Abstract
Eukaryotic translation termination is mediated by two interacting release factors, eRF1 and eRF3, which act cooperatively to ensure efficient stop codon recognition and fast polypeptide release. The crystal structures of human and Schizosaccharomyces pombe full-length eRF1 in complex with eRF3 lacking the GTPase domain revealed details of the interaction between these two factors and marked conformational changes in eRF1 that occur upon binding to eRF3, leading eRF1 to resemble a tRNA molecule. Small-angle X-ray scattering analysis of the eRF1/eRF3/GTP complex suggested that eRF1's M domain contacts eRF3's GTPase domain. Consistently, mutation of Arg192, which is predicted to come in close contact with the switch regions of eRF3, revealed its important role for eRF1's stimulatory effect on eRF3's GTPase activity. An ATP molecule used as a crystallization additive was bound in eRF1's putative decoding area. Mutational analysis of the ATP-binding site shed light on the mechanism of stop codon recognition by eRF1.
Keywords: eRF1, eRF3, protein biosynthesis, stop codon recognition, translation termination, translational GTPase
Termination of protein synthesis occurs when the stop codon of an mRNA enters the ribosomal A site. Stop codons are recognized by class I release factors (RFs), which also promote hydrolysis of the ester bond in peptidyl-tRNA in the peptidyl transfer center (PTC) of the large ribosomal subunit (Nakamura and Ito 2003). The bacterial class-I RFs, RF1 and RF2, recognize UAG/UAA and UGA/UAA stop codons, respectively, whereas the single eukaryotic class-I RF eRF1 recognizes all three stop codons (Kisselev et al. 2003). Crystal structures of the Thermus thermophilus 70S ribosome in complex with tRNA, mRNA containing a cognate stop codon, and either RF1 or RF2 (Laurberg et al. 2008; Weixlbaumer et al. 2008), indicate that stop codon recognition involves conserved elements in each RF, including the tripeptide recognition motif (PxT in RF1 and SPF in RF2) (Ito et al. 2000) and 16S ribosomal RNA.
The crystal structure of human eRF1 showed that it consists of three domains (N, M, and C), with domain N involved in stop codon recognition and domain M containing the universally conserved GGQ motif that is required to trigger peptidyl-tRNA hydrolysis (Song et al. 2000). The interaction between eRF1 and stop codons remains poorly understood. Various amino acids in the N domain, including the conserved NIKS and YxCxxxF sequence motifs, are implicated in codon recognition (Chavatte et al. 2002; Frolova et al. 2002; Seit-Nebi et al. 2002; Kolosov et al. 2005; Lekomtsev et al. 2007), suggesting that, in contrast to RF1/RF2, eRF1 recognizes stop codons through a complex three-dimensional network formed by conserved residues.
Translation termination also requires class II RFs, RF3 in prokaryotes (Grentzmann et al. 1994; Mikuni et al. 1994), and eRF3 in eukaryotes (Stansfield et al. 1995; Zhouravleva et al. 1995). Both RF3 and eRF3 are translational GTPases with limited homology that is restricted to their GTP-binding domains (Kisselev and Buckingham 2000). The functional C-terminal region of eRF3 comprises GTP-binding domain (G domain) and the β-barrel domains 2 and 3 that are similar to the respective domains of elongation factors EF-Tu and eEF1A, but with a different orientation of domain G relative to domains 2 and 3 (Song et al. 1999; Andersen et al. 2000; Kong et al. 2004). RF3 and eRF3 have entirely different functions in the termination process. The role of prokaryotic RF3 is to mediate recycling of RF1/RF2 from the post-termination complexes (Zavialov et al. 2001; Gao et al. 2007), whereas the GTPase activity of eukaryotic eRF3 couples codon recognition and peptidyl-tRNA hydrolysis mediated by eRF1 to ensure rapid and efficient peptide release (Salas-Marco and Bedwell 2004; Alkalaeva et al. 2006). Thus, eRF3 strongly enhances peptide release by eRF1 in the presence of GTP and abrogates it in the presence of the nonhydrolyzable GTP analog GDPNP, even when recycling of eRF1 is not required (Alkalaeva et al. 2006).
The mutual interdependence of eRF1 and eRF3 in termination involves not only eRF3's stimulation of peptide release by eRF1, but also stimulation by eRF1 of GTP binding to eRF3 (Hauryliuk et al. 2006; Mitkevich et al. 2006; Pisareva et al. 2006) and of eRF3's ribosome-dependent GTPase activity (Frolova et al. 1996). A distinguishing feature of eukaryotic RFs is that they form a stable complex (Stansfield et al. 1995; Zhouravleva et al. 1995) through interaction of their C-terminal domains (Ito et al. 1998; Ebihara and Nakamura 1999; Merkulova et al. 1999), and this physical interaction is required for (1) stimulation of GTP binding to eRF3 by eRF1 (Hauryliuk et al. 2006; Mitkevich et al. 2006; Pisareva et al. 2006), (2) induction of eRF3's GTPase activity by eRF1 on the ribosome (Frolova et al. 2000), and (3) stimulation by eRF3 of peptide release mediated by eRF1 (Alkalaeva et al. 2006).
The exact mechanism by which eRF3 stimulates peptide release by eRF1 is not known. One of the proposed roles of eRF3 is to promote binding of eRF1 to ribosomal pretermination complexes containing a stop codon in the ribosomal A-site, in analogy with the function of EF-Tu to increase the affinity of the aminoacyl-tRNA to the A-site programmed with a sense codon (Nakamura et al. 1996); this is supported by the finding that eRF1, eRF3, and GTP form a relatively stable long-lived complex (Hauryliuk et al. 2006; Mitkevich et al. 2006; Pisareva et al. 2006). The molecular mechanisms by which eRF1 specifically promotes binding of GTP (but not GDP) to eRF3 and by which it stimulates eRF3's ribosome-dependent GTPase activity are also not understood. Importantly, the C-terminal domain of eRF1, which is mainly responsible for the eRF1/eRF3 interaction, is not sufficient for stimulation of eRF3's GTP-binding and hydrolysis activities, and the M domain of eRF1 is also required for both processes (Kononenko et al. 2008).
To obtain insights into molecular mechanisms of interplay between eRF1 and eRF3 during eukaryotic translation termination, we determined the crystal structure of human and Schizosaccharomyces pombe full-length eRF1 in complex with domains 2 and 3 of eRF3. These structures revealed the details of interaction between eRF1 and eRF3 and a large conformational change in eRF1 that occur upon binding to eRF3. Molecular modeling, small-angle X-ray scattering (SAXS) analysis, and mutagenesis suggested that R192 in the M domain of human eRF1 is important for stimulating the GTPase activity of eRF3 by eRF1 and the ribosome.
Results
Overall architecture of the eRF1/eRF3-23 complexes
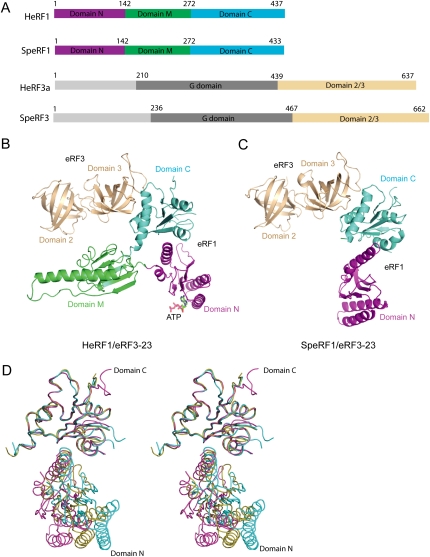
The crystal structures of full-length eRF1 in complex with domains 2 and 3 (residues 439–637) (Fig. 1A) of eRF3a from human (designated as HeRF1/eRF3-23) and full-length eRF1 (Sup45) in complex with domains 2 and 3 (residues 467–662) of eRF3 (Sup35) from S. pombe (designated as SpeRF1/eRF3-23) have been determined at resolutions of 3.8 Å and 3.5 Å, respectively. The final model of the HeRF1/eRF3-23 complex contains four complexes in the asymmetric unit (AU), which can be categorized into two groups (named as forms I and II) by the noncrystallographic symmetry (NCS) axis. The GGQ motif and most of the long helix adjacent to it are disordered in form II. Since no substantial differences are observed between the structures of forms I and II, and form I is more ordered, all subsequent analyses referred to the coordinates of form I. The final model of the SpeRF1/eRF3-23 complex also contains four copies of the complexes related by NCS in the AU. The model of the SpeRF1/eRF3-23 complex is less complete than that of the human complex, with domain M in all copies and domain N in two copies of the former being disordered. Statistics of structure determination and refinement are summarized in Table 1 (see the Materials and Methods).
Figure 1.
Structures of the human and S. pombe eRF1/eRF3-23 complexes. (A) Schematic representation of the domain organization of eRF1 and eRF3 from human and S. pombe. Domains N, M, and C of eRF1 are colored in magenta, limegreen, and cyan, respectively. Domains 2 and 3 of eRF3 are colored in wheat and domain G is shown in gray. (B) The structure of the HeRF1/eRF3-23 complex. The ribbon diagram is drawn with the orientation of domains 2 and 3 of eRF3 as in the SpeRF1/eRF3-23 complex. The coloring scheme is as in A. (C) The structure of the SpeRF1/eRF3-23 complex. Domain M of eRF1 is disordered. Color coding of each domain is as in B. (D) Stereo view of the superimposed domains N and C of eRF1 in free form (cyan), the human complex (yellow), and the S. pombe complex (magenta).
Table 1.
Data collection and refinement statistics
Values in parentheses indicate the specific values in the highest resolution shell.
aRmerge = ∑|Ij − <I>|/∑Ij, where Ij is the intensity of an individual reflection, and <I> is the average intensity of that reflection.
bRcryst = ∑||Fo| − |Fc||/∑|Fc|, where Fo denotes the observed structure factor amplitude, and Fc denotes the structure factor amplitude calculated from the model.
cRfree is as for Rcryst, but calculated with 5.0% of randomly chosen reflections omitted from the refinement.
The ribbon diagrams of the structures of HeRF1/eRF3-23 and SpeRF1/eRF3-23 are shown in Figure 1, B and C. In both complexes, eRF1 interacts with eRF3 exclusively via their C-terminal domains (eRF1: domain C; eRF3: domain 3). The orientation of domain N of eRF1 in the S. pombe complex is different from its counterpart in the human complex (Fig. 1). When domain C of eRF1 in free eRF1 and in different complexes is superimposed, the orientations of domain N relative to domain C differ by 15, 16, and 30° for human complex versus free, human complex versus S. pombe complex. and S. pombe complex versus free, respectively (Fig. 1D). A possible explanation for the difference in domain N's position is due to incomplete rearrangement of either S. pombe or human eRF1 (or of both, but to different extents) upon binding to domains 2 and 3 of eRF3 in the absence of the G domain. Intriguingly, an ATP molecule used as a crystallization additive was found to be situated in the putative decoding center of each eRF1 molecule in form I but not in form II in the human complex (Supplemental Fig. S1).
The eRF1/eRF3 interface
The interface between eRF1 and eRF3 involves domain C of eRF1 and domain 3 of eRF3 in both the human and S. pombe complexes (Fig. 1B,C). Since the interface of eRF1 and eRF3 is well conserved between human and S. pombe complexes (root-mean-square deviation [RMSD] of 0.9 Å for all the equivalent Cα atoms by superposition of domain C of eRF1 together with domain 3 of eRF3), for simplicity, only the interface of S. pombe eRF1 and eRF3 is discussed.
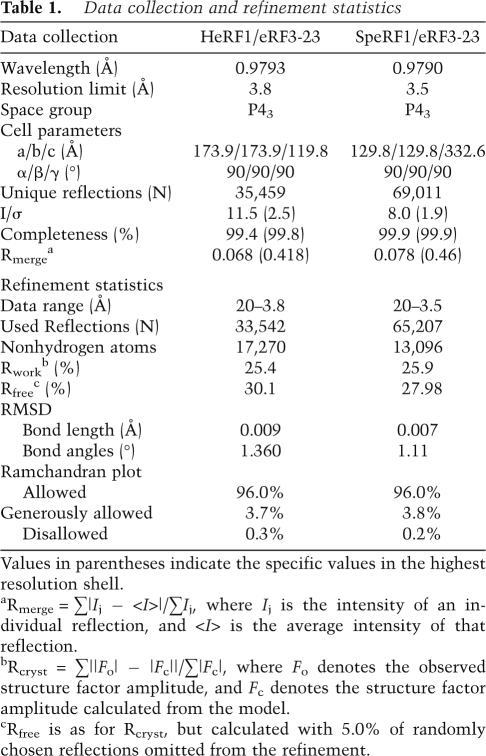
The interaction between domain C of eRF1 and domain 3 of eRF3 buries a pairwise accessible surface area of 974.0 Å2. Domain C of eRF1 interacts with eRF3 via helices α8, α11, and strand β10 (Fig. 2A). Domain 3 of eRF3 interacts with domain C of eRF1, mainly through its loop regions connecting β15–β16, β16–β17, β18–β19, and β21–β22 (numbering as in Kong et al. 2004). The surface groove on domain C of eRF1 that interacts with domain 3 of eRF3 is composed mainly of hydrophobic amino acids (Phe288, Ile291, Tyr298, Phe300, and Phe405). These residues are located in helices α8, α11, and strand β10, and form a hydrophobic patch that interacts with two hydrophobic residues, Ile572 and Phe612 of domain 3 of eRF3 (Fig. 2A). In addition to these predominant hydrophobic interactions, residues Gln396 and Gln400 of domain C in eRF1 contact Ile572 and Asp647 of domain 3 in eRF3 through van der Waals interactions. The residues involved in the eRF1/eRF3 interface are strongly conserved from yeast to human (Supplemental Fig. S2).
Figure 2.
Interaction between eRF1 and eRF3. (A, left) The SpeRF1/eRF3-23 complex shown as in Figure 1B. (Right) Stereo view of the interface of eRF1/eRF3. Residues involved in the interactions are labeled and shown in yellow stick model. (B) Yeast two-hybrid assays for S. pombe eRF1 and eRF3. (Top) eRF1 wild-type and interface mutations in AD (δ-N2 construct of eRF1) vector against eRF3 in BD vector. (Bottom) eRF3 wild-type and interface mutations in BD vector against eRF1 in AD vector (δ-N2 construct). The first mutation is indicated at the top and additional mutation(s) are indicated to the left in both panels.
To test the role of the interface in the S. pombe complex, we mutated residues in the interface of eRF1 and eRF3 and examined the effects of these mutations on their interactions and the ability to restore cell growth (i.e., complementation test). Isothermal Titration Calorimetry (ITC) showed that all five single mutations in eRF3, S571A, I572A, Y577A, F612A, and D647A reduced its binding to eRF1 by at least ninefold (Supplemental Table S1; Supplemental Fig. S3). Consistent with this observation, residues Tyr577 and Asp647 of eRF3 have been identified as being involved in the eRF1–eRF3 interaction (Ebihara and Nakamura 1999). Similarly, the eRF1 mutations F288A, I291A, Q400A, and F405A reduced its binding to eRF3 by at least sixfold, whereas Y298A only moderately reduced binding, and mutations S292A and F300A had very little effect on binding to eRF3 (Supplemental Table S1).
In the yeast two-hybrid assays, the only single mutations that diminished the eRF1–eRF3 interaction were F288A in eRF1 and I572A in eRF3. However, systematic combinations of two, three, or four mutations in most instances led to a reduction in the eRF1–eRF3 interaction, which correlated with complementation defects (Fig. 2B; Supplemental Table S2), showing that the defect in translation termination was specifically caused by mutational defects in eRF1–eRF3 binding. Taken together, these results documented that the residues in the interface are required for the interaction of eRF1 and eRF3 and are important for cell growth.
Comparison of the eRF1/eRF3-23 complex with EF-Tu–GTP–tRNA: modeling of the eRF1/eRF3/GTP ternary complex
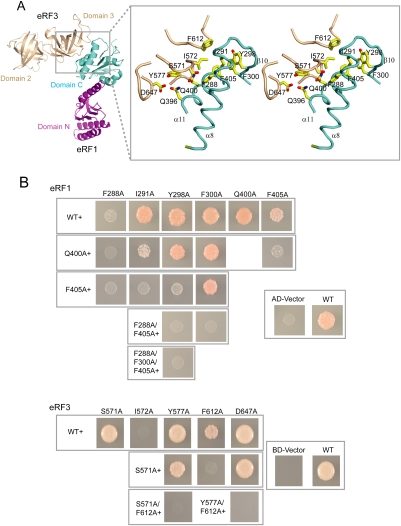
The relative orientation of domain M of eRF1 with respect to domain N differs significantly between the free form (Song et al. 2000) and form I of the human complex. Superimposition of domain N in the free form with that in form I revealed that upon binding to eRF3, domain M shifts and rotates, moving the tip of this domain by ∼44 Å from its position in the free form (Fig. 3A), causing eRF1 to adopt a bent conformation. This conformational change of eRF1 reduces the distance between the GGQ motif and the putative decoding site from 80 Å to 75 Å, and leads eRF1 to resemble a tRNA molecule (Fig. 3B).
Figure 3.
Structural comparison of the eRF1/eRF3-23 complexes with EF-Tu–GTP–tRNA, and the modeled eRF1/eRF3/GTP complex. (A) Relocation of the GGQ motif in domain M of eRF1 upon binding to eRF3. The GGQ motif is shown in sphere. Domains N and M of eRF1 in form I and in free state are shown in limegreen and lemon, respectively. (B) Structural comparison of eRF1 in form I with tRNA in the EF-Tu–GTP–tRNA complex. Both are shown in solid deep-olive surface model. (C,D) Superposition of the HeRF1/eRF3-23 complex (C) and the SpeRF1/eRF3-23 complex (D) with the EF-Tu–GTP–tRNA complex. The ribbon diagrams are generated by superposition of domains 2 and 3 of eRF3 with those of EF-Tu. The tRNA is shown in orange, EF-Tu in green, and the human and S. pombe eRF1/eRF3-23 complexes in magenta and salmon, respectively. The switch I and switch II regions of EF-Tu are shown in yellow and red, respectively, Mg2+ ion in blue sphere, and GTP in stick model. (E) The model of the eRF1/eRF3/GTP complex. G domain and domains 2 and 3 of eRF3 are colored in gray and wheat, respectively. The coloring scheme for the final modeled eRF1 is the same as in Figure 1. Superimposed domains M and N of eRF1 in the human complex as a rigid body with domain N of eRF1 in the S. pombe complex is shown in light pink. Arg192 and Arg203 in human eRF1 are shown in stick models. (F) Crystal structure of Cdc42/RhoGDI. Cdc42 is shown in gray. The regulatory arm of RhoGDI is shown in green and the rest of the molecule in cyan, Mg2+ ion in blue sphere, and switches I and II in yellow and red.
Superposition of domains 2 and 3 of human eRF3 and S. pombe eRF3 in the eRF1/eRF3-23 complexes with those of EF-Tu in the EF-Tu–GTP–tRNA complex (Nissen et al. 1995) gives an RMSD for equivalent Cα atoms of 1.2 Å and 1.3 Å, respectively, indicating that domains 2 and 3 are very similar in these structures. Since eRF1 in the HeRF1/eRF3-23 complex resembles a tRNA molecule (see above), the high structural similarity of domains 2 and 3 in eRF3 and EF-Tu suggests that eRF1 and tRNA could interact in a similar manner with these domains of eRF3 and of EF-Tu, respectively. As shown in Figure 3, C and D, domain C of eRF1 interacts with domain 3 of eRF3 in the eRF1/eRF3-23 complexes in an analogous manner to the interaction of domain 3 of EF-Tu with the T-stem of tRNA in the EF-Tu–GTP–tRNA complex. Despite these common features, there are some differences between the human and S. pombe complexes. In the human complex, domain M, which corresponds to the aminoacyl stem of tRNA, has no contacts with domains 2 and 3 of eRF3 arising from the orientation differences between eRF1 and the tRNA molecule (Fig. 3C), possibly due to loss of strong positive cooperativity in the absence of the G domain (Hauryliuk et al. 2006). In the S. pombe complex, domain M is disordered, and the orientation of domain N is different from that of the human complex (see above). However, when domains 2 and 3 of eRF3 are superimposed on corresponding domains in EF-Tu, domains N and C of eRF1 are almost perfectly matched with the anti-codon and T-stems of tRNA, respectively (Fig. 3D), suggesting that the orientations of these two domains in the S. pombe complex might represent those in the eRF1/eRF3/GTP complex.
eRF3 is structurally very similar to EF-Tu, albeit with quite different domain arrangements, and free eRF3 is unable to bind GTP due to the disorder of switch I and switch II regions (Kong et al. 2004). The observation that eRF3 gains the ability to bind GTP in the presence of eRF1 (Hauryliuk et al. 2006; Mitkevich et al. 2006; Pisareva et al. 2006) suggests that eRF3 undergoes conformational changes upon binding to eRF1. These conformational changes in eRF3 might be analogous to those of EF-Tu upon binding to GTP/tRNA (Berchtold et al. 1993), and involve the change in orientation of domain G relative to domains 2 and 3, as well as ordering of the switch regions. This hypothesis, together with the structural similarity between the EF-Tu–GTP–tRNA complex and the SpeRF1/eRF3-23 complex (Fig. 3D), allows us to model the eRF1/eRF3/GTP complex using the EF-Tu–GTP–tRNA complex as a template. We first modeled the structure of the G domain of human eRF3 in the GTP form by using the EF-Tu structure in the EF-Tu–GTP–tRNA complex (Nissen et al. 1995) as a template. Connection of this modeled G domain of eRF3 with domains 2 and 3 of eRF3 in the human complex gave a complete structure of human eRF3 in the GTP form. Given the high structural similarity between the S. pombe complex and EF-Tu–GTP–tRNA, superposition of this modeled eRF3–GTP with domains 2 and 3 of eRF3 in the S. pombe complex determines the orientation of eRF3–GTP in the eRF1/eRF3/GTP complex. The eRF1 structure in the eRF1/eRF3/GTP complex was generated in three steps. First, domain C of eRF1 was copied from the human complex. Second, superposition of domains M and N of eRF1 in the human complex as a rigid body with domain N of eRF1 in the S. pombe complex gave the positions of domain N, and domain M, which is in close proximity, but makes no contacts with domain G of eRF3 (Fig. 3E). The final position of domain M of eRF1 was determined by manually rigid body fitting of this domain to the aminoacyl stem of tRNA in the EF-Tu–GTP–tRNA complex. The final model of the eRF1/eRF3/GTP complex (Fig. 3E) predicted that domain M of eRF1 would contact the GTPase domain of eRF3. Consistent with our modeling result, domain M of eRF1 has been shown to be involved in interaction with eRF3 (Kononenko et al. 2008). Interestingly, the predicted interaction between domain M of eRF1 and the switch regions of eRF3 is strikingly similar to the interaction between the regulatory domains of RhoGDI and Cdc42 (Hoffman et al. 2000), suggesting that domain M of eRF1 may regulate the GTPase activity of eRF3 (Fig. 3F).
Validation of the model of eRF1/eRF3/GTP ternary complex by SAXS analysis
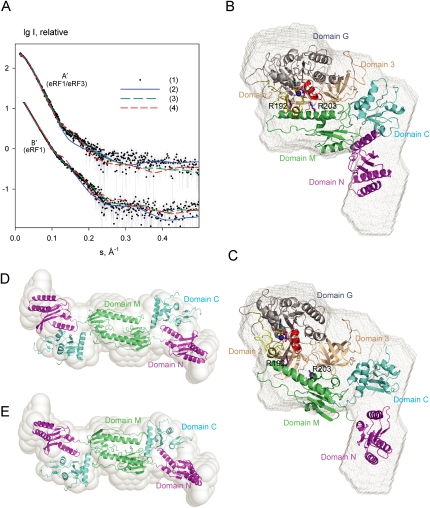
To validate our model of the eRF1/eRF3/GTP complex, SAXS was used. The molecular mass (MM) derived from the scattering data in Figure 4A (90 ± 8 kDa) agrees well with that expected for a 1:1 eRF1/eRF3 complex, indicating that the complex is monomeric in solution. The experimental radius of gyration (Rg =39 ± 1 Å) somewhat exceeds that of the model (34.8 Å) and the scattering pattern computed from the model, while yielding a reasonable fit with discrepancy χ = 1.80, displays systematic deviations from the experimental SAXS data (Fig. 4A, eRF1/eRF3 curve 2). The molecular model appears similar but somewhat more compact than the low-resolution shape of the complex reconstructed ab initio by DAMMIN (Svergun 1999), as displayed in Figure 4B. The molecular model was therefore refined against the SAXS pattern by rigid body movements and rotations of the eRF1 and eRF3 domains defined in the above molecular modeling section using SASREF (Petoukhov and Svergun 2005). Several simulated annealing (SA) refinement runs yielded reproducible results, and a typical rigid body model displayed in Figure 4C neatly fits the experimental data with χ = 1.03 (Fig. 4A, eRF1/eRF3 curve 3), and also matches the ab initio shape of the complex (Fig. 4C). The SAXS-refined models kept the structural organization of the molecular model (within RMSD of ∼9 Å). There is no steric clash between domain 2 of eRF3 and the M domain of eRF1, as these close contacts are monitored during the refinement. Importantly, although the refined model displayed an overall more extended conformation of the complex (with Rg ∼37.5 Å), eRF1 maintained a bent configuration similar to that in the crystal structure of the complex. Alternative rigid body modeling of the complex was performed by fixing eRF1 in the open conformation as observed in free eRF1 (PDB code 1dt9) (Song et al. 2000). Under this restraint, no good fit to the experimental data could be obtained (the best model yielding χ = 1.5 still displayed systematic deviations) (Fig. 4A, eRF1/eRF3 curve 4). These results suggest that the modeled eRF1/eRF3/GTP complex agrees reasonably well with the SAXS model that was obtained independently, and that eRF1 has a bent conformation in the complex.
Figure 4.
SAXS data and shape reconstructions. (A) Experimental and computed SAXS scattering data. (A′) the eRF1/eRF3/GTP complex, (B′) free eRF1. The logarithm of the scattering intensity is plotted against the momentum transfer s = 4π sinθ/λ, where 2θ is the scattering angle and λ = 1.5 Å is the X-ray wavelength. The plots are displaced along the ordinate for better visualization. (1) Experimental scattering. (2) Computed scattering from the molecular model of eRF1/eRF3 (in A′) and from the crystallographic dimer of eRF1 (in B′). (3) Computed scattering from these two models after rigid body refinement. (4) Computed scattering from the rigid body model of eRF1/eRF3 using extended eRF1 (A′) and from the rigid body model of eRF1 dimer using a compact eRF1 monomer (B′). (B,C) The modeled eRF1/eRF3/GTP complex (B) and the rigid body refined model (C) are superimposed onto the ab initio shape reconstruction (gray mesh). (D,E) The crystallographic dimer of extended eRF1 (D) and the rigid body model of the dimer from extended monomers (E) are superimposed onto the ab initio shape reconstruction (semitransparent beads). The coloring scheme for eRF1 and eRF3 is as in Figure 3E.
To further verify whether the bent conformation of eRF1 is induced by forming a complex with eRF3, SAXS from free eRF1 was measured. The experimental MM and Rg (85 ± 8 kDa and 48 ± 1 Å, respectively) significantly exceeded the values of the monomer and indicated that the protein is dimeric in solution. Among the possible crystallographic dimers, the extended one with the dimerization interface involving the M domain (Rg = 49.9 Å) (Fig. 4D) best fits the experimental SAXS pattern with χ = 1.7 (Fig. 4A, eRF1 curve 2) and also matches the ab initio shape of dimeric eRF1 (Fig. 4D). A rigid body refinement of this model using SASREF in terms of monomeric eRF1 (Song et al. 2000) (1DT9) yields a model (Fig. 4E), which is close to the crystallographic dimer (RMSD 8.8 Å), and neatly fits the SAXS pattern (χ = 1.13) (Fig. 4A, eRF1 curve 3). The refined model has Rg = 47.1 Å and also fits well with the ab initio shape (Fig. 4E). At the same time, attempts to construct a rigid body model of dimeric eRF1 from the bent monomers extracted from the eRF1/eRF3 complex yielded poor fits to the experimental SAXS data (typical Rg = 46 Å and χ = 2.0) (Fig. 4A, eRF1 curve 4). These results indicated that free eRF1 in solution is a dimer of the extended monomers observed in the crystal structure of its free form (Song et al. 2000), and that its compaction is only induced during complex formation with eRF3.
The interaction of the M domain of eRF1 with eRF3 is important for stimulation of eRF3's GTPase activity by eRF1 and the ribosome
The C-terminal domain of eRF1 is not sufficient for stimulation of eRF3's GTPase activity, and domain M of eRF1 is also required for this process (Kononenko et al. 2008). Whereas domain C of eRF1 interacts only with domain 3 of eRF3, our molecular modeling combined with SAXS analysis predict that domain M of eRF1 contacts domains 2 and 3 of eRF3 and the switch regions of its GTPase domain (Fig. 4B,C). The mechanism by which eRF1 stimulates eRF3's GTPase activity is not known, but the possibility that it involves an “arginine finger” cannot be excluded. In this respect, R192 and R203 in domain M of human eRF1 (R189 and R200 in S. pombe), which according to our model would contact the switch regions of eRF3 in the eRF1/eRF3/GTP complex (Fig. 4B,C), are of particular interest.
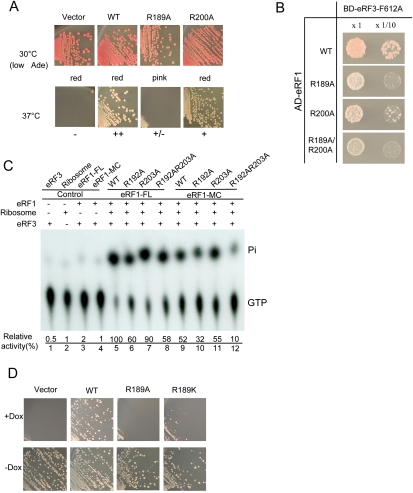
To examine the role of these residues in eRF1-mediated stimulation of eRF3's GTPase activity, we tested the abilities of single or double Ala-substituted mutants of S. pombe eRF1 to restore cell growth and to bind to eRF3, and their activities in stimulating eRF3's GTPase activity. Of these mutants, R189A exhibited a strong defect, and R200A showed a moderate defect in cell growth, whereas the double mutant R189A/R200A had a similar defect to that of R189A (data not shown) in the temperature-sensitive eRF1 yeast strain (Fig. 5A). Interestingly, eRF1(R189A) inhibited cell growth dominantly when supplied under the control of the strongest promoter (GPD) (data not shown). GGQ motif mutations in domain M of eRF1 that abolish the peptide hydrolysis reaction on the ribosome exhibit a similar effect (Song et al. 2000). Consistently, yeast transformed with the eRF1(R189A) plasmid showed a slight, but significant reduction in red pigment in the ade1-101(UAA) strain (Fig. 5A), suggesting nonsense suppression caused by dominant inhibition of translation termination. The dominant nonsense suppression effect of the R189A mutant was also confirmed by an in vivo readthrough assay performed at 25°C, which showed an increase of ∼20% in translation readthrough activity (data not shown). Two-hybrid assays showed substantially reduced binding of eRF1 R189A, R200A, and R189A/R200A mutants to eRF3 in the context of the F612A mutation (Fig. 5B). These results are consistent with the observation that domain M of eRF1 is involved in eRF3 binding (Kononenko et al. 2008), and validated the biological relevance of our eRF1/eRF3/GTP model.
Figure 5.
The M domain of eRF1 harbors an arginine residue important for GTPase activation of eRF3. (A) Growth complementation of eRF1 ts strains. (Top panel) Growth of transformants of wild-type and eRF1-M domain mutant expression plasmids at permissive temperature (30°C). Transformants formed on SC-Ura plates were streaked on low adenine SC-Ura plates to monitor nonsense suppression by red pigment due to insufficiency of full-length Ade2 protein. The colony color is indicated below. (Bottom panel) Growth of transformants of wild-type and eRF1-M domain mutant expression plasmids at nonpermissive temperature (37°C). (B) Yeast two-hybrid assays for S. pombe domain M mutant of eRF1 and eRF3 (F612A). (C) The stimulatory effects of wild-type human eRF1 (both full-length and MC domain) and its mutants on the GTPase activity of human eRF3. A PhosphorImager was used to quantify the released free phosphate (Pi) and the residual GTPr, which had not been hydrolyzed. The percentage of GTP hydrolyzed was calculated as Pi divided by the sum of Pi and GTPr. The numbers shown here are the relative GTPase activities of eRF3 stimulated by wild-type (assigned as 100) and mutant eRF1 proteins. (D) Growth complementation of Tet-OFF eRF1 strain by R189 substitutions. Tet-off eRF1 strains transformed with wild-type, R189A, R189K, and empty vector were streaked on the marker selective plates with (+Dox)/without (−Dox) 10 μg/mL Doxycyclin, a tetracycline derivative, and growth was monitored for 3 d.
To examine the importance of residues R189 and R200 for the ability of eRF1 to stimulate eRF3's GTPase activity, we created single and double Ala substitutions of the equivalent residues R192 and R203 of human eRF1. As expected, in the presence of ribosomes, wild-type human eRF1 strongly stimulated the GTPase activity of human eRF3 (Fig. 5C). Mutation of R192 to Ala (R192A) significantly reduced eRF1's stimulatory effect on eRF3's GTPase activity, whereas the R203A mutation had little influence. The R192A/R203A double mutations had a similar defect to that of R192A alone. Moreover, the same set of mutants created in the context of the MC domain of human eRF1 showed similar defects in stimulating eRF3's GTPase activity. The reduced stimulatory effect on GTPase activity is not due to reduced GTP-binding affinity as the recognition specificity of the bound GTP is conferred solely by the residues from eRF3 (Kong et al. 2004), and R192 may stabilize only the transition state, as in the case of GAPs for small GTPases (Bos et al. 2007). Consistently, our ITC titration experiments showed that wild-type eRF1-MC/eRF3 and eRF1-MC-R189A/eRF3 bind to GTP with similar affinities (Kd values of 0.49 μM and 0.55 μM, respectively) (Supplemental Table S1), indicating that the binding of GTP to eRF1/eRF3 is not affected by the R189A (R192A in human) mutation in eRF1.
To test whether R192 can be substituted by Lys, the R192K mutation in human eRF1 and the R189K mutation in S. pombe eRF1 were generated. A growth complementation assay performed in the Tet-OFF eRF1 strain showed that the R189K mutant was much less active than the wild type (Fig. 5D), and other substitutions, including those with basic or acidic side chains, poorly restored growth (data not shown). Consistent with this result, the GTPase assay showed that R192A and R192K substitutions in eRF1 reduced its ability to stimulate GTP hydrolysis to a similar extent (data not shown). Although the fact that R192 could not be functionally substituted by Lys resembles the situation for the GAPs (p120GAP and neurofibromin) of the small GTPase Ras, wherein the catalytic arginine finger cannot be replaced by a lysine residue (Ahmadian et al. 1997), the moderate reduction in GTP hydrolysis caused by the R192A and R192K mutations suggest that R192 (R189 in S. pombe) is likely not a bona fide arginine finger. However, these results indicate that R192 in domain M of eRF1 is nevertheless important for its stimulatory effect on eRF3's GTPase activity.
Molecular dissection of the stop codon decoding site
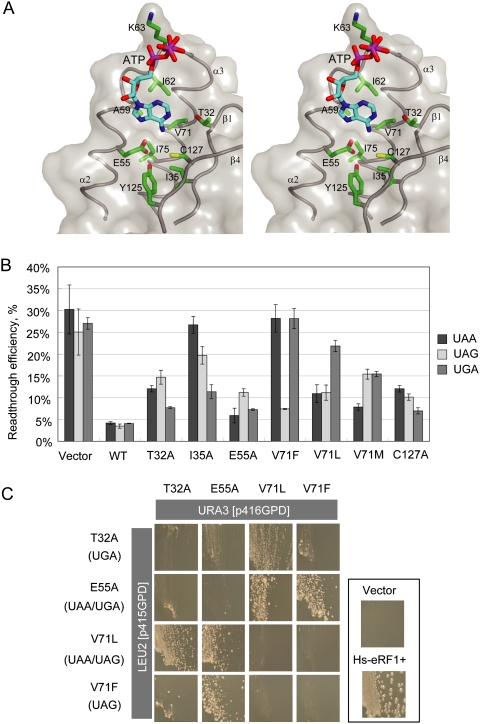
In form I of the human complex, an ATP molecule is located in a pocket formed by helices α2, α3, and the central anti-parallel β-sheet in domain N of eRF1 (Fig. 6A). The adenine base of the ATP dips into this pocket, making multiple contacts with the protein residues, while the γ-phosphate group is exposed to the solvent region (Fig. 6A), and the α- and β-phosphate group contacts Lys63 in the “NIKS” motif. The adenine base interacts with hydrophobic residues Ala59, Ile62, Val71, and Ile75 from α2–loop–α3, and Ile35 from β1. In addition, the adenine base appears to form several hydrogen bonds with the residues in the pocket (Fig. 6A). The N1 and N6 atoms could be hydrogen bonded to the side chains of Thr32 and Cys127, respectively, while the N7 atom could form hydrogen bonds with residues Glu55 and Tyr125. The bound ATP is further stabilized by the loop (residues 118–121) between β3 and β4 of the NCS-related eRF1 molecule in the AU (Supplemental Fig. S1). Consistent with this region being involved in ATP binding, the tripeptide QFM just preceding the YxCxxxF motif in Stylonychia eRF1, which corresponds to residues 118–121 in human eRF1, plays a crucial role in restricting its specificity toward UGA (Lekomtsev et al. 2007). Since ATP was not included as an additive for crystal growth of the S. pombe complex, and the ATP-binding pocket lacks the contribution from residues 118–121 of a neighboring symmetry-related eRF1 molecule in form II of the human complex, no ATP is observed in eRF1 in either the S. pombe complex or form II of the human complex.
Figure 6.
The binding of ATP to a putative decoding site on domain N of eRF1. (A) Stereo view of the binding pocket. Domain N of eRF1 is shown in transparent gray surface. Residues involved in this binding pocket are labeled and drawn in the green stick model. ATP molecule is shown in the cyan stick model. (B) In vivo data bar graph for selected mutations. (C) Dual-eRF1 complementation assay: Human eRF1 mutant genes that showed prominent single or double stop codon specificities (indicated in parentheses on the left; i.e., loss of omnipotency) were cloned into yeast expression vectors with different selection markers (p416GPD for URA3, p415GPD for LEU2). Tet-off eRF1 strains transformed with dual combinations of vectors were streaked on the marker selective plates supplemented with 10 μg/mL Doxycyclin, a tetracycline derivative, and growth was monitored for 5 d.
The pocket in which the adenine base is situated is near the anticipated site for stop codon recognition (Song et al. 2000). Among critical residues mentioned above, mutations on Glu55, Val71, Tyr125, and Cys127 were shown to be codon-specific from various analyses (Bertram et al. 2000; Seit-Nebi et al. 2002; Kolosov et al. 2005). Thus, it is tempting to propose that the ATP binding to domain N of eRF1 could mimic the base interaction with the eRF1 decoding site, the nature of which has been a long-standing unresolved question.
To test this possibility, we conducted a systematic mutational analysis of residues of human eRF1 involved in ATP binding. The overall translation termination activities of the mutated human eRF1s were monitored by growth complementation of a conditional eRF1 knockout strain (tet-off eRF1), and codon specificity was characterized using the Dual-Luciferase reporter readthrough assay (Supplemental Table S3). In the complementation test, at least one mutation from a series of amino acid substitutions at predicted critical residues showed a severe growth defect, suggesting their crucial role in translation termination. In the readthrough assay, many of the mutated eRF1s showed deviant stop codon recognition (Fig. 6B; Supplemental Table S3). For example, the T32A eRF1 mutant exhibited much higher readthrough at both UAA and UAG versus UGA codons, thus tending to UGA unipotency, like eRF1 of certain Ciliates (Lekomtsev et al. 2007). eRF1 with mutations at Thr32 (T32A, T32V), Ile35 (I35A), and Cys127 (C127A) tended to UGA unipotency. On the other hand, eRF1 containing mutation at Glu55 (E55A) exhibited UAA and UGA dual potency. Intriguingly, mutations at Val71 in eRF1 generated three different codon recognition patterns: V71A, V71F, V71Y, and V71G mutations tended toward UAG unipotency, V71M made eRF1 tend toward UAA unipotency, and V71L tended to UAA/UAG dual potency. Neither UAG nor UAA unipotent eRF1 variants have been reported previously.
In order to confirm the codon specificity of mutated eRF1s, we devised a new assay system, designated the “Dual eRF1 assay system,” in which two eRF1 variants are coexpressed in eRF1 knockout strains. In this system the combination of eRF1s with a tendency for specificity for different codons suppresses the conditional lethality of the eRF1 knockout strain by reconstituting omnipotency via complementation of each other's defective recognition of specific codons, just like bacterial codon-specific class I RFs RF1 and RF2. As briefly shown with four typical human eRF1 variants created in this study (Fig. 6C), only those pairs with reconstituted omnipotent activity could maintain cell growth. The codon specificity of mutations at Thr32, Ile35, Glu55, Val71, and Cys127 strongly suggested that these residues are crucial for stop recognition and/or discrimination. To our knowledge, this is the first experimental evidence that like in prokaryotes, termination codons could be recognized by multiple eukaryotic eRFs, each with restricted specificities.
Discussion
The crystal structures of the eRF1/eRF3-23 complexes, combined with molecular modeling and SAXS analysis, show that eRF1 interacts with eRF3 in a highly specific manner with their C-terminal domains conferring the main specificity, and the M domain of eRF1 providing the additional affinity in the presence of GTP. The observation that eRF1 interacts with eRF3 predominantly via their respective C-terminal domains in the eRF1/eRF3-23 complexes is consistent with the results of previous deletion studies (Ito et al. 1998; Ebihara and Nakamura 1999; Eurwilaichitr et al. 1999; Merkulova et al. 1999). Importantly, the binding of eRF3 to eRF1 does not shield the subdomain of domain C (residues 338–381; human numbering) that has been implicated in PP2A binding (Andjelkovic et al. 1996), suggesting that eRF1 may also use this subdomain to associate with other proteins regulating translation termination (e.g., Upf1). Domain M of eRF1 enhances the overall affinity between eRF1 and eRF3 (Kononenko et al. 2008), and under certain conditions, the interaction between eRF1 and eRF3 requires the presence of GTP (Kobayashi et al. 2004; Ivanov et al. 2008). These observations are in agreement with our modeling and SAXS analysis, which predict a direct interaction between domain M of eRF1 and the GTPase domain of eRF3 in the presence of GTP. Interestingly, as predicted by this model, domain M of eRF1 would interact with the switch regions of eRF3 in a manner similar to the interaction of the regulatory domains of RhoGDI with Cdc42 (Hoffman et al. 2000). The modeling data therefore argue strongly that in addition to its role in triggering polypeptide release with its GGQ motif, domain M is also involved in stimulation of the GTPase activity of eRF3. Consistent with the latter role, our mutagenesis and GTPase assays showed that R192 in the M domain of eRF1, which would contact the switch regions of eRF3's GTPase domain, contributes to eRF1's stimulation of the GTPase activity of eRF3.
Recently, a model for translation termination in eukaryotes has been proposed based on an in vitro reconstitution system (Alkalaeva et al. 2006). In this model, eRF1 and eRF3 act in a cooperative manner to ensure fast peptide release, and this cooperativity between eRF1 and eRF3 required the binding of eRF3 to the C-terminal domain of eRF1. However, the molecular basis of this cooperativity remains obscure. Our structural and mutagenesis data offer a possible mechanism by which eRF1 acts in concert with eRF3 for rapid peptide release in the termination process. In this model, prior to GTP binding, eRF1 forms a complex either with free eRF3 or with eRF3–GDP via the interaction of its domain C with domain 3 of eRF3. Since the cellular GTP concentration is at least 10 times higher than GDP, and the eRF1/eRF3 complex has six times higher affinity for GTP than for GDP at physiological Mg2+ concentration, either the eRF1/eRF3 or eRF1/eRF3-GDP complex will be converted to the eRF1/eRF3/GTP complex by anchoring domain C of eRF1 onto domain 3 of eRF3 and moving the M domain of eRF1 toward the G domain of eRF3. The interaction of domain M of eRF1 with the G domain of eRF3 would stabilize the switch regions of eRF3, which are disordered in free eRF3 (Kong et al. 2004), therefore enabling them to acquire the ability to bind the γ-phosphate and Mg2+. Our molecular modeling and SAXS analysis suggested that the eRF1/eRF3/GTP complex adopts a conformation similar to that of the EF-Tu–GTP–tRNA complex (Nissen et al. 1995), which would likely result in increased affinity to the ribosome of eRF1 in the eRF1/eRF3 complex compared with free eRF1. The high similarity in shape between eRF1 and tRNA would facilitate positioning of eRF1's decoding site in the N domain close to the stop codon and the GGQ motif pointing to the PTC. Binding of the eRF1/eRF3/GTP complex to the ribosome would induce large conformational changes, as evidenced by the toeprint shift in a reconstituted in vitro system (Alkalaeva et al. 2006), which might lead to optimal stop codon recognition by eRF1 and/or enable eRF1 together with the GTPase activation center (GAC) in the ribosome to stimulate the GTPase activity of eRF3. Upon GTP hydrolysis, the conformational changes in the switch regions, on one hand, would relieve the interaction of the M domain with the G domain of eRF3, consistent with the data that GDP has no effect on eRF1/eRF3 interaction (Mitkevich et al. 2006; Pisareva et al. 2006), and on the other hand, would change the orientation of the M domain relative to the N domain to such a way that the GGQ motif is positioned optimally in the PTC for peptide release. The mode of action of eRF1 in the cycles of the nucleotide exchange of eRF3 presented here would argue that eRF1 promotes/stabilizes the binding of GTP to eRF3 prior to binding to the ribosome, and acts as a GAP on the ribosome. The dual role of the M domain would allow eRF1 to regulate the GTP binding and hydrolysis of eRF3, and to use the energy of GTP hydrolysis to couple stop codon recognition with efficient peptide release. Consistent with this view, mutations affecting the rate of GTP hydrolysis of eRF3 have been found to influence stop signal decoding and, consequently, efficient peptide release (Salas-Marco and Bedwell 2004). Although previously reported results indicate that eRF1's stimulation of eRF3's ribosome-dependent GTPase activity can occur in the absence of the A-site stop codon (Frolova et al. 1996), the kinetic comparison of GTP hydrolysis by eRF3 on individual ribosomes and on functional pretermination complexes has not been performed. It is therefore possible that the interaction between eRF1 and a stop codon might accelerate GTP hydrolysis by eRF3. Resolving this question would require further kinetic, biochemical, and genetic studies of translation termination, and determination of the atomic structures of the eRF1/eRF3 complex at different nucleotide states with and without a bound ribosome.
Our structure showed that the pocket defined by residues Glu55, Val71, Cys127, Tyr125, Ile75, and Thr32 is occupied by the adenine base of ATP. This pocket is unlikely the first U-binding site, since the mutants associated with ATP binding exhibited readthrough activities for some, but not all stop codons (Supplemental Table S3). Among mutations analyzed, those at residues Thr32, Val71, and Cys127, which interact with six-membered rings of the adenine base, strongly affected codon specificity. Substitution of Thr32 to hydrophobic amino acids such as alanine and valine restricted the omnipotent eRF1 activity to UGA unipotent, whereas substitution to polar amino acids such as serine, cysteine, or lysine did not restrict omnipotency. The C127A substitution also led to UGA unipotency. Consistently, a recent genetic screen for mutations affecting stop codon recognition in yeast identified a C127S (C124 in yeast) mutation that specifically enhances UGA recognition (Fan-Minogue et al. 2008). The polarity of these residues is therefore essential for fitting and/or accepting the adenine base at the second position of stop codons. On the other hand, intriguingly, defective mutants at residue 71 commonly neglected UGA, indicating the importance of valine at this position for recognition of guanine at the second base. These residues are in close contact with N1 and N6 atoms of the adenine base. If the adenine base is replaced with a guanine base, these residues would contact atoms N1, O6, and N2 in the guanine base (Supplemental Fig. S1). These structural features explain the observed mutational effects on selection of the second base, and suggest that the bound ATP molecule is most likely mimicking the second base of the stop codon.
Meanwhile, E55A and Y125F compromised only UAG recognition in vivo, as previously reported in vitro (Kolosov et al. 2005). Therefore, whereas Thr32, Val71, and Cys127 are crucial for recognition of the second base, Glu55 and Tyr125, which contacted the adenine base of ATP from the opposite side, are critical for recognition of adenine of the third base of stop codons. Thus, mutation of residues in close contact with the ATP molecule affected recognition of the second and/or the third base. These results suggested that residues involved in recognition of either base influence selection of the other one; i.e., the recognition of the second and the third bases of stop codons are coordinated events. It is tempting to speculate that eRF1 excludes only the UGG sense codon out of the four purine(2nd)–purine(3rd) combinations through such a molecular interplay mechanism. Consistent with this notion, a series of Val71 variants showed transitions in codon specificity pattern; i.e., V71M, V71L to V71G (and also A, F, and Y) successfully traced A(2nd)–A(3rd), A(2nd)–A/G(3rd) to A(2nd)–G(3rd) shift in a stepwise manner. It is apparent that these transitions by Val71 mutants are closely related with their side-chain characteristics, suggesting that Val71 is critical for coordination of second- and third-base recognition.
Materials and methods
X-ray data collection
Details of protein expression, purification, and crystallization are described in the Supplemental Material. For data collection, crystals of the HeRF1/eRF3-23 complex were dehydrated by gradually increasing the concentration of sucrose in crystallization solution, and fast frozen in liquid nitrogen. Crystals belong to the space group P43 with cell parameters a = b = 174.5 Å, c = 120.2 Å, and contain four complexes per AU. X-ray data were collected at ESRF, ID29, and processed with Mosflm (Collaborative Computational Project, Number 4. 1994). Crystals of the SpeRF1/eRF3-23 complex were transferred to cryo-buffer (50 mM HEPES at pH 7.0, 400 mM KCl, 10% PEG400, 15% MPD) and fast frozen in liquid nitrogen. Crystals belong to the space group P43 with the cell parameters a = b = 129.8 Å, c = 332.6 Å, and contain four complexes per AU. X-ray data were collected at DESY, BW7A, and processed with Mosflm (Collaborative Computational Project, Number 4. 1994).
Structure determination
Details of structure determination and refinement are described in the Supplemental Material. Briefly, the structures of HeRF1/eRF3-23 and SpeRF1/eRF3-23 were determined by molecular replacement using program PHASER (McCoy et al. 2007) using human eRF1 (accession code 1DT9) (Song et al. 2000) and S. pombe eRF3c (accession code 1R5B) (Kong et al. 2004) as search models. Crystallographic refinement was carried out by REFMAC5 (Murshudov et al. 1997) and model building was performed by Coot (Emsley and Cowtan 2004) and O (Jones et al. 1991). The final refinement statistics are summarized in Table 1.
Strains for yeast in vivo complementation
For in vivo complementation test, two types of S. cerevisiae strains, Tet-OFF and temperature-sensitive, were used in this study. Tet-OFF eRF1 (SUP45) and eRF3 (SUP35) strains were constructed according to the same procedure described in Gari et al. (1997) in the BY4727 and BY4805 strain background (Brachmann et al. 1998), respectively. The genotypes were MAT α, his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 tTA-tetO (YBR143C/SUP45)∷ kanMX4 for eRF1-Tet-OFF and MAT α, ade2Δ∷ hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 SUP35∷ HIS3 HO∷tTA-tetO (YDR172W/SUP35)∷ hphMX4 for eRF3-Tet-OFF, respectively. These Tet-OFF strains hardly grew in the medium supplemented with 10 μg/mL Doxycyclin, a tetracycline derivative.
Accession numbers
The coordinates and structure-factor amplitudes of HeRF1/eRF3-23 and SpeRF1/eRF3-23 have been deposited in the Protein Data Bank with accession codes 3E1Y and 3E20, respectively.
Acknowledgments
We thank V. Hauryliuk and M. Ehrenberg for providing the yeast ribosomes, and C.U. Hellen for reading the manuscript. This work is financially supported by the Biomedical Research Council of A*STAR (Agency for Science, Technology, and Research) (H.S.). D.I.S. and T.V.P. acknowledge financial support from the Human Frontier Science Program, Research Grant Reference RGP 55/2006. D.I.S. also acknowledges support from the EU Framework 6 Programme (Design Study SAXIER, RIDS 011934). T.V.P. also acknowledges support from NIH grant R01 GM 80623 and thanks L.L. Kisselev and L. Yu. Frolova for the gift of eRF1 and eRF3 expression vectors.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1770109.
Supplemental material is available at http://www.genesdev.org.
References
- Ahmadian M.R., Stege P., Scheffzek K., Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 1997;4:686–689. doi: 10.1038/nsb0997-686. [DOI] [PubMed] [Google Scholar]
- Alkalaeva E.Z., Pisarev A.V., Frolova L.Y., Kisselev L.L., Pestova T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Andersen G.R., Pedersen L., Valente L., Chatterjee I., Kinzy T.G., Kjeldgaard M., Nyborg J. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Bα. Mol. Cell. 2000;6:1261–1266. doi: 10.1016/s1097-2765(00)00122-2. [DOI] [PubMed] [Google Scholar]
- Andjelkovic N., Zolnierowicz S., Van Hoof C., Goris J., Hemmings B.A. The catalytic subunit of protein phosphatase 2A associates with the translation termination factor eRF1. EMBO J. 1996;15:7156–7167. [PMC free article] [PubMed] [Google Scholar]
- Berchtold H., Reshetnikova L., Reiser C.O., Schirmer N.K., Sprinzl M., Hilgenfeld R. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993;365:126–132. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- Bertram G., Bell H.A., Ritchie D.W., Fullerton G., Stansfield I. Terminating eukaryote translation: Domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6:1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.L., Rehmann H., Wittinghofer A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Chavatte L., Seit-Nebi A., Dubovaya V., Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Ebihara K., Nakamura Y. C-terminal interaction of translational release factors eRF1 and eRF3 of fission yeast: G-domain uncoupled binding and the role of conserved amino acids. RNA. 1999;5:739–750. doi: 10.1017/s135583829998216x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Eurwilaichitr L., Graves F.M., Stansfield I., Tuite M.F. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol. 1999;32:485–496. doi: 10.1046/j.1365-2958.1999.01346.x. [DOI] [PubMed] [Google Scholar]
- Fan-Minogue H., Du M., Pisarev A.V., Kallmeyer A.K., Salas-Marco J., Keeling K.M., Thompson S.R., Pestova T.V., Bedwell D.M. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol. Cell. 2008;30:599–609. doi: 10.1016/j.molcel.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L., Le G X, Zhouravleva G., Davydova E., Philippe M., Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- Frolova L.Y., Merkulova T.I., Kisselev L.L. Translation termination in eukaryotes: Polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA. 2000;6:381–390. doi: 10.1017/s135583820099143x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L., Seit-Nebi A., Kisselev L. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 2002;8:129–136. doi: 10.1017/s1355838202013262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Zhou Z., Rawat U., Huang C., Bouakaz L., Wang C., Cheng Z., Liu Y., Zavialov A., Gursky R., et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Gari E., Piedrafita L., Aldea M., Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Grentzmann G., Brechemier-Baey D., Heurgue V., Mora L., Buckingham R.H. Localization and characterization of the gene encoding release factor RF3 in Escherichia coli. Proc. Natl. Acad. Sci. 1994;91:5848–5852. doi: 10.1073/pnas.91.13.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauryliuk V., Zavialov A., Kisselev L., Ehrenberg M. Class-1 release factor eRF1 promotes GTP binding by class-2 release factor eRF3. Biochimie. 2006;88:747–757. doi: 10.1016/j.biochi.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Hoffman G.R., Nassar N., Cerione R.A. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- Ito K., Ebihara K., Nakamura Y. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. Nature. 1998;4:958–972. doi: 10.1017/s1355838298971874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Uno M., Nakamura Y. A tripeptide ‘anticodon’ deciphers stop codon in messenger RNA. Nature. 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- Ivanov P.V., Gehring N.H., Kunz J.B., Hentze M.W., Kulozik A.E. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kisselev L.L., Buckingham R.H. Translational termination comes of age. Trends Biochem. Sci. 2000;25:561–566. doi: 10.1016/s0968-0004(00)01669-8. [DOI] [PubMed] [Google Scholar]
- Kisselev L., Ehrenberg M., Frolova L. Termination of translation: Interplay of mRNA, rRNAs and release factors? EMBO J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Funakoshi Y., Hoshino S., Katada T. The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. J. Biol. Chem. 2004;279:45693–45700. doi: 10.1074/jbc.M405163200. [DOI] [PubMed] [Google Scholar]
- Kolosov P., Frolova L., Seit-Nebi A., Dubovaya V., Kononenko A., Oparina N., Justesen J., Efimov A., Kisselev L. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res. 2005;33:6418–6425. doi: 10.1093/nar/gki927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong C., Ito K., Walsh M.A., Wada M., Liu Y., Kumar S., Barford D., Nakamura Y., Song H. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- Kononenko A.V., Mitkevich V.A., Dubovaya V.I., Kolosov P.M., Makarov A.A., Kisselev L.L. Role of the individual domains of translation termination factor eRF1 in GTP binding to eRF3. Proteins. 2008;70:388–393. doi: 10.1002/prot.21544. [DOI] [PubMed] [Google Scholar]
- Laurberg M., Asahara H., Korostelev A., Zhu J., Trakhanov S., Noller H.F. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- Lekomtsev S., Kolosov P., Bidou L., Frolova L., Rousset J.P., Kisselev L. Different modes of stop codon restriction by the Stylonychia and Paramecium eRF1 translation termination factors. Proc. Natl. Acad. Sci. 2007;104:10824–10829. doi: 10.1073/pnas.0703887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkulova T.I., Frolova L.Y., Lazar M., Camonis J., Kisselev L.L. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 1999;443:41–47. doi: 10.1016/s0014-5793(98)01669-x. [DOI] [PubMed] [Google Scholar]
- Mikuni O., Ito K., Moffat J., Matsumura K., McCaughan K., Nobukuni T., Tate W., Nakamura Y. Identification of the prfC gene, which encodes peptide-chain-release factor 3 of Escherichia coli. Proc. Natl. Acad. Sci. 1994;91:5798–5802. doi: 10.1073/pnas.91.13.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitkevich V.A., Kononenko A.V., Petrushanko I.Y., Yanvarev D.V., Makarov A.A., Kisselev L.L. Termination of translation in eukaryotes is mediated by the quaternary eRF1*eRF3*GTP*Mg2+ complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Res. 2006;34:3947–3954. doi: 10.1093/nar/gkl549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Ito K. Making sense of mimic in translation termination. Trends Biochem. Sci. 2003;28:99–105. doi: 10.1016/S0968-0004(03)00006-9. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Ito K., Isaksson L.A. Emerging understanding of translation termination. Cell. 1996;87:147–150. doi: 10.1016/s0092-8674(00)81331-8. [DOI] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard M., Thirup S., Polekhina G., Reshetnikova L., Clark B.F., Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- Petoukhov M.V., Svergun D.I. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva V.P., Pisarev A.V., Hellen C.U., Rodnina M.V., Pestova T.V. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J. Biol. Chem. 2006;281:40224–40235. doi: 10.1074/jbc.M607461200. [DOI] [PubMed] [Google Scholar]
- Salas-Marco J., Bedwell D.M. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seit-Nebi A., Frolova L., Kisselev L. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002;3:881–886. doi: 10.1093/embo-reports/kvf178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Parsons M.R., Rowsell S., Leonard G., Phillips S.E. Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 A resolution. J. Mol. Biol. 1999;285:1245–1256. doi: 10.1006/jmbi.1998.2387. [DOI] [PubMed] [Google Scholar]
- Song H., Mugnier P., Das A.K., Webb H.M., Evans D.R., Tuite M.F., Hemmings B.A., Barford D. The crystal structure of human eukaryotic release factor eRF1–mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- Stansfield I., Jones K.M., Kushnirov V.V., Dagkesamanskaya A.R., Poznyakovski A.I., Paushkin S.V., Nierras C.R., Cox B.S., Ter Avanesyan M.D., Tuite M.F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A., Jin H., Neubauer C., Voorhees R.M., Petry S., Kelley A.C., Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov A.V., Buckingham R.H., Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Zhouravleva G., Frolova L., Le G X., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]