Abstract
The cytoskeleton is a key regulator of cell morphogenesis. Crescentin, a bacterial intermediate filament-like protein, is required for the curved shape of Caulobacter crescentus and localizes to the inner cell curvature. Here, we show that crescentin forms a single filamentous structure that collapses into a helix when detached from the cell membrane, suggesting that it is normally maintained in a stretched configuration. Crescentin causes an elongation rate gradient around the circumference of the sidewall, creating a longitudinal cell length differential and hence curvature. Such curvature can be produced by physical force alone when cells are grown in circular microchambers. Production of crescentin in Escherichia coli is sufficient to generate cell curvature. Our data argue for a model in which physical strain borne by the crescentin structure anisotropically alters the kinetics of cell wall insertion to produce curved growth. Our study suggests that bacteria may use the cytoskeleton for mechanical control of growth to alter morphology.
Keywords: bacterial cell morphogenesis, cell curvature, crescentin, cytoskeleton, peptidoglycan
Introduction
The cytoskeleton has a central function in the morphogenesis of eukaryotes and prokaryotes alike. It can achieve this function in two main ways. One is by directly deforming the cell membrane, as in lamella (Pollard and Borisy, 2003), and the other is by affecting cell growth (Smith and Oppenheimer, 2005; Cabeen and Jacobs-Wagner, 2007; Fischer et al, 2008). In bacteria, as in plants and fungi, cell growth depends on cell wall growth. The bacterial peptidoglycan cell wall is a covalently closed meshwork of rigid glycan strands cross-linked by relatively flexible peptide bridges (Vollmer et al, 2008). It thereby forms a strong but elastic fabric that resists internal turgor pressure and prevents cell bursting. Under normal growth conditions, cells are turgid, as the force exerted by turgor pressure maintains the peptidoglycan in a stretched configuration (Koch, 1984; Koch et al, 1987; Baldwin et al, 1988). To expand, bonds in the peptidoglycan must be hydrolyzed to generate insertion sites for new wall material. The stretching from turgor pressure helps overcome molecular cohesion, facilitating bond hydrolysis and thereby providing a source of energy for cell wall growth (Koch et al, 1982; Harold, 2002). To preserve the integrity of the peptidoglycan fabric, cell wall hydrolases and synthases coordinate their activities by forming complexes (Romeis and Höltje, 1994; von Rechenberg et al, 1996; Höltje, 1998; Schiffer and Höltje, 1999; Vollmer et al, 1999; Bertsche et al, 2006).
The location and timing of new wall insertion is critical for generating nonspherical shapes, as turgor pressure is nondirectional (Harold, 2007; den Blaauwen et al, 2008). In most rod-shaped bacteria, elongation occurs by insertion of peptidoglycan material along the sidewall of the cell, with little to no insertion at the poles (de Pedro et al, 1997; Daniel and Errington, 2003). Cytoskeletal proteins of the actin family (MreB and its homologs), which form helical structures along the long axis of the cell beneath the cytoplasmic membrane (Jones et al, 2001; Shih et al, 2003; Figge et al, 2004), are thought to spatially restrict insertion of new peptidoglycan to the sidewall (Daniel and Errington, 2003; Figge et al, 2004; Dye et al, 2005; Kruse et al, 2005; Carballido-Lopez et al, 2006; Divakaruni et al, 2007).
Vibrioid (curved rod) or helical morphologies are also common among bacteria, but their morphogenetic mechanisms are largely unresolved. It is known that the intermediate filament-like protein crescentin is required for the crescent shape of Caulobacter crescentus as in its absence, the cells are straight rod shaped (Ausmees et al, 2003). Crescentin localizes along the inner cell curvature, beneath the cytoplasmic membrane, but how crescentin confers cell curvature remains unclear.
Physical modelling has proposed that filaments attached to the cell wall could produce a tension leading to curved or helical morphology (Wolgemuth et al, 2005). Our findings suggest a model in which the attachment of a crescentin structure at the cell envelope results in a compressive force that sets up a gradient of growth rate across the short cell axis to contribute to cell curvature.
Results
Crescentin assembles into a single coiled structure that is maintained in an extended configuration along the membrane
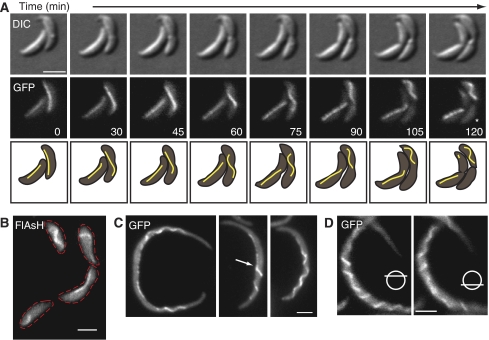
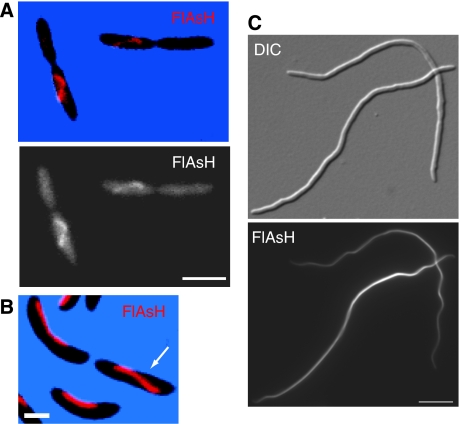
Although crescentin polymerizes into filaments in vitro (Ausmees et al, 2003), it was unclear whether it also formed filamentous structures in cells. Thus, we attempted to dislodge crescentin from its fixed position along the membrane by weakening the cell wall and widening the cell. We used a strain (CJW2203) co-expressing wild-type crescentin and crescentin-GFP, which form functional hybrid structures (Ausmees et al, 2003), and deleted for the β-lactamase encoding-gene (Δbla), allowing us to use the β-lactam antibiotic mecillinam, which inhibits peptidoglycan cross-linking (Seitz and Brun, 1998). Time-lapse imaging revealed that over time, the GFP signal began to detach from its static localization along the membrane and move as a single structure in some cells. The crescentin structure appeared to be anchored to the cell envelope continuously along its length, as detachment tended to occur gradually and progressively (Figure 1A; see also below and Figure 1C, arrow). Strikingly, as crescentin structures detached, they collapsed into curved or helical shapes (Figure 1A; Supplementary Movie 1). This often resulted in only one daughter cell inheriting the entire detached crescentin structure after division (Figure 1A, asterisk). Detachment of crescentin structures was also seen by FlAsH staining of a strain (CJW2995) producing tetracysteine (TC)-tagged crescentin rather than crescentin-GFP (Figure 1B). Crescentin-TC is fully functional, as plasmid-encoded creS-tc restores curvature to ΔcreS cells (Supplementary Figure S1). The motion of detached structures inside the cells showed their loss of cell envelope attachment and their flexibility (Supplementary Movie 1), consistent with the known flexibility of intermediate filaments (Herrmann et al, 2007). Continued mecillinam treatment eventually caused cell death and the disappearance of filamentous crescentin structures (not shown).
Figure 1.
Crescentin forms a single filamentous structure that collapses into a helical structure when released from the membrane. (A) Time lapse of log-phase CJW2203 cells (CB15N Δbla creS∷pBGST18creS-gfp∷pBGENTcreS) grown on an agarose-M2G+ pad containing 40 μg/ml mecillinam. Asterisk represents cell lacking crescentin-GFP signal after retraction of the crescentin structure into only one daughter cell. DIC, differential interference contrast. (B) CJW2995 cells (CB15N Δbla/pMR20creS-tc) treated in liquid with 40 μg/ml mecillinam for 4.5 h and stained with FlAsH dye. Dashed lines indicate cell outlines. (C) CJW2592 cells (CB15N Δbla ftsZ∷pBGENTPxylftsZ creS∷pBGST18creS-gfp∷pBGENTcreS) depleted for FtsZ for 2.5 h, then grown in liquid for 4.3 h after the addition of 10 μg/ml mecillinam to induce detachment of crescentin structures. Some crescentin structures displayed partial detachment (arrow). (D) Two optical sections of a detached crescentin structure, showing left-handed helicity. CJW2592 cells were depleted of FtsZ as in panel C for 3 h, then grown for another 4 h with 5 μg/ml mecillinam in liquid culture. Bars, 2 μm.
The shape of the detached crescentin structures made us wonder whether they adopt a preferential configuration when unanchored from the envelope. We reasoned that this would be clearer if the cells and crescentin structures were longer. Thus, we created a strain (CJW2592) to deplete the cell division protein FtsZ and create elongated cells before treating with mecillinam. Elongated FtsZ-depleted cells before treatment displayed crescentin at their inner curvatures (not shown). After about 4 h of growth with mecillinam, a portion of the elongated cells had released part or all of the crescentin structure (Figure 1C). Remarkably, these unanchored structures were helical (Figure 1C) and always left handed (Figure 1D; n=116 cells), with a helical pitch of 1.4±0.15 μm (n=128 helical turns) that was consistent among different cells and experiments. This helicity was also observed with cells producing crescentin-TC (CJW2612) or when cells (CJW2046) were treated with a different drug, phosphomycin, which inhibits an early step in peptidoglycan precursor synthesis (not shown). These findings suggest that the crescentin structure is helical and that this structure is normally held at a fixed position along the membrane in a less favourable stretched configuration.
Disruption of the crescentin structure results in slow, growth-dependent shape straightening
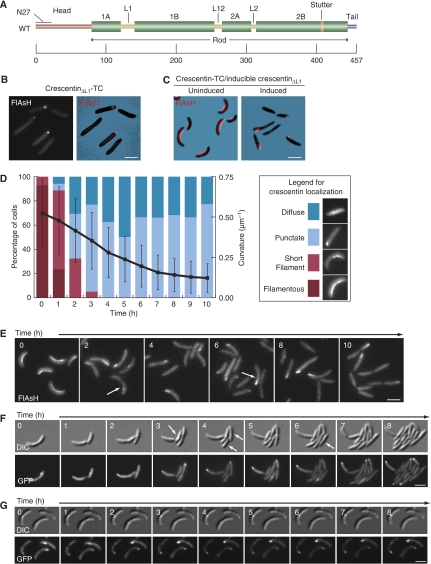
To hold the crescentin structure in an extended configuration, the cell would need to absorb a compressive force exerted by the crescentin structure. Isolated, hydrated peptidoglycan molecules are flexible and elastic (Koch and Woeste, 1992; Yao et al, 1999; Boulbitch et al, 2000), and experiments using atomic force microscopy on live bacteria have demonstrated the elasticity of the cell envelope of several bacterial species (Penegar et al, 1999; Arnoldi et al, 2000; Yao et al, 2002; Gaboriaud et al, 2005; Vadillo-Rodriguez et al, 2008). Therefore, if the crescentin structure applied a great enough force, it could in principle bend the cell into a curved shape. In this case, disruption of the crescentin structure (i.e. release of the force) should cause growth-independent elastic relaxation of cell curvature. Although mecillinam treatment eventually causes crescentin structure detachment, the cells widen during growth, complicating interpretation. Therefore, we made a dominant-negative mutant, an approach successfully used to disrupt intermediate filament networks in eukaryotes (Chen et al, 2003; Schietke et al, 2006). As dominant-negative mutations in intermediate filament proteins are often assembly defective, we generated a mutant similar to an assembly-defective vimentin mutant (Herrmann et al, 1999). The mutant, crescentinΔL1, had a 20-residue deletion (122–142) corresponding to the L1 linker in the rod domain (Figure 2A). CrescentinΔL1-TC did not curve crescentin-null (ΔcreS) cells and often appeared diffuse (51% of cells) or in a polar focus (49% of cells; n=207) rather than filamentous (Figure 2B), consistent with an assembly defect. To test its dominant-negative activity, we produced xylose-inducible untagged crescentinΔL1 from the chromosome in a ΔcreS strain producing wild-type crescentin-TC from a low-copy plasmid (CJW2788). Before induction of crescentinΔL1 synthesis, cells were curved and crescentin-TC displayed its normal filamentous structure at the inner cell curvatures (Figure 2C, left panel). However, after 10 h of induction, cells were straight, and FlAsH staining, which labelled only the functional protein, revealed that it localized diffusely or in a focus (Figure 2C, right panel), showing that filamentous structure formation is essential for crescentin function and that crescentinΔL1 efficiently disrupts crescentin structures.
Figure 2.
Cell straightening upon dominant-negative crescentinΔL1 production is gradual and growth dependent. (A) Crescentin domain organization. Amino acid positions are shown at the bottom. Green bars indicate coiled-coil forming regions. The N-terminal head, C-terminal tail, linkers, stutter (interruption in coiled-coil repeat) and N27 region are marked. (B) FlAsH-stained cells expressing crescentinΔL1-TC in a ΔcreS background (CJW1521; CB15N ΔcreS pMR20creSΔL1-tc). (C) FlAsH-stained cells (CJW2788; CB15N ΔcreS xylX∷pHL23PxylcreSΔL1/pMR20creS-tc) expressing wild-type crescentin-TC (red), without (left) or with (right) co-expression of untagged crescentinΔL1 for 10 h. (D) Graphical representation of cell curvature loss and crescentin localization during crescentin structure disruption. Coloured bars show the fraction of cells (left axis) with each crescentin localization pattern at each time point. The black line shows mean cell curvature in μm−1 for a population of cells (713<n<1919) at each time point (right axis); error bars indicate standard deviation. (E) Representative FlAsH-stained cells of CJW2788 strain taken at the noted intervals after addition of the xylose inducer of crescentinΔL1 synthesis. Only wild-type crescentin-TC is visualized with FlAsH. White arrows denote cells in which crescentin displays punctate localization pattern but that still retain some curvature. (F) CJW2778 cells preincubated in M2G+ with 0.3% xylose for 2 h, then mounted on M2G+ agarose pads with 0.3% xylose and imaged at the indicated time intervals. Arrows indicate examples of straight daughter cells produced by growth in the absence of an intact crescentin structure. (G) Experiment as in (F), but with 10 μg/ml chloramphenicol added to the agarose pad to halt cell growth. Bars, 2 μm.
Using this strain, we induced crescentinΔL1 synthesis in liquid cultures, scoring wild-type crescentin-TC localization (n>450 cells for each time point) and analysing cell curvature hourly. To quantify cell curvature, we developed a Matlab-based numerical method (see Materials and methods; Supplementary Figure S2). The majority of filamentous crescentin structures had been replaced by punctate foci or diffuse signal within 3 h of induction (Figure 2D). In contrast, cell curvature (713<n<1919) was lost gradually over 8 h (Figure 2D). Moreover, we often observed cells with punctate or diffuse crescentin localization but which retained curvature (Figure 2E, arrows).
The gradual loss of cell curvature after crescentin structure disruption suggested growth dependence. However, the bacterial wall exhibits a small amount of viscoelastic creep (time-dependent deformation during constant applied force) on a short-time scale in response to a short-lived force application (Vadillo-Rodriguez et al, 2008). This raised the possibility that, if the crescentin structure mechanically pulled the cell into a curved shape over a long period of time, it might produce a viscoelastic response requiring a longer recovery time. We tested this possibility by blocking cell growth after crescentin structure disruption and observing cell curvature over time. We used a creS/creS-gfp chromosomal merodiploid with plasmid-encoded xylose-inducible crescentinΔL1 (CJW2778) rather than crescentin-TC because GFP has greater photostability than FlAsH, allowing us to perform long time-lapse experiments. We preincubated cells with xylose for 2 h in liquid to disrupt most crescentin structures before substantial loss of cell curvature occurred. We then imaged cells for 8 h with or without chloramphenicol, which arrests protein synthesis and cell growth (Figure 2F and G). Without chloramphenicol, cells grew, divided and became progressively straighter (Figure 2F, arrows). In stark contrast, cells with chloramphenicol exhibited no discernable cell curvature change, even after 8 h, despite disruption of the crescentin structure (Figure 2G). To ensure that immobilization on the agarose pad was not an obstacle to relaxation of cell curvature, we performed the same experiment in liquid with a strain (CJW2788) carrying wild-type crescentin-TC and xylose-inducible, untagged crescentinΔL1. Cell curvature analysis before addition of chloramphenicol and 5 and 10 h thereafter (522<n<1327) revealed no apparent cell curvature change, whereas cell populations without chloramphenicol exhibited a drastic loss in cell curvature (Supplementary Figure S3). Thus, the crescentin structure affects cell curvature in a growth-dependent fashion.
Crescentin creates a gradient of peptidoglycan insertion rates around the cell circumference
The growth dependence of cell curvature loss suggested that the crescentin structure alters peptidoglycan structure or synthesis. Therefore, we analysed the shape and composition of isolated peptidoglycan (sacculi), taking advantage of the straight ΔcreS strain and the hypercurvature of a crescentin-overproducing strain with the creS gene on a medium-copy plasmid (CJW1430; CB15N/pJS14creS) (Figure 3A). Immunofluorescence showed that a thicker crescentin structure retained its characteristic localization at the inner curvature of these hypercurved crescentin-overproducing cells (Figure 3B). The sacculi from hypercurved cells were clearly more curved than the slightly curved wild-type sacculi, and sacculi from ΔcreS cells exhibited no curvature along their long axis (Figure 3C). We also noticed that sacculi from curved cells were generally straighter than whole cells (compare Figure 3A with C), an effect likely due to turgor pressure loss and flattening of the sacculi on the electron microscope (EM) grid. No variations in peptidoglycan thickness between inner and outer curvatures were apparent in sacculi, in agreement with electron cryotomography studies of C. crescentus (Briegel et al, 2006).
Figure 3.
Cell curvature is the result of a peptidoglycan growth differential around the cell circumference. (A) Strains lacking crescentin (CB15N ΔcreS), with wild-type levels of crescentin (CB15N), or overproducing crescentin (CB15N/pJS14creS). Bar, 2 μm. (B) DAPI staining (blue) overlaid with anti-crescentin immunofluorescence (red) of wild-type CB15N cells and cells overproducing crescentin (CB15N/pJS14creS). The line scan gives the fluorescence values along the yellow lines in the micrographs. Bar, 2 μm. (C) Transmission EM of uranyl acetate-contrasted peptidoglycan sacculi isolated from the three differently curved strains in panel A. As observed earlier (Poindexter and Hagenzieker, 1982), the isolated sacculi contained polyhydroxybutyrate (PHB) granules (arrow). Bar, 1 μm. (D) After digestion of the sacculi, muropeptides were subjected to HPLC to determine their relative composition. For the hypercurved CB15N/pJS14creS strain (CJW1430), which carries a plasmid encoding chloramphenicol resistance, we used a strain containing the empty vector (CB15N/pJS14; CJW1534) as a control; this strain exhibits wild-type curvature (data not shown). Major peaks 1–6 are proposed to have the following structures based on elution similarity to known E. coli muropeptides and to the published muropeptide profile from C. crescentus (Markiewicz et al, 1983): (1) disaccharide tetrapeptide, (2) disaccharide pentapeptide(Gly5), (3) disaccharide pentapeptide, (4) bis-disaccharide tetrapentapeptide(Gly5), (5) bis-disaccharide tetratetrapeptide and (6) bis-disaccharide tetrapentapeptide. (E) Schematic of how a cell length gradient along the long axis of the sidewall constitutes cell curvature. The sidewall displays a smooth gradient of lengths from the longest line, at the middle of the outer curvature (line a), through line b, at the middle of the gradient, to the shortest line, at the middle of the inner curvature (line c). (F) Transmission EM of peptidoglycan sacculi from a creS-overexpressing strain (CB15N/pJS14creS; CJW1430) and a ΔcreS strain carrying an empty vector (CB15N ΔcreS/pJS14; CJW2855) chased for 90 min after growth in D-Cys to reveal regions of new peptidoglycan insertion. D-Cys residues were biotinylated, then the sacculi were labelled with anti-biotin and gold-coupled anti-rabbit antibodies. Areas devoid of gold label show regions where new peptidoglycan material was incorporated during the chase. The antibodies used to detect the biotinylated D-Cys residues appeared to clump together. Measured lengths of the cleared region on each side of the sacculus are shown with broken lines. PHB, polyhydroxybutyrate granules. Bar, 0.5 μm.
Computational modelling has shown that cell curvature can be produced by decreasing the number of peptide cross-bridges along one side of the cell cylinder (Huang et al, 2008). We used high-performance liquid chromatography (HPLC) of digested sacculi to resolve and quantify differently cross-linked muropeptide species. This analysis showed that there were no noticeable differences in the degree of cross-linking or relative abundance of each particular muropeptide species among wild-type, crescentin-null (ΔcreS and creS∷Tn5) and crescentin-overproducing strains (Figure 3D), strongly arguing that curvature is not a product of alterations in peptidoglycan structure. Although this method cannot rule out a structural difference between inner and outer curvature sides that precisely cancels out, it is highly unlikely that the crescentin structure, located at the inner curvature only, could produce a perfectly compensatory effect at the outer curvature.
The fact that curved sacculi lack observable variations in thickness, cross-linking or composition compared with straight sacculi, therefore, argues that the length differential between inner and outer curvatures is a result of differential growth. A curved cell (or sacculus) has its longest length at the outer curvature (line a in Figure 3E), and its shortest length at the inner curvature (line c in Figure 3E), with a gradient of length in between (i.e. length increasing from line c to line a through line b in Figure 3E). Accordingly, the crescentin structure would not only reduce peptidoglycan insertion at the side where crescentin is located but would also generate a gradient of increasing peptidoglycan growth rates from its side (inner curvature) to the opposite side of the cell (outer curvature). To test this hypothesis, we used D-cysteine pulse-chase labelling of the peptidoglycan (de Pedro et al, 1997) and compared the peptidoglycan growth of straight and curved strains, using ΔcreS and hypercurved, crescentin-overproducing strains to accentuate any differences. Cells were grown with D-Cys for four generations, then the D-Cys was washed out and the cells were grown for 90 min. The D-Cys provides thiol groups that can be labelled and detected, distinguishing peptidoglycan newly synthesized during the chase period by its lack of label. In C. crescentus, there are two different modes of peptidoglycan insertion during cell elongation (Aaron et al, 2007). A dispersed/helical insertion phase early in the cell cycle is followed by a period of zonal elongation at midcell preceding cell division. Unlike the dispersed insertion during the first phase, insertion during the zonal phase produces regions of new synthesis at midcell that are clearly discernable by the D-Cys labelling, providing a suitable method for visualizing any differential growth. We found that hypercurved sacculi showed trapezoidal regions of zonal midcell clearing, with longer cleared spaces at the outer curvature (Figure 3F; Supplementary Figure S4), indicating a gradient of peptidoglycan insertion between inner and outer curvatures. Meanwhile, straight sacculi displayed rectangular cleared regions with comparable lengths along both sides (Figure 3F; Supplementary Figure S4). These results show that the crescentin structure caused differential peptidoglycan insertion rates around the cell circumference to produce cell curvature.
Force alone causes growth-dependent curvature in C. crescentus
Although a compressive force generated by the crescentin structure is not sufficient to physically bend the cells to their observed curvature, we wondered whether a force could affect cell growth rate. As our results indicated that the crescentin structure produces a gradient of growth rates to cause curvature, we reasoned that perhaps an external force could do likewise. It has been shown that when growing Escherichia coli cells are confined in circular agarose microchambers, they become curved (Takeuchi et al, 2005). As cell curvature is retained after release of E. coli from the chambers (Takeuchi et al, 2005), it might be the product of a force-induced growth rate gradient along the short cell axis, or possibly the result of a slow viscoelastic response (hours of confinement requiring hours of recovery).
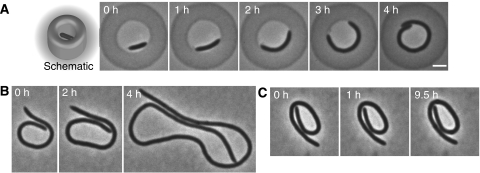
To examine these questions, we performed a series of experiments with C. crescentus cells (CJW1819), in which cell division could be blocked by FtsZ depletion. These ΔcreS cells, which are genetically unable to produce curvature, were placed in circular agarose microchambers (Figure 4A, schematic) and depleted for FtsZ, causing continued cell elongation. Eventually, the cells contacted the chamber walls, at which point the cells started curving typically along the chamber wall (Figure 4A; Supplementary Movie 2). Upon release onto an agarose pad, curvature was lost not immediately but progressively over time (Figure 4B), consistent with the E. coli data (Takeuchi et al, 2005). As with the E. coli experiments, it remained unclear whether the curvature loss represented slow relaxation or whether it was growth dependent. However, blocking cell growth with chloramphenicol after release from the chambers abolished curvature loss (Figure 4C), implying that physical force produced by contact with the chamber walls produced cell curvature by affecting cell growth.
Figure 4.
Physical force can alter growth to produce cell curvature. (A) Time-lapse series of a ΔcreS cell (CJW1819; CB15N ΔcreS ftsZ∷pBJM1) grown in a circular agarose microchamber. Cell division was blocked by FtsZ depletion to promote cell elongation and contact with the chamber wall. Bar, 2 μm. (B) A CJW1819 cell released from a circular chamber, placed on an agarose pad (containing medium), and imaged just after release and then after 2 or 4 h of growth. (C) A cell released from a circular chamber and placed on an agarose pad containing 40 μg/ml chloramphenicol to block cell growth. Images show cell just after release and 1 and 9.5 h later.
Synthesis of crescentin is sufficient to produce curvature in E. coli
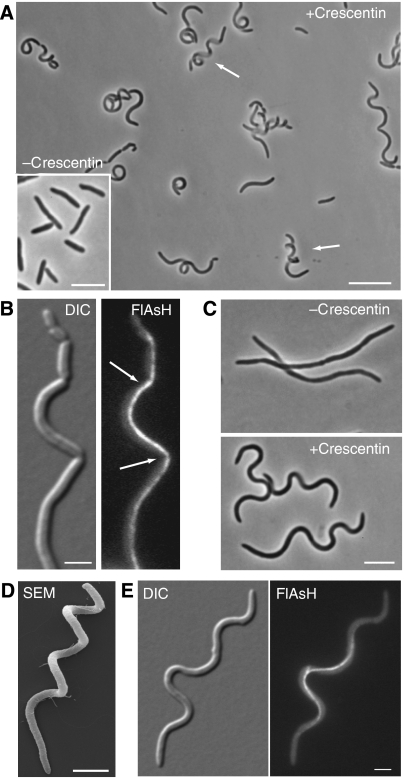
The results of the microchamber experiments raised the possibility that a force derived from the crescentin structure could produce growth-dependent curvature. As a force-based mechanism should operate in vastly divergent bacterial species, we tested whether producing crescentin in E. coli is sufficient to cause cell curvature. Crescentin or crescentin-TC was produced in wild-type E. coli strain MC1000 under arabinose-inducible control (strains CJW2193 or CJW2194, respectively). When crescentin synthesis was repressed, E. coli cells exhibited wild-type rod morphology (Figure 5A, inset). Strikingly, a curved morphology developed upon induction (Figure 5A). Crescentin production also caused cell chaining and filamentation, leading to the appearance of helical cells (Figure 5A, arrows). We attributed these additional phenotypes to interference with cell division in E. coli (Figure 5B, arrows), which is probably not equipped to handle the filamentous crescentin structure during cytokinesis. Therefore, to better visualize cell curvature, we blocked cell division with cephalexin while repressing or inducing crescentin synthesis.
Figure 5.
Crescentin can produce cell curvature in E. coli. (A) Composite phase-contrast image (assembled from overlapping frames) of CJW2193 (E. coli MC1000/pBAD18[Cm]creS) cells grown in M9-glycerol supplemented with 0.2% arabinose (to induce crescentin production) for 4 h. The arrows indicate elongated, helical cells. The inset (bar 5 μm) shows the morphology of CJW2193 cells when grown in the presence of 0.2% glucose, which represses crescentin production. Bar, 10 μm. (B) CJW2194 cells (MC1000/pBAD18creS-tc) grown for 4 h in M9-glycerol with 0.2% arabinose and stained with FlAsH to visualize the crescentin-TC structure. Arrows show cell chaining, presumably caused by interference at cell division sites by the crescentin structure. Bar, 2 μm. (C) CJW2193 cells (E. coli MC1000/pBAD18[Cm]creS) grown in M9-glycerol with 0.2% glucose for 3.5 h (to repress crescentin production) or 0.2% arabinose for 4 h (to induce crescentin synthesis) in the presence of cephalexin (to block cell division). Bar, 5 μm. (D) SEM of a cephalexin-treated CJW2193 E. coli cell producing crescentin for 4.5 h. (E) Cephalexin-treated CJW2194 (MC1000/pBAD18[Cm]creS-tc) grown in M9-glycerol with arabinose for 4.5 h and stained with FlAsH to visualize the crescentin-TC structure. Bars, 2 μm.
Cephalexin-treated E. coli cells producing crescentin commonly displayed a helical morphology, whereas without crescentin, they only produced random bends due to cell flexibility (Figure 5C). Scanning electron microscopy (SEM; Figure 5D) revealed that helical E. coli cells producing crescentin were invariably left handed (100%, n=196 cells), mimicking the invariable (n=200) left-handed helicity of C. crescentus cells blocked for cell division (not shown). The E. coli cells displayed a filamentous crescentin-TC structure along their inner cell curvatures (Figure 5E). Cells occasionally displayed multiple crescentin-TC structures (Supplementary Figure S5, arrows), suggesting that control of crescentin assembly in E. coli is not as highly tuned as in C. crescentus. However, by and large, crescentin assembled in E. coli into filamentous structures at the cell periphery that robustly generated cell curvature.
Crescentin requires its basic N-terminus for cell envelope attachment and function
To hold the crescentin structure in an extended configuration and at a fixed position along the membrane, a connection between the crescentin structure and the peptidoglycan is required, as the peptidoglycan is the only element with sufficient size and structural rigidity to perform such a function. The ability of crescentin to curve E. coli suggests that this connection would be mediated by one or more proteins that are not unique to C. crescentus. As E. coli is not under any selective pressure to maintain crescentin-interacting proteins, cell envelope association is most likely mediated by relatively nonspecific interactions that are shared among divergent bacterial species. With this in mind, we sought primary sequence features of crescentin that might be required for such interactions.
The first 27 amino acids of crescentin (N27) have a calculated isoelectric point (pI) of 9.7, compared with 5.3 for the entire protein and 5.5 for the head domain (first 79 residues) alone (see Figure 2A). We hypothesized that charged residues in the N27 region (which is out of the way of the coiled-coil forming rod domains and potentially more accessible) might be important for cell wall attachment of the crescentin structure. A mutant lacking the basic N27 region (crescentinΔN27) failed to complement the straight-rod shape of ΔcreS cells (CJW2927), indicating that this N-terminal region is essential for function (Figure 6A). CrescentinΔN27-TC formed arcs and S-shaped filamentous structures (Figure 6A) that moved within the cell (Supplementary Movie 3), indicating detachment from the cell envelope. Moreover, crescentinΔN27-TC failed to pellet with the membrane fraction in biochemical fractionation experiments while a portion of wild-type crescentin was present in this fraction (Supplementary Figure S6). Therefore, the N27 region is dispensable for filamentous structure formation but required for cell envelope attachment. Substitution of a lysine with a glutamine at position 12 (K12Q) lowers the predicted pI of the N27 region from 9.7 to 8.3. Although largely functional, this loss of a single basic residue within the N27 region caused a five-fold increase in crescentin structure detachment over wild type (1.6 versus 0.3%, n=716 and 689, respectively; Figure 6B).
Figure 6.
Crescentin requires its basic N-terminus for cell envelope attachment. (A) FlAsH-stained crescentinΔN27-TC (red, laid over phase contrast, cyan) produced in a ΔcreS background (CJW2927; CB15N ΔcreS/pMR20creSΔN27-tc). A FlAsH-only micrograph is also provided for better visualization of the fluorescent signal. Bar, 2 μm. (B) FlAsH-stained crescentinK12Q-TC (red, laid over phase contrast, cyan) produced in a ΔcreS background (CJW2818; CB15N ΔcreS/pMR10creSK12Q-tc). Arrow indicates a cell in which the crescentinK12Q-TC structure has detached from the cell membrane. Bar, 1 μm. (C) Cephalexin-treated CJW2199 cells (MC1000/pBAD18creSΔN27-tc) producing crescentinΔN27-TC induced for 4 h. Bar, 5 μm.
This N27 region was required for function in E. coli as well. Cephalexin-treated E. coli cells producing crescentinΔN27-TC (CJW2199) had only the random and transient curvatures due to cell flexibility (Figure 6C), in contrast to the helical morphology of E. coli cells expressing wild-type crescentin (Figure 5C). CrescentinΔN27-TC formed filamentous structures in these cells that showed no consistent localization patterns.
Discussion
Walled bacterial cells are pressurized by the force of turgor pressure, which is maintained in a wide variety of conditions by sophisticated osmoregulatory systems (Wood, 1999). The strong but elastic peptidoglycan restrains this pressure, causing the bonds within the peptidoglycan meshwork to stretch (Koch, 1984; Koch et al, 1987; Baldwin et al, 1988). Although the orientation of the stiff glycan strands relative to the membrane and the cell axes is still being debated (Höltje, 1996, 1998; Koch, 1998, 2000; Dmitriev et al, 2003; Vollmer and Höltje, 2004; Meroueh et al, 2006), it is agreed that peptide bridges within a peptidoglycan layer are parallel to the membrane, where they stretch under the stress of turgor pressure. Stretched peptide bridges are easier to break, as energy added to the bonds by stretching forces helps overcome molecular cohesion and lowers the energy barrier of breakage (Bustamante et al, 2004). This stretch thus facilitates the activity both of hydrolytic enzymes and new wall insertion, through the coordinated activity of hydrolysis and insertion (Romeis and Höltje, 1994; Vollmer et al, 1996, 1999; Höltje, 1998; Schiffer and Höltje, 1999; Bertsche et al, 2006).
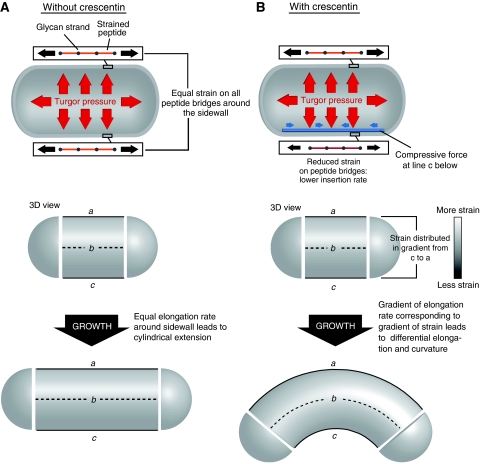
Most rod-shaped bacteria elongate by inserting new peptidoglycan along the cylindrical sidewall, with little or no insertion at the poles (de Pedro et al, 1997; Daniel and Errington, 2003; Tiyanont et al, 2006). As the contribution of turgor pressure to peptide bridge stretch is uniform around the circumference, new insertion is equally favourable all around the cell cylinder (Figure 7A). Our results show that the crescentin structure produces a bias in new wall insertion. The crescentin structure is stably anchored to the cell envelope and lengthens from the structure ends only (Charbon et al, in preparation), while cells elongate not from the ends but from the sidewall, either along its entire length or from a zone at midcell (Aaron et al, 2007). Therefore, any tension produced in the crescentin structure by virtue of its being extended as a cell elongates along its length or from the midcell zone would produce an equal compressive force in the cell wall. Such a compressive force, even if very small in magnitude, necessarily opposes turgor-induced peptide bridge stretch (Figure 7B). Physical modelling experiments demonstrate that a cell can be physically bent into a curved shape if a single filament applies a compressive force on pressurized elastic peptidoglycan (Wolgemuth et al, 2005). In our proposed model, the compressive force is much smaller, so that it does not substantially bend the cell but rather alters the kinetics of peptidoglycan hydrolysis and insertion. By reducing the stretching on the peptide bridges, the crescentin structure makes bond hydrolysis slightly less favourable, reducing the probability of hydrolysis and insertion (Figure 7B). A key advantage of this mechanism is that an elastic fabric like the peptidoglycan can propagate stress over distance, distributing the compressive force away from the point of crescentin structure attachment (line c, Figure 7B) in a gradient around the circumference of the sidewall (Figure 7B). This results in a gradient of cell wall elongation rates, which translates over time to a gradient of longitudinal cell lengths around the sidewall (Figure 7B). Notably, this mechanism is not based on differential localization of cell wall enzymes but only on their differential activity. If crescentin contributes to cell curvature in additional ways, for example by positioning a peptidoglycan synthesis inhibitor or by producing inhibition through some other path, this inhibitor or pathway would have to be present in E. coli and would require a way to distribute the inhibitory activity in a gradient away from the site of crescentin structure attachment.
Figure 7.
Model for crescentin action. (A) A rod-shaped cell lacking crescentin. Turgor pressure strains peptide bridges (red), which favours hydrolysis and new wall insertion. The glycan strands are represented as perpendicular to the long axis of the cell according to the prevailing view, but the model holds irrespective of the orientation of the glycan strands, as the peptide bridges are the extensible elements of the peptidoglycan. In such a cell, turgor pressure produces equal stress all around the circumference of the sidewall (3D view, a, b, c), and thus wall insertion is equally favoured all around the sidewall circumference. Cell elongation then causes straight elongation into a longer rod. (B) A cell with a crescentin structure (blue), which is affixed to the cell membrane in a stretched configuration. This in turn produces a compressive force on the cell wall (blue arrows), which slightly reduces local peptide bridge strain in a line corresponding to c in the 3D view. This small reduction in peptide bridge strain at line c is distributed in a smooth gradient of strain around the circumference of the sidewall because of the elasticity of the peptidoglycan fabric. Although it is too small a force to produce discernable morphological effects, the gradient of strain sets up a gradient of sidewall growth rates. Cell elongation under these conditions produces a gradient of cell lengths (from line a to line c), and thus curved morphology.
The model we propose is fully consistent with force alone (as in a microchamber) altering cell wall elongation rates to produce curved morphology. Similarly, the sufficiency of crescentin alone to produce curvature in E. coli argues against a requirement for additional species-specific regulatory factors. The linear-to-helical configuration shift of crescentin structures when released from their static location along the membrane shows that these structures are flexible, stretchable and elastic. These properties, which distinguish intermediate filaments from other cytoskeletal elements (Kreplak and Fudge, 2007), are ideal for resisting strain and generating a compressive force to modulate cell wall growth.
The cell envelope attachment mediated by the basic N27 region is essential for crescentin function, and the crescentin structure appears to be attached continuously along its length. This is notable, as even weak (presumably electrostatic) individual interactions along the length of the crescentin structure could be quite substantial when considered cumulatively. Many proteins interact directly or indirectly with the peptidoglycan (Cascales and Lloubes, 2004; Buist et al, 2008; Sauvage et al, 2008), and they provide a potential connection between the crescentin structure and the cell wall. The function of one such protein, MreB, has been recently shown to be required for crescentin structure attachment (Charbon et al, in preparation).
In order to grow, C. crescentus faces the same challenges as other walled organisms, such as plants, fungi and other bacteria—the cell wall must expand under turgor pressure. Wall expansion is therefore subject to forces cooperating with or opposing turgor pressure. Cytoskeletal elements that can bear such forces might be a common strategy to control growth in space and time to achieve specific shapes in walled cells. Recently, it has been shown that the bacterial tubulin homolog FtsZ can produce a constricting force (Osawa et al, 2008) in addition to its established role in the recruitment of cell division proteins (Margolin, 2005; den Blaauwen et al, 2008). It is conceivable that this constricting force may influence the direction of cell wall growth or help to mitigate the outward force of turgor pressure. Mathematical modelling in fact predicts that a very small force (∼8 pN) can promote peptidoglycan cross-bridge cleavage to direct growth at the division site (Lan et al, 2007). Thus, the conversion of mechanical forces into biochemical responses may be a morphogenetic strategy in bacteria, as it is in eukaryotes (Vogel and Sheetz, 2006).
Materials and methods
Strains, plasmids, media and growth conditions
C. crescentus was grown in PYE (peptone-yeast extract) or M2G supplemented with 1% PYE (M2G+) at 30°C. Plasmids were mobilized from E. coli strain S17-1 into C. crescentus by conjugation (Ely, 1991). Plasmids and strains are listed in Supplementary Table S1, and their construction and primers used are described in supplemental text. Log-phase cultures were used for all experiments. The xylose-inducible promoter (Pxyl) was induced by adding 0.3 or 0.03% xylose to M2G+. Mecillinam was used with C. crescentus Δbla strains at 5-40 μg/ml. E. coli MC1000 was grown in M9 medium with 0.2% glycerol; the arabinose promoter was repressed or induced with 0.2% glucose or arabinose, respectively. Cephalexin was used at 40 μg/ml for E. coli MC1000 strains.
Preparation of anti-crescentin antibodies and immunoblottling
His6-tagged crescentin was purified as described (Ausmees et al, 2003) and used to make polyclonal rabbit antibodies (Pocono Rabbit Farm & Laboratory). Antibodies were affinity purified from serum using His6-crescentin bound to nitrocellulose membrane. Immunoblotting was performed with anti-crescentin at a dilution of 1:10 000.
Sacculus isolation, electron microscopy and muropeptide analysis
Sacculi for EM and muropeptide analysis were isolated as described (Glauner, 1988). Briefly, cells were chilled, harvested by centrifugation, resuspended in water, dropped into boiling 8% SDS and boiled for 30 min, ultracentrifuged and washed free of SDS by repeated resuspension in water and ultracentrifugation. The sacculi were successively treated with amylase and pronase. After addition of SDS to 1%, the sacculi were heated at 80°C for 20 min, and SDS was removed as described earlier.
EM was performed as described (de Pedro et al, 1997). The isolated sacculi were immobilized on glow discharged carbon-pioloform-coated copper grids (400 mesh) and incubated 10 min with PBG (0.5% bovine serum albumin, 0.2% gelatin in phosphate-buffered saline). After two brief washes with PBS and four brief washes with water, the sacculi were contrasted with 1% (W/V) uranyl acetate for 1 min. The grids were briefly washed with water and air dried and were visualized using a Philips CM10 transmission EM at 60 kV.
Muropeptides were obtained by digestion of isolated sacculi with cellosyl, followed by reduction with sodium borohydride. HPLC analysis was performed as published (Markiewicz et al, 1983; Glauner et al, 1988).
D-cysteine labelling
Log-phase cells were grown for about four doublings in PYE medium supplemented with 125 μg/ml D-cysteine, then they were pelleted, resuspended in PYE alone and grown for 90 min. Sacculi were purified, reduced and biotinylated as described (de Pedro et al, 1997). The biotinylated sacculi were diluted in dH2O and applied to glow-discharged carbon-coated nickel EM grids (400 mesh). Sacculi were blocked for 15 min with 2% BSA in PBS, then probed with 1:50 rabbit anti-biotin antibodies (Abcam) for 1 h, rinsed thrice in PBS, probed with 1:10 or 1:25 nanogold-coupled goat anti-rabbit antibody (Nanoprobes) for 1 h, rinsed thrice in PBS, postfixed with 1% glutaraldehyde in PBS for 3 min, rinsed in dH2O for 5 min twice, silver enhanced (Nanoprobes) for 4 min, rinsed with dH2O thrice, stained with 2% uranyl acetate 5 min and rinsed in dH2O thrice before drying and microscopy. Images were taken with a Hamamatsu Orca-HR camera with AMT Image Capture software on a JEOL JEM-1230 transmission EM at 80 kV.
Light microscopy
Cells were imaged at room temperature (∼22°C) or with an objective heater at 31°C using either a Nikon E1000 microscope fitted with 100 × DIC or phase-contrast objectives and a Hamamatsu Orca-ER LCD camera or a Nikon Eclipse 80i microscope equipped with 100 × DIC or phase-contrast objectives and an Andor iXonEM+ (DU-897E) EMCCD camera. Cells were immobilized on 1% agarose-M2G+ or M9-glycerol pads supplemented with 0.3% xylose or 0.2% arabinose when appropriate. Immunofluorescence microscopy was performed as described earlier (Ausmees et al, 2003) using anti-crescentin antibodies at 1:200 or 1:400. Images were taken and analysed with Metamorph software (Molecular Devices). Composite images were assembled using Photoshop CS3 (Adobe Systems).
Cells producing TC-tagged crescentin were stained in M2G+ (C. crescentus) or M9-glycerol (E. coli) with 5 μM FlAsH reagent (Griffin et al, 1998) (Invitrogen or made in-house by DBW) and 20 μM ethanedithiol (EDT) (plus inducers when desired) for 30 min at 30°C (C. crescentus) or 37°C (E. coli) in an Eppendorf Thermomixer. Stained cells were then washed and resuspended in FlAsH-free medium (with inducer when desired) before mounting on slides.
Scanning electron microscopy
E. coli cultures in M9-glycerol with 0.2% arabinose were harvested and fixed in 5% glutaraldehyde, 4% formaldehyde in 0.08 M sodium phosphate buffer pH 7.2 for 30 min at room temperature. Cells were washed twice with 1 × PBS, applied to poly-L-lysine-coated glass coverslips, dehydrated in a series of ethanol baths, critical-point dried and gold coated. Images were captured in an FEI XL-30 ESEM FEG microscope (10 kV, spot size 3, working distance 7.5–10 mm).
Microchamber experiments
Agarose microchamber arrays were made from a PDMS mold (made in-house by DBW) by pouring liquid 3% agarose-PYE with 0.05% BSA at 65°C onto a mold and placing a microscope slide on top. After polymerization, excess agarose was removed, 0.5 μl of cell suspension was placed on the array, and a cover slip was placed over the array for light microscopy. For release of confined cells after growth, the coverslip was removed from the array and immediately transferred to a fresh 1% agarose-PYE (with 40 μg/ml chloramphenicol to arrest growth when required) pad and imaged.
Cell shape analysis
To detect the cells, a home-made program written in Matlab language was used. Cell outlines were obtained from phase-contrast images by thresholding and subsequence smoothing. The centerline (skeleton) of each cell was then detected by finding points along the long axis of the cell equidistant from opposite edges of the cell outline. Occasional cells where the centerline shape was not representative of cell curvature (e.g. predivisional cells with a kink at the division plane) were removed manually before analysis. To find the curvature of the cells, multiple points located equidistantly along the centerline were fitted to an arc of a circle, minimizing the sum of the squared distances to the centerline points from their projections to the arc. The reciprocal of the radius of the best-fit arc is the curvature value. Statistical calculations were done with Matlab.
Biochemical fractionation
Cells (500 ml) were grown to mid-log phase in PYE medium, washed in CBB buffer (20 mM Tris–Cl pH 8.0, 25 mM NaCl, 5 mM EDTA, 3.6 mM β-mercaptoethanol), and resuspended in 10 ml CBB buffer with a protease inhibitor cocktail tablet (Roche). Cells were lysed by two passes through a French press (∼1500 psi) and the lysate was cleared by centrifugation at 20 000 g for 15 min at 4°C. The cleared lysate was then centrifuged at 150 000 g for 2 h at 4°C, and a sample of the supernatant was taken for analysis. The pellet was resuspended in 1 ml CBB buffer and spun at 105 000 g for 1 h. The supernatant was discarded and the washed pellet was resuspended in 1 ml CBB with 1% SDS and used for analysis.
Supplementary Material
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Information
Acknowledgments
We thank O Sliusarenko and T Emonet for help with the automated cell shape analysis and for helpful discussions; Z Jiang for help with the SEM; G Ebersbach for advice about E. coli experiments; J Beckwith for the pBAD vectors; H Schwarz for help with transmission EM; E Dufresne for helpful discussions; and the Jacobs-Wagner laboratory for critical reading of the manuscript. MTC was supported by the NSF GRFP and the Mustard Seed Foundation. This work was supported by the European Commission (LSHM-CT-2004-512138 to WV), the German Research Council DFG (FOR449 to WV), the Searle Scholars Program (to DW), 3M Corporation National (DW), the National Institutes of Health (GM076698 to CJ-W) and the Pew Charitable Trusts (to CJ-W). Author contributions: MTC, GC, NA, CJ-W designed research; MTC, WV, PB, DBW performed experiments; MTC, WV, CJ-W analysed the data; MTC and CJ-W wrote the paper.
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C (2007) The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol 64: 938–952 [DOI] [PubMed] [Google Scholar]
- Arnoldi M, Fritz M, Bauerlein E, Radmacher M, Sackmann E, Boulbitch A (2000) Bacterial turgor pressure can be measured by atomic force microscopy. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics 62 (1 Part B): 1034–1044 [DOI] [PubMed] [Google Scholar]
- Ausmees N, Kuhn JR, Jacobs-Wagner C (2003) The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115: 705–713 [DOI] [PubMed] [Google Scholar]
- Baldwin WW, Sheu MJ, Bankston PW, Woldringh CL (1988) Changes in buoyant density and cell size of Escherichia coli in response to osmotic shocks. J Bacteriol 170: 452–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsche U, Kast T, Wolf B, Fraipont C, Aarsman ME, Kannenberg K, von Rechenberg M, Nguyen-Disteche M, den Blaauwen T, Höltje JV, Vollmer W (2006) Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol Microbiol 61: 675–690 [DOI] [PubMed] [Google Scholar]
- Boulbitch A, Quinn B, Pink D (2000) Elasticity of the rod-shaped gram-negative eubacteria. Phys Rev Lett 85: 5246–5249 [DOI] [PubMed] [Google Scholar]
- Briegel A, Dias DP, Li Z, Jensen RB, Frangakis AS, Jensen GJ (2006) Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol Microbiol 62: 5–14 [DOI] [PubMed] [Google Scholar]
- Buist G, Steen A, Kok J, Kuipers OP (2008) LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68: 838–847 [DOI] [PubMed] [Google Scholar]
- Bustamante C, Chemla YR, Forde NR, Izhaky D (2004) Mechanical processes in biochemistry. Annu Rev Biochem 73: 705–748 [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C (2007) Skin and bones: the bacterial cytoskeleton, cell wall, and cell morphogenesis. J Cell Biol 179: 381–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-Lopez R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J (2006) Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell 11: 399–409 [DOI] [PubMed] [Google Scholar]
- Cascales E, Lloubes R (2004) Deletion analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol Microbiol 51: 873–885 [DOI] [PubMed] [Google Scholar]
- Chen F, Chang R, Trivedi M, Capetanaki Y, Cryns VL (2003) Caspase proteolysis of desmin produces a dominant-negative inhibitor of intermediate filaments and promotes apoptosis. J Biol Chem 278: 6848–6853 [DOI] [PubMed] [Google Scholar]
- Daniel RA, Errington J (2003) Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113: 767–776 [DOI] [PubMed] [Google Scholar]
- de Pedro MA, Quintela JC, Höltje JV, Schwarz H (1997) Murein segregation in Escherichia coli. J Bacteriol 179: 2823–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA (2008) Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev 32: 321–344 [DOI] [PubMed] [Google Scholar]
- Divakaruni AV, Baida C, White CL, Gober JW (2007) The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol Microbiol 66: 174–188 [DOI] [PubMed] [Google Scholar]
- Dmitriev BA, Toukach FV, Schaper KJ, Holst O, Rietschel ET, Ehlers S (2003) Tertiary structure of bacterial murein: the scaffold model. J Bacteriol 185: 3458–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z (2005) Two independent spiral structures control cell shape in Caulobacter. Proc Natl Acad Sci USA 102: 18608–18613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B (1991) Genetics of Caulobacter crescentus. Methods Enzymol 204: 372–384 [DOI] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW (2004) MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol 51: 1321–1332 [DOI] [PubMed] [Google Scholar]
- Fischer R, Zekert N, Takeshita N (2008) Polarized growth in fungi—interplay between the cytoskeleton, positional markers and membrane domains. Mol Microbiol 68: 813–826 [DOI] [PubMed] [Google Scholar]
- Gaboriaud F, Bailet S, Dague E, Jorand F (2005) Surface structure and nanomechanical properties of Shewanella putrefaciens bacteria at two pH values (4 and 10) determined by atomic force microscopy. J Bacteriol 187: 3864–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauner B (1988) Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem 172: 451–464 [DOI] [PubMed] [Google Scholar]
- Glauner B, Höltje JV, Schwarz U (1988) The composition of the murein of Escherichia coli. J Biol Chem 263: 10088–10095 [PubMed] [Google Scholar]
- Griffin BA, Adams SR, Tsien RY (1998) Specific covalent labeling of recombinant protein molecules inside live cells. Science 281: 269–272 [DOI] [PubMed] [Google Scholar]
- Harold FM (2002) Force and compliance: rethinking morphogenesis in walled cells. Fungal Genet Biol 37: 271–282 [DOI] [PubMed] [Google Scholar]
- Harold FM (2007) Bacterial morphogenesis: learning how cells make cells. Curr Opin Microbiol 10: 591–595 [DOI] [PubMed] [Google Scholar]
- Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U (2007) Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol 8: 562–573 [DOI] [PubMed] [Google Scholar]
- Herrmann H, Haner M, Brettel M, Ku NO, Aebi U (1999) Characterization of distinct early assembly units of different intermediate filament proteins. J Mol Biol 286: 1403–1420 [DOI] [PubMed] [Google Scholar]
- Höltje JV (1996) A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology 142 (Part 8): 1911–1918 [DOI] [PubMed] [Google Scholar]
- Höltje JV (1998) Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev 62: 181–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS (2008) Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci USA 105: 19282–19287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J (2001) Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104: 913–922 [DOI] [PubMed] [Google Scholar]
- Koch AL (1984) Shrinkage of growing Escherichia coli cells by osmotic challenge. J Bacteriol 159: 919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AL (1998) Orientation of the peptidoglycan chains in the sacculus of Escherichia coli. Res Microbiol 149: 689–701 [DOI] [PubMed] [Google Scholar]
- Koch AL (2000) Length distribution of the peptidoglycan chains in the sacculus of Escherichia coli. J Theor Biol 204: 533–541 [DOI] [PubMed] [Google Scholar]
- Koch AL, Higgins ML, Doyle RJ (1982) The role of surface stress in the morphology of microbes. J Gen Microbiol 128: 927–945 [DOI] [PubMed] [Google Scholar]
- Koch AL, Lane SL, Miller JA, Nickens DG (1987) Contraction of filaments of Escherichia coli after disruption of cell membrane by detergent. J Bacteriol 169: 1979–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AL, Woeste S (1992) Elasticity of the sacculus of Escherichia coli. J Bacteriol 174: 4811–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreplak L, Fudge D (2007) Biomechanical properties of intermediate filaments: from tissues to single filaments and back. Bioessays 29: 26–35 [DOI] [PubMed] [Google Scholar]
- Kruse T, Bork-Jensen J, Gerdes K (2005) The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol 55: 78–89 [DOI] [PubMed] [Google Scholar]
- Lan G, Wolgemuth CW, Sun SX (2007) Z-ring force and cell shape during division in rod-like bacteria. Proc Natl Acad Sci USA 104: 16110–16115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W (2005) FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol 6: 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz Z, Glauner B, Schwarz U (1983) Murein structure and lack of DD- and LD-carboxypeptidase activities in Caulobacter crescentus. J Bacteriol 156: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S (2006) Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc Natl Acad Sci USA 103: 4404–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP (2008) Reconstitution of contractile FtsZ rings in liposomes. Science 320: 792–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penegar I, Toque C, Cornell SDA, Smith JR, Campbell SA (1999) Nano-indentation measurements of the marine bacteria Sphingomonas paucimobilis using the atomic force microscope. 10th International Congress on Marine Corrosion Fouling; Melbourne, Australia
- Poindexter JS, Hagenzieker JG (1982) Novel peptidoglycans in Caulobacter and Asticcacaulis spp. J Bacteriol 150: 332–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112: 453–465 [DOI] [PubMed] [Google Scholar]
- Romeis T, Höltje JV (1994) Specific interaction of penicillin-binding proteins 3 and 7/8 with soluble lytic transglycosylase in Escherichia coli. J Biol Chem 269: 21603–21607 [PubMed] [Google Scholar]
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P (2008) The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32: 556. [DOI] [PubMed] [Google Scholar]
- Schietke R, Brohl D, Wedig T, Mucke N, Herrmann H, Magin TM (2006) Mutations in vimentin disrupt the cytoskeleton in fibroblasts and delay execution of apoptosis. Eur J Cell Biol 85: 1–10 [DOI] [PubMed] [Google Scholar]
- Schiffer G, Höltje JV (1999) Cloning and characterization of PBP 1C, a third member of the multimodular class A penicillin-binding proteins of Escherichia coli. J Biol Chem 274: 32031–32039 [DOI] [PubMed] [Google Scholar]
- Seitz LC, Brun YV (1998) Genetic analysis of mecillinam-resistant mutants of Caulobacter crescentus deficient in stalk biosynthesis. J Bacteriol 180: 5235–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih YL, Le T, Rothfield L (2003) Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA 100: 7865–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Oppenheimer DG (2005) Spatial control of cell expansion by the plant cytoskeleton. Annu Rev Cell Dev Biol 21: 271–295 [DOI] [PubMed] [Google Scholar]
- Takeuchi S, DiLuzio WR, Weibel DB, Whitesides GM (2005) Controlling the shape of filamentous cells of Escherichia coli. Nano Lett 5: 1819–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiyanont K, Doan T, Lazarus MB, Fang X, Rudner DZ, Walker S (2006) Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc Natl Acad Sci USA 103: 11033–11038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadillo-Rodriguez V, Beveridge TJ, Dutcher JR (2008) Surface viscoelasticity of individual gram-negative bacterial cells measured using atomic force microscopy. J Bacteriol 190: 4225–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V, Sheetz M (2006) Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol 7: 265–275 [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA (2008) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32: 149–167 [DOI] [PubMed] [Google Scholar]
- Vollmer W, Höltje JV (2004) The architecture of the murein (peptidoglycan) in gram-negative bacteria: vertical scaffold or horizontal layer(s)? J Bacteriol 186: 5978–5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, von Rechenberg M, Höltje JV (1999) Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J Biol Chem 274: 6726–6734 [DOI] [PubMed] [Google Scholar]
- von Rechenberg M, Ursinus A, Höltje JV (1996) Affinity chromatography as a means to study multienzyme complexes involved in murein synthesis. Microb Drug Resist 2: 155–157 [DOI] [PubMed] [Google Scholar]
- Wolgemuth CW, Inclan YF, Quan J, Mukherjee S, Oster G, Koehl MA (2005) How to make a spiral bacterium. Phys Biol 2: 189–199 [DOI] [PubMed] [Google Scholar]
- Wood JM (1999) Osmosensing by bacteria: signals and membrane-based sensors. Microbiol Mol Biol Rev 63: 230–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Jericho M, Pink D, Beveridge T (1999) Thickness and elasticity of gram-negative murein sacculi measured by atomic force microscopy. J Bacteriol 181: 6865–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Walter J, Burke S, Stewart S, Jericho MH, Pink D, Hunter R, Beveridge TJ (2002) Atomic force microscopy and theoretical considerations of surface properties and turgor pressures of bacteria. Colloids Surf B Biointerfaces 23: 213–230 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Information