Abstract
Background: Five genes—UNG, SMUG1, MBD4, TDG, and DUT—are involved in the repair or prevention of uracil misincorporation into DNA, an anomaly that can cause mutagenic events that lead to cancer. Little is known about the determinants of uracil misincorporation, including the effects of single nucleotide polymorphisms (SNPs) in the abovementioned genes. Because of their metabolic function, folate and other one-carbon micronutrients may be important factors in the control of uracil misincorporation.
Objectives: We sought to identify polymorphisms in uracil-processing genes that are determinants of DNA uracil concentration and to establish whether one-carbon nutrient status can further modify their effects.
Design: We examined the relations between 23 selected variants in the 5 uracil-processing genes, uracil concentrations in whole-blood DNA, and one-carbon nutrient (folate, vitamins B-6 and B-12, and riboflavin) status in 431 participants of the Boston Puerto Rican Health Study.
Results: Four SNPs in DUT, UNG, and SMUG1 showed a significant association with DNA uracil concentration. The SNPs in SMUG1 (rs2029166 and rs7296239) and UNG (rs34259) were associated with increased uracil concentrations in the variant genotypes (P = 0.011, 0.022, and 0.045, respectively), whereas the DUT SNP (rs4775748) was associated with a decrease (P = 0.023). In this population, one-carbon nutrient status was not associated with DNA uracil concentration, and it did not modify the effect of these 4 identified SNPs.
Conclusion: Because elevated uracil misincorporation may induce mutagenic lesions, possibly leading to cancer, we propose that the 4 characterized SNPs in DUT, UNG, and SMUG1 may influence cancer risk and therefore deserve further investigation.
INTRODUCTION
The maintenance of DNA integrity through DNA repair is critical in the prevention of cancer. Several conditions are associated with an increased risk of cancer due to germline mutations in genes encoding DNA-repair enzymes (1), and single nucleotide polymorphisms (SNPs) in DNA-repair genes have been related to cancer risk as well (2, 3). Among the different enzymes involved in DNA repair are those that prevent or correct the misincorporation of uracil in DNA: these enzymes either excise uracil from the DNA molecule (uracil glycosylases) or degrade uridylate before it is incorporated into DNA (UTPases) (4). Uracil misincorporation can occur either during DNA replication as a result of uracil substitution for thymidine, resulting in a U:A mispair, or is formed from the spontaneous hydrolytic deamination of cytosine, leading to U:G mispairs (4). If unrepaired, U:G mispairs may give rise to C:G to T:A transition mutations on DNA replication (4). Moreover, in the process of excising uracil residues, transient breaks in the phosphodiester backbone of DNA are created, and double-strand breaks can appear if uracil residues are closely spaced on opposite DNA strands (5). Accumulation of the latter is particularly mutagenic and is a putative risk factor for cancer (6, 7). For reasons that are not yet understood, specific regions of the genome are particularly susceptible to strand breakage and such breaks are associated with impaired expression of those genes contained within susceptible regions, as was previously observed by our laboratory for the p53 and Apc genes (8, 9).
Four uracil glycosylases excise uracil from DNA in humans: the UNG- and SMUG1-encoded uracil-DNA glycosylases and the MBD4- and TDG-encoded thymine-DNA glycosylases (4). One UTPase, encoded by the DUT gene, has been identified in humans. This enzyme minimizes intracellular dUTP concentrations, thus lowering the probability of uracil misincorporation (4). Although many SNPs have been identified for these 5 uracil-processing genes, there is very little information on their associated phenotypes. Notably, their effects on the uracil concentration in DNA of individuals are totally unknown. Three studies have examined an SNP in MBD4 and observed an association with an altered risk of adenocarcinoma of the lung (10, 11) and squamous carcinoma of the esophagus (12); however, no data exists on the effect of this SNP on DNA uracil concentrations.
Another determinant of DNA integrity is the B vitamin folate, in part because it is a critical cofactor in the enzymatic conversion of uridylate to thymidylate. Studies in human cells, intact humans, and animals have shown that inadequate folate availability promotes the misincorporation of uracil into DNA, which results in DNA strand breakage and chromosome aberrations (13–16). Also, because one-carbon nutrients are highly interdependent in their biochemical functions, vitamins B-12, vitamin B-6, and riboflavin may also affect DNA uracil. For instance, vitamin B-12 depletion alone produces a marked increase in uracil incorporation into the colonic DNA of rodents (17). Additionally, because alcohol consumption and chronic cigarette smoking are known to inhibit aspects of one-carbon metabolism (18, 19), these factors should also be taken into account when defining the relation between one-carbon nutrients and uracil misincorporation in DNA.
The results of many observational studies have generally been consistent with these concepts, which demonstrates an inverse association between high dietary intakes or blood concentrations of folate, vitamins B-6 and B-12, and the risk of cancers such as those of the colorectum and breast, especially among individuals consuming alcohol (20–22). Nevertheless, a few observational studies have failed to observe an association between folate and cancer (23–26), and results from randomized trials of folate alone or in combination with other B vitamins and cancer risk are inconsistent (27–30) and one trial has even raised concerns about deleterious effects (28). Some investigators have suggested that this lack of consistency may be explained by other factors such as timing and genetic background, which might modulate the interaction between folate status and cancer (31, 32).
The goal of the current study was to identify common genetic polymorphisms that may determine the degree to which an individual accumulates uracil in DNA and to define how one-carbon nutrient status may modify these relations. Our a priori hypothesis was that the mutant alleles of selected SNPs in the 5 uracil-processing genes mentioned above would result in altered uracil incorporation in DNA. We further hypothesized that the effects of such polymorphisms might be influenced by the availability of one-carbon nutrients, by tobacco use, and by alcohol consumption. Finally, to verify the hypothesis that elevated uracil incorporation in DNA results in DNA strand breakage, we also sought to determine whether uracil concentrations predict excess strand breakage in the hypermutable region of the p53 gene. These associations were tested in a subpopulation of the ongoing Boston Puerto Rican Health Study (BPRHS).
SUBJECTS AND METHODS
Subjects and study design
The current study was performed in a nested fashion within the BPRHS—a prospective cohort study of men and women of self-reported Puerto Rican origin aged 45–75 y living in the Boston, MA, metropolitan area (initial recruitment date: May 2004). Details on the BPRHS have been published (33). In brief, home interviews were conducted by a bilingual staff to collect health-related and anthropometric data as well as dietary intake information by using a validated ethnic-specific food-frequency questionnaire (34). Fasting blood samples were collected on the morning of the health interviews for biochemical and genetic analyses. Approval for the BPRHS was obtained from the Institutional Review Board of Tufts Medical Center. The current nested study was declared exempt by the institutional review board because of the use of deidentified samples and data (exemption category 4 as defined by the US Department of Health and Human Services).
A subset of 500 subjects were randomly selected from the cohort after the exclusion of individuals taking medications that might potentially affect DNA uracil content: antipurines (eg, azathioprine), antifolate chemotherapy drugs (eg, methotrexate), and sulfonamide antibiotics. DNA was extracted from 1 mL frozen whole blood by using a standard phenol:choloroform extraction after proteinase K and RNase treatment. Sufficient DNA for uracil analysis was recovered from 431 individuals, who were then genotyped for the SNPs of interest. Demographic data, energy intakes, and plasma B vitamin concentrations of those 431 study participants are presented in Table 1.
TABLE 1.
Demographic characteristics, dietary intakes, and blood variables of the participants by sex1
| Men | Women | P | |
| Age (y) | 57.2 ± 7.52 (111) | 58.3 ± 7.1 (320) | 0.160 |
| Weight (kg) | 83.7 ± 18.0 (110) | 79.1 ± 18.0 (316) | 0.0203 |
| BMI (kg/m2) | 29.8 ± 5.8 (111) | 33.2 ± 7.0 (316) | <0.00013 |
| Energy intake (kcal/d) | 2951 ± 1645 (108) | 2257 ± 1223 (317) | <0.00013 |
| Riboflavin intake (mg/d) | 3.2 ± 1.7 (108) | 2.7 ± 1.5 (317) | 0.0033 |
| Multivitamin use (%) | 16.4 (18) | 18.8 (60) | <0.566 |
| Smoking status (%) | <0.00014 | ||
| Never | 18.4 (20) | 49.1 (156) | |
| Past | 43.1 (47) | 28.9 (92) | |
| Current | 38.5 (42) | 22 (70) | |
| Alcohol consumer (%) | <0.00014 | ||
| Never | 6.5 (7) | 40.6 (130) | |
| Past | 36.1 (39) | 26.9 (86) | |
| Current | 57.4 (62) | 32.5 (104) | |
| Plasma folate (ng/mL) | 17.7 ± 7.9 (110) | 20.2 ± 9.3 (318) | 0.0143 |
| Plasma vitamin B-12 (pg/mL) | 493 ± 234 (110) | 538 ± 273 (318) | 0.125 |
| Plasma pyridoxal phosphate (nmol/L) | 54.2 ± 56.4 (110) | 57.3 ± 58.3 (319) | 0.623 |
| Plasma homocysteine (μmol/L) | 10.8 ± 5.8 (110) | 9.0 ± 4.9 (319) | 0.0023 |
| Plasma creatinine (mg/dL) | 0.99 ± 0.48 (110) | 0.79 ± 0.43 (319) | <0.00013 |
| Blood cell DNA uracil (pg/μg DNA) | 1.07 ± 1.16 (111) | 1.14 ± 1.99 (320) | 0.141 |
n in parentheses.
Mean ± SD (all such values).
Student's t test.
Chi-square test.
SNP selection
We obtained a list of SNPs reported within and mapping near to each of the 5 genes of interest from the National Center for Biotechnology Information (NCBI) database (2008). In brief, the strategy for SNP selection was a balanced approach that integrated the results of published studies with bioinformatics-based predictions of the putative functional consequence of the 2 alleles and with linkage disequilibrium (LD) analyses that enabled us to diversely and economically sample different genetic blocks.
First, to have sufficient statistical power to detect potential differences in DNA uracil concentrations between different genotypes, we preferentially selected SNPs with a reported minor allele frequency ≥0.2 in European or African populations (NCBI database). Puerto Ricans are a mixture of 3 population groups: West African, European, and Taíno Native American (35). Because no genotype data are available for Puerto Rican or Taíno populations, we used the data available for European and African populations in our SNP selection. Second, LD data retrieved for each gene of interest from the International HapMap project database (36, 37) was used to select TagSNPs and to avoid selection of more than one SNP from the same LD block. More precisely, we used TAGGER (38) with a pairwise LD correlation coefficient of r2 = 0.80 and a minor allele frequency ≥0.05 in the CEU (Utah residents with northern and Western European ancestry) or in the YRI (Yoruva in Ibadan, Nigeria) populations. Third, we searched the literature to identify SNPs that had been previously linked to uracil concentrations or studied for their association with cancer risk. Finally, putative allele-specific functions were assessed according to position within the gene. SNPs in upstream regions and introns were assessed for their potential to alter transcription factor binding sites (MAPPER; 39). Intronic SNPs could also affect mRNA splicing. Nonsynonymous SNPs within coding sequences of exons could alter protein sequence, structure and function, whereas synonymous SNPs could call for codons of a different frequency, the effects of which can be similar to nonsynonymous SNPs. Last, 3′-untranslated region SNPs can alter secondary structures, which can affect mRNA stability or interactions with small RNAs (eg, miRNAs). These approaches were detailed previously (40).
A total of 26 autosomal, diallelic SNPs from the 5 genes of interest were selected. For each SNP, data for the SNP location on the gene and chromosome, function, and accession number were obtained from databases published by the NCBI (2008). Note that we did not find any examples in the literature on SNPs linked to DNA uracil concentration. A list of all genotyped SNPs, with information on chromosome and gene location, accession number, and function, is shown in Table 2.
TABLE 2.
Characteristics of the 26 selected single nucleotide polymorphisms (SNPs)1
| Gene name, mRNA accession, cytogenetic locus | SNP ID no. | SNP alleles | Gene region | Location relative to TSS (bp) | Justification for selection2 | Literature |
| DUT, NM_001025248.1, 15q15-q21.1 | rs8042136 | C/T | Upstream | −6272 | H; TagYRI; M: site Sry-delta abolished with allele C | Resides within a predicted regulatory region3 |
| rs11637235 | C/T | Intron 4 | 9533 | H; TagCEU; M: site PTF1-beta abolished with allele C | Genotype published (41) | |
| rs4775748 | G/T | Downstream | 15175 | H, TagYRI | ||
| rs8033556 | A/G | Downstream | 18686 | H | Undergoes positive selection in whites (42) | |
| rs12592330 | C/T | Downstream | 21660 | H | ||
| MBD4, NM_003925.1, 3q21-q22 | rs10470431 | A/G | Upstream | −8390 | H; TagYRI; M: site Pbx abolished with allele G | |
| rs2311394 | C/T | Upstream | −986 | H; TagYRI; M: none | ||
| rs3138355 | A/G | Intron 5 | 6623 | H; tagYRI; M: sites Ubx and Bsap abolished with allele A | ||
| rs140696 | C/T | Exon 6 | 6764 | H; TagYRI; S | ||
| rs4128190 | C/G | Downstream | 15902 | H; TagCEU and YRI | ||
| SMUG1, NM_014311.1, 12q13.11-q13.3 | rs7296239 | C/T | Upstream | −8946 | H; TagYRI; M: none | |
| rs2029166 | C/T | Upstream | −7341 | H; TagCEU and YRI; M: sites CDP and Sp3 abolished with allele C | ||
| rs3136392 | A/G | 3′-UTR, exon 4 | 7212 | H; TagYRI | ||
| rs1994356 | A/G | Downstream | 11593 | H; TagYRI | ||
| TDG, NM_003211.3, 12q24.1 | rs703657 | A/T | Upstream | −8765 | H; TagCEU and YRI; M: none | |
| rs172814 | A/G | 5′ near gene | −1802 | H; TagCEU and YRI; M: site GAGA factor abolished with allele A | Genotype published (41) | |
| rs4135050 | A/T | Intron 1 | 2860 | H; TagCEU; M: none | Genotype published (41) | |
| rs4135054 | C/T | Intron 1 | 3994 | H; TagCEU and YRI; M: site E2F abolished with allele T, site SRF abolished with allele C | Genotype published (41) | |
| rs4135063 | C/T | Intron 1 | 6046 | H; TagYRI; M: site SPI-B and Brn-2 abolished with allele T | Genotype published (41) | |
| rs2723877 | C/T | Intron 1 | 9510 | H; TagCEU; M: multiple TFBPs are affected by this SNP (eg, sites ISGF-3 and IRF-1 abolished with allele C) | ||
| rs10861152 | A/G | Intron 2 | 13040 | H; TagCEU and YRI; M: site Zic1 abolished with allele A | Genotype published (41) | |
| rs1866074 | C/T | Intron 3 | 14826 | H; TagCEU and YR; M: none | Genotype published (41): P = 0.032 for association with risk of meningioma | |
| UNG, NM_080911.1, 12q23-q24.1 | rs1018784 | C/T | 5′-UTR, exon 1 | 10 | H; TagYRI; M: none | |
| rs3219266 | C/T | Intron 5 | 10914 | H; TagYRI; M: none | ||
| rs34259 | C/G | Downstream | 15227 | H; TagYRI | ||
| rs246085 | C/T | Downstream | 22191 | H; TagCEU and YRI |
TSS, transcription start site; bp, base pairs; UTR, untranslated region.
H, listed in the HapMap project; TagYRI, TagSNP in the YRI (Yoruba in Ibadan, Nigeria) population using the genome browser from the HapMap project and TAGGER (with a pairwise linkage disequilibrium correlation coefficient of r2 = 0.80 and a minor allele frequency ≥ 0.05) (38); TagCEU, TagSNP in the CEU (Utah residents with ancestry from Northern and Western Europe) population using the genome browser from the HapMap project and TAGGER (with a pairwise linkage disequilibrium correlation coefficient of r2 = 0.80 and a minor allele frequency ≥ 0.05) (38); M, analyzed for altering transcription factor binding sites (MAPPER; 39); S, synonymous.
Observation made based on sequence conservation seen in an alignment of genomic sequence from 7 species using the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu/).
Genotyping
SNPs were genotyped with the Applied Biosystems TaqMan SNP genotyping system (43). For all genotyping, blinded no-template controls and replicates of DNA samples were incorporated in each of the DNA sample plates, which were routinely checked by laboratory personnel. On the basis of our internal quality control as well as that estimated independently by external laboratories, the genotyping error rate was <1%.
Biochemical measurements
Plasma folate and vitamin B-12 concentrations were measured with Immulite chemiluminescent kits according to the manufacturer's instructions (Diagnostic Products Corporation/Siemens, Los Angeles, CA). Between-run and within-run CVs were 4% for plasma folate and 6% and 5% for plasma vitamin B-12, respectively. Plasma concentrations of pyridoxal phosphate (PLP, vitamin B-6) were measured with the radioenzymatic method of Camp et al (44). Total plasma homocysteine (Hcy) concentrations were measured by HPLC with fluorescence detection, as described previously (45). Between-run and within-run CVs were 6% and 5% for plasma vitamin B-6, respectively, and 6% for plasma Hcy.
Measurement of DNA uracil concentrations
DNA (5–10 μg) was used to determine the uracil concentration after uracil DNA glycosylase (New England Biolabs, Ipswich, MA) treatment according to the gas chromatography–mass spectrometry method of Blount and Ames (46), with modifications (47). Between-run and within-run CVs were 7% and 5%, respectively.
Determination of strand breaks within the p53 hypermutable region
Exons 6 and 8 of the p53 gene were chosen to assess DNA strand breakage, because these loci are within the most frequently mutated region of the gene and because previous animal studies have shown that this region is particularly susceptible to strand breakage due to folate depletion (8, 48, 49). The detection of p53 exon-specific strand breaks was determined with a polymerase chain reaction (PCR) method, previously used by both us and others (49), which is based on the principle that preexisting lesions in DNA halt the progression of Taq polymerase during PCR amplification. The following sets of primers were used for the real-time PCR reaction: 5′-GTCTGGCCCCTCCTCAGC-3′ (forward) and 5′-TCCCAGAGACCCCAGTTGC-3′ (reverse) for p53 exon 6; 5′-TAGTGGTAATCTACTGGGACGGAAC-3′ (forward) and 5′-TGCTTGCTTACCTCGCTTAGTG-3′ (reverse) for p53 exon 8. A 41-bp segment of GAPDH was amplified as a control region for the real-time PCR reaction by using the following set of primers: 5′-ACCGGGAAGGAAATGAATGG−3′(forward) and 5′-GCAGGCTTTCCTAACGGCT −3′(reverse). Breaks in p53 exons 6 and 8 are reported as a ΔCt value (Ctp53exon6-8 – CtGAPDH), with a higher ΔCt indicating a lower template integrity. Although the specificity of this assay is not absolute, because other aberrations in the structure of DNA, such as adducts, can also interfere with amplification, we previously showed that incremental increases in gaps in the phosphodiester backbone leads to progressive increases in the readout of this assay (49).
Statistical analyses
Statistical analyses were performed by using SAS 9.1 (SAS Institute, Cary, NC) and SYSTAT 11 (San Jose, CA). Chi-square tests were used to determine whether genotype frequencies of the selected SNPs were in Hardy-Weinberg equilibrium. Because the DNA uracil concentration was not normally distributed, this variable was log transformed to achieve normality before fitting the statistical models. Likely covariates of DNA uracil concentration (plasma concentrations of folate, PLP, and vitamin B-12; riboflavin intakes; alcohol consumption; tobacco use; sex; and age) were assessed by using multiple linear regression, as were the associations between SNP genotypes and DNA uracil concentrations. In the latter analyses, the dependent variable was DNA uracil concentration, and the independent variables were the genotypes of each selected SNP. These analyses also allowed us to adjust for putative covariates and to test for interactions between each SNP genotype and B vitamin status. A general genetic model (ie, testing for the 3 possible genotypes) was used as a first step in our analyses, and then a specific genetic model (ie, a recessive or an additive model) was used as appropriate. The association between DNA uracil content and p53 strand breaks was examined with Pearson correlation coefficients. All data are reported as means ± SEMs, except when specified otherwise, using the untransformed data. P values ≤ 0.05 were considered statistically significant. When a Bonferroni correction for multiple comparisons was applied, a P value ≤ 0.0023 was considered significant (α was divided by 22 because one pair of SNPs among the 23 successfully assayed SNPs was in strong LD, see Results).
Population admixture and linkage disequilibrium analyses
Individual ancestry was calculated based on the genotypes of 100 informative ancestral markers in the BPRHS population by using the program STRUCTURE 2.2 with reference to the 3 ancestral populations: West African, European, and Taíno (50). Using the estimated admixture of each subject, we adjusted for population admixture in all statistical analyses. Pairwise LD among all successfully genotyped SNPs within each gene of interest was estimated as correlation coefficients by using the Haploview 4.1 program (51).
RESULTS
Characteristics of participants
Several demographic and biochemical characteristics differed significantly by sex (Table 1). In particular, BMI and plasma folate were significantly higher in women than in men (P < 0.05). In contrast, plasma Hcy and creatinine concentrations and energy and riboflavin intakes were higher in men than in women (P < 0.05). The percentage of subjects who reported smoking or drinking was also significantly higher in men than in women (P < 0.0001). Moreover, only 0.5% of the population had inadequate folate status, as defined by plasma folate <5 ng/mL (52). Similarly, the frequencies of subjects with deficient status in vitamins B-12 and B-6 were 3.3% and 10.7%, respectively [defined as plasma vitamin B-12 < 200 pg/mL (53) and as plasma PLP <20 nmol/L (54)] (data not shown).
Genotyping results
Of the 26 candidate SNPs, 23 were successfully assayed: 3 assays did not adequately amplify and distinguish both alleles. Minor allele frequencies of the 23 successfully genotyped SNPs are presented in Table 3 and ranged from 0.04 to 0.50. Six SNPs were not in Hardy-Weinberg equilibrium: 3 in DUT, 1 in MBD4, and 2 in UNG (Table 3). A total of 27 pairs of SNPs appeared to be either in strong (r2 ≥ 0.8) or in intermediate (0.1 ≤ r2 < 0.8) LD in this subset of the BPRHS population (see Supplemental Table 1 under “Supplemental data” in the online issue; within-gene LD only). In particular, 2 SNPs in DUT (rs4775748 and rs12592330) and 2 SNPs in UNG (rs1018784 and rs3219266) appeared to be in strong LD in this population (r2 = 0.83 and 0.86, respectively). Because the latter pairwise LD was >0.85, the corresponding pair of SNPs was studied together in subsequent analyses.
TABLE 3.
Observed minor allele frequencies (MAFs) and genotype counts for the 23 successfully genotyped single nucleotide polymorphisms (SNPs)
| Allele1 |
Genotype count |
|||||||
| Gene and SNP ID no. | 1 | 2 | 11 | 12 | 22 | Total | MAFs | HWE P2 |
| DUT | ||||||||
| rs8042136 | T | C | 381 | 48 | 0 | 429 | 0.06 | >0.05 |
| rs11637235 | C | T | 129 | 193 | 107 | 429 | 0.47 | 0.04 |
| rs4775748 | T | G | 275 | 131 | 25 | 431 | 0.21 | >0.05 |
| rs8033556 | G | A | 257 | 139 | 34 | 430 | 0.24 | 0.02 |
| rs12592330 | T | C | 295 | 115 | 20 | 430 | 0.18 | 0.05 |
| MBD4 | ||||||||
| rs10470431 | A | G | 138 | 214 | 76 | 428 | 0.43 | >0.05 |
| rs3138355 | G | A | 337 | 94 | 0 | 431 | 0.11 | 0.01 |
| rs140696 | C | T | 303 | 111 | 15 | 429 | 0.16 | >0.05 |
| rs4128190 | C | G | 360 | 67 | 2 | 429 | 0.08 | >0.05 |
| SMUG1 | ||||||||
| rs7296239 | T | C | 127 | 227 | 77 | 431 | 0.44 | >0.05 |
| rs2029166 | C | T | 172 | 206 | 50 | 428 | 0.36 | >0.05 |
| rs1994356 | G | A | 195 | 199 | 35 | 429 | 0.31 | >0.05 |
| TDG | ||||||||
| rs703657 | A | T | 156 | 217 | 56 | 429 | 0.38 | >0.05 |
| rs172814 | A | G | 288 | 124 | 15 | 427 | 0.18 | >0.05 |
| rs4135050 | T | A | 211 | 174 | 46 | 431 | 0.31 | >0.05 |
| rs4135054 | C | T | 315 | 105 | 8 | 428 | 0.14 | >0.05 |
| rs4135063 | C | T | 186 | 193 | 51 | 430 | 0.34 | >0.05 |
| rs2723877 | C | T | 331 | 96 | 4 | 431 | 0.12 | >0.05 |
| rs1866074 | T | C | 102 | 226 | 100 | 428 | 0.50 | >0.05 |
| UNG | ||||||||
| rs1018784 | C | T | 395 | 32 | 3 | 430 | 0.04 | 0.01 |
| rs3219266 | T | C | 389 | 38 | 3 | 430 | 0.05 | >0.05 |
| rs34259 | G | C | 224 | 169 | 33 | 426 | 0.28 | >0.05 |
| rs246085 | T | C | 346 | 76 | 11 | 430 | 0.11 | 0.01 |
Alleles 1 and 2 refer to the major and minor alleles, respectively, in the 431 participants of the Boston Puerto Rican Health Study population.
Test for Hardy-Weinberg equilibrium (HWE; P values, chi-square test). Genotypes are not in HWE when P < 0.05.
Effect of B vitamin status, alcohol/tobacco consumption, age, and sex on DNA uracil concentration
Plasma concentrations of folate, vitamin B-12, and vitamin B-6 and riboflavin intake were not correlated with uracil concentration in blood DNA (P > 0.05; data not shown). Age, alcohol consumption, and tobacco use were also not associated with uracil content (P > 0.05; data not shown). DNA uracil concentrations did not differ between men and women (Table 1, P = 0.141).
Association between variations in the uracil-processing genes and DNA uracil concentrations
Four of the tested SNPs in 3 of the uracil-processing genes showed significant associations with blood DNA uracil concentration at α = 0.05 in the BPRHS population: rs4775748, located in the downstream region of the DUT gene; rs2029166 and rs7296239, both located in the promoter region of the SMUG1 gene; and rs34259, located in the downstream region of the UNG gene (Figures 1–3). None of these associations remained significant when the Bonferroni correction was used at the α = 0.0023 level. The SNPs in DUT and SMUG1 conformed best to a recessive model (ie, the phenotype of heterozygous individuals did not differ from the phenotype of homozygous wild-type individuals). For the SNP in UNG, an additive model appeared to be a better fit, the variant allele showing an additive effect on the phenotype of interest. These 4 SNPs were all in Hardy-Weinberg equilibrium.
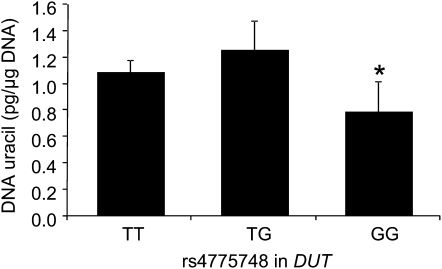
FIGURE 1.
Mean (±SEM) blood DNA uracil concentrations by TT (n = 275), TG (n = 131), and GG (n = 25) genotypes for the rs4775748 single nucleotide polymorphism in DUT. *Log uracil was significantly different (P = 0.029, general linear model) from the other 2 genotypes combined (recessive model). The P values were 0.023 after adjustment for age, sex, riboflavin intake, and plasma folate, vitamin B-12, and vitamin B-6 and 0.027 after further adjustment for population admixture.
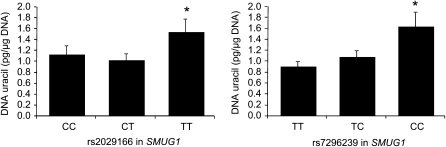
FIGURE 2.
Mean (±SEM) blood DNA uracil concentrations by genotype for the rs2029166 and rs7296239 single nucleotide polymorphisms (SNPs) in SMUG1. SNP rs2029166: n = 172, 206, and 50 for the CC, CT, and TT genotypes, respectively. SNP rs7296239: n = 127, 227, and 77 for the TT, TC, and CC genotypes, respectively. *Log uracil was significantly different (P < 0.05, general linear model) from the other 2 genotypes combined (recessive model). The unadjusted P values were 0.022 and 0.017 for SNPs rs2029166 and rs7296239, respectively. The P values were 0.011 and 0.022, respectively, after adjustment for age, sex, riboflavin intake, and plasma folate, vitamin B-12, and vitamin B-6 and 0.013 and 0.022, respectively, after further adjustment for population admixture.
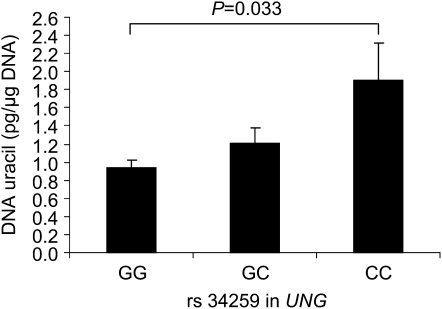
FIGURE 3.
Mean (±SEM) blood DNA uracil concentrations by GG (n = 224), GC (n = 169), and CC (n = 33) genotypes for the rs34259 single nucleotide polymorphism in UNG. Log uracil was significantly different (P for trend = 0.033, general linear model) between the 3 genotypes (additive model). The P values were 0.045 after adjustment for age, sex, riboflavin intake, and plasma folate, vitamin B-12, and vitamin B-6 and 0.064 after further adjustment for population admixture.
Homozygosity for the minor allele (G) of the rs4775748 SNP in DUT was associated with significantly lower uracil concentrations in blood DNA when compared with the pooled wild-type and heterozygote groups (Figure 1). More precisely, the uracil concentration was 31% lower (P = 0.029) in the GG subjects than in the pooled TT and TG subjects. This association remained significant after adjustment for age, sex, riboflavin intake, and plasma folate, vitamin B-12, and vitamin B-6 (P = 0.023) and after further adjustment for population admixture (P = 0.027). In strong LD with the SNP rs4775748 and 6.5 kb downstream, the SNP rs12592330 showed a similar association with DNA uracil concentration: ie, homozygotes for the minor allele C tended to have a lower uracil concentration (0.83 ± 0.28 compared with 1.12 ± 0.09 pg/μg DNA) than did carriers of the common allele (TC + TT), although it was not significant (P = 0.119 after adjustment for age, sex, riboflavin intake, and plasma folate, vitamin B-12, and vitamin B-6) (data not shown).
The variant genotypes for the 2 SNPs in SMUG1 were associated with a significant increase in the DNA uracil concentration (Figure 2). Individuals with the TT genotype for the rs2029166 SNP had a 43% increase (P = 0.022) in DNA uracil concentration compared with those with the CC or CT genotypes. For the rs7296239 SNP, the uracil concentration was 62% higher (P = 0.017) in the CC subjects than in the pooled TT and TC subjects. Both associations remained significant after correction for age, sex, riboflavin intake, and plasma folate, vitamin B-12, and vitamin B-6 (P = 0.011 and P = 0.022, respectively) and after further correction for population admixture (P = 0.013 and P = 0.022, respectively). These 2 SNPs in SMUG1 were in intermediate LD in the BPRHS population (r2 = 0.56; see Supplemental Table 1 under “Supplemental data” in the online issue).
DNA uracil concentration was also significantly different between the genotypes for the rs34259 SNP in the UNG gene (Figure 3). In comparison with homozygous individuals for the major allele (G), heterozygotes and homozygotes for the minor allele (C) had 28% and 103% higher uracil concentrations, respectively (P for trend = 0.033). As for the other SNPs, this association remained significant after adjustment for age, sex, riboflavin intake, and plasma folate, vitamin B-12, and vitamin B-6 (P = 0.045). However, the association was attenuated after further adjustment for population admixture (P = 0.064).
No significant association was found between any other SNP genotypes and uracil concentration in blood DNA in this population. We did not observe a significant effect of an interaction between B vitamin status and genotypes in any of the 4 identified SNPs on the uracil phenotype (data not shown).
Association between uracil concentrations in blood DNA and strand breaks within the p53 hypermutable region
We observed a significant positive correlation between uracil concentration in blood DNA and strand breaks measured in exon 8 of the p53 gene. Greater DNA uracil concentration was associated with a higher ΔCt value, which is indicative of lower template integrity (P = 0.012 with untransformed uracil and P = 0.035 with log-transformed uracil; data not shown). No correlation was found between DNA uracil concentration and strand breaks measured in exon 6 of the p53 gene (P > 0.05; data not shown).
DISCUSSION
The primary objective of the present study was to determine whether polymorphisms in the 5 uracil-processing genes are associated with DNA uracil concentrations, and furthermore, to determine whether the status of folate and other B vitamins can modify these associations.
To the best of our knowledge, this is the largest study of the determinants of DNA uracil content to date. The mean (± SD) amount of uracil detected in all of the DNA blood samples from our study was 1.12 ± 1.81 pg/μg DNA (range: ≈0.04–21.9 pg/μg DNA). The extremely large range of values did not seem to be due to analytic error because the between-run and within-run CVs were small (7% and 5%, respectively). It may rather be explained by the large interindividual variation of uracil among our population. This range of uracil values approximates previously reported values: 0.35–3.29 pg/μg DNA in human white blood cells (55) and 1–10.8 pg/μg DNA in cultured human lymphocytes (47, 56).
The primary observation in this study is the characterization of 4 SNPs at the DUT, UNG, and SMUG1 loci that are significantly associated with altered uracil concentrations in blood DNA. None of these associations passed the highly stringent Bonferroni correction test (P < 0.0023), but they were all significant at the α =0.05 level, and this was true despite the relatively modest sample size of our study. Moreover, these associations remained significant, or borderline significant, after further adjustment for population admixture, which is of particular importance in this genetically mixed population (35, 50).
We observed that the variant genotype for SNP rs4775748, located in the downstream region of the DUT gene, was associated with a decreased DNA uracil concentration (Figure 1). The DUT gene encodes for the human UTPase (4). Although functional data regarding this SNP is lacking, we can hypothesize that because it maps within the 3′-untranslated region, this SNP may modify the secondary structure of the mRNA, affecting mRNA stability or interactions with small RNAs. In this case, the variant genotype would result in increased synthesis of the UTPase and a reduction in uracil misincorporation. A similar mechanism may underlie the observed association between SNP rs34259 at the UNG locus and DNA uracil content, because this SNP is also located in the downstream region of the gene. We found that the variant allele (C) for this SNP was associated with significant stepwise increases in DNA uracil concentrations for each additional C allele (Figure 3). We propose that the variant allele of SNP rs34259 may modify the secondary structure of the UNG mRNA, leading in this case to a decreased synthesis of the UNG protein.
We also observed that 2 SNPs in the promoter region of the SMUG1 gene, rs2029166 and rs7296239, were associated with a significantly elevated DNA uracil concentration in the homozygous variant genotypes (Figure 2). The observation that the SNPs in SMUG1 were in intermediate LD (r2 = 0.56) is consistent with the fact that both variants were associated with uracil concentrations and reinforces our findings. Of the 4 uracil glycosylases that are known in humans, UNG and SMUG1 are thought to be the most efficient at the repair of misincorporated uracil, which is consistent with our observation that it was variants in these 2 enzymes that displayed phenotypes (3). Because the 2 SNPs in SMUG1 are located in the promoter region, we suggest that one or both of them may modify the degree of gene expression, which leads to a decreased expression of the SMUG1 protein with the variant genotype. Of particular interest, computational analysis suggests that SNP rs2029166 alters putative transcription factor binding sites for CDP and Sp3.
Although the mechanisms proposed to explain how the 4 characterized SNPs can affect the DNA uracil content are biologically plausible, we cannot exclude the possibility that these SNPs are nonfunctional but are in strong LD with other functional SNPs not selected for study, especially because they were all identified to be TagSNPs. Nevertheless, the observation that polymorphisms in the UNG, SMUG1, and DUT genes are associated with alterations in DNA uracil concentrations is potentially of great import, and it warrants confirmation in a larger cohort and definitive demonstration of the functional nature of the identified SNPs. If the 4 SNPs identified in this study are proven to be nonfunctional, the search for those SNPs that are genuinely causal should focus on SNPs that are tagged by those characterized in our study and that also have a high likelihood of being functional.
Contrary to one of our a priori hypotheses, we did not observe any associations between the systemic status of folate, vitamins B-6 and B-12, or riboflavin and DNA uracil concentrations. Nor did status of these B vitamins modify the effects of the 4 identified SNPs on the uracil phenotype. In retrospect, there are compelling reasons for this paucity of nutrient-uracil and nutrient-gene interactions. First, several recent human studies have failed to confirm a relation between folate status and uracil misincorporation (55, 57, 58), which suggests that the relation initially reported by Blount et al (14) was only possible because of the flagrant degree of folate deficiency evident in their subjects (RBC folate < 140 ng/mL) and because they exclusively used splenectomized subjects. As recently shown (59), such an RBC folate concentration exists in <1% of the US population; therefore, it is highly unlikely that the low folate status necessary to produce an elevation in uracil will be observed nowadays in a population-based study in the United States. Moreover, even in the context of nationwide folic acid fortification, our cohort had an exceptionally high B vitamin status. For instance, the mean concentration of plasma folate in our cohort was >60% higher than the median value of serum folate for adults in the United States in 2004 (59). It is entirely likely that the effect of B vitamins on uracil misincorporation is only detectable in a population with a significant number of individuals with lower B vitamin status. From our perspective, perhaps the most significant message conveyed by these results is that the effects of SNPs are sufficiently robust by themselves to create a biochemical phenotype, even within a study population that clearly had no significant limitation in the availability of B vitamins.
We also observed no association between alcohol consumption or tobacco use—2 lifestyle factors known to have a negative effect on one-carbon metabolism (18, 19)—and DNA uracil concentrations in this population. Although these factors might truly not affect uracil incorporation, our findings might instead indicate that the effects of alcohol and tobacco are minor enough to require a larger sample size to demonstrate an association or, alternatively, the effects might only be observed in the context of a population with more limited availability of one-carbon nutrients.
We also observed that DNA uracil concentration and strand breaks within exon 8 of the p53 gene were significantly correlated in this Puerto Rican population. This observation confirms earlier preclinical and human studies (13, 15, 16), which indicate that excess uracil in DNA is associated with a variety of metrics that reflect genetic instability, including strand breaks. Because increased DNA breakage is considered a mutagenic event, our results add evidence that increased uracil misincorporation in DNA may increase cancer risk.
Despite the many strengths of this study, several limitations existed that one must be mindful of when interpreting the data. First, individuals from the BPRHS population had a very high B vitamin status, which may have impeded our ability to observe an effect of B vitamins on the uracil phenotype. Second, most of the individuals in this cohort were women who had a very high prevalence of obesity. This questions the extent to which we can extrapolate these observations to other populations. Finally, because the current uracil assay is not conducive to high throughput analyses, the sample size of our study was relatively small for the purposes of molecular epidemiology. Nevertheless, these findings introduce some very novel and potentially important concepts and therefore merit further investigation.
In conclusion, we showed that 4 of 23 studied SNPs in 3 of the 5 uracil-processing genes were associated with altered DNA uracil concentrations and that the associations lacked any apparent interactions with the one-carbon nutrient status of individuals. This represents the first study to examine the biochemical phenotypes of SNPs in the 5 human uracil-processing genes. Future studies should be directed toward confirming the associations between the 4 SNPs and DNA uracil concentrations in a larger population, establishing the functional nature of the SNPs and further defining the relation between elevated DNA uracil content and the risk of cancer.
Supplementary Material
Acknowledgments
We thank Chao-Qiang Lai for his help with the population admixture and LD analyses, Jose Ordovas for facilitating the SNPs analyses, Yu-Chi Lee and Christine Gary for their technical support, and Gayle Petty and the Nutrition Evaluation Laboratory for the blood analyses.
The authors’ responsibilities were as follows—AC and EDC: genotyping reactions and uracil measurements; AC and LDP: SNP selection; ZL: p53 strand break measurements; AC, JWC, and JBM: manuscript preparation; AC and ZL: statistics; and KLT: human study. None of the authors had a conflict of interest.
REFERENCES
- 1.Mohrenweiser HW, Jones IM. Variation in DNA repair is a factor in cancer susceptibility: a paradigm for the promises and perils of individual and population risk estimation? Mutat Res 1998;400:15–24 [DOI] [PubMed] [Google Scholar]
- 2.Hung RJ, Hall J, Brennan P, Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol 2005;162:925–42 [DOI] [PubMed] [Google Scholar]
- 3.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev 2002;11:1513–30 [PubMed] [Google Scholar]
- 4.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet 2004;38:445–76 [DOI] [PubMed] [Google Scholar]
- 5.Dianov GL, Timchenko TV, Sinitsina OI, Kuzminov AV, Medvedev OA, Salganik RI. Repair of uracil residues closely spaced on the opposite strands of plasmid DNA results in double-strand break and deletion formation. Mol Gen Genet 1991;225:448–52 [DOI] [PubMed] [Google Scholar]
- 6.Moynahan ME, Jasin M. Loss of heterozygosity induced by a chromosomal double-strand break. Proc Natl Acad Sci USA 1997;94:8988–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott B, Jasin M. Double-strand breaks and translocations in cancer. Cell Mol Life Sci 2002;59:373–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YI, Shirwadkar S, Choi SW, Puchyr M, Wang Y, Mason JB. Effects of dietary folate on DNA strand breaks within mutation-prone exons of the p53 gene in rat colon. Gastroenterology 2000;119:151–61 [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Choi SW, Crott JW, et al. Mild depletion of dietary folate combined with other B vitamins alters multiple components of the Wnt pathway in mouse colon. J Nutr 2007;137:2701–8 [DOI] [PubMed] [Google Scholar]
- 10.Shin MC, Lee SJ, Choi JE, et al. Glu346Lys polymorphism in the methyl-CpG binding domain 4 gene and the risk of primary lung cancer. Jpn J Clin Oncol 2006;36:483–8 [DOI] [PubMed] [Google Scholar]
- 11.Miao R, Gu H, Liu H, et al. Tagging single nucleotide polymorphisms in MBD4 are associated with risk of lung cancer in a Chinese population. Lung Cancer 2008;62:281–6 [DOI] [PubMed] [Google Scholar]
- 12.Hao B, Wang H, Zhou K, et al. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res 2004;64:4378–84 [DOI] [PubMed] [Google Scholar]
- 13.Crott JW, Mashiyama ST, Ames BN, Fenech M. The effect of folic acid deficiency and MTHFR C677T polymorphism on chromosome damage in human lymphocytes in vitro. Cancer Epidemiol Biomarkers Prev 2001;10:1089–96 [PubMed] [Google Scholar]
- 14.Blount BC, Mack MM, Wehr CM, et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci USA 1997;94:3290–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duthie SJ, Grant G, Narayanan S. Increased uracil misincorporation in lymphocytes from folate-deficient rats. Br J Cancer 2000;83:1532–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linhart HG, Troen A, Bell GW, et al. Folate deficiency induces genomic uracil misincorporation and hypomethylation but does not increase DNA point mutations. Gastroenterology 2009;136:227–235 e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi SW, Friso S, Ghandour H, Bagley PJ, Selhub J, Mason JB. Vitamin B-12 deficiency induces anomalies of base substitution and methylation in the DNA of rat colonic epithelium. J Nutr 2004;134:750–5 [DOI] [PubMed] [Google Scholar]
- 18.Mason JB, Choi SW. Effects of alcohol on folate metabolism: implications for carcinogenesis. Alcohol 2005;35:235–41 [DOI] [PubMed] [Google Scholar]
- 19.Gabriel HE, Crott JW, Ghandour H, et al. Chronic cigarette smoking is associated with diminished folate status, altered folate form distribution, and increased genetic damage in the buccal mucosa of healthy adults. Am J Clin Nutr 2006;83:835–41 [DOI] [PubMed] [Google Scholar]
- 20.Choi SW, Mason JB. Folate and carcinogenesis: an integrated scheme. J Nutr 2000;130:129–32 [DOI] [PubMed] [Google Scholar]
- 21.Giovannucci E. Epidemiologic studies of folate and colorectal neoplasia: a review. J Nutr 2002;132:2350S–5S [DOI] [PubMed] [Google Scholar]
- 22.Zhang SM, Willett WC, Selhub J, et al. Plasma folate, vitamin B6, vitamin B12, homocysteine, and risk of breast cancer. J Natl Cancer Inst 2003;95:373–80 [DOI] [PubMed] [Google Scholar]
- 23.Rohan TE, Jain MG, Howe GR, Miller AB. Dietary folate consumption and breast cancer risk. J Natl Cancer Inst 2000;92:266–9 [DOI] [PubMed] [Google Scholar]
- 24.Baglietto L, English DR, Gertig DM, Hopper JL, Giles GG. Does dietary folate intake modify effect of alcohol consumption on breast cancer risk? Prospective cohort study. BMJ 2005;331:807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinstein SJ, Albanes D, Selhub J, et al. One-carbon metabolism biomarkers and risk of colon and rectal cancers. Cancer Epidemiol Biomarkers Prev 2008;17:3233–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J, Lee IM, Cook NR, et al. Plasma folate, vitamin B-6, vitamin B-12, and risk of breast cancer in women. Am J Clin Nutr 2008;87:734–43 [DOI] [PubMed] [Google Scholar]
- 27.Paspatis GA, Karamanolis DG. Folate supplementation and adenomatous colonic polyps. Dis Colon Rectum 1994;37:1340–1 [DOI] [PubMed] [Google Scholar]
- 28.Cole BF, Baron JA, Sandler RS, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA 2007;297:2351–9 [DOI] [PubMed] [Google Scholar]
- 29.Logan RF, Grainge MJ, Shepherd VC, Armitage NC, Muir KR. Aspirin and folic acid for the prevention of recurrent colorectal adenomas. Gastroenterology 2008;134:29–38 [DOI] [PubMed] [Google Scholar]
- 30.Jaszewski R, Misra S, Tobi M, et al. Folic acid supplementation inhibits recurrence of colorectal adenomas: a randomized chemoprevention trial. World J Gastroenterol 2008;14:4492–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulrich CM, Potter JD. Folate and cancer–timing is everything. JAMA 2007;297:2408–9 [DOI] [PubMed] [Google Scholar]
- 32.Han J, Hankinson SE, Zhang SM, De Vivo I, Hunter DJ. Interaction between genetic variations in DNA repair genes and plasma folate on breast cancer risk. Cancer Epidemiol Biomarkers Prev 2004;13:520–4 [PubMed] [Google Scholar]
- 33.Tucker KL. Stress and nutrition in relation to excess development of chronic disease in Puerto Rican adults living in the Northeastern USA. J Med Invest 2005;52(suppl):252–8 [DOI] [PubMed] [Google Scholar]
- 34.Tucker KL, Bianchi LA, Maras J, Bermudez OI. Adaptation of a food frequency questionnaire to assess diets of Puerto Rican and non-Hispanic adults. Am J Epidemiol 1998;148:507–18 [DOI] [PubMed] [Google Scholar]
- 35.Choudhry S, Coyle NE, Tang H, et al. Population stratification confounds genetic association studies among Latinos. Hum Genet 2006;118:652–64 [DOI] [PubMed] [Google Scholar]
- 36.The International HapMap Consortium A haplotype map of the human genome. Nature 2005;437:1299–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Internation HapMap Project. Homepage. Available from: http://www.hapmap.org (cited June2008).
- 38.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet 2005;37:1217–23 [DOI] [PubMed] [Google Scholar]
- 39.Marinescu VD, Kohane IS, Riva A. The MAPPER database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res 2005;33:D91–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai CQ, Tucker KL, Parnell LD, et al. PPARGC1A variation associated with DNA damage, diabetes, and cardiovascular diseases: the Boston Puerto Rican Health Study. Diabetes 2008;57:809–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bethke L, Murray A, Webb E, et al. Comprehensive analysis of DNA repair gene variants and risk of meningioma. J Natl Cancer Inst 2008;100:270–6 [DOI] [PubMed] [Google Scholar]
- 42.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol 2006;4:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal 1999;14:143–9 [DOI] [PubMed] [Google Scholar]
- 44.Camp VM, Chipponi J, Faraj BA. Radioenzymatic assay for direct measurement of plasma pyridoxal 5′-phosphate. Clin Chem 1983;29:642–4 [PubMed] [Google Scholar]
- 45.Araki A, Sako Y. Determination of free and total homocysteine in human plasma by high-performance liquid chromatography with fluorescence detection. J Chromatogr 1987;422:43–52 [DOI] [PubMed] [Google Scholar]
- 46.Blount BC, Ames BN. Analysis of uracil in DNA by gas chromatography-mass spectrometry. Anal Biochem 1994;219:195–200 [DOI] [PubMed] [Google Scholar]
- 47.Mashiyama ST, Courtemanche C, Elson-Schwab I, et al. Uracil in DNA, determined by an improved assay, is increased when deoxynucleosides are added to folate-deficient cultured human lymphocytes. Anal Biochem 2004;330:58–69 [DOI] [PubMed] [Google Scholar]
- 48.Pogribny IP, Basnakian AG, Miller BJ, Lopatina NG, Poirier LA, James SJ. Breaks in genomic DNA and within the p53 gene are associated with hypomethylation in livers of folate/methyl-deficient rats. Cancer Res 1995;55:1894–901 [PubMed] [Google Scholar]
- 49.Crott JW, Liu Z, Choi SW, Mason JB. Folate depletion in human lymphocytes up-regulates p53 expression despite marked induction of strand breaks in exons 5-8 of the gene. Mutat Res 2007;626:171–9 [DOI] [PubMed] [Google Scholar]
- 50.Lai CQ, Tucker KL, Choudhry S, et al. Population admixture associated with disease prevalence in the Boston Puerto Rican Health Study. Hum Genet 2009;125:199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–5 [DOI] [PubMed] [Google Scholar]
- 52.Selhub J, Jacques PF, Wilson PW, Rush D, Rosenberg IH. Vitamin status and intake as primary determinants of homocysteinemia in an elderly population. JAMA 1993;270:2693–8 [DOI] [PubMed] [Google Scholar]
- 53.Tucker KL, Rich S, Rosenberg I, et al. Plasma vitamin B-12 concentrations relate to intake source in the Framingham Offspring study. Am J Clin Nutr 2000;71:514–22 [DOI] [PubMed] [Google Scholar]
- 54.Haller J, Lowik MR, Ferry M, Ferro-Luzzi A. Nutritional status: blood vitamins A, E, B6, B12, folic acid and carotene. Euronut SENECA investigators. Eur J Clin Nutr 1991;45(suppl 3):63–82 [PubMed] [Google Scholar]
- 55.Ren J, Ulvik A, Refsum H, Ueland PM. Uracil in human DNA from subjects with normal and impaired folate status as determined by high-performance liquid chromatography-tandem mass spectrometry. Anal Chem 2002;74:295–9 [DOI] [PubMed] [Google Scholar]
- 56.Crott JW, Mashiyama ST, Ames BN, Fenech MF. Methylenetetrahydrofolate reductase C677T polymorphism does not alter folic acid deficiency-induced uracil incorporation into primary human lymphocyte DNA in vitro. Carcinogenesis 2001;22:1019–25 [DOI] [PubMed] [Google Scholar]
- 57.Kapiszewska M, Kalemba M, Wojciech U, Milewicz T. Uracil misincorporation into DNA of leukocytes of young women with positive folate balance depends on plasma vitamin B12 concentrations and methylenetetrahydrofolate reductase polymorphisms: a pilot study. J Nutr Biochem 2005;16:467–78 [DOI] [PubMed] [Google Scholar]
- 58.Narayanan S, McConnell J, Little J, et al. Associations between two common variants C677T and A1298C in the methylenetetrahydrofolate reductase gene and measures of folate metabolism and DNA stability (strand breaks, misincorporated uracil, and DNA methylation status) in human lymphocytes in vivo. Cancer Epidemiol Biomarkers Prev 2004;13:1436–43 [PubMed] [Google Scholar]
- 59.Pfeiffer CM, Johnson CL, Jain RB, et al. Trends in blood folate and vitamin B-12 concentrations in the United States, 1988 2004. Am J Clin Nutr 2007;86:718–27 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.