Abstract
The early Caenorhabditis elegans embryo contains abundant transcripts for two α- and two β-tubulins, raising the question of whether each isoform performs specialized functions or simply contributes to total tubulin levels. Our identification of two recessive, complementing alleles of a β-tubulin that disrupt nuclear-centrosome centration and rotation in the early embryo originally suggested that this tubulin, tbb-2, has specialized functions. However, embryos from tbb-2 deletion worms do not have defects in nuclear-centrosome centration and rotation suggesting that the complementing alleles are not null mutations. Both complementing alleles have distinct effects on microtubule dynamics and show allele-specific interactions with the two embryonically expressed α-tubulins: One of the alleles causes microtubules to be cold stable and resistant to the microtubule-depolymerizing drug benomyl, whereas the other causes cell cycle-specific defects in microtubule polymerization. Gene-specific RNA interference targeting all four embryonically expressed tubulin genes singly and in all double combinations showed that the tubulin isoforms in the early embryo are largely functionally redundant with the exception of tbb-2. tbb-2 is required for centrosome stabilization during anaphase of the first cell division, suggesting that tbb-2 may be specialized for interactions with the cell cortex.
INTRODUCTION
Most organisms have multiple α- and β-tubulins, which are the proteins that heterodimerize to form microtubules (MTs). The existence of different tubulin isoforms is attributed to both functional divergence between the isoforms and the need to maintain protein levels (Luduena, 1998). In yeast, investigators showed that the two α-tubulins in the genome are functionally interchangeable in the laboratory environment (Schatz et al., 1986), whereas in Drosophila, one β-tubulin isoform is unable to fully substitute for another, indicating specialization in function (Hoyle and Raff, 1990). In Caenorhabditis elegans, a β-tubulin, MEC-7, is required in six touch-receptor neurons to generate special 15 protofilament MTs needed for sensory transduction in touch cell processes (Savage et al., 1989), whereas worms mutant for ben-1, another β-tubulin, have no apparent phenotype except for resistance to the neurological phenotypes produced by exposure to the MT-depolymerizing drug benomyl (Driscoll et al., 1989). The expression of two α- and two β-tubulin genes (Baugh et al., 2003) and the diversity of MT-dependent events in the early C. elegans embryo (including meiosis, spindle positioning, and mitosis) raises the question whether there is any functional specification associated with the tubulin isoforms expressed in the embryo.
In specific blastomeres of the early embryo, spindle alignment with the axis of anteroposterior polarization is accomplished by active rotation of the nuclear-centrosome complex before spindle assembly. Perfusion of MT-depolymerizing drugs into early embryos inhibits nuclear-centrosome rotation in the one-cell embryo, P0, and the posterior blastomere of the two-cell embryo, P1, suggesting that MTs are essential for these rotations (Strome and Wood, 1983; Hyman and White, 1987). Using a laser to disrupt MTs in P1, Hyman (1989) discovered that MTs connecting the forward rotating centrosome and the cell cortex are important for rotation. Additionally, RNA interference (RNAi) experiments have demonstrated that dynein and components of the associated dynactin complex are required for centrosome rotation in P0 (Skop and White, 1998; Gonczy et al., 1999a). These observations precipitated the development of a theory that MT motors shorten MTs connecting the centrosome to the cell cortex, creating a force that causes the rotation of the nuclear-centrosome complex. LET-99, a protein asymmetrically localized in a band near the posterior of blastomeres where rotation occurs, may also contribute to centrosome rotation by modulating MT stability (Tsou et al., 2002).
Similar processes are required for nuclear movement in Saccharomyces cerevisiae during budding. Spindle orientation toward the bud before and during mitosis involves astral MTs contacting a cortical site in the bud, and the action of dynein and kinesin-like MT motor proteins (Schuyler and Pellman, 2001; Segal and Bloom, 2001). Furthermore, mutations in these motor proteins cause spindle orientation defects in part because they disrupt MT dynamics (Carminati and Stearns, 1997; Cottingham and Hoyt, 1997; DeZwaan et al., 1997). Consistent with these observations, a yeast tubulin mutant with nondynamic MTs fails to maintain correct spindle orientation (Gupta et al., 2002). These studies have focused attention on the importance of precise control of MT dynamics in spindle orientation.
Here, we describe the analysis of two C. elegans β-tubulin mutants. One allele of tbb-2 eliminates specific nuclear-centrosome rotations in the early embryo, whereas the other disrupts a wider range of MT-dependent events. This study of these tbb-2 mutants concentrated on two related issues. The first concerned how the altered β-tubulins affect MT function and the second on the apparent specialization of tubulin isoforms in the early embryo. We found that the two mutant β-tubulins differentially affect MT dynamics, cold stability, and sensitivity to MT-depolymerizing drugs and show allele-specific interactions with α-tubulins. And despite what was suggested by the tbb-2 mutant phenotypes, depleting specific α- and β-tubulin isoforms with RNAi revealed considerable functional redundancy in the early embryo, with the exception that tbb-2 but not tbb-1, seems to be specifically required to stabilize centrosomes during anaphase of the first cell division.
METHODS AND MATERIALS
Strains and Culture Conditions
Nematode strains were grown under standard culture conditions (Brenner, 1974) at 15°Cor25°C as needed. The following strains were used in this study: N2, wild-type Bristol; HC48, tbb-2(qt1) III; GE2255, tbb-2(t1623) unc32(e189)/qC1[dpy-19(e1259) glp-1(q339)] III; him-3(e1147) IV; CB4856, wild-type CB sub-clone of HA-8; MT696, nDf12/unc-93(e1500) dpy-17(e164) III; BC4697, sDf121(s2098) unc-32(e189) III; sDp3 (III;f); BC4637, sDf130(s2427) unc-32(e189) III; sDp3 (III;f); SP471, dpy-17(e164) unc-32(e189) III; HC55 tbb-2(qt1) dpy-17(e164) unc-32(e189) III; HC73, daf-2(e1370) tbb-2(qt1) dpy-17(e163) III; VC167, tbb-2(gk130) III; VC169, tbb-2(gk129)III; VC364, tbb-1(gk207)III, EU995, tbb-2(or362ts) dpy-17(e164) III. All HC strains were created in the course of this study, tbb-2(gk129), tbb-2(gk130), and tbb-1(gk207) were isolated by the C. elegans Knockout Consortium; EU995 was provided by G. Ellis and B. Bowerman (University of Oregon, Eugene, OR), whereas the remaining strains were acquired from the Caenorhabditis Genetics Center.
Isolation, Mapping, and Cloning of tbb-2(qt1) and tbb-2(t1623)
tbb-2(qt1) is an EMS-induced mutation isolated in a F2 clonal screen for temperature-sensitive, embryonic lethal mutations and was outcrossed two times before analysis. tbb-2(qt1) is a strict maternal effect lethal mutation because crossing wild-type male worms to tbb-2(qt1) hermaphrodites did not rescue the embryonic lethality (91% dead, n = 1599). tbb-2(qt1) mapped to the left of dpy-17 on linkage group III by using standard meiotic recombination methods. Deficiency mapping showed that tbb-2(qt1) complemented the deficiencies nDf12 and sDf121 and failed to complement Df130, which confined tbb-2(qt1) to a 1.5 map unit region. We used single nucleotide polymorphisms to further refine the map position to a 30-gene interval flanked by single nucleotide polymorphisms C03D11 (9767) and F37A8 (11354) (Wicks et al., 2001). We sequenced a β-tubulin gene, tbb-2 (C36E8.5), in the interval and found mutations in the qt1 and t1623 strains. tbb-2(t1623) was formerly called rot-2(t1623) (Gonczy et al., 1999b). tbb-2(qt1) and tbb-2(t1623) are complementing alleles (Table 1), but another allele of tbb-2, or326ts, fails to complement both qt1 and t1623 (our unpublished data; Ellis and Bowerman, personal communication).
Table 1.
RNAi primer sequences
| Primer name | Primer sequence |
|---|---|
| tba-1for | T7-CGATGGAACTATGCCATCAG |
| tba-1rev | T3-GGCTCAAGATCTACAAAAATCG |
| tba-1utrfor | T7-TCGTATACAACACAAGCGATG |
| tba-1utrrev | T3-AAACGAGGAGGAAGGAGAAG |
| tba-2for | T7-CGATGGAACCATGCCAACTC |
| tba-2rev | T3-GCTCGAGATCGACGAAGATG |
| tba-2utrfor | T7-TGTTTATGACGATCGAACTGC |
| tba-2utrrev | T3-AGGAGAAGAGGAGGGAGAG |
| tbb-1for | T7-CGACGAGACTTACTGTATCG |
| tbb-1rev | T3-AGCCAACTTGCGAAGATCAGC |
| tbb-1utrfor | T7-TTAATCATCGTTCATTCGAGTC |
| tbb-1utrrev | T3-CGAGCCACTCGACGAGTTC |
| tbb-2for | T7-ACCTACTGCATTGACAACG |
| tbb-2rev | T3-CTTGCGGAGATCGGCATTG |
| tbb-2utrfor | T7-ATGGAATTGAAGAGAGATTGTG |
| tbb-2utrrev | T3-CTGCCGAAGACGACGTCG |
T7-taatacgactcactataggg; T3-attaaccctcactaaaggga.
Embryo Observation and Centrosome Position Measurements
The percentage of embryonic lethality was determined by placing L4 hermaphrodites on individual plates at the appropriate temperature and determining the percentage of embryos they produced that failed to hatch. The complete genotype of worms referred to as tbb-2(t1623) is tbb-2(t1623) unc-32(e189). Observed tbb-2(t1623)/+ embryos were from tbb-2(t1623) unc-32(e189)/qC1[dpy-19(e1259) glp-1(q339)] adults, whereas the percentage of embryonic lethality was determined using tbb-2(t1623) unc32(e189)/+ adults. The genotype of the adults in the tbb-2(t1623)/tbb-2(qt1) complementation test was either tbb-2(t1623) unc-32(e189)/tbb-2(qt1)or tbb-2(t1623) unc-32(e189)/tbb-2(qt1) dpy-17(e164) unc-32(e189). Observed embryos were obtained from adults of the later genotype.
Unless otherwise noted, embryos were dissected from adults grown at the indicated temperature, mounted on 2% agarose pads, overlaid with a coverslip, and observed or recorded at 23°C on an Axiophot or Axioskop (Carl Zeiss, Thornwood, NY). To observe without pressure, some embryos were placed on a poly-l-lysine-coated coverslip and inverted over the space created by adhering two coverslips 0.5 inch apart. Differential interference contrast (DIC) time-lapse recordings (1 frame every 5 s) of select embryos were obtained using OpenLab 3.0 software (Improvision, Coventry, United Kingdom). For the centrosome position measurements, DIC time-lapse movies (1 frame every 1 s) of select embryos were captured during mitosis. After the spindle began to move to the posterior during anaphase, the position of the anterior and posterior centrosomes in every frame was determined with respect to a Cartesian coordinate grid using the OpenLab 3.0 software (Improvision). Centrosome position along the y-axis was translated into percentage of egg width and graphed over time. The difference in value (in percentage of egg width) between the minimum and maximum position along the y-axis for the anterior and posterior centrosomes for each embryo was calculated and averaged for all embryos of a similar genotype.
RNAi of Tubulin Genes
For each of the four tubulin genes targeted by RNAi (Fire et al., 1998), a fragment of DNA was amplified from embryonic cDNA for two distinct regions in the gene; one in the coding region and one in the 3′ untranslated region (UTR). We picked regions of each gene where exact nucleotide matches to other tubulin genes were no longer than 14 bp to reduce cross-interference. Primer sequences (Table 1) were also restricted to these regions. After amplification, polymerase chain reaction products were transcribed to produce single-stranded RNAs, which were annealed to form dsRNAs that were injected (1 μg/μl) into the gonad. The unique coding region and 3′ UTR RNAs produced the same phenotype for each gene. The coding region RNA was injected for most of the β-tubulin experiments, whereas the coding and 3′ UTR RNAs were coinjected (1 μg/μl each) for most of the α-tubulin experiments. For all double RNAi experiments, the coding region RNA for each gene was coinjected (1 μg/μl each). Worms were typically placed at 25°C after injection, and embryos were observed 23-34 h postinjection.
Tubulin Staining
Embryos in water on a poly-l-lysine slide were slightly squashed with pressure from a coverslip, permeabilized by freeze cracking, fixed for 10 min in room temperature methanol, and then air dried. To visualize MTs, embryos were stained with an α-tubulin antibody (T9026; Sigma-Aldrich, St. Louis, MO) diluted 1:200 in phosphate-buffered saline (100 mM phosphate, pH 9.9, 500 mM NaCl) with 0.1% Tween 20 and a rhodamine goat anti-mouse secondary antibody diluted 1:100. Hoechst 33258 was used to visualize DNA. Images of tubulin and Hoechst-stained embryos were collected on an Olympus IX70 and deconvolved using Delta Vision Soft Works 2.10 software (Applied Precision, Issaquah, WA).
Cold Destabilization Assays
This method was first described by Hannak et al., 2002. Briefly, embryos were extruded from 20 adults in 4 μl of water on a poly-l-lysine-coated slide by squashing the adults with an 18 × 18-mm coverslip. MTs were depolymerized by placing the slides on an aluminum plate in a water and ice bath for 0.5, 1, 2, 3, or 5 min, and in some cases, repolymerized by returning the slides to room temperature (∼22°C) for 0, 15, 30, or 90 s. After the specified amount of time, the slides were placed in liquid nitrogen and processed for α-tubulin staining as described above. A set of embryos incubated at room temperature served as a control. Images of tubulin and Hoechst stained embryos were collected on an Axiophot or Axioskop (Carl Zeiss) with OpenLab 3.0 software (Improvision).
Microtubule Inhibitor Experiments
Adults were cut open to extrude embryos into embryonic culture media (ECM; 4% sucrose, 0.1 M NaCl) alone or ECM with 10 μg/ml benomyl (gift from A. Murray, Harvard University, Cambridge, MA), 10 μg/ml vinblastine (V1377; Sigma-Aldrich), or 50 μg/ml colchicine (C3915; Sigma-Aldrich). The benomyl and ECM alone solutions also contained 0.01% dimethyl sulfoxide. Embryos were transferred to a poly-l-lysine-coated coverslip that was inverted over a space formed by adhering two coverslips 0.5 inch apart to a microscope slide. The appropriate culture media was added to the space. A hole was then punched in the coverslip above a two- or “three-” cell embryo by using a laser. The glass shards from the coverslip punctured the embryonic eggshell, causing extrusion of one cell and allowing the culture media containing the drug to enter the embryo. The effect of the drug on the nuclear division of the intact cells was scored. Temperature refers to both the temperature at which adult worms were grown and the temperature at which the experiments were done.
RESULTS
tbb-2(qt1) Embryos Have Defects in Nuclear-Centrosome Centration and Rotation
tbb-2(qt1) was isolated in a genetic screen for temperature-sensitive, embryonic-lethal mutants that mislocalize the cell fate determinant PAL-1 in the early embryo. Because tbb-2(qt1) is a recessive (Table 2), maternal effect mutation, embryos from homozygous mothers are referred to as tbb-2(qt1) embryos for simplicity. At 15°C, 5% of tbb-2(qt1) embryos are inviable, whereas at 25°C, 96% of the embryos are inviable (Table 2). Observations of early cell divisions in tbb-2(qt1) embryos indicated that the PAL-1 mislocalization phenotype results from a major disruption of the first cell division. After fertilization in wild-type embryos, the maternal pronucleus migrates to meet the paternal pronucleus and its associated centrosomes in the posterior of the embryo (Figure 1, A and B). Initially the centrosomes are aligned in a transverse orientation, but after pronuclear meeting, the pronuclear-centrosome complex centers and undergoes a 90° rotation so that the centrosomes and eventually the first mitotic spindle are aligned longitudinally along the anterior-posterior axis (Figure 1, B-E; Albertson, 1984). In the tbb-2(qt1) embryos, the pronuclear-centrosome complex fails to center and rotate causing the first spindle to set up in a transverse orientation in the posterior of the embryo (Figure 1, F-I, and Table 2). During cytokinesis, two cleavage planes ingress, one bisecting the transverse spindle and the other in the normal orientation but anteriorly displaced. The result is a three-cell embryo with two nucleated cells in the posterior and an anucleate anterior cytoplast (Figure 1J). One of the posterior cells often fuses with the anterior cytoplast to produce a normal-looking “two-cell” embryo. In one-third of tbb-2(qt1) embryos, the maternal pronucleus fails to migrate to meet the paternal pronucleus (Table 2). However, the centrosomes associated with the haploid paternal pronucleus still assemble a transverse spindle in the posterior. The maternal chromosomes either join the spindle late or segregate randomly to produce multinucleated cells.
Table 2.
Effects of tbb-2 mutations on the first cell division
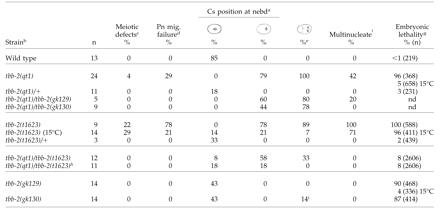
Cs, centrosome; nebd, nuclear envelope breakdown; pn, pronuclear; na, not applicable; nd, not determined.
As nebd occurred, the orientation of the centrosomes (transverse, diagonal, longitudinal) and extent of centrosome/pronuclear complex centering (center, rear) in each embryo was noted. Only the percentage of the embryos that completely centered and rotated (4th column) or completely failed to center and rotate (5th column) are shown
All embryos are from animals placed at 25°C at least 24 hrs before observation and were observed slightly squashed by a coverslip unless otherwise noted
Percentage of embryos with meiotic defects as determined by the presence of additional pronuclei
Percentage of embryos in which the maternal pronucleus failed to complete migration prior to ncbd
Percentage of embryos that divided transversely to form a three-cell embryo
Percentage of embryos that had multinucleated cells after the first cell division due to defects in meiosis, pronuclear migration, and/or chromosomal segregation
Embryonic lethality was determined at 25°C unless otherwise noted
Embryos observed without coverslip pressure (see Materials and Methods)
Vigorous rocking turned the spindle transverse during anaphase in these two embryos
Figure 1.
tbb-2(qt1) and tbb-2(t1623) embryos have defects in P0 nuclear-centrosome centration and rotation. DIC images of the first cell division in wild-type and mutant embryos. (A-E) Wild-type embryo from an adult raised at 25°C. During pseudocleavage, the maternal pronucleus (M) migrates to meet the paternal pronucleus (P) in the posterior (A). After pronuclear meeting (B), the pronuclear-centrosome complex moves to the center of the embryo and undergoes a 90° rotation so that the first mitotic spindle is aligned longitudinally (C). A cleavage plane (D) divides the cell into a larger anterior cell, AB, and a smaller posterior cell, P1 (E). (F-J) tbb-2(qt1) embryo from an adult raised at 25°C. Pronuclear migration (F) and pronuclear meeting (G) are normal, but the pronuclear-centrosome complex fails to center and rotate, resulting in a transverse mitotic spindle in the rear of the embryo (H). Two cleavage planes bisect the embryo (I), creating an anterior anucleate cytoplast and two posterior nucleated cells (J). (K-O) tbb-2(t1623) embryo from an adult raised at 15°C. Pronuclear migration (K) and pronuclear meeting (L) are normal, but the pronuclear-centrosome complex fails to center and completely rotate, resulting in a diagonal mitotic spindle in the rear of the embryo (M) that subsequently skews onto the longitudinal axis during anaphase (N). A cleavage plane (N) divides the cell (O). Note the two nuclei in the anterior cell, indicating chromosomal segregation defects. (P-T) tbb-2(t1623) embryo from an adult raised at 25°C. Pronuclear migration and meeting fail (P and Q) and the paternal pronuclear-centrosome complex fails to center and rotate resulting in a transverse mitotic spindle in the rear of the embryo (R). Two cleavage planes bisect the embryo (S) creating an anterior cytoplast and two posterior cells (T). In this embryo, maternal chromosomes were randomly segregated to the anterior cell. Anterior is left. The arrowheads indicate visible centrosomes and spindle poles. Bar, 10 μm.
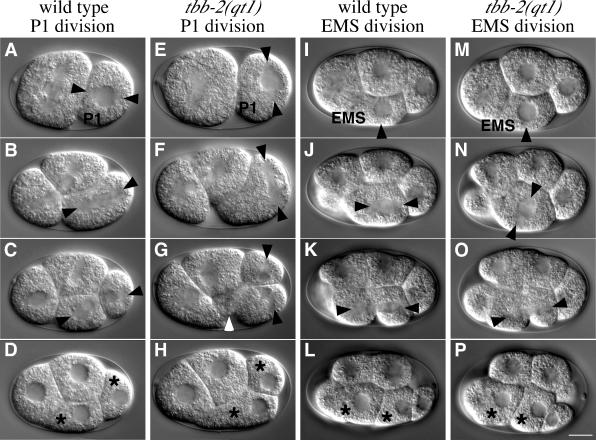
In wild-type embryos, nuclear-centrosome rotation also occurs in the P1, EMS, P2, and P3 blastomeres (Figure 2,A-D, I-L; Hyman and White, 1987). Because tbb-2(qt1) is temperature sensitive, it is possible to examine whether these later rotation events are perturbed in the mutant embryos. When two-cell tbb-2(qt1) embryos were shifted to the restrictive temperature, 100% of the embryos failed to undergo nuclear-centrosome rotation in P1, resulting in a transverse spindle and embryonic lethality (n = 11) (Figure 2, E-H). Similar to the P0 division, the spindle was positioned in the extreme posterior of P1, and 18% of the P1 cells produced an anterior cytoplast (n = 11) (Figure 2G). A similar experiment was performed to investigate nuclear-centrosome rotation in EMS by shifting four-cell embryos to the restrictive temperature. Nuclear-centrosome rotation in EMS failed in 79% of the embryos (n = 14) (Figure 2, M and N). However, as the transverse spindle elongated during anaphase, it skewed, apparently due to eggshell constraints, to assume a longitudinal orientation that resulted in viable embryos (Figure 2, O and P). Nuclear-centrosome rotation in P2 is subtle, and we could not discern any defects in the P2 divisions of up-shifted tbb-2(qt1) embryos. We did not examine rotation in the P3 blastomere. Consistent with the tbb-2(qt1) mutation not compromising development at or after the four-cell stage, 100% of tbb-2(qt1) embryos shifted to the restrictive temperature at the four-cell stage produced viable L1s (n = 22), and L1s shifted to the restrictive temperature produced normal, fertile adults. This suggests that the effects of the tbb-2(qt1) mutation are limited to the early, large blastomeres.
Figure 2.
tbb-2(qt1) embryos have defects in nuclear-centrosome rotation in P1 and EMS blastomeres. DIC images of P1 and EMS blastomere divisions in wild-type and tbb-2(qt1) embryos. P1 is the posterior blastomere in the two-cell embryo and EMS is the ventral blastomere in the four-cell embryo. tbb-2(qt1) embryos were shifted to the restrictive temperature at the two-cell stage before examining P1 divisions and at the four-cell stage before examining EMS divisions. (A-D) Wild-type P2 division. The centrosomes have rotated (A) and produce a longitudinal mitotic spindle (B). P1 is asymmetrically cleaved into a larger ventral cell, EMS, and a smaller posterior cell, P2 (C and D). (E-H) tbb-2(qt1) P2 division. The centrosomes failed to rotate (E) and produce a transverse mitotic spindle in the posterior of the cell (F). Two cleavage planes initially create two nucleated cells and an anterior cytoplast (white arrowhead) (G) that is later reabsorbed into the ventral most cell (H). (I-L) Wild-type EMS division. Transversely aligned centrosomes (I) rotate to align the subsequent spindle longitudinally (J). A cleavage furrow (K) divides the cell into two daughter cells (L). (M-P) tbb-2(qt1) EMS division. Transversely aligned centrosomes (M) fail to rotate producing a transversely aligned mitotic spindle (N). However, during anaphase, the spindle skews onto the longitudinal axis (O) and a cleavage plane divides the cell into two daughter cells (P). This embryo was viable. Anterior is left, ventral is down. Resulting daughter cells are indicated with stars, visible centrosomes and spindle poles are indicated with arrowheads. Bar, 10 μm.
tbb-2(t1623) Disrupts Additional Tubulin-dependent Functions in the Early Embryo
We obtained another allele of tbb-2, t1623 (Gonczy et al., 1999b). Like tbb-2(qt1), tbb-2(t1623) is a recessive (Table 2), maternal effect mutation, and embryos from homozygous tbb-2(t1623) mothers will be referred to as tbb-2(t1623) embryos. At 15 and 25°C, 96 and 100% of tbb-2(t1623) embryos are inviable, respectively. The defects in one-cell embryos from adults raised at 25°C are more severe than those observed in embryos from adults raised at 15°C (Table 2 and Figure 1, K-T). For example, 21% of 15°C tbb-2(t1623) embryos fail to undergo pronuclear migration, whereas 78% of the 25°C embryos fail to undergo pronuclear migration (Figure 1, P and Q). Nuclear-centrosome centration and rotation fail to occur normally at either temperature, but the spindles of embryos from adults grown at 15°C orient longitudinally during anaphase (Figure 1, M and N). Thus, most of the 15°C tbb-2(t1623) embryos divide longitudinally into two-cell embryos (Figure 1O), whereas almost all the 25°C tbb-2(t1623) embryos divide transversely into three-cell embryos (Figure 1T). In addition, tbb-2(t1623) embryos at both temperatures have meiotic defects and extensive chromosomal segregation defects (Table 2). Because the first division is defective in most tbb-2(t1623) embryos due to rotation failure or chromosome segregation defects, nuclear-centrosome rotation in P1 and EMS could not be evaluated. Compared with the tbb-2(qt1) mutation, the tbb-2(t1623) mutation affects more MT-dependent events and more strongly compromises MT function.
tbb-2(qt1) and tbb-2(t1623) Are Complementing, Missense Mutations in a β-Tubulin
We mapped qt1 to a small region on linkage group III that contains a β-tubulin gene, tbb-2 (see MATERIALS AND METHODS). We sequenced the tbb-2 open reading frame in the tbb-2(qt1) strain, and found a G-to-A transversion at position 593 that changes amino acid 198 from glutamic acid to lysine. In the tbb-2(t1623) strain, we found a G-to-A transversion at position 937 that changes amino acid 313 from valine to methionine. Although tbb-2(qt1) and tbb-2(t1623) are both alleles of tbb-2, worms heterozygous for both mutations produce viable progeny, indicating intragenic complementation (Table 2). When observing embryos from the double heterozygotes, we saw no defects in meiosis or pronuclear migration, but the nuclear-centrosome complex failed to undergo complete centration and rotation in many embryos with the spindle skewing onto the diagonal or longitudinal axis during anaphase to allow a longitudinal division. Under normal observation conditions, when embryos are squashed slightly by the coverslip, 33% of the double heterozygote embryos divided transversely, which contradicts the 8% lethality rate (Table 2). However, when the embryos were observed without squashing, all the embryos eventually divided longitudinally indicating that reorientation during anaphase is sensitive to coverslip pressure (Table 2).
tbb-2(qt1) and tbb-2(t1623) Are Not Null Mutations
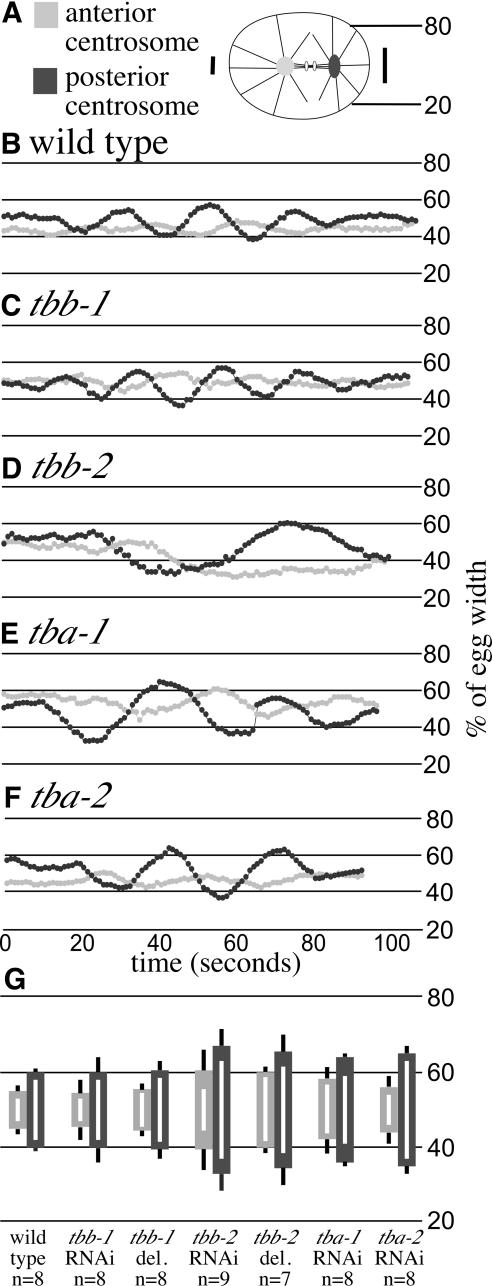
To investigate whether the tbb-2(qt1) or tbb-2(t1623) mutations cause a null or strong loss-of-function phenotype, we used RNAi (Fire et al., 1998) to deplete the tbb-2 gene product from wild-type embryos. Because β-tubulin genes are highly conserved at the nucleotide level, we chose regions of sequence unique to tbb-2 for RNAi (see MATERIALS AND METHODS). Previous RNAi of tbb-2 in C. elegans resulted in embryos with no spindles (Gonczy et al., 2000), likely due to RNAi cross-interference with other β-tubulin genes. We found the tbb-2(RNAi) animals produced embryos with fairly normal early divisions, yet these embryos failed to hatch. All MT-dependent processes disrupted in tbb-2(qt1)or tbb-2(t1623) embryos were wild-type in the tbb-2(RNAi) embryos (Table 3). However, a majority of the tbb-2(RNAi) embryos showed defects in centrosome stabilization during anaphase of the first mitotic division. Centrosome movement in wild-type and RNAi embryos was analyzed by translating centrosome position to percentage of egg width and plotting it over time (Figure 3). During anaphase in wild-type embryos, the anterior centrosome is relatively stable, whereas the posterior centrosome oscillates transversely (Albertson, 1984; Figure 3, A and B; Video 1). Compared with wild type, the anterior centrosome of tbb-2-depleted embryos moved, on average, through 100% more embryo space, whereas the posterior centrosome moved through 70% more embryo space (Figure 3G). In addition, the periodicity of the oscillations was lacking in >50% of the tbb-2-depleted embryos.
Table 3.
Effects of RNAi of α- and β-tubulin genes on the first cell division of wild-type embryos
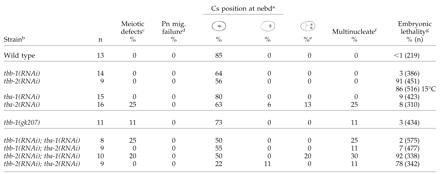
See Table 2 for all footnote clarifications
See Table 2 for all footnote clarifications
See Table 2 for all footnote clarifications
See Table 2 for all footnote clarifications
See Table 2 for all footnote clarifications
See Table 2 for all footnote clarifications
See Table 2 for all footnote clarifications
Figure 3.
tbb-2 RNAi and deletion embryos have defects in spindle pole stabilization during anaphase of the first cell division. (A) Schematic of anaphase during the first cell division in a wild-type embryo with 20 and 80% egg width marked. Vertical lines indicate the relative movement of the anterior and posterior centrosomes during anaphase. (B-F) Position of the anterior and posterior centrosomes in representative wild-type, mutant, and RNAi embryos during anaphase. Centrosome position on the y-axis was translated into percentage of egg width and plotted over time. This allows pictorial representation of the oscillation of the spindle poles in wild-type (B), tbb-1(RNAi) (C), tbb-2 deletion (D), tba-1(RNAi) (E), and tbb-2(RNAi) (F) embryos. The movies that the measurements were taken from correspond to Videos 1, 2, 3, 4, and 5. (G) The difference (in percentage of egg width) between the most extreme positions of each spindle pole on the y-axis within a genotype was calculated, averaged, and graphed centered on 50% egg width. The thick light gray bars represent the anterior centrosome average, whereas the dark gray bars represent the posterior centrosomes average. The black-and-white lines indicate the highest and lowest differences observed for the anterior and posterior centrosomes within each genotype, respectively.
The lack of tbb-2(RNAi) phenotypes similar to those caused by the tbb-2(qt1) or tbb-2(t1623) mutations led us to question whether the RNAi treatment was effective. Therefore, we injected the tbb-2 double-stranded RNA into tbb-2(qt1) and tbb-2(t1623) hermaphrodites. The early MT-dependent events in most progeny from the injected hermaphrodites were normal, but the embryos still displayed the centrosome stabilization defect characteristic of tbb-2(RNAi) embryos and died during embryogenesis (Table 4). Thus, we presume that tbb-2(qt1) and tbb-2(t1623) are gain-of-function mutations and the centrosome stabilization phenotype is the tbb-2 null phenotype.
Table 4.
Effects of RNAi of α- and β-tubulin genes on the first cell division of tbb-2 mutant embryos
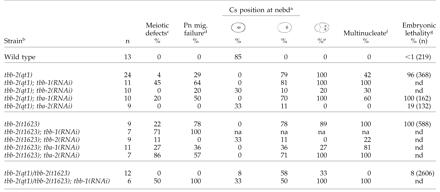
See Table 2 for all footnote clarifications
See Table 2 for all footnote clarifications
See Table 2 for all footnote clarifications
See Table 2 for all footnote clarifications
See Table 2 for all footnote clarifications
See Table 2 for all footnote clarifications
See Table 2 for all footnote clarifications
To confirm the tbb-2 null phenotype, deletion alleles of tbb-2 were obtained from the C. elegans Knockout Consortium. Both deletions, tbb-2(gk129) and tbb-2(gk130) remove the start codon of TBB-2 and TBB-2 protein is not detectable by Western analysis (Ellis and Bowerman, personal communication). The deletion strains show a temperature-sensitive embryonic lethality. At 15°C, hermaphrodites homozygous for either tbb-2 deletion produce viable embryos (Table 2). At 25°C, a large portion of the embryos fail to hatch, but similar to the tbb-2(RNAi), the first cell division seems normal except for the centrosome stabilization phenotype (Table 2 and Figure 3, D and G; Video 3).
tbb-2(t1623) Disrupts a Variety of MT Structures
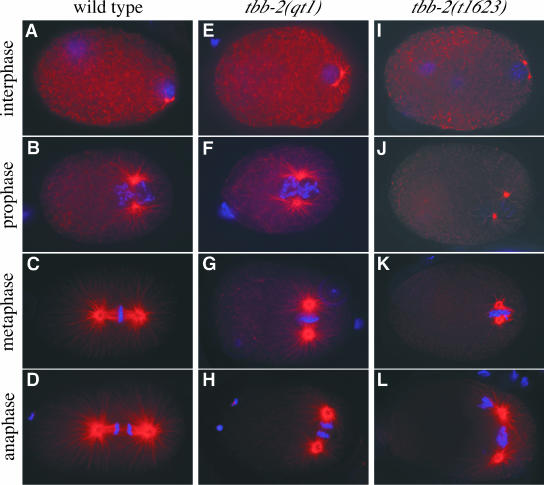
We visualized MTs in wild-type and mutant embryos by immunofluorescence by using an α-tubulin antibody (Figure 4). In tbb-2(qt1) embryos, MT organization looked normal except for the transverse orientation of the mitotic spindle (Figure 4, E-H). The maximum length of astral MTs during metaphase and anaphase was shorter in tbb-2(qt1) embryos (Figure 4, G and H) than in wild-type embryos, but this may be a direct result of reduced available space, because the spindle is aligned along the shortest axis of the embryo. MT structure and organization in tbb-2(qt1) embryos did not suggest an obvious reason for the failure of nuclear-centrosome rotation.
Figure 4.
tbb-2(t1623) embryos have defects in astral and spindle microtubules. α-Tubulin immunofluorescence (orange) and DNA staining (blue) of wild-type (A-D), tbb-2(qt1) (E-H), and tbb-2(t1623) embryos (I-L). (A, E, and I) Interphase. Duplication of the centrosomes associated with the paternal pronucleus. Note the two maternal pronuclei in the tbb-2(t1623) embryo (I). (B, F, and J) Prophase. Asters continue to enlarge after pronuclear meeting in wild-type (B) and tbb-2(qt1) (F) embryos, but not to the same degree in tbb-2(t1623) embryos (J). Note in J that the centrosomes seem disassociated from the pronuclei. (C, G, and K) Metaphase. In wild-type (C) and tbb-2(qt1) (G) embryos, the chromosomes have congressed on the metaphase plate, although the smaller tbb-2(qt1) spindle is orientated incorrectly. The MTs in the tbb-2(t1623) embryo (K) remain short and chromosomes are trapped, as opposed to aligned, between the centrosomes. tbb-2(t1623) embryos at this stage often have peripherally positioned chromosomes. (D, H, and L) Anaphase. In wild-type (D) and tbb-2(qt1) (H) embryos, the chromosomes have segregated into discrete bundles. In the tbb-2(t1623) embryo (L), the MTs have dramatically increased in length since metaphase, but the resulting spindle is disorganized and contains both anaphase bridges and DNA excluded from the spindle.
In contrast, tbb-2(t1623) embryos showed extensive defects in MT organization. In α-tubulin stained tbb-2(t1623) embryos from adults raised at 15 and 25°C, we saw similar patterns of MT organization. The meiotic spindle seemed wild type, but we observed embryos with multiple pronuclei (Figure 4I) and large polar bodies suggestive of meiotic defects. Although the interphase MT mesh was intact and the centrosome duplicated as expected (Figure 4I), the centrosomes were occasionally observed to be disassociated from the nucleus/chromosomes later in the cell cycle (Figure 4J). Determining subsequent cell-cycle stages was difficult because of the extensive defects in MT organization and spindle formation. In contrast to wild-type and tbb-2(qt1) embryos, where the asters are enlarged in prophase (Figure 4, B and F) and spindles are fully developed by metaphase (Figure 4, C and G), in tbb-2(t1623) embryos, MTs were significantly shorter and sparser in prophase and “metaphase” (Figure 4, J and K). Surprisingly, embryos at “anaphase” had large asters that extended to the cell cortex, suggesting either a delay in MT polymerization or a cell cycle-dependent defect. Spindles associated with these large asters were disorganized and contained varying amounts of kinetochore MTs, such that chromosomes were often excluded from the central spindle and anaphase bridges formed (Figure 4L). The increase in astral MT length between metaphase and anaphase was confirmed by time-lapse observation of tbb-2(t1623) embryos expressing green fluorescent protein:tubulin (our unpublished data). The abnormally short MTs at prophase when nuclear-centrosome rotation normally occurs may directly contribute to the tbb-2(t1623) phenotype.
tbb-2(qt1) Affects MT Dynamics
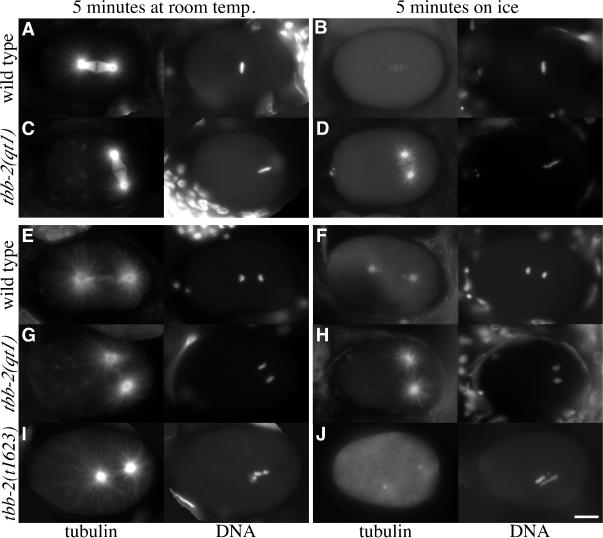
To assay for other alterations in MT behavior, we observed the effect of cold-induced depolymerization and subsequent repolymerization on wild-type and mutant embryos. Embryos were incubated on ice to depolymerize most MTs and then fixed and stained for α-tubulin at intervals after return to room temperature. Astral MTs in wild-type embryos at metaphase were undetectable after 5 min on ice (Figure 5B) but grew back rapidly when returned to room temperature (our unpublished data; Hannak et al., 2002). In contrast, in metaphase tbb-2(qt1) embryos, astral and kinetochore MTs associated with the centrosomes were obvious after cold destabilization (Figure 5D). At anaphase, short kinetochore MTs and occasionally short, sparse astral MTs were visible in wild-type embryos (Figure 5F), whereas the tbb-2(qt1) embryos had kinetochore and extensive astral MTs (Figure 5H). These results suggest that the tbb-2(qt1) mutation stabilizes MTs. In contrast, MTs in tbb-2(t1623) mutant embryos were indistinguishable from wild-type after cold destabilization (Figure 5J). On MT reassembly at room temperature, the tbb-2(qt1) spindle returns to full size, whereas two classes of tbb-2(t1623) embryos are apparent that mirror the phenotypes observed during normal growth: embryos at anaphase that contain larger but mispositioned MT structures and embryos at prophase or metaphase that contain very short MTs (our unpublished data).
Figure 5.
tbb-2(qt1) microtubules are present after cold destabilization. Tubulin and DNA staining of wild-type (A and B) and tbb-2(qt1) (C and D) embryos at metaphase and wild-type (E and F), tbb-2(qt1) (G and H), and tbb-2(t1623) (I and J) embryos at anaphase with and without cold destabilization. After 5 min on ice, only centrosomes and very short kinetochore MTs are visible in metaphase-stage wild-type embryos (B). In contrast, astral MTs as well as kinetochore MTs are visible in metaphase-stage tbb-2(qt1) embryos (D). In the anaphase stage wild-type embryo, short kinetochore MTs and very short, sparse astral MTs are visible after cold destabilization (F), whereas nearly full-length astral and kinetochore MTs are present in the anaphase stage tbb-2(qt1) embryo (H). In the tbb-2(t1623) anaphase embryo (J), the MTs have depolymerized so that only the centrosomes and some faint astral MTs remain.
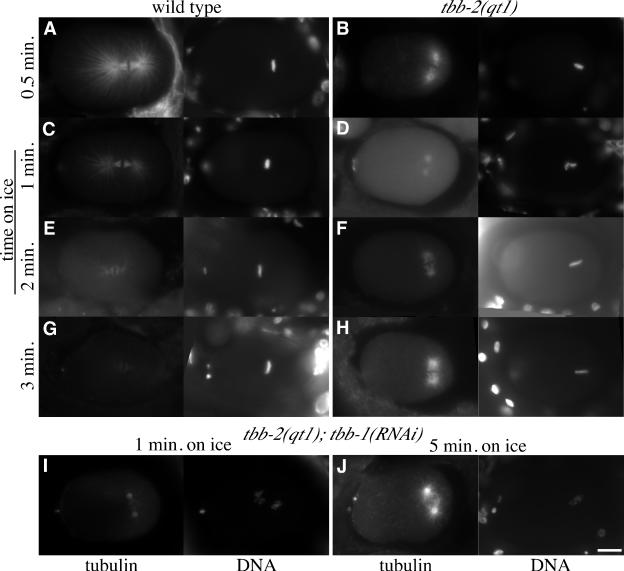
Because tbb-2(qt1) seems to cause MTs to be partially cold stable, we investigated the kinetics of depolymerization. MTs were examined after the embryos had been on ice for 0.5, 1, 2, or 3 min. We found that MTs in metaphase stage tbb-2(qt1) embryos initially depolymerized almost completely and then reassembled to form the asters visible after 5 min on ice (Figure 6, B, D, F, and H). In contrast, MTs in metaphase stage wild-type embryos only depolymerized (Figure 6, A, C, E, and G). MTs in anaphase stage embryos did not depolymerize to the extent seen in metaphase stage embryos in either genotype. We infer that after initial depolymerization, the tbb-2(qt1) MTs are capable of repolymerizing in the cold. It is possible that the initial MT disassembly reflects a requirement that the reassembled, cold-stable MTs contain predominantly qt1 β-tubulin. To test this, we asked whether MTs that already contained predominantly qt1 β-tubulin would be cold-stable without a reassembly step by repeating the experiment with tbb-2(qt1);tbb-1(RNAi) embryos. We found that after 1 and 5 min on ice, MTs in metaphase tbb-2(qt1); tbb-1(RNAi) embryos were indistinguishable from the MTs of tbb-2(qt1) embryos (Figure 6, I and J). (RNAi of tbb-1 in wild-type worms will be discussed below). This suggests that the assembly-disassembly process is not required to change the composition of the MTs but that the observed cold stability partially reflects an assembly-dependent modification in MT structure.
Figure 6.
Microtubules in tbb-2(qt1) embryos on ice initially depolymerize then reassemble. (A-H) Tubulin and DNA staining of wild-type and tbb-2(qt1) embryos incubated on ice for 0.5, 1, 2, or 3 min. With increasing time on ice, astral MTs in wild-type embryos (A, C, E, and G) get progressively shorter until they are essentially gone and only short kinetochore MTs remain (G). See Figure 5B for an image of a wild-type embryo incubated on ice for 5 min. In tbb-2(qt1) embryos, astral MTs rapidly decrease in length, with minimal astral MT length occurring after 1 min on ice (D), and then the MTs regrow, achieving a maximal length just short of control embryos after 5 min on ice. See Figure 5D for an image of a tbb-2(qt1) embryo incubated on ice for 5 min. (I-J) Tubulin and DNA staining of tbb-2(qt1);tbb-1(RNAi) embryos incubated on ice for 1 and 5 min. tbb-2(qt1);tbb-1(RNAi) embryos behave like tbb-2(qt1) embryos with astral MTs depolymerizing up to 1 min on ice (I), and then repolymerizing (J).
tbb-2(qt1) Embryos Are Resistant to Benomyl
The tbb-2(qt1) mutation changes the highly conserved glutamic acid at position 198 to lysine. Mutations that change this glutamic acid to lysine, alanine, glycine, or asperagine have been documented in 16 different fungi, including Aspergillus, Neurospora, Penicillum, and Saccharomyces (Fujimura et al., 1992; Jung et al., 1992; Koenraadt et al., 1992; Yarden and Katan, 1993; Buhr and Dickman, 1994; Hollomon et al., 1998; Richards et al., 2000). The only phenotype associated with any of these mutations is resistance to the MT-depolymerizing drug benomyl, which binds β-tubulin. To determine whether the tbb-2(qt1) mutation confers benomyl resistance, we introduced benomyl through a laser-induced hole in the eggshell of two/three-cell embryos and assayed for completion of nuclear division (Table 5; see MATERIALS AND METHODS). Whereas nuclei from wild-type embryos failed to divide when exposed to benomyl, 93% of the nuclei in tbb-2(qt1) embryos at the restrictive temperature did divide, indicating the tbb-2(qt1) mutation causes benomyl resistance. tbb-2(qt1) cells at the permissive temperature and tbb-2(t1623) cells showed a low frequency of division. Wild-type and tbb-2(qt1) embryos showed similar sensitivity to two other MT-inhibiting drugs vinblastine and colchicine (Table 5; Strome and Wood, 1983).
Table 5.
Effects of MT-depolymerizing drugs on wild-type, tbb-2(qt1), and tbb-2(t1623) embryos
| Genotype | Temp. (°C) | Drug | % of Nuclei divided (n) |
|---|---|---|---|
| N2 | 23 | None | 100 (9) |
| tbb-2(qt1) | 23 | None | 85 (13) |
| tbb-2(t1623) | 23 | None | 100 (3) |
| N2 | 23 | Benomyl | 0 (16) |
| N2 | 15 | Benomyl | 0 (14) |
| tbb-2(qt1) | 23 | Benomyl | 93 (14) |
| tbb-2(qt1) | 15 | Benomyl | 35 (23) |
| tbb-2(t1623) | 23 | Benomyl | 22 (9) |
| N2 | 23 | Vinblastine | 25 (12) |
| tbb-2(qt1) | 23 | Vinblastine | 36 (14) |
| N2 | 23 | Colchicine | 29 (7) |
| tbb-2(qt1) | 23 | Colchicine | 0 (4) |
RNAi of tba-1, tba-2, and tbb-1 in Wild-Type Embryos
The C. elegans genome contains genes for nine α-tubulins, six β-tubulins, and one γ-tubulin. In addition to tbb-2 (C36E8.5), the early C. elegans embryo expresses one other β-tubulin gene, tbb-1 (K01G5.7), and two α-tubulin genes, tba-1 (F26E4.8) and tba-2 (C47B2.3), at high levels (Baugh et al., 2003). In addition to their expression in the early embryo, these tubulins are among the most highly expressed in larval and adult C. elegans, and they are the only tubulins consistently expressed throughout the worm's life cycle (Hill et al., 2000).
RNAi of these three tubulin genes separately in wild-type animals did not result in significant embryonic lethality (Table 3). However, if tba-1 and tba-2 or tbb-1 and tbb-2 are depleted by RNAi in the same animal, its progeny fail to make spindle structures, indicating that each individual double-stranded RNA preparation is active. Previous RNAi of tba-1 and tba-2 (Fraser et al., 2000) and tbb-1 (Gonczy et al., 2000) that did result in embryonic lethality was likely due to cross-interference. Early cleavages in tbb-1(RNAi), tba-1(RNAi), and tba-2(RNAi) embryos were essentially wild type, with the exception of low frequency meiotic defects in the tba-2(RNAi) and one tba-2(RNAi) embryo that failed to undergo nuclear-centrosome rotation (Table 3). Importantly, the tbb-1(RNAi) embryos did not show the centrosome stabilization phenotype seen in the tbb-2 deletion and RNAi embryos (Figure 3C; Video 2). However, centrosome stabilization during anaphase was affected in the tba-1(RNAi) and tba-2(RNAi) embryos (Figure 3, E and F; Videos 4 and 5). In the tba-1(RNAi) embryos, the anterior and posterior centrosomes both move through 50% more embryo space than wild-type centrosomes, whereas in tba-2 (RNAi) embryos only the movement of the posterior centrosome was more exaggerated (Figure 3G). Unlike the tbb-2 deletion and RNAi embryos, the regular periodicity of the centrosome oscillations was not disrupted in the α-tubulin RNAi embryos. Overall, qualitatively the disruption in centrosome behavior in the tbb-2 deletion and RNAi embryos is much more severe than in the tba-1 and tba-2 RNAi embryos. To confirm that depletion of tbb-1 does not affect centrosome stabilization, a deletion strain of tbb-1 was obtained from the C. elegans Knockout Consortium. By Western analysis, TBB-1 protein is not detected in these worms (our unpublished data) and similar to the tbb-1(RNAi) worms, the first cell division was identical to wild-type in all respects (Table 3 and Figure 3G).
From the results of the single and double RNAi experiments, we conclude that the α-tubulins are functionally redundant in the early embryo and that an embryo can survive with either TBA-1 or TBA-2; however, both α-tubulins seem to have a role in centrosome stabilization. The β-tubulins are also redundant for most MT-dependent functions in the early embryo, but the TBB-2 protein seems to have a specific role in centrosome stabilization as well as later in embryonic development at 25°C because embryonic lethality results when the gene product is removed. RNAi of tba-1, tba-2, and tbb-1 also results in variable levels of larval lethality and adult phenotypes that include uncoordination and vulval defects that were not explored further.
RNAi of α- and β-Tubulin in tbb-2(qt1) and tbb-2(t1623) Worms
To investigate the functional capacity of the qt1 and t1623 TBB-2 proteins, we used RNAi to deplete wild-type α- and β-tubulins in the mutant strains. Depletion of tbb-1 gave contrasting results in the two strains. The tbb-2(qt1); tbb-1(RNAi) embryos had a higher incidence of meiotic defects and pronuclear migration failure than the tbb-2(qt1) embryos, but the embryos were still capable of forming spindle structures and undergoing mitosis, even though the predominant β-tubulin in the embryo was the defective qt1 TBB-2 (Table 4). RNAi of tbb-1 in tbb-2(t1623) animals produced embryos with no spindle structures (Table 4). Thus, the t1623 tubulin alone is not capable of forming functional MTs, supporting the idea that the tbb-2(t1623) mutation is more deleterious than the tbb-2(qt1) mutation.
RNAi of each α-tubulin in both mutants produced an intriguing result. Depleting TBA-1 from tbb-2(qt1) embryos increased the percentage of embryos with defects in meiosis and pronuclear migration (Table 4). However, depleting TBA-2 from tbb-2(qt1) embryos suppressed the pronuclear migration and nuclear-centrosome rotation phenotypes as well as the embryonic lethality (Table 4), suggesting that the qt1 TBB-2 protein may form abnormal heterodimers exclusively with TBA-2. In contrast, depleting TBA-2 from tbb-2(t1623) embryos increased the level of meiotic defects while depleting TBA-1 partially rescued the pronuclear migration and transverse spindle orientation phenotypes (Table 4). This rescue is not as dramatic as the rescue of tbb-2(qt1) embryos by tba-2(RNAi) because suppression of the centration and rotation defect was limited; only during anaphase did the spindle shift to the correct orientation. However, it may indicate a preference for t1623 TBB-2 to form heterodimers with TBA-1.
RNAi of α- and β-Tubulin Combinations in Wild-Type Embryos
The observation that the mutant β-tubulins may preferentially heterodimerize with different α-tubulins, raises the question of whether the wild-type α- and β-tubulins have specific heterodimer partners. We therefore used RNAi to remove all four possible α- and β-tubulin combinations. Depleting one α- and one β-tubulin leaves the embryo capable of making predominantly one kind of heterodimer. If any of the tubulins have absolute heterodimer partner specificity, some of the experimental combinations would likely give rise to embryos that produced no spindle structures because the remaining α- and β-tubulins would be unable to form heterodimers. The results of the RNAi experiments indicated that there are not specific α- and β-tubulin heterodimer partners; each of the four possible RNAi combinations produced embryos with mostly normal MT function (Table 3). Defects in meiosis and/or nuclear-centrosome rotation were detected at a low frequency in most double tubulin combinations (Table 3). This is not surprising since the composition of the tubulin pool in the RNAi embryos is severely altered.
DISCUSSION
tbb-2(qt1) and tbb-2(t1623) Mutations Differentially Affect MTs
Our experiments indicate that the tbb-2(qt1) and tbb-2(t1623) mutations differentially affect MT stability. Generally, MT stability is determined by the frequency with which α- and β-tubulin heterodimers are added or removed from MTs, a process termed dynamic instability (Desai and Mitchinson, 1997). This process can be affected by various factors, including the primary sequence of tubulin, MT binding proteins, and the environment of the cell. Thus, the changes we observe in MT behavior could be caused by the mutations affecting the structure of TBB-2 or altering a protein-binding site. Incorporation of mutated TBB-2 tubulin into MTs then leads to the observed phenotypes.
MTs containing qt1 TBB-2 subunits are more stable than wild-type MTs because they are cold stable and resistant to the MT-depolymerizing drug benomyl. However, in vitro experiments with fungal tubulins indicate that mutations affecting aa 198, as the tbb-2(qt1) mutation does, prevents benomyl from binding to tubulin, which could account for the observed resistance (Hollomon et al., 1998). Amino acid 198 is highly conserved among all eukaryotes (Burns and Surridge, 1994) and contributes to a pocket on the inside the tubulin subunit located near the lumen of the assembled MT (Richards et al., 2000) at the juncture between the GTP binding domain and the central taxol binding domain of the tubulin subunit (Nogales et al., 1998). Examination of β-tubulin sequences from Antarctic fishes, whose MTs polymerize at 4°C, revealed that several have an amino acid change at aa 200. It has been proposed that alterations to this juncture may promote the stabilization of MTs by causing the β-tubulin subunit to take on a conformation similar to the more stable GTP bound form (Detrich et al., 2000). The tbb-2(qt1) mutation could have a similar effect. Alternatively the mutation may disrupt a binding site for a protein involved in MT destabilization.
MTs in tbb-2(t1623) embryos fail to lengthen substantially during prophase and metaphase. At anaphase, MT length increases until the MTs contact the cortex of the cell. The tbb-2(t1623) mutation affects aa 313, which is conserved among metazoan (Burns and Surridge, 1994). Amino acid 313 is located on the exterior of the tubulin protein and the assembled MT within a large domain that has been described as the assembly domain, but aa 313 is not in any of the areas that have been linked to lateral contacts, longitudinal contacts, or GTP hydrolysis (Nogales et al., 1998). The observation that MT repolymerizaton after depolymerization on ice results in two classes of embryos, depending on the stage in the cell cycle suggests that the short MT phenotype is cell cycle specific as opposed to a generalized delay in MT polymerization. The correlation of short MTs with the cell cycle and the location of the affected amino acid near the outside of assembled MTs suggests that the tbb-2(t1623) mutation may disrupt the interaction of MTs with a MT-associated protein involved in cell cycle-dependent MT stabilization. One candidate is ZYG-9, an MT-stabilizing protein that localizes to centrosomes until telophase and spindle MTs until early anaphase (Matthews et al., 1998). The human homolog of ZYG-9, TOGp, binds to tubulin and MTs but not through the C-terminal tail where many proteins associate with MTs (Spittle et al., 2000).
An interesting observation is the rescue and partial rescue of the tbb-2(qt1) and tbb-2(1623) phenotypes by RNAi of different α-tubulins. Rescue of β-tubulin mutation phenotype by depletion of an α-tubulin was documented in fission yeast; however, researchers were unable to determine whether the rescue was isoform specific as observed here (Radcliffe et al., 1998). The isoform-specific rescue is hard to understand because the two α-tubulins are 98% identical and only one of the 12 amino acid differences between TBA-1 and TBA-2 is predicted to lie near the α-/β-tubulin interface (Nogales et al., 1998). The tbb-2(qt1) and tbb-2(t1623) mutations, although not located directly at the α-/β-tubulin interface, are both nearby. Perhaps conformational changes caused by the tbb-2(qt1) and tbb-2(t1623) mutations are transmitted to the dimer interface, which would allow for differential α-tubulin interactions and create heterodimer specificity.
Altered MT Dynamics Disrupt Centrosome Rotation
The discovery that the tbb-2(qt1) and tbb-2(t1623) mutations affect tubulin confirm previous results indicating that MTs are required for centrosome rotation. Mutations in zyg-9, mei-1, and mel-26 prevent the formation of long astral MTs and cause P0 rotation phenotypes similar to those seen in tbb-2(qt1) and tbb-2(t1623) embryos (Albertson, 1984; Matthews et al., 1998; Mains et al., 1990; Dow and Mains, 1998; Srayko et al., 2000).
Defective MTs most likely account for the failure of nuclear-centrosome complex rotation in tbb-2(t1623) and tbb-2(qt1) embryos. Astral MTs in tbb-2(t1623) embryos remain short until after nuclear-centrosome rotation normally occurs; thus, the failure of astral MTs to interact with the cortex could explain the rotation failure. The short MTs are also likely responsible for the pronuclear migration and chromosome segregation defects. In contrast, tbb-2(qt1) embryos do not show gross defects in MT organization and astral MTs are apparently in contact with the cortex. It is possible that the potentially more stable, thus less dynamic, nature of the qt1 TBB-2 containing MTs prevents MT shortening to a degree that disrupts rotation in the larger blastomeres. It is notable that treatments and mutations that destabilize MTs [nocodozale, zyg-9, mei-1, tbb-2(t1623)] have the same effect on nuclear-centrosome centration and rotation as the tbb-2(qt1) mutation, which stabilizes MTs, indicating that the rotation process is very sensitive to changes in MT dynamics.
tbb-2(qt1) and tbb-2(t1623) Complement Because of Their Different Effects on MTs
Although the tbb-2 alleles are recessive, they seem to be gain-of-function mutations because removal of the defective TBB-2 subunits by RNAi rescues the mutant phenotypes and when tbb-2(qt1) is hemizygous the phenotype is slightly improved (Table 2). It is likely that both mutations produce abnormal tubulins that interfere with MT function when incorporated into MTs. Depletion of the affected tubulin by RNAi simply removes the interfering activity. Because both alleles are recessive, presumably in heterozygotes the wild-type TBB-1 and TBB-2 subunits are able to compensate for the defective qt1 TBB-2 and t1623 TBB-2 subunits, allowing MTs to function normally. However, when either mutation is homozygous, the amount of wild-type β-tubulin in MTs is no longer sufficient to compensate for the mutant TBB-2. This idea is supported by the observation that the mutant phenotypes worsen when tbb-1 is depleted (Table 4). Given the difference in the two phenotypes, one explanation for the complementation is that the effects of the tbb-2(qt1) and tbb-2(t1623) mutations are independent. This idea suggests that qt1 TBB-2 is able to compensate for the function affected by the tbb-2(t1623) mutation and vice versa in tbb-2(qt1)/tbb-2(t1623) embryos. Another explanation is that the effects of the mutations are additive and the tendency for stable MTs caused by the tbb-2(qt1) mutation is counterbalanced by the tbb-2(t1623) mutation's effect of making MTs less stable. Either way, wild-type TBB-1 is absolutely required for complementation because when it is depleted by RNAi, tbb-2(qt1) and tbb-2(t1623) fail to complement (Table 4).
One issue that complicates our understanding of the nature of the tbb-2(t1623) and tbb-2(qt1) mutant phenotypes is tubulin homeostasis, an autoregulatory mechanism that serves to maintain constant tubulin protein levels (Cleveland and Theodorakis, 1994). Data generated by Ellis and Bowerman (personal communication) suggest that constant β-tubulin levels are maintained in the early C. elegans embryo because deletion of tbb-2 leads to increased levels of TBB-1. However, in tbb-2(t1623) mutants, TBB-1 protein levels are not elevated. Thus the tbb-2(t1623) mutant phenotype may reflect a loss of TBB-2 function compounded by the lack of increased expression of TBB-1. Therefore, the suppression of tbb-2(t1623) by tbb-2(RNAi) may be due to up-regulation of TBB-1 in addition to removal of mutant TBB-2. This hypothesis still implies that incorporation of qt1 and t1623 TBB-2 is detrimental to MT function, which is the only explanation for the distinctive phenotypes, but suggests that the phenotypes may be exacerbated by a failure to upregulate TBB-1 (at least for the t1623 embryos).
TBB-2 Is Required for Centrosome Stabilization during Anaphase
The only consistent tubulin RNAi phenotypes we observed were defects in centrosome stabilization during spindle rocking in anaphase of the first cell division. The rocking was mildly affected in both α-tubulin RNAi embryos but severely affected whenever tbb-2 was depleted or deleted. Grill et al. (2001) proposed that the rocking of the posterior centrosome in the wild-type P0 blastomere is caused by destabilization of posterior astral MTs at the cortex relative to anterior astral MTs. An explanation for the phenotype of the tbb-2(RNAi) and deletion embryos might be that TBB-1 is not sufficient to interact with the cortical protein(s) required for MT stabilization. PAR-3, PAR-1, and Gα proteins have been implicated in regulating MT stability (Labbe et al., 2003) and are candidates to show preferential β-tubulin interactions. Although TBB-1 and TBB-2 are 98% identical, 7 of the 11 amino acid differences between the two β-tubulins are found in the last 19 amino acids of the protein. These seven amino acid differences are conserved in the TBB-1 and TBB-2 homologs of the related nematode C. briggsae. The carboxy terminus is where α- and β-tubulins most often interact with MT binding proteins, supporting the idea that TBB-1 and TBB-2 may differentially interact with cortical proteins. An alternate explanation for the centrosome stabilization phenotypes is that depletion of the various tubulins may disrupt total tubulin dimer levels, causing the phenotypes. The observation that β-tubulin levels are regulated argues against that, but subtle alternations could still be disruptive.
Overall, our results indicate a specific requirement for TBB-2 in centrosome stabilization and demonstrate that in the early embryo TBB-1 and TBB-2 are not functionally equivalent. Conversely, we were not able to detect a specific requirement for TBA-1 or TBA-2 in early embryonic development because depletion of either slightly affects centrosome stabilization. Thus, we suggest that in the early C. elegans embryo, β-tubulins, but not α-tubulins, have functional specification.
Supplementary Material
Acknowledgments
We thank G. Ellis, J. Phillips, B. Bowerman, C. Lu, and P. Mains for communicating unpublished results, sharing reagents, and reviewing this manuscript, T. Stiernagle at the Caenorhabditis Genetics Center (supported by the National Institutes of Health National Center for Research Support) for supplying worm strains, and D. Mootz for critical review of this manuscript. Deletion mutations used in this work were provided by the C. elegans Reverse Genetics Core Facility at the University of British Columbia which is funded by the Canadian Institute for Health Research, Genome Canada, and Genome BC. Funding was provided by a grant from National Science Foundation IBN 0110480 to C.P.H.
Abbreviations used: DIC, differential interference contrast; MT, microtubule; RNAi, RNA interference.
Online version of this article contains video material for some figures. Online version is available at www.molbiolcell.org.
References
- Albertson, D.G. (1984). Formation of the first cleavage spindle in nematode embryos. Dev. Biol. 101, 61-72. [DOI] [PubMed] [Google Scholar]
- Baugh, L.R., Hill, A.A., Slonim, D.K., Brown, E.L., and Hunter, C.P. (2003). Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130, 889-900. [DOI] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr, T.L., and Dickman, M.B. (1994). Isolation, characterization, and expression of a second β-tubulin-encoding gene from Colletotrichum gloeosporioides f. sp. aeschynomene. Appl. Environ. Microbiol. 60, 4155-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, R.G., and Surridge, C.D. (1994). Tubulin: conservation and structure. In: Microtubules, ed. J.S. Hyams and C.W. Lloyd, New York: Wiley-Liss, 3-31.
- Carminati, J.L., and Stearns, T. (1997). Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138, 629-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland, D.W., and Theodorakis, N.G. (1994). Regulation of tubulin synthesis. In: Microtubules, ed. J.S. Hyams and C.W. Lloyd, New York: Wiley-Liss, 47-58.
- Cottingham, F.R., and Hoyt, M.A. (1997). Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 138, 1041-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., and Mitchinson, T.J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83-117. [DOI] [PubMed] [Google Scholar]
- Detrich, H.W. 3rd, Parker, S.K., Williams, R.C., Jr., Nogales, E., and Downing, K.H. (2000). Cold adaptation of microtubule assembly and dynamics. J. Biol. Chem. 275, 37038-37047. [DOI] [PubMed] [Google Scholar]
- DeZwaan, T.M., Ellingson, E., Pellman, D., and Roof, D.M. (1997). Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J. Cell Biol. 138, 1023-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow, M.R., and Mains, P.E. (1998). Genetic and molecular characterization of the Caenorhabditis elegans gene, mel-26, a negative postmeiotic regulator of MEI-1, a meitotic-specific spindle component. Genetics 150, 119-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll, M., Dean, E., Reilly, E., Bergholz, E., and Chalfie, M. (1989). Genetic and molecular analysis of a Caenorhabditis elegans β-tubulin that conveys benzimidazole sensitivity. J. Cell Biol. 109, 2993-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- Fraser, A.G., Kamath, R.S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M., and Ahringer, J. (2000). Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325-330. [DOI] [PubMed] [Google Scholar]
- Fujimura, M., Oeda, K., Inoue, H., and Kato, T. (1992). A single amino-acid substitution in the beta-tubulin gene of Neurospora confers both carbendazim resistance and diethofencarb sensitivity. Curr. Genet. 21, 399-404. [DOI] [PubMed] [Google Scholar]
- Gonczy, P., Pichler, S., Kirkham, M., and Hyman, A.A. (1999a). Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 147, 135-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy, P., Schnabel, H., Kaletta, T., Amores, A.D., Hyman, T., and Schnabel, R. (1999b). Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J. Cell Biol. 144, 927-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy, P., et al. (2000). Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408, 331-336. [DOI] [PubMed] [Google Scholar]
- Grill, S.W., Gonczy, P., Stelzer, E.H.K., and Hyman, A.A. (2001). Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature 409, 630-633. [DOI] [PubMed] [Google Scholar]
- Gupta, M.L., Jr., Bode, C.J., Thrower, D.A., Pearson, C.G., Suprenant, K.A., Bloom, K.S., and Hines, R.H. (2002). β-Tubulin C354 mutations that severely decrease microtubule dynamics do not prevent nuclear migration in yeast. Mol. Biol. Cell 13, 2919-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak, E., Oegema, K., Kirkham, M., Gonczy, P., Habermann, B., and Hyman, A.A. (2002). The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is γ-tubulin dependent. J. Cell Biol. 157, 591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, A.A., Hunter, C.P., Tsung, B.T., Tucker-Kellogg, G., and Brown, E.L. (2000). Genomic analysis of gene expression in C. elegans. Science 290, 809-812. [DOI] [PubMed] [Google Scholar]
- Hollomon, D.W., Butters, J.A., H. Barker, L. Hall. (1998). Fungal β-tubulin, expressed as a fusion protein, binds benzimidazole and phenylcarbamate fungicides. Antimicrob. Agents Chemother. 42, 2171-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle, H.D., and Raff, E.C. (1990). Two Drosophila beta tubulin isoforms are not functionally equivalent. J. Cell Biol. 111, 1009-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman, A.A. (1989). Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J. Cell Biol. 109, 1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman, A.A., and White, J.G. (1987). Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. J. Cell Biol. 105, 2123-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, K.M., Wilder, I.B., and Oakley, B.R. (1992). Amino acid alterations in the benA (β-tubulin) gene of Aspergillus nidulans that confer benomyl resistance. Cell Motil. Cytoskeleton 22, 170-174. [DOI] [PubMed] [Google Scholar]
- Koenraadt, H., Somerville, S.C., and Jones, A.L. (1992). Characterization of mutations in the beta-tubulin gene of benomyl-resistant field strains of Venturia inaequalis and other plant pathogenic fungi. Phytopathology 82, 1348-1354. [Google Scholar]
- Labbe, J.C., Maddox, P.S., Salmon, E.D., and Goldstien, B. (2003) PAR proteins regulate microtubule dynamics at the cell cortex in C. elegans. Curr. Biol. 13, 707-714. [DOI] [PubMed] [Google Scholar]
- Luduena, R.F. (1998). Multiple forms of tubulin: different gene products and covalent modifications. Int. Rev. Cytol. 178, 207-75. [DOI] [PubMed] [Google Scholar]
- Mains, P.E., Kemphues, K.J., Sprunger, S.A., Sulston, I.A., and Wood, W.B. (1990). Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics 126, 593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, L.R., Carter, P., Thierry-Mieg, D., and Kemphues, K. (1998). ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J. Cell Biol. 141, 1159-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales, E., Wolf, S.G., and Downing, K.H. (1998). Structure of the αβ tubulin dimer by electron crystallography. Nature 391, 199-203. [DOI] [PubMed] [Google Scholar]
- Radcliffe, P., Hirata, D., Childs, D., Vardy, L., and Toda, T. (1998). Identification of novel temperature-sensitive lethal alleles in essential β-tubulin and nonessential α2-tubulin genes as fission yeast polarity mutants. Mol. Biol. Cell 9, 1757-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, K.L., Anders, K.R., Nogales, E., Schwartz, K., Downing, K.H., and Botstein, D. (2000). Structure-function relationships in yeast tubulins. Mol. Biol. Cell 11, 1887-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage, C., Hamelin, M., Culotti, J.G., Coulson, A., Albertson, D.G., and Chalfie, M. (1989). mec-7 is a β-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 3, 870-881. [DOI] [PubMed] [Google Scholar]
- Schatz, P.J., Solomon, F., and Botstein, D. (1986). Genetically essential and nonessential α-tubulin genes specify functionally interchangeable proteins. Mol. Cell. Biol. 6, 3722-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler, S.C., and Pellman, D. (2001). Search capture and signal: games microtubules and centrosomes play. J. Cell Sci. 114, 247-55. [DOI] [PubMed] [Google Scholar]
- Segal, M., and Bloom, K. (2001). Control of spindle polarity and orientation in Saccharomyces cerevisiae. Trends Cell Biol. 11, 160-166. [DOI] [PubMed] [Google Scholar]
- Skop, A.R., and White, J.G. (1998). The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr. Biol. 8, 1110-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spittle, C., Charrasse, S., Larroque, C., and Cassimeris, L. (2000). The interaction of TOGp with microtubules and tubulin. J. Biol. Chem. 275, 20748-20753. [DOI] [PubMed] [Google Scholar]
- Srayko, M., Buster, D.W., Bazirgan, O.A., McNally, F.J., and Mains, P.E. (2000). MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14, 1072-1084. [PMC free article] [PubMed] [Google Scholar]
- Strome, S., and Wood, W.B. (1983). Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35, 15-25. [DOI] [PubMed] [Google Scholar]
- Tsou, M.F., Hayashi, A., DeBella, L.R., McGrath, G., and Rose, L.S. (2002). LET-99 determines spindle position and is asymmetrically enriched in response to PAR polarity cues in C. elegans embryos. Development 129, 4469-4481. [DOI] [PubMed] [Google Scholar]
- Wicks, S.R., Yeh, R.T., Gish, W.R., Waterson, R.H., and Plasterk, R.H. (2001). Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28, 160-164. [DOI] [PubMed] [Google Scholar]
- Yarden, O., and Katan, T. (1993). Mutations leading to substitutions at amino acids 198 and 200 of beta-tubulin that correlate with benomyl-resistance phenotypes of field strains of Botrytis cinerea. Phytopathology 83, 1478-1483. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.