Abstract
Heterodimeric integrin adhesion receptors regulate cell migration, survival and differentiation in metazoa by communicating signals bi-directionally across the plasma membrane. Protein engineering and mutagenesis studies have suggested that the dissociation of a complex formed by the single-pass transmembrane (TM) segments of the α and β subunits is central to these signalling events. Here, we report the structure of the integrin αIIbβ3 TM complex, structure-based site-directed mutagenesis and lipid embedding estimates to reveal the structural event that underlies the transition from associated to dissociated states, that is, TM signalling. The complex is stabilized by glycine-packing mediated TM helix crossing within the extracellular membrane leaflet, and by unique hydrophobic and electrostatic bridges in the intracellular leaflet that mediate an unusual, asymmetric association of the 24- and 29-residue αIIb and β3 TM helices. The structurally unique, highly conserved integrin αIIbβ3 TM complex rationalizes bi-directional signalling and represents the first structure of a heterodimeric TM receptor complex.
Keywords: cell adhesion, integrin receptors, membrane proteins, transmembrane signalling
Introduction
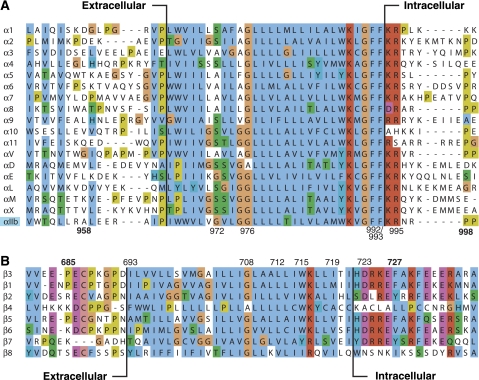
The family of integrin cell adhesion receptors regulates cell migration, survival and differentiation in metazoa by responding to intracellular and extracellular signals in inside-out and outside-in signalling pathways, respectively (Hynes, 2002; Askari et al, 2009; Harburger and Calderwood, 2009). Integrins are Type I heterodimeric receptors that consist of large extracellular domains (>700 residues), single-pass transmembrane (TM) segments and generally short cytosolic tails (<70 residues). Integrin αIIbβ3, the primary adhesion receptor of blood platelets, mediates platelet aggregation by binding to the multivalent blood protein fibrinogen (Hynes, 2002; Bennett, 2005). It is essential to both the arrest of bleeding at sites of vascular injury and pathological thrombosis culminating in heart attack and stroke. The sequences of the TM segments of the 18 α and 8 β human subunits are well conserved (Figure 1), and the TM region of integrin αIIbβ3 closely resembles many of the 18 α and 8 β human subunits, in particular the distinct, hydrophobic sequences C-terminal to the first charged residues on the intracellular side, αIIb(K989) and β3(K716). Integrin bi-directional TM signalling involves the dissociation of a complex formed by the α–β TM segments (Hughes et al, 1996; Kim et al, 2003; Li et al, 2005; Partridge et al, 2005; Zhu et al, 2007), which is accompanied by large rearrangements of its extracellular domains (Xiao et al, 2004; Adair et al, 2005; Rocco et al, 2008; Ye et al, 2008; Zhu et al, 2008). Recently, we have determined the structures and lipid embedding of the monomeric integrin αIIb and β3 TM segments in phospholipid bicelles (Lau et al, 2008a, 2008b), laying the foundation for understanding the structural transition between dissociated and associated TM states, and thus integrin αIIbβ3 TM signalling. Although the monomeric β3 TM segment forms a 29-residue α-helix that is tilted in the membrane and immerses β3(K716), the αIIb segment adopts a straight, 24-residue TM helix that terminates at αIIb(K989). C-terminal to the αIIb helix an unusual Gly-Phe-Phe backbone reversal is observed that immerses the two fully conserved Phe residues (Figure 1) back into the intracellular membrane leaflet (Lau et al, 2008a).
Figure 1.
Sequence alignment of the transmembrane segments of all human integrin subunits. (A) The 18 α subunits and (B) 8 β subunits are depicted. Proposed minimal lipid tail-to-headgroup borders for monomeric and heterodimeric α and β subunits are depicted (c.f. Figure 7) (Armulik et al, 1999; Stefansson et al, 2004; Lau et al, 2008a, 2008b). Conserved amino acids are coloured by the Jalview multiple alignment editor (Clamp et al, 2004) using the ClustalX colour scheme.
Many structural models of the signalling events in the membrane have been proposed and the structural information on the monomeric, dissociated αIIb and β3 TM states increases the possible structural transitions between the associated and dissociated state even further. In general, changes in the TM segment membrane crossing angles accompanying lateral separation of the TM segments pictured in helical conformation are predicted (Luo et al, 2004; Gottschalk, 2005; Li et al, 2005; Partridge et al, 2005; Wegener et al, 2007). In such an event, the Gly-Phe-Phe motif may convert to helical conformation, as suggested by reported cytosolic αIIbβ3 tail complex structures (Vinogradova et al, 2002; Weljie et al, 2002) and induce an αIIb TM helix elongation and tilt that may favour a modelled Glycophorin A-like αIIbβ3 association (Gottschalk, 2005). Alternatively, the five-residue hydrophobic stretch C-terminal to the first charged residues on the intracellular side of the β3 subunit, K716, may repartition into the cytosol, leading to a straight β3 TM helix and a more parallel αIIbβ3 TM association. Moreover, membrane embedding of the TM helices may also change at the extracellular face in a piston-like manner and thereby transmit a force to the integrin ectodomain. Finally, a direct association between the monomeric αIIb–β3 TM structures may occur, which would be the first example of an asymmetric, heterodimeric TM association. Here, we report the structure of the noncovalently associated αIIbβ3 TM complex, structure-based site-directed mutagenesis and lipid embedding estimates to differentiate the plethora of integrin TM signalling possibilities. Aside from providing insight into integrin biology, the integrin αIIbβ3 TM complex structure also advances the understanding of dimeric cell receptor TM complexes of which only homodimeric structures have been reported (MacKenzie et al, 1997; Call et al, 2006; Bocharov et al, 2007, 2008).
Results and discussion
Integrin αIIb–β3 heterodimerization and structure determination
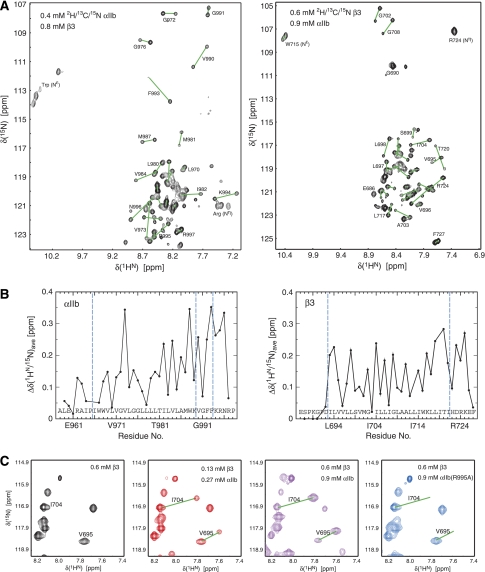
The structure of the noncovalently associated αIIbβ3 TM complex was determined in the phospholipid bilayer environment of small bicelles (Lee et al, 2008); this is essential for obtaining accurate integrin TM conformations (Lau et al, 2008b) and was achieved by solution NMR spectroscopy using peptides of several different 2H/13C/15N labelling patterns. Random mutagenesis of the β3 TM segment and the entire cytosolic tail shows that the used peptides, encompassing αIIb(A958-P998) and β3(P685-F727), contain all TM and cytosolic αIIbβ3 heterodimerization elements (Partridge et al, 2005). Under the chosen experimental conditions, the mixing of αIIb and β3 peptides resulted in the presence of the monomeric NMR resonances obtained earlier (Lau et al, 2008a, 2008b) and a new set of resonances, corresponding to the associated αIIbβ3 state (Figure 2A and B). All backbone resonance positions and, hence, the structures and chemical environment of the co-existing monomers and heterodimer were independent of αIIb-to-β3 peptide ratio and lipid-to-peptide ratio (Figure 2C). In analogy to the monomeric αIIb and β3 TM structures (Lau et al, 2008a, 2008b), no significant variation in backbone chemical shifts and, hence, αIIbβ3 complex structure was found between the herein used lipids of 16/18 and 14/14 carbon atom tails, and phosphocholine and serine headgroups (data not shown), which represent the major platelet lipids (GarciaGuerra et al, 1996). Incomplete, but dominant, heterodimerization in bicelles (Figure 2C) agrees with a relatively weak αIIbβ3 TM association detected in Escherichia coli membranes (Li et al, 2004; Schneider & Engelman, 2004), although the truncated TM segments used in these earlier studies may have unduly favoured homodimerization over heterodimerization. The αIIbβ3 association was sensitive to the integrity of the proposed αIIb(R995)–β3(D723) salt bridge (Hughes et al, 1996), as the introduction of the αIIb(R995A) substitution essentially led to the loss of heterodimeric resonances (Figure 2C; Supplementary Figure 2A and B). However, αIIb(R995A) did not abrogate the heterodimeric association completely. The signal intensities of the monomeric resonances still experienced significant reductions in the presence of αIIb(R995A) peptide (Supplementary Figure 2C). This indicates the presence of additional heterodimerization elements and, on the timescale of the NMR measurements, reflects a transition from the slow to the fast exchange limit. In conclusion, the αIIb and β3 TM segments associate quantitatively and specifically in phospholipid bicelles with contributions from αIIb(R995)–β3(D723) electrostatic interactions and additional heterodimerization elements.
Figure 2.
Illustration of αIIb–β3 heterodimerization at the protein backbone level. (A) H–N correlation spectra for each labelled subunit in the presence of its unlabelled partnering subunit. For each transmembrane resonance, a second one is obtained, as indicated by connecting lines for resonances in well-resolved spectral regions. For comparison, spectra for monomeric αIIb and β3 peptides are shown in Supplementary Figure 1. Spectra were recorded at 23°C and a 1H frequency of 700 MHz using bicelles composed of 385 mM 1,2-dihexanoyl-sn-glycero-3-phosphocholine, 83 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 41 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-L-serine]. (B) Average H–N chemical shift difference for a given residue between monomeric and heterodimeric resonances. The shift difference for a residue i is calculated as {[Δδi(1HN)2 + (Δδi(15N)/5)2]/2}1/2 (Grzesiek et al, 1997). (C) Verification of specific αIIb–β3 heterodimerization. The signal intensities of the monomeric and heterodimeric peaks, for example, V695 and I704 of the 2H/13C/15N-labelled β3 subunit, correlate with peptide concentrations, but their positions are independent of peptide concentrations, showing slow exchange on the NMR timescale. Moreover, a mutant αIIb(R995A) peptide does not induce any significant second set of resonances, verifying the specificity of the αIIbβ3 heterodimeric association in bicelles and corroborating strong αIIb(R995)–β3(D723) electrostatic interaction (c.f. main text).
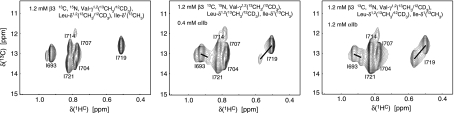
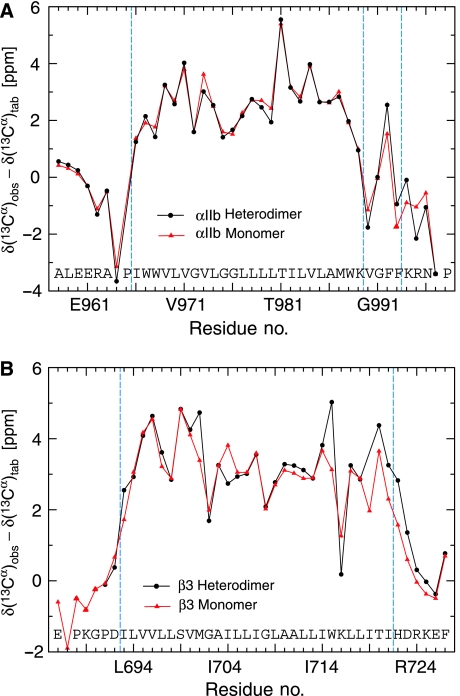
The observation of separate, nonaveraged signals for monomeric and heterodimeric protein backbone resonances (slow exchange limit; Figure 2) allows for the direct backbone structure determination of these co-existing species and a simultaneous comparison of their lipid embedding. It shows as well that complex dissociation is relatively slow in bicelles, taking place on the millisecond timescale, but association is also slow as heterodimerization is incomplete. Slow exchange behaviour was also observed at the level of side chain resonances, but, more often, monomeric and heterodimeric signals were averaged, resulting in only one resonance (fast exchange limit; Figure 3). The behaviour of a resonance to exhibit fast or slow exchange depends, in addition to the underlying exchange kinetics, on the chemical shift (frequency) difference between monomeric and heterodimeric states (Cavanagh et al, 1996). As the monomeric αIIb and β3 TM structures are already known (Lau et al, 2008a, 2008b), they can serve as a convenient reference for assessing any possible change in backbone conformation on heterodimerization. Secondary 13Cα chemical shifts exhibit a high correlation with backbone conformation and low dependence on the chemical environment (Spera and Bax, 1991; Xu and Case, 2001), which makes them the most sensitive NMR parameter for assessing backbone structural changes. The secondary 13Cα chemical shifts between monomeric and heterodimeric states are essentially superimposable (Figure 4). Minor 13Cα chemical shift differences at the intracellular membrane face are well within expected margins from the stabilization of secondary structures, particularly the two C-terminal β3 helix turns, by heterodimerization involving the newly formed αIIb(R995)–β3(D723) salt bridge. Thus, a transition of the distinct αIIb(G991-F992-F993) backbone reversal to helical conformations can be excluded. H–N chemical shifts, on the other hand, exhibit a low correlation with backbone conformation and high dependence on the chemical environment (Wagner et al, 1983; Wang and Jardetzky, 2004), which results in widespread HN–N shift changes from direct and indirect effects for all TM residues (Figure 2B). Further analysis of Hα, 15N, 13Cα and 13C′ backbone chemical shifts (Cornilescu et al, 1999) and HN–HN NOEs confirms that backbone secondary structures are indeed indistinguishable between monomeric and heterodimeric states (Supplementary Figure 3 and Table I). To reliably define the heterodimerization interface, extensive use of selective methyl labelling (Tugarinov and Kay, 2003) was necessary to identify close αIIb–β3 interproton distances, while suppressing the otherwise dominating intrasubunit NOE signals. Including the αIIb(R995)–β3(D723) salt bridge verified here by mutagenesis, 27 intersubunit distance restraints were thus unambiguously identified (Supplementary Figure 4), yielding appropriate coordinate precisions of 0.68 Å for the backbone-heavy atoms of the ensemble of 20 calculated simulated annealing structures (Figure 5A; Supplementary Table II). The salt bridge may also be excluded without adverse effect on the dimer structure, which independently juxtaposes αIIb(R995) and β3(D723) (Figure 5E and F). Residual dipolar couplings for the heterodimeric state could be measured with only limited accuracy, which negated their inclusion in the structure calculations.
Figure 3.
Illustration of αIIb–β3 heterodimerization at the protein side chain level. H–C correlation spectra of the β3 isoleucine methyl groups are shown for β3 alone, and for β3 in the presence of increasing concentrations of its partnering αIIb subunit. For I693 and I719, a second heterodimeric resonance is obtained, as indicated by the connecting lines (slow exchange limit). For the remaining four isoleucines, small or no shifts of only one resonance are detected (fast exchange limit). The behaviour of a resonance to exhibit fast or slow exchange depends, among other factors, on the chemical shift difference between monomeric and heterodimeric states.
Figure 4.
Comparison of 13Cα secondary chemical shifts between monomeric and heterodimeric integrin αIIb–β3 transmembrane states. (A, B) Secondary 13Cα chemical shifts, defined as the difference between the observed and tabulated random-coil 13Cα shift of a residue, correlate with the underlying backbone conformation but are also sensitive to local backbone dynamics (Spera and Bax, 1991; Ulmer et al, 2005). The minor differences between monomeric and heterodimeric shifts indicate the absence of significant backbone rearrangements on heterodimerization and the stabilization of secondary structure at the intracellular side in the presence of αIIb(R995)–β3(D723) electrostatic interactions (Figure 2C). Chemical shifts were measured in bicelles composed of 385 mM 1,2-dihexanoyl-sn-glycero-3-phosphocholine, 83 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 41 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-L-serine].
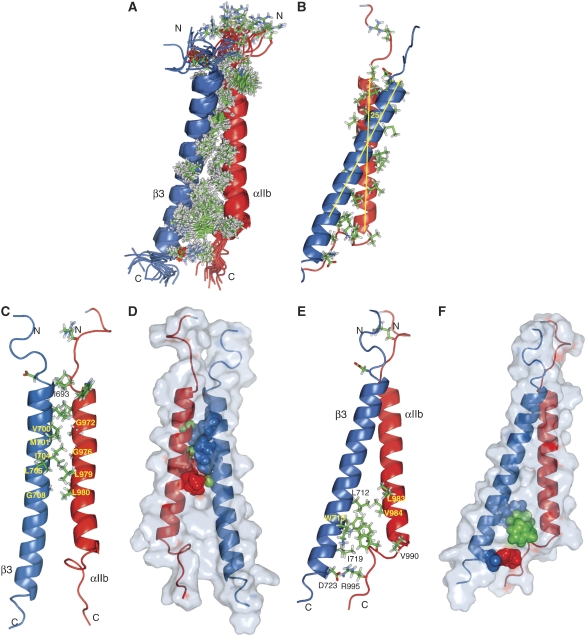
Figure 5.
Structure of the integrin αIIbβ3 transmembrane complex. (A) Superposition of the ensemble of 20 calculated simulated annealing structures. αIIb(I966-R995) and β3(I693-D723) adopt well-structured conformations. (B, C, E) Selected views of the energy minimized, average structure. (D) The outer membrane clasp: illustration of glycine packing. αIIb(G972), αIIb(G976) and β3(G708) are shown in green spheres, with their β3 and αIIb packing residues shown in blue and red spheres, respectively. (F) The inner membrane clasp: stabilization of the α–β TM helix arrangement by αIIb(F992-F993)-mediated interhelical packing and αIIb(R995)–β3(D723) electrostatic interaction. β3(W715) and β3(D723) are shown in blue, αIIb(R995) in red, and αIIb(F992-F993) in green spheres.
Structure of the αIIbβ3 TM complex and structure-based mutagenesis
The αIIb and β3 TM helices commence in close proximity at the extracellular membrane border, with close side chain distances detectable (Figure 5C) in agreement with previous disulfide cross-linking studies in the native receptor (Luo et al, 2004). Guided by packing interactions with three distinct glycine residues, αIIb(G972), αIIb(G976) and β3(G708), the TM helices cross within their N-terminal halves at an angle of 25±3° (Figure 5B–D). This and the differing length of the αIIbβ3 TM helices would result in a loss of interhelical contacts C-terminal to β3(L712) on the intracellular side (Figure 5E). Such a loss is apparently compensated by the placement of αIIb(F992-F993) between the TM helices, which results from the distinct αIIb backbone reversal C-terminal to its TM helix (Figure 5E and F). The aromatic rings of αIIb(F992-F993) are in proximity to that of β3(W715), and the hydrophobic assembly is augmented by contacts between β3(I719) and αIIb(F992-F993) as well as the hydrophobic moiety of αIIb(R995)'s side chain (Figure 5E and F; Supplementary Figure 4A). This assembly enables the strong electrostatic attractions detected by mutagenesis between αIIb(R995) and β3(D723) (Figure 2C) within the relatively low dielectric environment of lipid headgroups, particularly compared with aqueous solution (Ulmer et al, 2001). Thus, integrin αIIbβ3 forms a TM dimer of unique structural complexity.
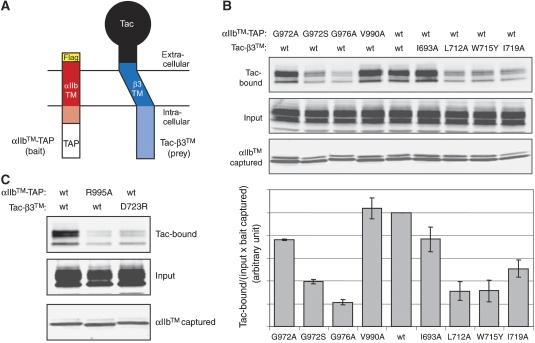
On the basis of the structure of the αIIbβ3 TM complex, two association elements are differentiated: an outer membrane association motif or clasp (OMC) characterized by packing interactions centred on αIIb(G972), αIIb(G976) and β3(G708) (Figure 5C and D), and an inner membrane association motif or clasp (IMC) based on the hydrophobic αIIb(F992-F993) and electrostatic αIIb(R995)–β3(D723) bridges (Figure 5E and F). It is noted that the IMC differs from a clasp that was defined earlier based on reported α-helical conformations for the αIIb(G991-F992-F993) motif (Vinogradova et al, 2002). The pivotal nature of the OMC and IMC residues is corroborated by the strongly activating, that is TM complex dissociating, nature of any of the αIIb(G972L), αIIb(G976L), β3(G708I/L), αIIb(F992A), αIIb(F993A), αIIb(R995D) and β3(D723R) point mutations in native receptors (Hughes et al, 1996; Luo et al, 2005; Partridge et al, 2005). Furthermore, these residues are highly conserved among human integrin α and β TM segments (Figure 1). A less strongly activating point mutation, αIIb(T981I/L) (Luo et al, 2005; Partridge et al, 2005), is also found peripheral to the dimer interface where newly introduced Ile/Leu may compete with native αIIb(L980)–β3(G708) interactions (Figure 5C) and thereby disturb the dimer interface. To learn more about the dimer interface, structure-based site-directed mutagenesis was performed in integrin αIIb and β3 TM constructs (Figure 6A) and their association in mammalian CHO cell membranes was evaluated. Reduced TM association was achieved by overpacking the OMC relative to the wild type by αIIb(G972S) and αIIb(G976A), but not by αIIb(G972A) (Figure 6B), in good agreement with the TM helix periodicities (Figure 5C and D). These substitutions, taken from other integrin TM segments (Figure 1), raise the possibility of an α–β TM affinity modulation by the OMC. It is also explicitly noted that not all occurring sequence variations have effects, for example, β3(I693A) underpacking at the extracellular membrane border is without consequence (Figure 6B). In addition to αIIb(F992A) or αIIb(F993A) (Hughes et al, 1996), destabilization of the IMC by underpacking was observed for β3(L712A), β3(W715Y) and β3(I719A), but not for αIIb(V990A) in excellent agreement with the complex structure (Figure 5E and F). Thus, two dominant integrin TM association motifs are discerned (IMC and OMC), and mutagenesis suggests that the α–β TM affinity of integrins, most notably the OMC, may vary among different subunit combinations.
Figure 6.
Site-directed mutagenesis of the αIIbβ3 dimer interface. (A) Selected mutations were introduced to an αIIb construct, αIIb™-TAP, consisting of the αIIb TM region plus cytosolic tail (Q954-E1008) with an N-terminal flag-tag, a C-terminal TAP (tandem affinity purification; calmodulin-binding domain and IgG-binding domain) tag, and a β3 construct, Tac-β3™, encompassing the β3 TM region plus cytosolic tail (V681-T762), with the extracellular domain of interleukin-2 receptor α (Tac) fused to the N-terminus (B) After co-transfection of αIIb™-TAP and Tac-β3™ constructs into CHO cells, their association was analysed by capturing αIIb™-TAP using calmodulin beads, and subsequently detecting bound Tac-β3™ through western blotting using an anti-Tac antibody (upper panels). Expression of Tac-β3™ (middle panels) and captured αIIb™-TAP (bottom panels) was verified by western blots using anti-Tac and anti-flag antibodies, respectively. The interaction was quantified by calculating the amount of bound Tac-β3™ divided by the amount of expressed Tac-β3™ and captured αIIb™-TAP. The mean and standard error from three independent experiments are depicted. (C) To verify the analogy of the assay with the NMR studies, the strongly TM-dissociating nature of the salt bridge mutations, αIIb™-TAP(R995A) and Tac-β3™(D723A), was confirmed.
Lipid embedding of the αIIbβ3 complex
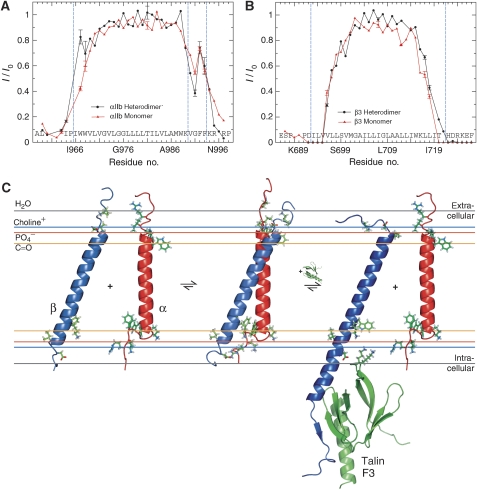
In addition to the αIIbβ3 complex structure, information about its lipid embedding is required to define the mechanism of integrin TM signalling. Protection from paramagnetic Mn2+EDDA2− present in the aqueous phase was quantified to examine the embedding of the αIIbβ3 complex in its bicelle lipid environment (Altenbach et al, 1994; Lee et al, 2008; Lau et al, 2008b) in comparison to the dissociated subunits. Protection between monomeric and heterodimeric states is quite similar overall (Figure 7A and B), excluding any large changes in lipid association on TM dissociation such as repartitioning the hydrophobic segments C-terminal to αIIb(K989) or β3(K716) on the intracellular side. This similarity agrees well with backbone conformations that are independent of the αIIbβ3 association state, and the αIIb protection pattern nicely illustrates the αIIb(G991-F992-F993) backbone reversal and lipid re-immersion of the two phenylalanine side chains in both monomeric and heterodimeric states. Compared with monomeric αIIb, the heterodimeric αIIb state showed a detectable increase in protection at the N-terminal TM helix side and a decrease in protection C-terminal to the TM helix (V990-G991). At the intracellular β3 TM helix terminus, an increase in protection is noted for the heterodimeric state (Figure 7B). These changes are compatible with small αIIbβ3 rearrangements relative to the bicelle lipids, and Figure 7C provides an estimate of integrin αIIbβ3 membrane embedding. The interhelical crossing angle in the αIIbβ3 complex suggests a straight αIIb helix and a β3 helix tilt of 25° in accordance with their differing TM helix length. At this point, it is noted that, in the absence of lipid environments, helical conformations for the αIIb(G991-F992-F993) sequence, which is fully conserved among all 18 integrin α subunits (Figure 1), were reported (Vinogradova et al, 2002; Weljie et al, 2002). In the absence of the unique environment created by the lipid tail-to-headgroup-to-solvent transition (Wiener and White, 1992), however, these structures may not be of physiological relevance. In conclusion, the structures, lengths and protection patterns of the integrin αIIb/β3 TM helices indicate an unusual straight/tilted combination in the membrane that is independent of their association state as is their backbone conformation.
Figure 7.
Model of integrin αIIbβ3 membrane embedding. (A, B) Signal broadening due to paramagnetic relaxation enhancement arising from the presence of net neutral Mn2+EDDA2− in the aqueous phase. The normalized ratio of H–N TROSY signal intensities, in the presence and absence of 1 mM Mn2+EDDA2−, I/I0, is used to quantify signal broadening for each monomeric and heterodimeric residue in slow exchange. Datasets were recorded at two temperatures (28 and 33°C) to verify the obtained broadening pattern. αIIb(I966-F993) and β3(I693-I721), marked by dashed lines, are considered membrane embedded (Lau et al, 2008a, 2008b). The αIIb TM helix border, αIIb(K989-V990), is also marked. Mn2+EDDA2− protection was evaluated in bicelles composed of 385 mM 1,2-dihexanoyl-sn-glycero-3-phosphocholine, 83 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 41 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-L-serine]. (C) Predicted orientations of the αIIb and β3 TM segments, shown in red and blue, in their monomeric and associated states, respectively, relative to the indicated functional groups of a lipid bilayer composed of 1,2-dioleoyl-sn-glycero-3-phosphocholine (Wiener and White, 1992), which is closely related to the long-chain lipids used herein. In addition, the activating β cytosolic tail–talin F3 complex (Wegener et al, 2007) (PDB ID 2h7e) has been fused to the monomeric β3 TM segment (PDB ID 2rmz). The depicted Lys322 and/or Lys324 side chains of talin are positioned to interfere with the IMC, particularly αIIb(R995)–β3(D723) electrostatic interactions.
Mechanism of integrin αIIbβ3 TM signalling
On the intracellular side, activating ligands can directly disrupt the IMC, which extends into the lipid headgroups (Figure 7C). For example, the activating talin F3 domain is likely to interfere with the IMC (Wegener et al, 2007). In fact, the unusual, asymmetric structure of the αIIbβ3 TM complex may relate to its openness to interference from cytosolic ligands and, compared with a purely helical coiled-coil-like assembly, offers additional structural elements amenable to regulatory intervention. The membrane re-immersion of αIIb(F992-F993) not only aids in juxtaposing αIIb(R995)–β3(D723), but also makes this central constraint readily accessible (Figure 7C). The highly conserved αIIb(Gly991-R995) sequence (Figure 1) suggests that this mode of inside-out activation is general to many integrin receptors. Interestingly, talin also stabilizes the helical propensity of the β tail on exiting the membrane (Ulmer et al, 2001; Li et al, 2002; Wegener et al, 2007), resulting in a stable, elongated helix for the β subunit (Figure 7C). In contrast, on the extracellular side, the agonist-binding site is far from the extracellular membrane border (c.f. Figure 9) and a direct, enthalpic disruption of the OMC is not possible. The structures of the extracellular domain of integrin αVβ3 and αIIbβ3 (Xiong et al, 2001; Adair et al, 2005; Zhu et al, 2008) show the C-terminal ends of the α–β ectodomain to be in close proximity (Figure 9B). Therefore, we suggest an indirect, entropic stabilization of the OMC by the associated extracellular lower leg assembly. If such stabilization were present, the weakly αIIbβ3 TM dissociating mutations identified in the absence of the ectodomain (Figure 6B) would be less dissociating in the native receptor. Indeed, all four of the experimentally amenable dissociating mutations were not able to activate native integrin αIIbβ3 in contrast to more disruptive substitutions (Figure 8). Thus, an entropic stabilization of the α–β TM dimer interface by the resting extracellular domains is present and may be lost as a consequence of conformational changes resulting from ligand binding (DiazGonzalez et al, 1996), resulting in outside-in signalling. By the symmetry of this arrangement, the integrity of the IMC and OMC stabilize the resting conformation of the extracellular domain and disruption of the IMC leads to inside-out signalling.
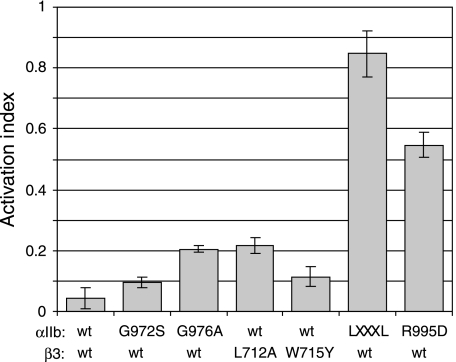
Figure 8.
Stabilization of the αIIbβ3 transmembrane complex by the resting integrin ectodomain. The αIIb(G972S), αIIb(G976A), β3(L712A) and β3(W715Y) substitutions, which were TM dissociating in the absence of the integrin ectodomain (Figure 6B), were evaluated to determine whether they were capable of activating full-length integrin αIIbβ3 receptors. The point mutation-bearing αIIb and β3 subunits were co-transfected into CHO cells. Subsequent to 24 h of transfection, cells were stained with PAC1 (an activation-specific αIIb–β3 antibody) to measure activation, and with D57 (an αIIb–β3 complex-specific antibody) to measure surface expression. D57-positive cells were analysed to calculate the Activation Index, defined as (F0–Fr)/(FMn–Fr), where F0 is the mean fluorescence intensity (MFI) of PAC1 binding, Fr is the MFI of PAC1 binding in the presence of competitive inhibitor (integrilin), and FMn is the MFI of PAC1 binding in the presence of 2 mM Mn2+. In contrast to activating control mutations, LXXXL denoting αIIb(G972 L)/αIIb(G976 L) and αIIb(R995D) (Hughes et al, 1996; Luo et al, 2005), the substitutions were unable to trigger significant integrin activation, showing the stabilization of the αIIbβ3 TM complex by the resting integrin ectodomain.
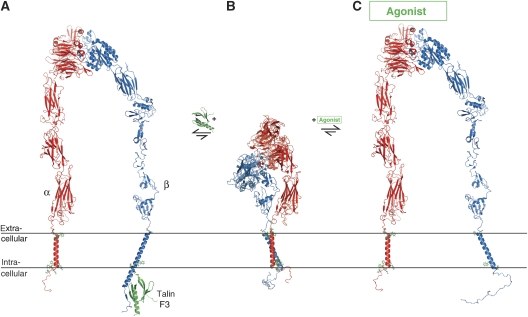
In summary, the integrin αIIbβ3 TM complex represents the first structure of a heterodimeric TM receptor and reveals a dimerization interface of intriguing complexity. The IMC stabilizes the integrin TM dimer interface on the intracellular side, the ectodomain and OMC do so on the extracellular side, and the transition between heterodimeric and monomeric TM states proceeds without large rearrangements of αIIbβ3 backbone structure or lipid association. On the basis of these observations, the notion of α–β TM complex dissociation on activation (Hughes et al, 1996; Kim et al, 2003; Li et al, 2005; Zhu et al, 2007), and the available structures of integrin domains, the following refined mechanism of integrin αIIbβ3 TM signalling is proposed. The resting state, with associated TM helices, is stabilized by the packing of C-terminal residues of the integrin ectodomain, as observed in the resting integrin αVβ3 and αIIbβ3 ectodomain structures (Xiong et al, 2001; Adair et al, 2005; Zhu et al, 2008), and the here described TM dimer interface (Figure 9B). Outside-in signalling is induced by the separation of the extracellular lower leg assembly (Xiao et al, 2004), after ectodomain interactions with agonists. This results in the loss of entropic stabilization of the TM complex, shifting the equilibrium to the dissociated state (Figure 9C) and transmitting a signal into the cell. For the extracellular domains, a transition from bent to extended conformations is depicted (Xiao et al, 2004; Zhu et al, 2008), but it is noted that alternative rearrangements have been also proposed (Arnaout et al, 2005; Rocco et al, 2008; Ye et al, 2008). Inside-out signalling is achieved by the direct disruption of the IMC by activating intracellular ligands such as talin, as suggested by cytosolic β tail–talin F3 structures (Figure 7C) (Garcia-Alvarez et al, 2003; Wegener et al, 2007). The enthalpic disruption of the IMC by talin leads to the collapse of the TM dimer interface, which destabilizes the ectodomain lower leg assembly and allows the extracellular domains to rearrange into high-affinity conformations (Figure 9A). Thus, we explain integrin αIIbβ3 bi-directional signalling by the unique structure of its TM complex, its ectodomain conformation-dependent stabilization and intracellular ligand-dependent structural interferences.
Figure 9.
Illustration of proposed integrin receptor functional states. (A) Inside-out activated integrin composed of an activated extracellular headpiece (Xiao et al, 2004) (PDB ID 2vdn), extended I-EGF1-2 domains (Shi et al, 2007) (PDB ID 2p28) and modelled tailpiece, fused to the monomeric αIIb and β3 TM structures (PDB ID 2k1a and 2mrz), and connected to the activating β cytosolic tail–talin F3 complex (Wegener et al, 2007) (PDB ID 2h7e). Talin F3 domain binding stabilizes α-helical structure subsequent to the β TM helix. (B) Resting integrin composed of the bent integrin αVβ3/αIIbβ3 structure (Xiong et al, 2001; Adair et al, 2005; Zhu et al, 2008) (PDB ID 1jv2), the αIIbβ3 TM complex (PDB ID 2k9j), and dynamically unstructured cytosolic tails (Ulmer et al, 2001; Li et al, 2002). (C) Outside-in activated integrin composed of an activated extracellular headpiece (Xiao et al, 2004) (PDB ID 2vdn), extended I-EGF1-2 domains (Shi et al, 2007) (PDB ID 2p28) and modelled tailpiece, fused to the monomeric αIIb and β3 TM structures (PDB ID 2k1a and 2mrz), and connected to dynamically unstructured cytosolic tails (Ulmer et al, 2001; Li et al, 2002). As no high-resolution structures of an entire, activated ectodomain exists, the depicted domain–domain orientations only serve to illustrate a presumed extended geometry (Adair and Yeager, 2002; Takagi et al, 2003; Zhu et al, 2008). In each functional transition, the TM helices have been rotated in addition to their dissociation. During both inside-out and outside-in signalling, additional ectodomain intermediates are likely to exist (Takagi et al, 2003; Xiao et al, 2004; Zhu et al, 2008) and some debate regarding the structural rearrangement of the ectodomain on activation remains (Arnaout et al, 2005; Rocco et al, 2008; Ye et al, 2008; Zhu et al, 2008).
Materials and methods
NMR sample preparation
Peptides encompassing human integrin αIIb(A958-P998) and β3(P685-F727), including β3(C687S), respectively, were produced as described earlier (Lau et al, 2008a, 2008b). In addition, a peptide incorporating αIIb(R995A) was produced analogously. Peptides were produced in four different isotope-labelling patterns: fully protonated, unlabelled (12C/14N) peptide; partially and highly deuterated, 13C/15N labelled peptide synthesized in 50 and 99% D2O solutions, respectively, using 1H/13C D-glucose/15N NH4Cl; and selectively Val-γ1,2 (13CH3/12CD3), Leu-δ1,2 (13CH3/12CD3), Ile-δ1 (13CH3)-labelled peptides within a perdeuterated background in combination with 13C/15N labelling synthesized by supplying the sodium salts of 2-keto-3-methyl-d3-butyric acid-1,2,3,4-13C4,3-d1 and 2-ketobutyric acid-13C4,3,3-d2 (Tugarinov and Kay, 2003), and 2H/13C D-glucose/15N NH4Cl in 99% D2O solution. For HN-detected experiments, the peptides were typically reconstituted at concentrations of 0.6 and 0.9 mM for 2H/13C/15N-labelled and unlabelled subunits, respectively, in 385 mM 1,2-dihexanoyl-sn-glycero-3-phosphocholine, 83 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 41 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-L-serine]. For HC-detected experiments, both peptides were reconstituted at concentrations of 1.2 mM in deuterated lipids, 350 mM 1,2-dihexanoyl(d22)-sn-glycero-3-phosphocholine-1,1,2,2-d4-N,N,N-trimethyl-d9, 105 mM 1,2-dimyristoyl-d54-sn-glycerol-3-phosphocholine-1,1,2,2-d4-N,N,N-trimethyl-d9. These samples were prepared in 25 mM HEPES·NaOH, pH 7.4, 6% D2O, 0.02% w/v NaN3 solutions and, if desired, freeze dried and exchanged into D2O. To measure residual dipolar couplings, the peptides were dissolved at concentrations of 0.3 mM and 0.4 mM for 2H/13C/15N-labelled and unlabelled subunits, respectively, in 25 mM KH2PO4/K2HPO4, pH 7.4, 75 mM KCl containing 1,2-dihexanoyl-sn-glycero-3-phosphocholine and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine at concentrations of 100/30 mM or 200/60 mM and G-tetrad liquid crystals formed from 25 mg/ml d(GpG) (Lorieau et al, 2008).
NMR spectroscopy and structure calculation
All NMR experiments were conducted on a cryoprobe-equipped Bruker Avance 700 spectrometer. Data were processed and analysed with the nmrPipe package (Delaglio et al, 1995) and CARA. Pulse sequences were applied with TROSY optimization (Pervushin et al, 1998). Starting from the monomeric αIIb and β3 resonances (Lau et al, 2008a, 2008b), HN, 15N, 13Cα backbone assignments of the heterodimeric set of resonances (Figure 2) were achieved from HSQC, HNCA, NOESY-TROSY and HSQC-NOESY-TROSY experiments using highly deuterated peptide in complex with fully protonated partner. Hα and H/C side chain assignments were achieved from (CT)-HSQC, HAHB(CBCACO)NH, (H)CCH-COSY, H(C)CH-COSY and NOESY-HSQC experiments (Gehring and Ekiel, 1998) using partially deuterated, 13C/15N-labelled peptide uncomplexed and in complex with fully protonated partner, respectively. Selectively, Val-γ1,2 (13CH3/12CD3), Leu-δ1,2 (13CH3/12CD3), Ile-δ1 (13CH3)-labelled peptides uncomplexed and in complex with fully protonated partner, respectively, were assigned by (H)CCC(CO)NH, H(C)CC(CO)NH and NOESY-HSQC experiments (Grzesiek et al, 1993). Spectral overlap was resolved and NOE intensities were optimized by recording experiments at different temperatures (23, 28 and 33 °C) and using different mixing times (75, 100 and 125 ms). H–N residual dipolar couplings were measured from 1JNH-scaled HNCO experiments (Kontaxis et al, 2000).
Backbone torsion angle restraints were derived from TALOS-based database searches (Cornilescu et al, 1999) using 1Hα, 15N, 13Cα and 13C′ chemical shifts. HN–HN distance constraints were obtained from 15N-edited NOESY experiments. To reliably detect interhelical NOEs selective methyl labelling was indispensable and from 13Cmethyl–1H, 13C-edited NOESY-HSQC experiments and the herein verified αIIb(R995)–β3(D723) salt bridge (Figure 2C), 27 unambiguous distance restraints were obtained. Representative NOE strips are provided in Supplementary Figure 4. Because of the complex relationship of individual NOE amplitudes and interproton distances in noncovalently associated complexes (Campbell and Sykes, 1993) and ensuing referencing ambiguities, only upper distance bounds were defined at ⩽5.5 Å for the detected methyl NOE connectivities, in analogy to protocols that do not require lower distance bounds (Kuszewski et al, 1999). Limited accuracy of the measured residual dipolar couplings negated their inclusion in the structure calculations. The structure of the αIIbβ3 TM complex was calculated by simulated annealing, starting at 3000 K, using the XPLOR-NIH program (Schwieters et al, 2003). In addition to standard force field terms for covalent geometry (bonds, angles and improper dihedrals) and nonbonded contacts (Van der Waals repulsion), dihedral angle and interproton distance restraints were implemented using quadratic square-well potentials, and a backbone–backbone hydrogen-bonding potential and torsion angle potential of mean force were used (Kuszewski et al, 1997; Grishaev and Bax, 2004). The final values for the force constants of the different terms in the simulated annealing target function are as follows: 1000 kcal mol−1 Å−2 for bond lengths; 500 kcal mol−1 rad−2 for angles and improper dihedrals, which serve to maintain planarity and chirality; 4 kcal mol−1 Å−4 for the quartic Van der Waals repulsion term; 30 kcal mol−1 Å−2 for interproton distance restraints; 200 kcal mol−1 rad−2 for dihedral angle restraints; 1.0 for the torsion angle potential; and a directional force of 0.20 and a linearity force of 0.05 for the hydrogen-bonding potential. Appropriate convergence for all 20 calculated structures to coordinate precisions of 0.68 and 1.05 Å relative to the mean coordinates was obtained for the backbone-heavy and all nonhydrogen atoms, respectively (Supplementary Table II). Moreover, the calculated structural ensemble is insensitive to the random removal of 10% of intermolecular distance restraints (data not shown). Structural statistics are summarized in Supplementary Table II. The atomic coordinates have been deposited in the Protein Data Bank with the accession number 2K9J.
Affinity capture of αIIb™-TAP/Tac-β3™ constructs
For the construction of αIIbTM-TAP (see Figure 6A), the sequences of the preprotrypsin leader sequence followed by three FLAG repeats (Sigma-Aldrich, Inc.) were cloned into pcDNA3.1 (Invitrogen, Inc.). Subsequently, PCR-generated fusion sequences coding for αIIb(Gln954-Glu1008) and TAP were ligated into the vector. Tac-β3TM (see Figure 6A) was generated by the ligation of PCR-generated fusion sequences coding for the extracellular Tac domain and β3(Val681-Thr762) into pcDNA3.1. Point mutations were performed using the Quick-change site-directed mutagenesis kit (Stratagene, Inc).
At 24 h after co-transfection of pcDNA3.1/αIIb™-TAP and pcDNA3.1/Tac-β3™, CHO cells were lysed with CHAPS lysis buffer (20 mM HEPES, pH 7.4, 1% CHAPS, 150 mM NaCl, 2 mM CaCl2, Roche EDTA-free protease inhibitor mixture) and were clarified by centrifugation at 14 000 rpm for 15 min. The clarified lysates were incubated with calmodulin sepharose (GE Healthcare, Inc.) for 2 h at 4 °C, and bound proteins were eluted with SDS reducing sample buffer, subjected to SDS–PAGE and analysed by western blotting.
Activation assay of functional integrin αIIbβ3
Flow cytometric activation assays were performed as described earlier (Feral et al, 2005; Han et al, 2006). In brief, αIIb and β3 subunits bearing point mutations in the TM segment (Figure 8) were co-transfected into CHO cells. At 24 h after transfection, cells were stained with PAC1 (an activation-specific αIIb–β3 antibody) to measure activation, and with D57 (an αIIb–β3 complex-specific antibody) to measure surface expression, in the presence and absence of the integrin-activating Mn2+ cation. After washing, cells were stained with fluorescein isothiocyanate-conjugated anti-mouse IgG and with R-phycoerythrin-conjugated anti-mouse IgM. Five minutes before analysis, propidium iodide (PI) was added, and PI-negative live cells were analysed on FACSCalibur (BD Biosciences). Data were analysed using WinMDI version 2.9 to generate dot plots (Supplementary Figure 5).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Diana Gegala for critically reading the manuscript. This work was supported by Grants from the US National Institutes of Health to TSU (HL089726) and MHG (HL70784 and AR27214). TSU is recipient of a Scientist Development Grant from the American Heart Association. Tong-Lay Lau and Chungho Kim are recipients of postdoctoral fellowships from the American Heart Association. The authors declare no competing financial interests.
References
- Adair BD, Xiong JP, Maddock C, Goodman SL, Arnaout MA, Yeager M (2005) Three-dimensional EM structure of the ectodomain of integrin alpha V beta 3 in a complex with fibronectin. J Cell Biol 168: 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair BD, Yeager M (2002) Three-dimensional model of the human platelet integrin alpha(llb)beta(3) based on electron cryomicroscopy and x-ray crystallography. Proc Natl Acad Sci USA 99: 14059–14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL (1994) A collision gradient-method to determine the immersion depth of nitroxides in lipid bilayers—application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci USA 91: 1667–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armulik A, Nilsson I, von Heijne G, Johansson S (1999) Determination of the border between the transmembrane and cytoplasmic domains of human integrin subunits. J Biol Chem 274: 37030–37034 [DOI] [PubMed] [Google Scholar]
- Arnaout MA, Mahalingam B, Xiong JP (2005) Integrin structure, allostery, and bidirectional signaling. Annu Rev Cell Dev Biol 21: 381–410 [DOI] [PubMed] [Google Scholar]
- Askari JA, Buckley PA, Mould AP, Humphries MJ (2009) Linking integrin conformation to function. J Cell Sci 122: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JS (2005) Structure and function of the platelet integrin alpha(IIb)beta(3). J Clin Invest 115: 3363–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharov EV, Mayzel ML, Volynsky PE, Goncharuk MV, Ermolyuk YS, Schulga AA, Artemenko EO, Efremov RG, Arseniev AS (2008) Spatial structure and pH-dependent conformational diversity of dimeric transmembrane domain of the receptor tyrosine kinase EphA1. J Biol Chem 283: 29385–29395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharov EV, Pustovalova YE, Pavlov KV, Volynsky PE, Goncharuk MV, Ermolyuk YS, Karpunin DV, Schulga AA, Kirpichnikov MP, Efremov RG, Maslennikov IV, Arseniev AS (2007) Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger. J Biol Chem 282: 16256–16266 [DOI] [PubMed] [Google Scholar]
- Call ME, Schnell JR, Xu CQ, Lutz RA, Chou JJ, Wucherpfennig KW (2006) The structure of the zeta zeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell 127: 355–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AP, Sykes BD (1993) The 2-dimensional transferred nuclear overhauser effect—theory and practice. Annu Rev Biophys Biomol Struct 22: 99–122 [DOI] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ (1996) Protein NMR Spectroscopy. San Diego: Academic Press [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20: 426–427 [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) Nmrpipe—a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- DiazGonzalez F, Forsyth J, Steiner B, Ginsberg MH (1996) Trans-dominant inhibition of integrin function. Mol Biol Cell 7: 1939–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feral CC, Nishiya N, Fenczik CA, Stuhlmann H, Slepak M, Ginsberg MH (2005) CD98hc (SLC3A2) mediates integrin signaling. Proc Natl Acad Sci USA 102: 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alvarez B, de Pereda JM, Calderwood DA, Ulmer TS, Critchley D, Campbell ID, Ginsberg MH, Liddington RC (2003) Structural determinants of integrin recognition by Talin. Mol Cell 11: 49–58 [DOI] [PubMed] [Google Scholar]
- GarciaGuerra R, GarciaDominguez JA, GonzalezRodriguez J (1996) A new look at the lipid composition of the plasma membrane of human blood platelets relative to the GPIIb/IIIa (integrin alpha IIb beta 3) content. Platelets 7: 195–205 [Google Scholar]
- Gehring K, Ekiel I (1998) H(C)CH-COSY and (H)CCH-COSY experiments for C-13-labeled proteins in H2O solution. J Magn Reson 135: 185–193 [DOI] [PubMed] [Google Scholar]
- Gottschalk KE (2005) A coiled-coil structure of the alpha llb beta 3 integrin transmembrane and cytoplasmic domains in its resting state. Structure 13: 703. [DOI] [PubMed] [Google Scholar]
- Grishaev A, Bax A (2004) An empirical backbone-backbone hydrogen-bonding potential in proteins and its applications to NMR structure refinement and validation. J Am Chem Soc 126: 7281–7292 [DOI] [PubMed] [Google Scholar]
- Grzesiek S, Anglister J, Bax A (1993) Correlation of backbone amide and aliphatic side-chain resonances in C-13/N-15-enriched proteins by isotropic mixing of C-13 magnetization. J Magn Reson Ser B 101: 114–119 [Google Scholar]
- Grzesiek S, Bax A, Hu JS, Kaufman J, Palmer I, Stahl SJ, Tjandra N, Wingfield PT (1997) Refined solution structure and backbone dynamics of HIV-1 Nef. Protein Sci 6: 1248–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JW, Lim CJ, Watanabe N, Soriani A, Ratnikov B, Calderwood DA, Puzon-McLaughlin W, Lafuente EM, Boussiotis VA, Shattil SJ, Ginsberg MH (2006) Reconstructing and deconstructing agonist-induced activation of integrin alpha IIb beta 3. Curr Biol 16: 1796–1806 [DOI] [PubMed] [Google Scholar]
- Harburger DS, Calderwood DA (2009) Integrin signalling at a glance. J Cell Sci 122: 159–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes PE, DiazGonzalez F, Leong L, Wu CY, McDonald JA, Shattil SJ, Ginsberg MH (1996) Breaking the integrin hinge—a defined structural constraint regulates integrin signaling. J Biol Chem 271: 6571–6574 [DOI] [PubMed] [Google Scholar]
- Hynes RO (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110: 673–687 [DOI] [PubMed] [Google Scholar]
- Kim M, Carman CV, Springer TA (2003) Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science 301: 1720–1725 [DOI] [PubMed] [Google Scholar]
- Kontaxis G, Clore GM, Bax A (2000) Evaluation of cross-correlation effects and measurement of one-bond couplings in proteins with short transverse relaxation times. J Magn Reson 143: 184–196 [DOI] [PubMed] [Google Scholar]
- Kuszewski J, Gronenborn AM, Clore GM (1997) Improvements and extensions in the conformational database potential for the refinement of NMR and X-ray structures of proteins and nucleic acids. J Magn Reson 125: 171–177 [DOI] [PubMed] [Google Scholar]
- Kuszewski J, Gronenborn AM, Clore GM (1999) Improving the packing and accuracy of NMR structures with a pseudopotential for the radius of gyration. J Am Chem Soc 121: 2337–2338 [Google Scholar]
- Lau T-L, Dua V, Ulmer TS (2008a) Structure of the integrin alphaIIb transmembrane segment. J Biol Chem 283: 16162–16168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau T-L, Partridge AP, Ginsberg MH, Ulmer TS (2008b) Structure of the integrin beta3 transmembrane segment in phospholipid bicelles and detergent micelles. Biochemistry 47: 4008–4016 [DOI] [PubMed] [Google Scholar]
- Lee D, Walter KFA, Bruckner A-K, Hilty C, Becker S, Griesinger C (2008) Bilayer in small bicelles revealed by lipid-protein interactions using NMR spectroscopy. J Am Chem Soc 130: 13822–13823 [DOI] [PubMed] [Google Scholar]
- Li RH, Babu CR, Valentine K, Lear JD, Wand AJ, Bennett JS, DeGrado WF (2002) Characterization of the monomeric form of the transmembrane and cytoplasmic domains of the integrin beta 3 subunit by NMR spectroscopy. Biochemistry 41: 15618–15624 [DOI] [PubMed] [Google Scholar]
- Li RH, Gorelik R, Nanda V, Law PB, Lear JD, DeGrado WF, Bennett JS (2004) Dimerization of the transmembrane domain of integrin alpha(IIb) subunit in cell membranes. J Biol Chem 279: 26666–26673 [DOI] [PubMed] [Google Scholar]
- Li W, Metcalf DG, Gorelik R, Li RH, Mitra N, Nanda V, Law PB, Lear JD, DeGrado WF, Bennett JS (2005) A push-pull mechanism for regulating integrin function. Proc Natl Acad Sci USA 102: 1424–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieau J, Yao LS, Bax A (2008) Liquid crystalline phase of G-tetrad DNA for NMR study of detergent-solubilized proteins. J Am Chem Soc 130: 7536–7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Carman CV, Takagi J, Springer TA (2005) Disrupting integrin transmembrane domain heterodimerization increases ligand binding affinity, not valency or clustering. Proc Natl Acad Sci USA 102: 3679–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Springer TA, Takagi J (2004) A specific interface between integrin transmembrane helices and affinity for ligand. PLoS Biol 2: 776–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie KR, Prestegard JH, Engelman DM (1997) A transmembrane helix dimer: structure and implications. Science 276: 131–133 [DOI] [PubMed] [Google Scholar]
- Partridge AW, Liu SC, Kim S, Bowie JU, Ginsberg MH (2005) Transmembrane domain helix packing stabilizes integrin aIIb beta 3 in the low affinity state. J Biol Chem 280: 7294–7300 [DOI] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K (1998) Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in C-13-labeled proteins. J Am Chem Soc 120: 6394–6400 [Google Scholar]
- Rocco M, Rosano C, Weisel JW, Horita DA, Hantgan RR (2008) Integrin conformational regulation: uncoupling extension/tail separation from changes in the head region by a multiresolution approach. Structure 16: 954–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Engelman DM (2004) Involvement of transmembrane domain interactions in signal transduction by alpha/beta integrins. J Biol Chem 279: 9840–9846 [DOI] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM (2003) The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160: 65–73 [DOI] [PubMed] [Google Scholar]
- Shi ML, Foo SY, Tan SM, Mitchell EP, Law SKA, Lescar J (2007) A structural hypothesis for the transition between bent and extended conformations of the leukocyte beta 2 integrins. J Biol Chem 282: 30198–30206 [DOI] [PubMed] [Google Scholar]
- Spera S, Bax A (1991) Empirical correlation between protein backbone conformation and C-alpha and C-beta C-13 nuclear-magnetic-resonance chemical-shifts. J Am Chem Soc 113: 5490–5492 [Google Scholar]
- Stefansson A, Armulik A, Nilsson IM, von Heijne G, Johansson S (2004) Determination of N- and C-terminal borders of the transmembrane domain of integrin subunits. J Biol Chem 279: 21200–21205 [DOI] [PubMed] [Google Scholar]
- Takagi J, Strokovich K, Springer TA, Walz T (2003) Structure of integrin alpha(5)beta(1) in complex with fibronectin. EMBO J 22: 4607–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugarinov V, Kay LE (2003) Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc 125: 13868–13878 [DOI] [PubMed] [Google Scholar]
- Ulmer TS, Bax A, Cole NB, Nussbaum RL (2005) Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem 280: 9595–9603 [DOI] [PubMed] [Google Scholar]
- Ulmer TS, Yaspan B, Ginsberg MH, Campbell ID (2001) NMR analysis of structure and dynamics of the cytosolic tails of integrin alpha IIb beta 3 in aqueous solution. Biochemistry 40: 7498–7508 [DOI] [PubMed] [Google Scholar]
- Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas TA, Plow EF, Qin J (2002) A structural mechanism of integrin alpha(IIb)beta(3) “inside-out” activation as regulated by its cytoplasmic face. Cell 110: 587–597 [DOI] [PubMed] [Google Scholar]
- Wagner G, Pardi A, Wuthrich K (1983) Hydrogen-bond length and H-1-NMR chemical-shifts in proteins. J Am Chem Soc 105: 5948–5949 [Google Scholar]
- Wang YJ, Jardetzky O (2004) Predicting N-15 chemical shifts in proteins using the preceding residue-specific individual shielding surfaces from phi, psi(i-1), and chi(1) torsion angles. J Biomol NMR 28: 327–340 [DOI] [PubMed] [Google Scholar]
- Wegener KL, Partridge AW, Han J, Pickford AR, Liddington RC, Ginsberg MH, Campbell ID (2007) Structural basis of integrin activation by talin. Cell 128: 171–182 [DOI] [PubMed] [Google Scholar]
- Weljie AM, Hwang PM, Vogel HJ (2002) Solution structures of the cytoplasmic tail complex from platelet integrin alpha IIb- and beta 3-subunits. Proc Natl Acad Sci USA 99: 5878–5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener MC, White SH (1992) Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of X-ray and neutron-diffraction data.3. Complete structure. Biophys J 61: 434–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Takagi J, Coller BS, Wang J-H, Springer TA (2004) Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature 432: 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Diefenbach B, Zhang RG, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA (2001) Crystal structure of the extracellular segment of integrin alpha V beta 3. Science 294: 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XP, Case DA (2001) Automated prediction of N-15, C-13(alpha), C-13(beta) and C-13 ‘ chemical shifts in proteins using a density functional database. J Biomol NMR 21: 321–333 [DOI] [PubMed] [Google Scholar]
- Ye F, Liu J, Winkler H, Taylor KA (2008) Integrin alpha(IIb)beta(3) in a membrane environment remains the same height after Mn2+ activation when observed by cryoelectron tomography. J Mol Biol 378: 976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Luo BH, Xiao T, Zhang C, Nishida N, Springer TA (2008) Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol Cell 32: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JQ, Carman CV, Kim M, Shimaoka M, Springer TA, Luo BH (2007) Requirement of alpha and beta subunit transmembrane helix separation for integrin outside-in signaling. Blood 110: 2475–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information