Abstract
In higher eukaryotes, heritable gene silencing is associated with histone deacetylation and late replication timing. In Saccharomyces cerevisiae, the histone deacetylase Rpd3 regulates gene expression and also modulates replication timing; however, these mechanisms have been suggested to be independent, and no global association has been found between replication timing and gene expression levels. Using 5-Bromo-2′-deoxyuridine (BrdU) incorporation to generate genome-wide replication profiles, we identified >100 late-firing replication origins that are regulated by Rpd3L, which is specifically targeted to promoters to silence transcription. Rpd3S, which recompacts chromatin after transcription, plays a primary role at only a handful of origins, but subtly influences initiation timing globally. The ability of these functionally distinct Rpd3 complexes to affect replication initiation timing supports the idea that histone deacetylation directly influences initiation timing. Accordingly, loss of Rpd3 function results in higher levels of histone H3 and H4 acetylation surrounding Rpd3-regulated origins, and these origins show a significant association with Rpd3 chromatin binding and gene regulation, supporting a general link between histone acetylation, replication timing, and control of gene expression in budding yeast. Our results also reveal a novel and complementary genomic map of Rpd3L- and Rpd3S-regulated chromosomal loci.
Keywords: DNA replication timing, replication origin, chromatin, histone deacetylase, 5-Bromo-2′-deoxyuridine (BrdU), S-phase checkpoint, microarrays
Chromosomal DNA replication initiates at sites distributed along chromosomes called origins, whose activity is regulated by cell cycle control and chromatin context. Origins are bound by the origin recognition complex (ORC), which in G1 phase recruits Cdc6 and Cdt1 to load the MCM proteins, thereby forming prereplicative complexes (pre-RCs) at each of these sequences (for review, see Bell and Dutta 2002). In the budding yeast, Saccharomyces cerevisiae, ∼500 loci distributed throughout the genome have been identified as confirmed or potential origins, which are denoted “ARSs,” for autonomously replicating sequences (for review, see MacAlpine and Bell 2005; Nieduszynski et al. 2007). As is generally the case in other eukaryotes, budding yeast replication origins vary in their initiation timing during S phase and in their efficiency of initiation, the latter of which is defined as the frequency of initiation events per cell cycle. Although the exact mechanisms that determine initiation timing and efficiency remain unclear, chromatin structure clearly plays an important role. For example, origins within heterochromatin, such as transcriptionally silenced regions and subtelomeric regions, typically are late-firing and/or inefficient. Whereas a general correlation exists between late replication timing and gene repression in most eukaryotes (for review, see Gilbert 2002; Weinreich et al. 2004), this relationship has not been demonstrated in the S. cerevisiae genome (with the exception of the subtelomeric regions).
Several factors that regulate gene expression through chromatin modification have been shown to influence initiation timing and/or efficiency. For example, Sir3, which interacts with the Sir2 histone deacetylase to silence genes within subtelomeric heterochromatin and at the silent mating-type loci, delays and/or suppresses initiation of subtelomeric origins (Stevenson and Gottschling 1999). The Sin3–Rpd3 histone deacetylase complex, known for its role as a gene-specific transcriptional repressor, regulates initiation timing. In budding yeast, deletion of RPD3 causes significantly earlier initiation of at least some nontelomeric, late-firing origins, along with increased acetylation of histones flanking these origins (Vogelauer et al. 2002; Aparicio et al. 2004). In direct support of the notion that local histone acetylation influences the timing of origin firing, targeting of a histone acetylase adjacent to a late-firing origin advances its time of initiation (Vogelauer et al. 2002; Goren et al. 2008).
The recent description of two functionally distinct Rpd3 complexes, large (Rpd3L) and small (Rpd3S), presented the opportunity to elucidate more clearly the mechanism of Rpd3's effect on replication timing (Carrozza et al. 2005b; Joshi and Struhl 2005; Keogh et al. 2005). Rpd3L represents the previously characterized transcriptional regulator, which is recruited by sequence-specific DNA-binding proteins such as Ume6 to promoters, where this complex typically represses gene expression by deacetylating proximal histones (Kadosh and Struhl 1998a,b; Rundlett et al. 1998). Unlike the large complex, Rpd3S is nonspecifically recruited to actively transcribed regions where it deacetylates chromatin in the wake of the transcription elongation machinery and suppresses spurious transcription initiation from cryptic start sites within ORFs (Carrozza et al. 2005b; Joshi and Struhl 2005; Li et al. 2007b). Conceivably, Sin3–Rpd3 may affect replication timing through Rpd3L's function as a promoter-specific regulator of gene expression, and/or through Rpd3S's function in condensing chromatin within transcribed regions that flank origins in the proximal intergenic regions. In fact, Set2, which recruits Rpd3S to chromatin, has been suggested to play a role in negatively regulating DNA replication (Biswas et al. 2008a).
Sequence-specific DNA-binding proteins normally target Rpd3 to specific promoters to regulate transcription; however, its mechanism of targeting to origins remains unclear. Deletion of the gene-specific repressor Ume6, which recruits Rpd3, does not advance the timing of late origin ARS603, which is proximal to a potential Ume6-binding site, as does deletion of RPD3 (Aparicio et al. 2004). Because of the apparent lack of correlation between gene expression levels and replication timing in yeast, and because deletion of RPD3 affects chromatin acetylation throughout extensive regions of the genome and is not restricted to promoters of regulated genes, it has been suggested that Rpd3 acts in an untargeted manner to affect origins (Vogelauer et al. 2002).
Previous studies have addressed the genome-wide function of Rpd3 in transcriptional control through analysis of gene expression levels in rpd3Δ and sin3Δ cells, and analysis of Rpd3 and Sin3 chromatin binding by chromatin immunoprecipitation (ChIP) (Bernstein et al. 2000; Fazzio et al. 2001; Kurdistani et al. 2002; Harbison et al. 2004; Robert et al. 2004; Sabet et al. 2004). In contrast, the genome-wide scope of origin deregulation in rpd3Δ cells is unknown. To enable comparison of Rpd3's role in regulating gene expression with its effects on origin timing, we carried out a genome-wide analysis of replication timing in rpd3Δ cells to identify the subset of origins whose activity is regulated by Rpd3. The application of newly developed statistical approaches for quantitative analyses of this type of data provided a robust foundation for examining the distinct roles of Rpd3L and Rpd3S in modulating origin function, and also enabled large-scale correlation studies between the role of Rpd3 at origins and its role in gene regulation. The findings show that Rpd3 delays initiation of >100 nontelomeric origins throughout the genome, with Rpd3L playing the predominant role at most of these origins. Rpd3S plays a major role at only six origins, but has a small effect globally, as might be expected from its general role in transcription. Our findings also show significant correlations between Rpd3 function in histone deacetylation, gene regulation, and regulation of origin firing-time, suggesting a common mechanism, although distinct factors are likely involved in targeting to different loci. These findings reiterate the importance of histone deacetylation in origin activity, support a general correlation between transcriptional control and replication timing, and provide a novel and complementary map of genomic loci targeted by Rpd3.
Results
Determination of genome-wide replication origin firing in hydroxyurea (HU)
Comparing chromosomal DNA replication dynamics on a genomic scale between different mutant strains requires a highly reproducible and quantifiable method for measurement of DNA synthesis. We chose BrdU-IP-chip, which involves in vivo incorporation of 5-Bromo-2′-deoxyuridine (BrdU) into newly replicated DNA, immunoprecipitation of the BrdU-labeled DNA, and identification of these DNA sequences by hybridization to a high-density, DNA oligonucleotide microarray. Wild-type cells blocked in G1 phase with α-factor were released synchronously into S phase in the presence of HU to impede replication forks from early-firing origins and inhibit replication initiation from late-firing origins; four such experimental replicates were performed to enable robust statistical analyses. Data normalization as well as peak identification and quantification were performed as described in (S.R.V. Knott, C.J. Viggiani, O.M. Aparicio, and S. Tavaré, in prep.) and briefly outlined in the Materials and Methods; an example of data prior to smoothing is also shown (Supplemental Fig. S1A).
In wild-type cells, significant peaks of BrdU incorporation were detected at 251 known or predicted origins, indicating that these origins initiated replication in the presence of HU (Supplemental Table S1). This number is comparable with the number of origins reported to initiate replication under similar conditions in two other genome-wide studies (247 by DNA copy number increase; 290 by density-shift analysis) (Yabuki et al. 2002; Alvino et al. 2007). Indeed, 238 of the 251 BrdU peaks have been described as replication origins in multiple studies (compiled in OriDB) (Nieduszynski et al. 2007), indicating that our approach accurately and sensitively identifies the firing of known origins. Underscoring the high sensitivity of the BrdU-IP-chip approach is the identification of BrdU peaks at 13 loci predicted to contain replication origins in only one previous study (and thus annotated as “dubious” origins in OriDB) (Fig. 1A, “Dub”), which also argues that these are bona fide origins that fire detectably in the presence of HU. An additional 16 significant BrdU peaks that did not align with any previously predicted origins are reported in Supplemental Table S2, but have not been analyzed further.
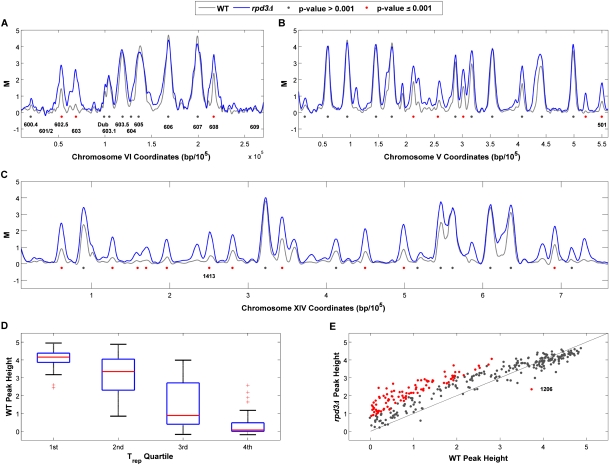
Figure 1.
Early S-phase replication profiles identify Rpd3-regulated origins. Wild-type (WT) and rpd3Δ cells blocked in G1 phase with α-factor were synchronously released into fresh medium containing HU plus BrdU for 1 h and harvested for BrdU-IP-chip analysis. Data were processed as described in the Materials and Methods; chromosomes VI (A), V (B), and XIV (C) are shown. Individual peaks are denoted with gray dots, and peaks that are significantly different in height (P ≤ 0.001) between the strains are denoted with red dots; origins discussed in the text are indicated. (D) Horizontal lines in the plots indicate the lower quartile, median (red line), and upper quartile of wild-type BrdU peak heights assigned to the corresponding Trep quartile (as indicated on the X-axis). Outliers are displayed with a red plus (+) sign. (E) BrdU peak heights determined for each origin in the rpd3Δ strain are plotted against the corresponding peak heights in the wild type; peaks that are significantly different in height in the rpd3Δ cells are indicated with red dots.
Examination of the BrdU profiles for chromosome VI origins, whose initiation timings and efficiencies have been carefully characterized (Friedman et al. 1997; Yamashita et al. 1997), shows that BrdU peak heights reflect origin characteristics such as replication timing and origin efficiency. For example, the early, efficient origins ARS603.5, ARS605, ARS606, and ARS607 exhibit large BrdU peaks that span up to ∼20 kb, reflecting that replication forks from early origins travel up to ∼10 kb before stalling in HU (Fig. 1A). In contrast, the late-firing origins ARS601/602, ARS603, and ARS609 incorporate much less or no BrdU (Fig. 1A), reflecting their inhibition by HU in most cells. The relatively early, but inefficient, origin ARS608 shows an intermediate-sized BrdU peak, and the very inefficient ARS604 exhibits no significant BrdU incorporation. ARS602.5, ARS603.1, and “Dub” were more recently identified and their timings and efficiencies have not been determined (Feng et al. 2006; Xu et al. 2006). In addition to these chromosome VI origins, large BrdU peaks are found at other well-characterized early-firing origins that fire efficiently in HU (e.g., ARS305 and ARS306) (Supplemental Fig. S2), and small BrdU peaks are found at well-characterized late-firing origins that are inhibited by HU (e.g., ARS501, ARS1413) (Fig. 1B,C; Aparicio et al. 1997, 2004; Santocanale and Diffley 1998). Thus, BrdU incorporation levels generally reflect the native origin efficiencies of early origins and the inefficient firing of late origins resulting from their inhibition by the checkpoint. However, it is impossible a priori, from these BrdU profiles alone, to distinguish whether small BrdU peaks reflect low efficiency or late replication timing.
To examine, quantitatively and systematically, the relationship between BrdU incorporation levels and replication timing, we compared BrdU peak heights with the mean replication initiation times (Trep) assigned to >300 origins in a previous study (Raghuraman et al. 2001). We aligned the wild-type BrdU profiles with the predicted origin coordinates from the previous study and found 147 BrdU peaks that correspond to a predicted origin (within 2 kb). The 147 origins were divided into quartiles based on Trep values and the respective BrdU peak heights were plotted (Fig. 1D). The data show that larger BrdU peaks are associated with lower Trep values that represent earlier firing origins, whereas smaller BrdU peaks are associated with higher Trep values that represent later firing origins. Additionally, a Spearman's rank correlation coefficient between BrdU peak height values and the Trep values assigned for these 147 origins demonstrates an anti-correlation between peak sizes and Trep values (ρ = −0.78). We also compared BrdU peak heights with late-replicating origins in Clb5-dependent regions (CDRs) (McCune et al. 2008); Clb5 is required for efficient activation of late-firing origins (Donaldson et al. 1998). The 251 BrdU peaks identified as origins were classified as belonging to one of three CDR classes (1, 2, or 3, with class 3 being the most stringent) or non-CDR and respective BrdU peak heights were plotted (Supplemental Fig. S1B). The results show that the set of origins within CDRs have significantly smaller BrdU peak heights than origins within non-CDRs (Wilcoxon rank sum test, P = 9.7 × 10−10), further indicating that smaller BrdU peaks are associated with late origins. Because origin efficiencies have not been reported on a large scale, a similar analysis of the association between peak sizes and origin efficiencies was not feasible. Nevertheless, our ability to distinguish origin-firing levels in HU based on BrdU peak heights (whether a reflection of differences in timing and/or efficiency) enables the quantitative comparison of replication profiles between strains, such as those lacking Rpd3 function or specifically lacking function of the Rpd3L or Rpd3S complex.
Rpd3 regulates initiation of many replication origins
We demonstrated previously that deletion of RPD3 advances the initiation timing of specific, late-firing origins, which allows these origins to escape inhibition by the HU-induced intra-S phase checkpoint (Aparicio et al. 2004). Our previous study also suggested that the effect of Rpd3 on replication timing might be quite broad across the genome, as deletion of RPD3 suppresses the defect in late origin activation and resulting slow S phase of clb5Δ cells. To explore the genomic scope of Rpd3's influence over origin function, we generated BrdU replication profiles in HU for rpd3Δ cells, in quadruplicate, as described above for wild-type cells. We identified 304 significant BrdU peaks, including 282 known origins as well as 22 peaks at loci previously predicted only in single studies according to OriDB. An additional 41 significant BrdU peaks that did not align with any previously predicted origins are reported in Supplemental Table S2, but have not been analyzed further. Whereas most large peaks representing early-firing origins are very similar in size in the two strains, there are numerous smaller (or absent) BrdU peaks in wild-type cells that are larger in rpd3Δ cells (Fig. 1A–C). For instance, we described previously the advanced firing, and resulting escape from HU-induced inhibition, of the late origin ARS603 in strains lacking RPD3. The wild-type replication profile shows slight, but significant incorporation of BrdU, consistent with this late origin firing inefficiently in HU (Fig. 1A); the rpd3Δ replication profile, however, shows a significantly greater BrdU peak height (empirical Bayes-moderated t-test, P ≤ 0.001). Similarly, the data show significantly larger BrdU peaks at other late origins previously identified as advanced-firing in rpd3Δ cells including ARS501 and ARS1413 (Fig. 1B,C). Thus, increases in BrdU peak heights at these origins in rpd3Δ cells reflect their advanced initiation resulting in more efficient firing in HU.
Global comparison of corresponding BrdU peak heights between strains reveals a total of 104 BrdU peaks that are significantly different in height (empirical Bayes-moderated t-test, P ≤ 0.001) in the rpd3Δ strain, including 53 origins that failed to initiate (or initiated below the level of detection) in the wild-type strain (Fig. 1E; Supplemental Table S3). Data points significantly above the diagonal reflect increased levels of origin firing in the rpd3Δ cells. Rpd3-regulated origins are overrepresented in CDRs versus non-CDRs; of the 104 Rpd3-regulated origins, 74 locate within CDRs (hypergeometric test, P = 3.8 × 10−5). Interestingly, the BrdU peak at ARS1206 is significantly smaller in the rpd3Δ strain suggesting that the deletion of RPD3 has a unique effect on this origin. Only two significant BrdU peaks are detected at telomere-proximal origins (defined here as within 20 kb of a telomere) in wild-type cells (one is deregulated rpd3Δ cells), consistent with the late and/or inefficient nature of subtelomeric origins. However, because of this small number and because probe coverage of origins annotated to these regions is poor (due to lack of sequence specificity), limiting the number of origins available for analysis, we cannot make a strong conclusion about the role of Rpd3 in subtelomeric regions. In summary, this analysis identifies 104 replication origins that incorporate BrdU to a significantly different degree in rpd3Δ cells, indicating that the Sin3–Rpd3 complex impacts the initiation timing and/or replication efficiencies of many origins genome wide.
Rpd3S significantly modulates the initiation of only a few select origins
Rpd3S deacetylates histones in the wake of the transcription elongation machinery (for review, see Lee and Shilatifard 2007). Disruption of Rpd3S results in histone hyperacetylation of much of the genome (Reid et al. 2004; Li et al. 2007b), suggesting that Rpd3S might be responsible for the Rpd3-dependent delay of replication initiation. To test the role of Rpd3S in modulating origin firing, we generated replication profiles in mutant strains that specifically disrupt the chromatin recruitment of Rpd3S (set2Δ, eaf3Δ, and rco1Δ) without affecting the function of Rpd3L. Visual examination of the replication profiles indicates that the set2Δ, eaf3Δ, and rco1Δ profiles more closely resemble wild-type than the rpd3Δ profiles (Figs. 2A; Supplemental Fig. S3A). This suggests that Rpd3S does not mediate most of the observed effects of Rpd3 on origin activity.
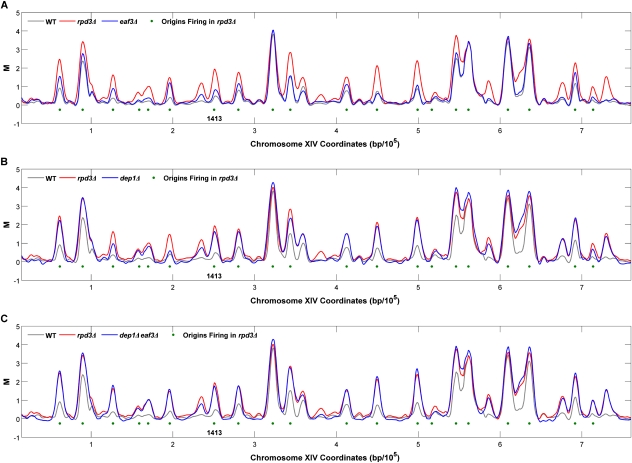
Figure 2.
Early S-phase replication profiles identify Rpd3S- and Rpd3L-regulated origins. eaf3Δ (A), dep1Δ (B), and dep1Δ eaf3Δ (C) cells were analyzed as described in the legend for Figure 1 and the resulting data for chromosome XIV are shown overlaid with the wild-type (WT) and rpd3Δ profiles. Peaks meeting significance criteria for initiation in rpd3Δ cells are indicated with green dots.
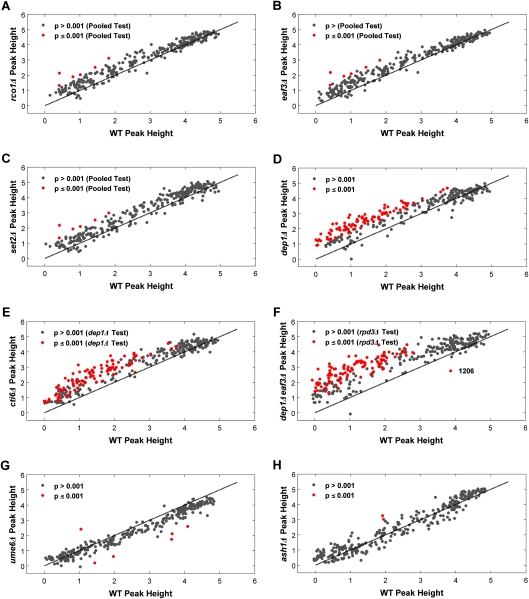
Global comparison of BrdU peak heights in each of the Rpd3S mutant strains versus wild-type confirms that disruption of Rpd3S function does not deregulate origin initiation like RPD3 deletion (cf. Figs. 3A–C and 1E). In contrast to the rpd3Δ mutant profile, only six BrdU peaks were significantly greater in the set2Δ, eaf3Δ, and rco1Δ strains (empirical Bayes-moderated t-test, P ≤ 0.001; t-tests were performed with data pooled from duplicate set2Δ, eaf3Δ, and rco1Δ experiments) (Supplemental Table S4A,B). To ensure that these individual mutations were fully inactivating Rpd3S function we also analyzed eaf3Δ set2Δ, and eaf3Δ rco1Δ strains. The results show strikingly similar replication profiles as the single mutants (see Fig. 4; Supplemental Fig. S3A). Taken together, these results indicate that Rpd3S modulates the activation of a handful of replication origins, but is not responsible for the much more extensive Rpd3-dependent regulation of origin firing demonstrated above.
Figure 3.
Global comparison of BrdU peak heights in Rpd3S and Rpd3L mutants with corresponding wild-type peaks. BrdU peak heights for each origin in rco1Δ (A), eaf3Δ (B), set2Δ (C), dep1Δ (D), cti6Δ (E), dep1Δ eaf3Δ (F), ume6Δ (G), and ash1Δ (H) mutant strains are plotted against the corresponding BrdU peak heights in wild-type (WT) cells; peaks that are significantly different in height from the wild type are indicated with red dots.
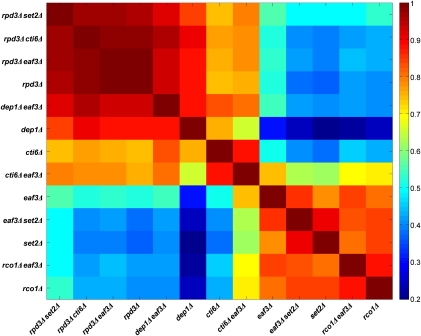
Figure 4.
Pairwise correlation analysis comparing Rpd3S and Rpd3L replication profiles. Wild-type peak heights were subtracted from the corresponding peak heights of each mutant to yield a “difference vector” for every strain. The full pairwise Pearson's correlation matrix for the set of mutant strains was calculated using these difference vectors.
Whereas disruption of Rpd3S function results in only a small number of origins that are significantly deregulated, our whole-genome analysis reveals slight, but reproducible increases in BrdU incorporation at many origins in these mutants (Figs. 2A, 3A–C; Supplemental Figs. S3A, S4A,B); a Wilcoxon signed rank test on the differences between BrdU peak heights in Rpd3S mutants and wild-type cells confirmed the significance of this global difference (P = 6.17 × 10−31). This indicates that in addition to the primary role in delaying a handful of late origins, the chromatin modifications by Rpd3S have a minor, global effect on origin timing or efficiency. Moreover, the well-studied late origin ARS603 is among the six origins significantly deregulated in the Rpd3S mutant strains (Supplemental Table S4). These findings underscore the value of employing whole-genome approaches to identify subtle, but pervasive differences in replication timing or efficiency, and reveal the limits of relying on “representative” origins (whose regulation may in fact be somewhat unique) as indicative of an entire class.
Rpd3L mediates the widespread Rpd3-dependent effect on origin firing
To determine Rpd3L's role in regulating origin initiation, we specifically disrupted Rpd3L by deleting DEP1 or CTI6, two components unique to the large complex, and performed BrdU-IP-chip analysis in HU as in the previous experiments. The replication profiles of dep1Δ and cti6Δ mutants closely resemble those of rpd3Δ cells, with significant increases in most of the same affected origins (Fig. 2B; Supplemental Fig. S3B). Global comparison of BrdU peak heights illustrates many significantly larger peaks in dep1Δ and cti6Δ cells, reminiscent of rpd3Δ cells (Fig. 3D,E); here the t-tests are based on quadruplicate experiments in the dep1Δ strain (Supplemental Table S5). For instance, 94 peaks are significantly larger in the dep1Δ strain than in the wild-type strain, and 81 of these are also deregulated in rpd3Δ cells (hypergeometric test, P = 7.96 × 10−14). These results strongly suggest that Rpd3L mediates the widespread effect on replication initiation timing or efficiency that we demonstrated for Rpd3.
To compare objectively the replication profiles for all mutant strains, we systematically determined how each mutant profile differed from the wild-type profile. Wild-type peak heights were subtracted from each of the corresponding mutant peak heights and Pearsons's correlation coefficient (ρ) was used to measure pairwise correlations between the resulting differences (Fig. 4). This analysis yields significant correlations between the Rpd3L mutant strains, dep1Δ and cti6Δ, and the rpd3Δ strain (ρ = 0.88 for dep1Δ vs. rpd3Δ; ρ = 0.74 for cti6Δ vs. rpd3Δ), demonstrating that replication patterns in these strains differ from wild-type in similar ways and that most of the same origins are deregulated in each. The slightly weaker correlation between cti6Δ and rpd3Δ probably reflects incomplete disruption of Rpd3L function by CTI6 deletion (M. Carrozza and J. Workman, pers. comm.). The data also show significant correlations (mean ρ = 0.87) between all of the Rpd3S targeting mutant strains—set2Δ, eaf3Δ, rco1Δ, set2Δ eaf3Δ, and eaf3Δ rco1Δ—indicating that these mutations all result in replication profiles that differ from the wild-type in similar ways. However, these mutant profiles do not correlate well with the rpd3Δ profile (ρ < 0.5), confirming that the Rpd3S mutant strains do not comparably deregulate origin firing like RPD3 deletion.
Deletion of both large and small complex subunits even more closely phenocopies rpd3Δ, as deletion of EAF3 in either dep1Δ or cti6Δ results in replication profiles that correlate more strongly with rpd3Δ than the respective Rpd3L single mutant strains (Figs. 2C, 4; Supplemental Fig. S3B). Strains with deletion of RPD3 together with an Rpd3S- (eaf3Δ rpd3Δ and set2Δ rpd3Δ) or Rpd3L- (cti6Δ rpd3Δ) specific gene show very high correlations with rpd3Δ, indicating that these genes function with RPD3, rather than independently (Fig. 4). In summary, these data demonstrate that Rpd3L plays the predominant role in modulating the initiation of Rpd3-regulated late origins, whereas Rpd3S plays the predominant role at very few sites, but has a minor repressive effect on origins globally.
Deletion of putative Rpd3L-targeting factors deregulates few Rpd3L-regulated origins
Our previous analysis of the Rpd3-targeting factor Ume6 indicated that UME6 deletion does not advance firing of ARS603, whereas deletion of RPD3 does (Aparicio et al. 2004). However, as mentioned above, ARS603 firing is delayed by Rpd3S in addition to Rpd3L, and hence, a role for Ume6 may have been obscured. Therefore, we re-examined the role of Ume6 in Rpd3L targeting by generating replication profiles in triplicate in ume6Δ cells for comparison with Rpd3L mutant profiles. Consistent with our previous study, deletion of UME6 did not alter late origin firing like RPD3 deletion (Fig. 3G; Supplemental Fig. S3D). In ume6Δ cells, only one BrdU peak is significantly increased relative to wild-type, whereas five BrdU peaks are significantly reduced (empirical Bayes-moderated t-test, P ≤ 0.001), suggesting that these five origins fire less efficiently or that their initiation timing is delayed when UME6 is deleted. These results suggest that Ume6 plays little or no role in recruiting Sin3–Rpd3, at least with respect to origin regulation. However, an alternative explanation is that Ume6 mediates additional factors that act in opposition to or independently of Rpd3. Indeed, Ume6 recruits both Isw2 and Rpd3 complexes to repress URS1-containing genes such that deletion of both regulators is required for full derepression (Goldmark et al. 2000). Thus, deletion of UME6 has effects distinct from RPD3 deletion on transcriptional repression, and this may explain the different effects on origin firing observed here.
We also tested the role of Ash1, a sequence-specific DNA-binding protein, which stably associates with Rpd3L and may participate in its recruitment to promoters (Carrozza et al. 2005a). We generated replication profiles in triplicate for ash1Δ strains and found little similarity between the ash1Δ and rpd3Δ (or dep1Δ) replication profiles (Fig. 3H; Supplemental Fig. S3D). In ash1Δ cells, two BrdU peaks (ARSXII-199 and ARSXIII-482) are significantly larger (empirical Bayes-moderated t-test, P ≤ 0.001) than the corresponding wild-type peaks. Interestingly, these origins are also significantly deregulated in the rpd3Δ and dep1Δ strains, and Ash1 appears to bind within the intergenic region occupied by ARSXII-199 (P = 0.072) (Harbison et al. 2004). These results are consistent with Ash1 recruiting Rpd3L to a subset of its targets, while Rpd3L is recruited to many sites by factors other than Ume6 and Ash1.
Rpd3L-regulated origins are locally associated with Sin3–Rpd3-regulated transcription and chromatin binding
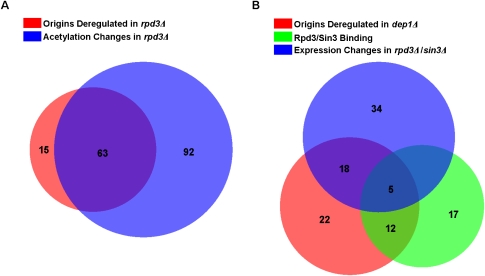
Our results suggest that the function of Rpd3L as a targeted regulator of gene expression through histone deacetylation generally delays or impedes the activity of proximal replication origins. To confirm that changes in histone acetylation were associated with origin deregulation in rpd3Δ cells, we compared the locations of deregulated origins in the rpd3Δ replication profile with published data on histone acetylation changes in rpd3Δ cells (Robyr et al. 2002). Of the 304 origins that fired in rpd3Δ cells, acetylation data were available for 229 origins (within 500 base pairs [bp] of the 5′ or 3′ end of the ARS as defined in OriDB), which included 78 origins that incorporated BrdU more robustly. Of these 78, 63 exhibited at least twofold increased acetylation of histone H3-K18, H4-K5, or H4-K12, indicating a significant association between locally increased histone acetylation and origin deregulation (hypergeometric test, P = 0.0016) (Fig. 5A; Supplemental Table S6).
Figure 5.
Rpd3L-regulated origins are associated with Rpd3-regulated genes. (A) Venn diagram of overlap between (1) origins deregulated in rpd3Δ cells and (2) regions that show increased acetylation in rpd3Δ cells. (B) Venn diagram of overlap between intergenic regions containing origins that are (1) deregulated in dep1Δ cells, (2) flanked by genes deregulated in rpd3Δ or sin3Δ cells, and (3) flanked by genes that bind Rpd3 or Sin3 in their promoter regions.
To examine the relationship of origin deregulation with transcription, we compared the locations of deregulated origins in the dep1Δ replication profile with published, genome-wide gene expression profiles of rpd3Δ and sin3Δ cells and genome-wide chromatin-binding maps of Rpd3 and Sin3 (Bernstein et al. 2000; Fazzio et al. 2001; Robert et al. 2004; Sabet et al. 2004). We focused this analysis on 241 confirmed origins whose precise intergenic locations are known (compiled at OriDB), allowing the unambiguous assignment of each origin to the flanking genes. The results of this analysis are depicted in Figure 5B. Of these 241 origins, 57 are significantly deregulated in dep1Δ cells. Rpd3 and/or Sin3 binding (P ≤ 0.05) has been reported at the promoters of genes adjacent to 45 of the 241 origins, and 17 of the 57 deregulated origins. A hypergeometric test indicates significant partial association (P = 0.014) of deregulated origins with proximal genes that bind Sin3–Rpd3 in their promoters. Next, we compared the locations of deregulated origins with the locations of Sin3–Rpd3-regulated genes. Of the 241 origins, 68 are adjacent to at least one gene whose expression level has been reported to increase (P ≤ 0.05, or twofold increased depending on the data source) in rpd3Δ or sin3Δ cells; 23 of these 68 origins are significantly deregulated in dep1Δ cells. A hypergeometric test reveals a significant partial association (P = 0.017) between deregulated origins and flanking genes whose expression levels are derepressed in rpd3Δ or sin3Δ cells.
Finally, in an effort to identify additional factors involved, potentially as Rpd3L-targeting factors, we examined whether other transcription factors were overrepresented near Rpd3L-regulated origins. We conducted Random Forest Regression analysis to identify factors associated with promoters of genes adjacent to Rpd3-regulated origins, using concatenated ChIP–chip data for 203 yeast transcription factors and six chromatin remodeling factors (including Rpd3 and Sin3) (Harbison et al. 2004; Robert et al. 2004). As the Rpd3 and Sin3 promoter binding reported in the ChIP–chip study likely reflects that of Rpd3L, we again chose the deregulated origins in the dep1Δ strain as our response variable to be compared against the DNA-binding factor data. For this analysis, we used 200 confirmed origins whose precise intergenic locations are known and that are present in the transcription factor data sets; 57 of these origins are deregulated in dep1Δ cells. Of the 209 DNA-binding factors examined, six are identified as significantly associated with origin deregulation (Supplemental Table S7). Importantly, this analysis identifies the binding of Rpd3 and Sin3 as most predictive of Dep1-dependent origin regulation, independently confirming the results of the previous association analysis. Rap1, Sum1, Smp1, and Swi6 also are overrepresented at genes adjacent to Dep1-regulated origins, suggesting that each of these factors may impact origin regulation. In addition, these findings suggest a functional link between each of these factors and Sin3–Rpd3, potentially as recruitment factors or coregulators, as discussed below.
Discussion
The global scope of Rpd3 in regulation of origin initiation
We used genomic DNA replication profiles to reveal a widespread role for Rpd3 in regulating replication origin activity. We identified 103 origins whose initiation timings are advanced and/or firing efficiencies increased in rpd3Δ cells using a stringent confidence level. Our previous analysis of origin function in rpd3Δ cells showed advanced initiation timing but no evidence for changes in origin efficiencies of late origins (necessarily measured in the absence of HU) or for disruption of the intra-S checkpoint; thus, we concluded that the increased origin activation in HU reflects advanced timing and resulting escape of intra-S checkpoint inhibition (Aparicio et al. 2004). We infer that the same relationship applies to most of the origins altered in rpd3Δ and related mutant strains in this study. Consistent with this, loss of Rpd3 function almost exclusively alters smaller BrdU peaks, which are globally correlated with late replication timing (Fig. 1D). Nevertheless, we cannot rule out that increased origin efficiencies account for some of the changes. Indeed, it has been suggested that differences in origin efficiencies underlie differences in timing (Rhind 2006).
The large number of origins affected by loss of Rpd3 function suggests that the delayed or diminished initiation of most late-replicating origins in the yeast genome can be attributed, at least in good measure, to Rpd3 function. In a different study, 104 potential replication origins (PROs) were identified as late-firing (out of 247 total), and were not detected in HU (Yabuki et al. 2002). A possible exception to the widespread effect of Rpd3 on late-firing origins is the subtelomeric origins, which may be unaffected by Rpd3 (Aparicio et al. 2004). Interestingly, deacetylation of subtelomeric regions depends mainly on the Hda1 and Sir2 histone deacetylases (Robyr et al. 2002), consistent with a reduced role of Rpd3 in subtelomeric histone deacetylation and accordant origin regulation.
Differential and shared roles of Rpd3L and Rpd3S in regulation of origin initiation
A main finding of this study is that Rpd3L is primarily responsible for modulating the initiation timing or efficiency of most Rpd3-regulated origins, whereas Rpd3S plays a primary role at only a handful of Rpd3-regulated origins. Rpd3S also has a minor, global influence on origin activity. Rpd3L-regulated origins are frequently associated with flanking genes whose expression is controlled by Rpd3. In addition, binding of Rpd3 and/or Sin3, as well as several other potentially related transcription factors, is overrepresented in promoter regions of genes adjacent to deregulated origins. These findings argue that Rpd3L's effect on replication origins is related to its role in regulating gene expression, and suggest that genes in the vicinity of Rpd3L-regulated origins are under Rpd3L control.
Whereas Rpd3L mediates the bulk of Rpd3's effect on origin firing, Rpd3S plays a distinct and independent role at a handful of origins. Rpd3L and Rpd3S function distinctly, with Rpd3L function largely limited to promoter regions to control transcription initiation, and Rpd3S function occurring over more extensive regions, being especially pronounced toward the 3′ end of transcribed regions, to suppress spurious initiation within the body of expressed genes. Thus, the one clearly shared effect of the two complexes is histone deacetylation, consistent with histone deacetylation directly modulating origin activation. Deletion of RPD3 increases histone acetylation surrounding Rpd3-regulated origins (Fig. 5A). Furthermore, artificial targeting of a histone acetylase near a late-replicating origin advanced its replication timing in yeast and mammalian cells, and targeting of Rpd3 protein to an actively firing origin in a Drosophila system diminished origin activity (Vogelauer et al. 2002; Aggarwal and Calvi 2004; Goren et al. 2008). In addition, Rpd3's effect on origins depends on its catalytic activity, arguing against Rpd3 acting through an alternative mechanism such as a recruitment factor or mediator of protein–protein interactions within chromatin (Aggarwal and Calvi 2004; Aparicio et al. 2004). It is conceivable that RPD3 deletion affects replication timing or efficiency by increasing the level of a limiting replication initiation factor. Therefore, we searched for genes with expression changes in at least two of the three available rpd3Δ gene expression data sets that were annotated with the gene ontology term “DNA replication” (GO: 0006260), but did not identify any likely candidates. We also considered the possibility that origin deregulation results from disruption of intra-S checkpoint signaling in cells lacking Rpd3 function. However, the available data suggest that rather than diminishing the checkpoint, deletion of RPD3 can suppress some checkpoint defects (Scott and Plon 2003; Lottersberger et al. 2007).
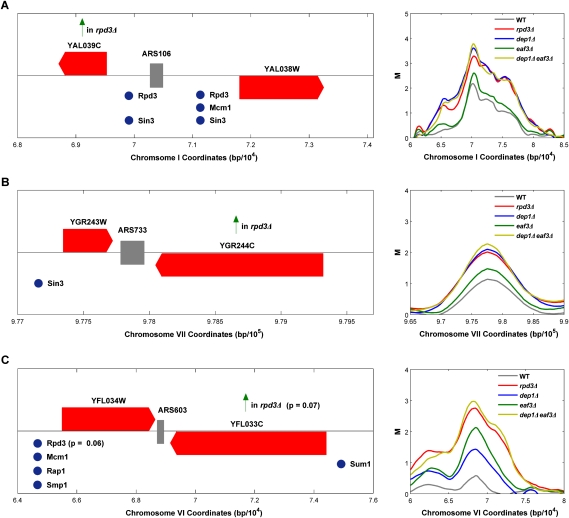
Closer examination of specific origins regulated by Rpd3L and Rpd3S offers further insight into the influence of histone deacetylation on origin function. Figure 6 illustrates distinct effects on three origins and their relationship to local gene organization and Rpd3-targeting mechanisms, which may serve as archetypes. First, we consider ARS106, whose activity Rpd3L modulates, and which lies between two diverging genes (Fig. 6A). Both flanking genes are reported to have Rpd3- and Sin3-binding sites in their promoter regions and YAL039C shows increased expression (greater than or equal to twofold) in the absence of RPD3. Here, the proximity of Sin3–Rpd3 binding to the origin likely has a direct effect on acetylation of histones bordering the origin (the sequence occupied by ORC is histone-free) (Thoma et al. 1984), delaying or suppressing origin activation.
Figure 6.
Different gene-origin configurations subject to regulation by Rpd3L and Rpd3S. See the Discussion. Red strips with points indicate ORF positions and direction of transcription, and gray strips indicate origin position. Vertical arrows indicate genes derepressed in rpd3Δ or sin3Δ cells, and spheres mark reported binding sites for the indicated proteins (P ≤ 0.05, unless indicated otherwise). Panels to the right show BrdU-incorporation tracings for the indicated strains.
Second, ARS733 (Fig. 6B) represents Rpd3L-regulated origins flanked by converging genes in which at least one of the flanking genes binds Sin3–Rpd3 and/or shows transcriptional derepression in strains lacking Sin3–Rpd3 function. Because increased transcription results in increased histone acetylation of transcribed regions (Li et al. 2007a), increased acetylation at the 3′ ends of genes derepressed by RPD3 deletion may influence proximal origins in a direct manner. In addition to increasing acetylation of histones, transcription may destabilize or remodel origin-proximal nucleosomes, increasing accessibility to the origin. Another possibility is that Rpd3 bound at the promoter can also act over a distance to deacetylate histones, particularly in cases where it is more stably bound.
Third, we consider ARS603 (Fig. 6C), which represents origins whose activity is significantly affected by disruption of Rpd3S function. Four of the six origins significantly altered in Rpd3S mutant strains occupy intergenic regions flanked by converging genes, and one of the six lies between tandem genes; the last of the six appears to reside between converging genes but has not been precisely mapped to the intergenic region, and is therefore excluded from further analysis below. Thus, all are proximal to 3′ ends of transcriptional units where deacetylation by Rpd3S is most pronounced. Deacetylation by Rpd3S is also more pronounced at longer transcriptional units (Li et al. 2007b), so we examined the lengths of genes flanking these five origins. The mean length of the converging genes flanking the Rpd3S-affected origins is ∼2.9 kb, compared with ∼1.6 kb for the entire collection of genes flanking converging intergenic origins. A permutation test on the mean demonstrates that the genes flanking the Rpd3S-regulated origins are significantly longer (P = 0.018) than the genes flanking four randomly selected origins between converging genes. The substantial effect of Rpd3S on this handful of origins strongly supports a direct effect of proximal histone deacetylation on origin initiation timing. Interestingly, Rpd3L also regulates four of the six Rpd3S-regulated origins, representing a nonrandom level of overlap (hypergeometric test, P = 0.057).
We noted that Rpd3S also exerts a subtle effect on a large number of origins. Perhaps this more limited effect of Rpd3S reflects the distance between the affected origins and the 3′ ends of transcripts where Rpd3S activity is concentrated. To address this, we tested whether disruption of Rpd3S affected BrdU peak heights of origins flanked by converging genes more than those flanked by diverging genes. Indeed, we find a significantly greater effect (P = 0.048, standard t-test) of Rpd3S on origins flanked by converging genes (mean log2 unit increase in BrdU peak size = 0.45) than on those flanked by diverging genes (mean log2 unit increase = 0.26). This finding further supports the idea that proximal histone deacetylation can influence origin activity; however, the contribution of Rpd3S is limited relative to Rpd3L.
Combinatorial gene regulation impacts DNA replication
The available data strongly suggest that histone deacetylation proximal to origins is sufficient to delay initiation timing. Histone deacetylation is associated with chromatin compaction, altered subnuclear localization, and the establishment of transcriptionally silenced chromatin, any of which may reduce the rate at which a replication initiation factor(s) gains access to a pre-RC. However, proximity to an Rpd3-regulated gene does not ensure late or inefficient initiation. For example, there are more origins adjacent to Rpd3-regulated genes that are not deregulated by loss of Rpd3L than those that are (Fig. 5B). This may partly reflect a limitation of our assay, which is designed to detect differences in timing or efficiency that affect the level of initiation in HU. For example, earlier firing of an already early-firing origin would not be detected. The Rpd3-regulated genes identified in the previous studies may be indirect targets of Rpd3, or may be influenced by competing factors that can override the effects of Rpd3. For example, elimination of Rpd3 binding may be insufficient to fully derepress transcription because overlapping mechanisms may contribute to repression (such as Ume6 and Isw2 as mentioned earlier), or because derepression requires positive activation, which is dependent on a specific signal or condition not elicited in our experiments. Similarly, many of the Rpd3-regulated origins we identified reside near genes that have not been demonstrated to bind or be regulated by Sin3–Rpd3. Again, this may reflect a failure to detect such interactions for technical reasons or because such interactions are limited to a particular cell cycle stage or growth condition.
Our search for transcription factor-binding sites overrepresented proximal to Rpd3L-regulated replication origins identified potential candidates. For example, Sum1, which enhances the initiation efficiencies of certain origins (Irlbacher et al. 2005), binds some of the same intergenic regions as Rpd3 and is overrepresented at origins deregulated in dep1Δ cells (Supplemental Table S7); eight origins whose timing is deregulated in Rpd3L mutant strains bind Sum1 and Rpd3 and/or Sin3 (data not shown). Thus, elimination of Rpd3 may facilitate binding of Sum1. Interestingly, Sum1 acts as a repressor of middle meiotic genes and is thought to recruit the histone deacetylase Hst1 for repression (Xie et al. 1999; Sutton et al. 2001; Weber et al. 2008). How Sum1 contributes to more efficient origin firing is unclear; however, transcriptional repression by Sum1 may protect against direct disruption of origin function by transcription.
Rap1 also is overrepresented at intergenic regions occupied by Rpd3L-regulated replication origins (Supplemental Table S7). Like Rpd3, Rap1 activates or represses transcription depending on the specific context by modulating chromatin structure and has been implicated in replication (for review, see Morse 2000). Rpd3 and Rap1 appear to play antagonistic roles in gene regulation, with Rap1 possibly recruiting the Esa1 histone acetylase for gene activation. Interestingly, Esa1 plays a role in recovery from replication stress, which may reflect a competition between Esa1 and Rpd3 in late origin firing (Lottersberger et al. 2007). Also, a recent study suggests that Esa1 functions in opposition to Rpd3S (Biswas et al. 2008b). Further future examination of the role of Esa1 in DNA replication is clearly warranted.
We also found overrepresentation of binding sites of the transcription factors Smp1 and Swi6 in proximity to Rpd3L-regulated replication origins (Supplemental Table S7). Although a direct interaction between Smp1 and Rpd3 has not been demonstrated, Smp1 and Rpd3 act downstream from Hog1 in the osmotic stress response, suggesting that Smp1 may recruit Rpd3 to activate stress response genes (de Nadal et al. 2003, 2004). Swi6 is a component of two transcription factor complexes, SBF and MBF, which activate late G1-/early S-phase-specific genes as well as meiotic genes (for review, see Breeden 2003). Swi6 binding is highly correlated with Rpd3 binding; and Rpd3 binding at a number of promoters is dependent on Swi6, suggesting a direct recruitment mechanism (Lee et al. 2002). We attempted to analyze replication profiles in swi6Δ cells but were unable to achieve satisfactory synchronization with α-factor. Future studies will investigate the possibility that replication timing is associated with the cell cycle regulated expression of genes, particularly genes expressed circa S phase.
Linking chromatin structure, gene expression, and DNA replication
The association of Rpd3L-regulated origins with Sin3–Rpd3 promoter binding and/or Sin3–Rpd3-dependent gene regulation leads us to conclude that a link between replication timing and gene expression, although more obscure in budding yeast, indeed exists. Together, these findings are consistent with the idea that localized histone deacetylation has generally comparable effects on transcription and replication—that is, reducing initiation rates—thereby coordinating these processes throughout much of the genome. However, combinatorial or competing controls also impinge significantly. Thus, the Rpd3L complex should serve as a model for studying the specification of gene expression and replication timing patterns that underlie cellular differentiation in higher eukaryotes. We also envision the possibility that changes in replication patterns may serve as useful indicators of altered transcription patterns (and even networks) that underlie normal cellular differentiation as well as oncogenesis.
Materials and methods
Yeast strains and methods
Yeast strains were constructed in the W303-1 background and are described in Supplemental Table S8. Gene deletions with KanMX and HIS5 cassettes were constructed and confirmed using PCR-based methods (Longtine et al. 1998). Strains contain a single integrated copy of p306-BrdU incorporation to enable BrdU uptake and incorporation into DNA (Viggiani and Aparicio 2006). YEPD medium was used for all experiments. Cultures were blocked in G1 phase using α-factor (5 nM) and released synchronously into fresh medium containing 0.2 mg/mL Pronase E (Sigma) as described previously (Aparicio et al. 2004). S-phase entry was monitored by analysis of budding morphology (Supplemental Fig. S5).
BrdU-IP-chip
Cultures were released from α-factor arrest into fresh medium containing 400 μg/mL BrdU (Sigma) plus 200 mM HU (Sigma), and harvested 1 h later with the addition of NaN3 to 0.1% final concentration. DNA isolation and immunoprecipitation were performed as described in Szyjka et al. (2005), except that anti-BrdU antibody (GE Healthcare) was used at 1:1000. Ten nanaograms of immunoprecipitated DNA and 10 ng of total genomic DNA (from a G1-arrested culture) were amplified separately using a GenomePlex Complete Whole Genome Amplification Kit (Sigma). One microgram of amplified immunoprecipitated DNA and 1 μg of amplified total DNA were labeled by Klenow extension using Cy5- and Cy3-conjugated random nonamers (Trilink Biotechnologies), respectively, according to the NimbleGen Arrays User's Guide, except that the Cy-labeled nonamers were used at OD = 0.1 in the indicated solution. Cy5-labeled immunoprecipitate and Cy3-labeled total DNA samples (1–5 μg each) were combined and hybridized for 12–16 h at 43°C to high-density oligonucleotide tiling microarrays representing the entire yeast genome (NimbleGen Systems, Inc., CGH 385k array, catalog no. B2436001; probe size 45–75 bp, with 12-bp median spacing), according to NimbleGen protocols, except that standard microarray hybridization chambers (Corning) and cover slips (Lifter Slip, Electron Microscopy Sciences) were used. Slides were washed according to NimbleGen protocols and scanned using a GenePix Personal 4100A Scanner (Axon Instruments). Slides were stripped and rehybridized up to three times according to NimbleChip protocols.
Array normalization
Array normalization was performed as described in S.R.V. Knott, C.J. Viggiani, O.M. Aparicio, and S. Tavaré (in prep.). Briefly, within-array normalization (for intensity bias) was performed with a novel dynamic programming algorithm—designed specifically for BrdU-IP-chip data sets—that applies principal component analysis to remove the intensity bias evident in probes plotted on the M = log2(IP/Total) versus A = [log2(IP) + log2(Total)]/2 plane. Between-array normalization (for location and scale) was performed by first removing the location differences between the arrays' empirical M distributions and then scale normalizing replicate arrays with a modified quantile normalization.
Peak identification and quantification
Replication peaks were identified in a three-step process as described in S.R.V. Knott, C.J. Viggiani, O.M. Aparicio, and S. Tavaré (in prep.). Briefly, a set of initial peaks was identified for each array based on signal intensity. The height of each peak was set as the median M value of probes within 500 bp of its apex. Next, each set of replicates was aligned based on initial peak locations, and only peaks that were present in all replicates were kept for further consideration. Finally, the remaining peaks were aligned with known/replicated origins in OriDB (Nieduszynski et al. 2007), and only peaks that aligned with origins were considered for subsequent analysis.
Strain comparisons
Individual peak height differences between wild-type and mutant strains were identified with an empirical Bayes-moderated t-test (Smyth 2004), using Benjamini-Hochberg correction for multiple testing (Benjamini and Hochberg 1995). The global similarity between each pair of strains was assessed with the Pearson's correlation coefficient (ρ) between the strains' difference vectors (as calculated by subtracting wild-type peak heights from the corresponding mutant peak heights) (see the Supplemental Material for details).
Acknowledgments
We thank Bing Li and Jerry Workman for helpful discussions, Michael Mehan for help with gene annotations, and Michelle Arbeitman for sharing equipment. This work was supported by NIH grants RO1 GM65494 (to O.M.A.) and P50 HG02790 (to S.K. and S.T.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1784309.
Supplemental material is available at http://www.genesdev.org.
References
- Aggarwal B.D., Calvi B.R. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- Alvino G.M., Collingwood D., Murphy J.M., Delrow J., Brewer B.J., Raghuraman M.K. Replication in hydroxyurea: It's a matter of time. Mol. Cell. Biol. 2007;27:6396–6406. doi: 10.1128/MCB.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O.M., Weinstein D.M., Bell S.P. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Aparicio J.G., Viggiani C.J., Gibson D.G., Aparicio O.M. The Rpd3–Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:4769–4780. doi: 10.1128/MCB.24.11.4769-4780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- Bernstein B.E., Tong J.K., Schreiber S.L. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D., Takahata S., Xin H., Dutta-Biswas R., Yu Y., Formosa T., Stillman D.J. A role for Chd1 and Set2 in negatively regulating DNA replication in Saccharomyces cerevisiae. Genetics. 2008a;178:649–659. doi: 10.1534/genetics.107.084202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D., Takahata S., Stillman D.J. Different genetic functions for the Rpd3(L) and Rpd3(S) complexes suggest competition between NuA4 and Rpd3(S) Mol. Cell. Biol. 2008b;28:4445–4458. doi: 10.1128/MCB.00164-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L.L. Periodic transcription: A cycle within a cycle. Curr. Biol. 2003;13:R31–R38. doi: 10.1016/S0960-9822(02)01386-6. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., Florens L., Swanson S.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Workman J.L. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta. 2005a;1731:77–87. doi: 10.1016/j.bbaexp.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005b;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- de Nadal E., Casadome L., Posas F. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol. Cell. Biol. 2003;23:229–237. doi: 10.1128/MCB.23.1.229-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E., Zapater M., Alepuz P.M., Sumoy L., Mas G., Posas F. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature. 2004;427:370–374. doi: 10.1038/nature02258. [DOI] [PubMed] [Google Scholar]
- Donaldson A.D., Raghuraman M.K., Friedman K.L., Cross F.R., Brewer B.J., Fangman W.L. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell. 1998;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- Fazzio T.G., Kooperberg C., Goldmark J.P., Neal C., Basom R., Delrow J., Tsukiyama T. Widespread collaboration of Isw2 and Sin3–Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 2001;21:6450–6460. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Collingwood D., Boeck M.E., Fox L.A., Alvino G.M., Fangman W.L., Raghuraman M.K., Brewer B.J. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat. Cell Biol. 2006;8:148–155. doi: 10.1038/ncb1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman K.L., Brewer B.J., Fangman W.L. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:667–678. doi: 10.1046/j.1365-2443.1997.1520350.x. [DOI] [PubMed] [Google Scholar]
- Gilbert D.M. Replication timing and transcriptional control: Beyond cause and effect. Curr. Opin. Cell Biol. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- Goldmark J.P., Fazzio T.G., Estep P.W., Church G.M., Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- Goren A., Tabib A., Hecht M., Cedar H. DNA replication timing of the human β-globin domain is controlled by histone modification at the origin. Genes & Dev. 2008;22:1319–1324. doi: 10.1101/gad.468308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irlbacher H., Franke J., Manke T., Vingron M., Ehrenhofer-Murray A.E. Control of replication initiation and heterochromatin formation in Saccharomyces cerevisiae by a regulator of meiotic gene expression. Genes & Dev. 2005;19:1811–1822. doi: 10.1101/gad.334805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A.A., Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Kadosh D., Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes & Dev. 1998a;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D., Struhl K. Targeted recruitment of the Sin3–Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 1998b;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh M.C., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kurdistani S.K., Robyr D., Tavazoie S., Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Shilatifard A. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat. Res. 2007;618:130–134. doi: 10.1016/j.mrfmmm.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Rinaldi N.J., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007a;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li B., Gogol M., Carey M., Pattenden S.G., Seidel C., Workman J.L. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes & Dev. 2007b;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., 3rd, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lottersberger F., Panza A., Lucchini G., Longhese M.P. Functional and physical interactions between yeast 14-3-3 proteins, acetyltransferases, and deacetylases in response to DNA replication perturbations. Mol. Cell. Biol. 2007;27:3266–3281. doi: 10.1128/MCB.01767-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine D.M., Bell S.P. A genomic view of eukaryotic DNA replication. Chromosome Res. 2005;13:309–326. doi: 10.1007/s10577-005-1508-1. [DOI] [PubMed] [Google Scholar]
- McCune H.J., Danielson L.S., Alvino G.M., Collingwood D., Delrow J.J., Fangman W.L., Brewer B.J., Raghuraman M.K. The temporal program of chromosome replication: Genomewide replication in clb5Δ Saccharomyces cerevisiae. Genetics. 2008;180:1833–1847. doi: 10.1534/genetics.108.094359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R.H. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 2000;16:51–53. doi: 10.1016/s0168-9525(99)01936-8. [DOI] [PubMed] [Google Scholar]
- Nieduszynski C.A., Hiraga S., Ak P., Benham C.J., Donaldson A.D. OriDB: A DNA replication origin database. Nucleic Acids Res. 2007;35:D40–D46. doi: 10.1093/nar/gkl758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman M.K., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Reid J.L., Moqtaderi Z., Struhl K. Eaf3 regulates the global pattern of histone acetylation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:757–764. doi: 10.1128/MCB.24.2.757-764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N. DNA replication timing: Random thoughts about origin firing. Nat. Cell Biol. 2006;8:1313–1316. doi: 10.1038/ncb1206-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F., Pokholok D.K., Hannett N.M., Rinaldi N.J., Chandy M., Rolfe A., Workman J.L., Gifford D.K., Young R.A. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D., Suka Y., Xenarios I., Kurdistani S.K., Wang A., Suka N., Grunstein M. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen A.A., Suka N., Turner B.M., Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- Sabet N., Volo S., Yu C., Madigan J.P., Morse R.H. Genome-wide analysis of the relationship between transcriptional regulation by Rpd3p and the histone H3 and H4 amino termini in budding yeast. Mol. Cell. Biol. 2004;24:8823–8833. doi: 10.1128/MCB.24.20.8823-8833.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C., Diffley J.F. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- Scott K.L., Plon S.E. Loss of Sin3/Rpd3 histone deacetylase restores the DNA damage response in checkpoint-deficient strains of Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:4522–4531. doi: 10.1128/MCB.23.13.4522-4531.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1444-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- Stevenson J.B., Gottschling D.E. Telomeric chromatin modulates replication timing near chromosome ends. Genes & Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A., Heller R.C., Landry J., Choy J.S., Sirko A., Sternglanz R. A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol. Cell. Biol. 2001;21:3514–3522. doi: 10.1128/MCB.21.10.3514-3522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyjka S.J., Viggiani C.J., Aparicio O.M. Mrc1 is required for normal progression of replication forks throughout chromatin in S. cerevisiae. Mol. Cell. 2005;19:691–697. doi: 10.1016/j.molcel.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Thoma F., Bergman L.W., Simpson R.T. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J. Mol. Biol. 1984;177:715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- Viggiani C.J., Aparicio O.M. New vectors for simplified construction of BrdU-Incorporating strains of Saccharomyces cerevisiae. Yeast. 2006;23:1045–1051. doi: 10.1002/yea.1406. [DOI] [PubMed] [Google Scholar]
- Vogelauer M., Rubbi L., Lucas I., Brewer B.J., Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol. Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- Weber J.M., Irlbacher H., Ehrenhofer-Murray A.E. Control of replication initiation by the Sum1/Rfm1/Hst1 histone deacetylase. BMC Mol. Biol. 2008;9:100. doi: 10.1186/1471-2199-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M, Palacios DeBeer M.A., Fox C.A., The activities of eukaryotic replication origins in chromatin. Biochim. Biophys. Acta. 2004;1677:142–157. doi: 10.1016/j.bbaexp.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Xie J., Pierce M., Gailus-Durner V., Wagner M., Winter E., Vershon A.K. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Aparicio J.G., Aparicio O.M., Tavaré S. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics. 2006;7:276. doi: 10.1186/1471-2164-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N., Terashima H., Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7:781–789. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]
- Yamashita M., Hori Y., Shinomiya T., Obuse C., Tsurimoto T., Yoshikawa H., Shirahige K. The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:655–665. doi: 10.1046/j.1365-2443.1997.1530351.x. [DOI] [PubMed] [Google Scholar]