholar]
- 51.Górski B, Cybulski C, Huzarski T, Byrski T, Gronwald J, Jakubowska A, Stawicka M, Gozdecka-Grodecka S, Szwiec M, Urbanski K, Mitus J, Marczyk E, Dziuba J, Wandzel P, Surdyka D, Haus O, Janiszewska H, Debniak T, Toloczko-Grabarek A, Medrek K, Masojc B, Mierzejewski M, Kowalska E, Narod SA, Lubinski J. Breast cancer predisposing alleles in Poland. Breast Cancer Res. Treat. 2005;92:19–24. doi: 10.1007/s10549-005-1409-1. [DOI] [PubMed] [Google Scholar]
- 52.Cybulski C, Górski B, Debniak T, Gliniewicz B, Mierzejewski M, Masojc B, Jakubowska A, Matyjasik J, Zlowocka E, Sikorski A, Narod SA, Lubinski J. NBS1 is a prostate cancer susceptibility gene. Cancer Res. 2004;64:1215–1219. doi: 10.1158/0008-5472.can-03-2502. [DOI] [PubMed] [Google Scholar]
- 53.Resnick IB, Kondratenko I, Pashanov E, Maschan AA, Karachunsky A, Togoev O, Timakov A, Polyakov A, Tverskaya S, Evgrafov O, Roumiantsev AG. 657del5 mutation in the gene for Nijmegen breakage syndrome (NBS1) in a cohort of Russian children with lymphoid tissue malignancies and controls. Am. J. Med. Genet. A. 2003;120:174–179. doi: 10.1002/ajmg.a.20188. [DOI] [PubMed] [Google Scholar]
- 54.Chrzanowska KH, Piekutowska-Abramczuk D, Popowska E, Gladkowska-Dura M, Maldyk J, Syczewska M, Krajewska-Walasek M, Goryluk-Kozakiewicz B, Bubala H, Gadomski A, Gaworczyk A, Kazanowska B, Koltan A, Kuzmicz M, Luszawska-Kutrzeba T, Maciejka-Kapuscinska L, Stolarska M, Stefanska K, Sznurkowska K, Wakulinska A, Wieczorek M, Szczepanski T, Kowalczyk J. Carrier frequency of mutation 657del5 in the NBS1 gene in a population of Polish pediatric patients with sporadic lymphoid malignancies. Int. J. Cancer. 2006;118:1269–74. doi: 10.1002/ijc.21439. [DOI] [PubMed] [Google Scholar]
- 55.Stanulla M, Stümm M, Dieckvoss BO, Seidemann K, Schemmel V, Müller Brechlin A, Schrappe M, Welte K, Reiter A. No evidence for a major role of heterozygous deletion 657del5 within the NBS1 gene in the pathogenesis of non-Hodgkin's lymphoma of childhood and adolescence. Br. J. Haematol. 2000;109:117–120. doi: 10.1046/j.1365-2141.2000.01973.x. [DOI] [PubMed] [Google Scholar]