Abstract
Oligopeptide-binding protein A (OppA) from Lactococcus lactis binds peptides of an exceptionally wide range of lengths (4–35 residues), with no apparent sequence preference. Here, we present the crystal structures of OppA in the open- and closed-liganded conformations. The structures directly explain the protein's phenomenal promiscuity. A huge cavity allows binding of very long peptides, and a lack of constraints for the position of the N and C termini of the ligand is compatible with binding of peptides with varying lengths. Unexpectedly, the peptide's amino-acid composition (but not the exact sequence) appears to have a function in selection, with a preference for proline-rich peptides containing at least one isoleucine. These properties can be related to the physiology of the organism: L. lactis is auxotrophic for branched chain amino acids and favours proline-rich caseins as a source of amino acids. We propose a new mechanism for peptide selection based on amino-acid composition rather than sequence.
Keywords: mass spectrometry, peptide binding, peptide transport, X-ray crystallography
Introduction
The Gram-positive lactic acid bacterium Lactococcus lactis is auxotrophic for several amino acids including the branched chain amino acids, leucine, isoleucine and valine. The organism can satisfy its demand for these amino acids by importing peptides, for example, when L. lactis grows in protein-containing media such as milk, it encounters an environment with low concentrations of free amino acids. To acquire the necessary amino acids, L. lactis proteolyses the abundant milk proteins (caseins) to peptides, which are taken up by peptide transporters, and further metabolized in the cytoplasm (Kunji et al, 1998).
The oligopeptide transport system OppABCDF is the major transporter used by the organism to import peptides. OppABCDF is an ATP-binding cassette (ABC) transporter consisting of five subunits: two homologous integral membrane proteins OppB and OppC, which together form the translocation pore; two homologous nucleotide-binding domains OppD and OppF, which fuel the transport by ATP binding and hydrolysis; and the receptor or substrate-binding protein (SBP) OppA (Tynkkynen et al, 1993) that determines the substrate specificity of the system (Doeven et al, 2004). OppA belongs to a large superfamily of SBPs associated with ABC transporters involved in nutrient uptake in prokaryotes (Monnet, 2003). These proteins consist of two domains with α/β-folds connected by a hinge. A rotation of the domains around the hinge allows the proteins to adopt closed and open conformations. Substrates bind between the two domains and shift the equilibrium towards a closed state, a process often referred to as a Venus flytrap mechanism (Mao et al, 1982). The closed, ligand-bound proteins associate with the membrane-embedded pore and deliver the cargo for translocation.
Although in Gram-negative bacteria SBPs are soluble proteins targeted to the periplasmic space, in L. lactis and other Gram-positive bacteria they are either anchored to the membrane through a lipid modification on the N-terminal cysteine (e.g. in OppA), or covalently linked to the translocation pore (Biemans-Oldehinkel et al, 2006). Crystal structures are known of the oligopeptide-binding proteins DppA from Escherichia coli (specific for dipeptides) (Nickitenko et al, 1995), OppA from Salmonella typhimurium (tri- to pentapeptides) (Tame et al, 1994), OppA from Yersinia pestis (unknown specificity) (Tanabe et al, 2007) and AppA from Bacillus subtilis (unknown specificity, possibly nonapeptides) (Levdikov et al, 2005). The structures revealed that the specificity for peptides is determined by hydrogen bonds with the ligand backbone. The side chains of the bound peptides are located in large and well-hydrated pockets. These pockets can accept any side chain, which is the basis for the well-documented lack of specificity for the ligand's amino-acid sequence. Specificity for peptides of a particular length is determined by salt bridges that constrain the positions of their N and C termini in the binding site. OppA of L. lactis is exceptional, because it accepts peptides of lengths from 4 to at least 35 residues and virtually any amino-acid composition (Detmers et al, 2000). To shed light on how lactococcal OppA achieves such broad specificity, we have determined its structures in the peptide-bound closed and open states.
Results
Crystallization and structure determination
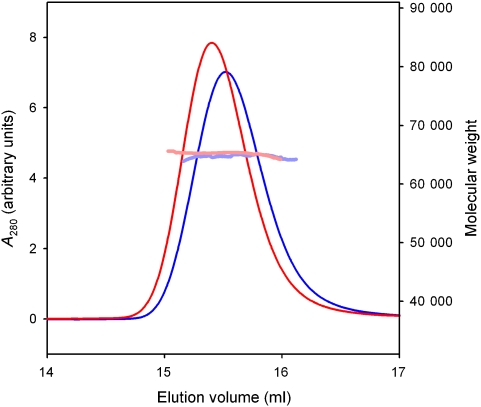
OppA without lipid anchor (termed OppA*) was produced in the cytoplasm of L. lactis where endogenous peptides are available for binding. These peptides bind so tightly that they remain associated with the protein throughout the purification (Lanfermeijer et al, 1999). The endogenous peptides can be removed from the protein only by guanidium chloride treatment, which generates ligand-free OppA* (Lanfermeijer et al, 1999). Removal of endogenous peptides was required to allow binding of defined peptides used for crystallization. Analytical ultracentrifugation and static light-scattering experiments showed that both ligand-free and ligand-bound OppA* was monomeric in solution (Figure 1; Supplementary Figure S1).
Figure 1.
Oligomeric state of OppA*. Molecular weight determination using size-exclusion chromatography coupled to static light scattering and refractive index measurements. The chromatograms are shown of OppA* with endogenous peptides bound (blue line) and without peptide (red line). The molecular mass was calculated throughout the eluting peaks and is indicated in pale blue and pale red for OppA* with endogenous peptides and without peptide, respectively (right-hand scale). The protein has the same molecular mass in both cases, but the elution volume of the gel filtration column is different, indicating that OppA* with bound peptide adopts a more compact conformation.
OppA* was crystallized (i) in complex with endogenous peptides in the closed-liganded conformation (space group P212121, diffracting to 1.3 Å resolution); (ii) bound to the high-affinity peptide bradykinin (RPPGFSPFR) in the closed-liganded conformation (space group P1, diffracting to 2.5 Å resolution) and (iii) in the presence of four different peptides (5, 8, 12 and 20 amino-acid long) in the open-liganded conformation (space group P21 diffracting to 1.5–1.8 Å) (see Supplementary Table I). To solve the structure of OppA* complexed with endogenous peptides, the phases were determined by multiple-wavelength anomalous diffraction (MAD) from bromide ions introduced in the crystals by soaking, combined with single-wavelength anomalous diffraction from sulphur atoms (sulphur-SAD). The other structures were solved by molecular replacement.
OppA* complexed with endogenous peptides
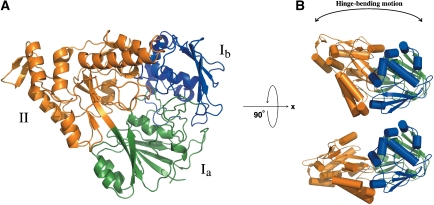
OppA* complexed with endogenous peptides consists of two α/β-domains (named I and II), which are in contact with each other and enclose a buried substrate-binding site, characteristic of SBPs in their closed conformation (Figure 2). The domains are connected by two segments, of which the central residues are Ala299 and Met542.
Figure 2.
The OppA* structure. (A) Ribbon representation of the domain organization of OppA* in the closed conformation (the structure with endogenous peptide, PDB code 3DRF). The refined model contains a continuous polypeptide trace from Gly32 to Val570. The two α/β-domains are made up of residues 32–300 and 543–572 (domain I, green and blue), and residues 301–542 (domain II, orange). Domain I has two subdomains: domain Ia (residues 32–82, 220–300 and 543–570, green), and domain Ib (residues 83–219, blue). Domain Ib is absent in most known substrate-binding proteins, but is present in the subfamily of oligopeptide-binding proteins. The hinge region corresponds to the segments that link domain Ia and domain II, that is, the green to orange transitions. (B) Differences in the relative orientation of the two α/β-domains between the closed (upper cartoon, PDB code 3DRF) and the open (lower cartoon, PDB code 3DRH) conformation. The colours of the domains are the same as in (A).
Despite limited sequence identities (Quiocho and Ledvina, 1996) (∼20%), the structure of OppA* is similar to other peptide-binding protein structures, such as AppA from B. subtilis (485 equivalent Cα atoms superimposed, RMSD of 2.5 Å), OppA from S. typhimurium, and DppA from E. coli. A sequence alignment of OppA* with AppA, DppA and OppA from S. typhimurium, based on their 3D structures is shown in Supplementary Figure S2.
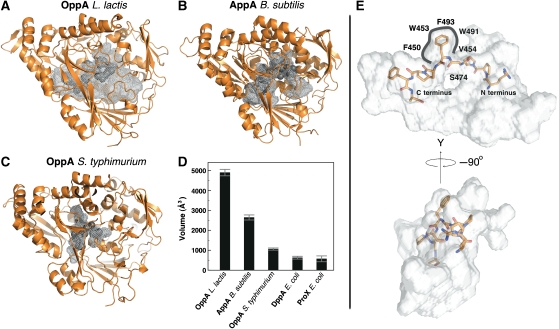
A striking feature of the OppA* structure is the enormous volume of the ligand-binding cavity buried between the two domains. Figure 3 shows a comparison of the volumes of the cavities of OppA*, AppA from B. subtilis, OppA from S. typhimurium, DppA from E. coli, and ProX, the proline-binding protein from E. coli (Figure 3D). The volumes of the binding cavities correlate well with the sizes of the substrates that are accepted. Accordingly, OppA*, which accepts peptides up to 35 residues long, has by far the largest binding cavity (∼4900 Å3), followed by the cavities in AppA (∼2600 Å3, probably specific for nonapeptides), OppA from S. typhimurium (∼1000 Å3, tri- to pentapeptides), DppA (∼700 Å3, dipeptides) and ProX (∼700 Å3, single amino acid). The greatly expanded cavity in OppA* is created mainly by two structural differences compared with AppA: The last loop before the C-terminal end (residues 557–564) as well as the loop containing residues 416–426 are shifted towards the surface of the protein.
Figure 3.
Peptide-binding sites of OppA* (A, PDB codes 3DRF), AppA from B. subtilis (B, PDB code 1XOC) and OppA from S. typhimurium (C, PDB code 1B9J), visualized as grey mesh; the bound peptide (endogenous peptides for OppA* and AppA, KLK for OppA from S. typhimurium) (dark grey) is shown in space filling representation. (D) Comparison of binding cavity volumes, including the cavities of DppA (PDB code 1DPE) and ProX (PDB code 1SW2) from E. coli. The volumes were calculated with the program Voidoo (Kleywegt and Jones, 1994) and the error is the standard deviation over 10 calculations. (E) The bound bradykinin visualized inside the binding cavity of OppA* reveals the extent of the unoccupied space. The cavity is shown as silver surface, the peptide is shown in stick representation, and the protein is not shown.
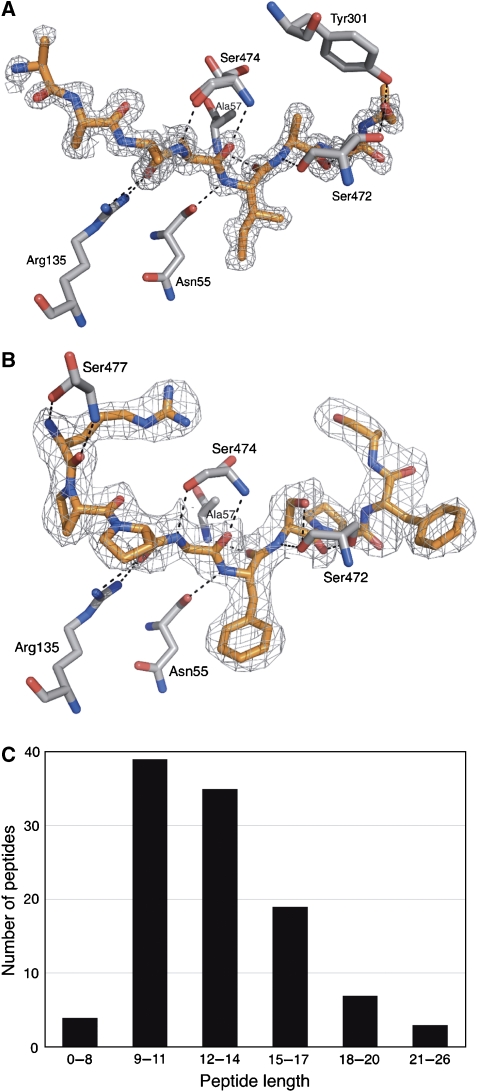
Residual electron density became visible between domains I and II during refinement and could account for bound peptide (Figure 4A). The bound peptide was completely buried with no access to the bulk solvent. The electron density allowed for straightforward model building of an octapeptide backbone with positions 1 and 8 partially occupied. In contrast to the backbone, the electron densities for the side chains of the peptide could not be readily interpreted except for the amino acid at position 5 that could be assigned unambiguously to an isoleucine. The observed electron density originates from tightly bound endogenous lactococcal peptides (see below for identification), which had co-purified with OppA* (Lanfermeijer et al, 1999). Because the endogenous peptides have different sequences, the electron density at the side chain positions is ambiguous. The peptide electron density becomes weaker towards both termini, indicating additional diversity in the lengths of the bound peptides, hence preventing the analysis of possible interactions of the N and C termini with the protein. At either side of the fading density (towards both the N- and the C-terminal end of the modelled peptide), the cavity expands dramatically, leaving space to accommodate long peptides.
Figure 4.
Bound ligands and their interactions. Electron density for the ligand (SigmaA-weighted 2Fo−Fc omit map) is shown for the endogenous mixture of peptides (A, PDB code 3DRF) and bradykinin (B, PDB code 3DRG). The ligands are shown in orange, protein residues that form hydrogen bonds to bradykinin are shown in grey. The density of both ligands is shown at a contour level of 1σ. (C) Distribution of lengths of endogenous peptides identified with tandem mass spectrometry.
The bound endogenous peptides were extracted from OppA* and identified by MALDI tandem mass spectrometry. In total, 107 different peptides originating from 14 proteins of L. lactis could be identified with a confidence level of >99% (see Supplementary Table II for a complete list of identified peptides). They ranged in length from 7 to 26 amino acids (Figure 4C), consistent with the known binding properties of OppA, which is able to bind peptides of a wide variety of lengths, including very long peptides. Strikingly, the bound peptides were rich in proline residues. The frequency of occurrence of proline residues was three-fold higher in the bound peptides than in the whole L. lactis proteome. Furthermore, 70% of the identified peptides contained at least one isoleucine, which could account for the observed electron density at position 5, but it must be noted that the relative abundance of the extracted peptides could not be determined based on the mass spectrometry analysis. No obvious correlations were found between the size of the peptides and their amino-acid composition or hydrophobicity.
OppA* complexed with the nonapeptide RPPGFSPFR
To analyse in detail the protein–ligand interactions, in particular with peptide side chains and termini, it was necessary to obtain a structure of OppA* in the closed conformation with a single defined peptide bound instead of a mixture. The structure of OppA* complexed with the high-affinity nonapeptide bradykinin (sequence RPPGFSPFR; Supplementary Figure S3) was in the desired closed conformation. The overall structure was very similar to the structure with endogenous peptides (RMSD of 0.4 Å), but now the peptide-binding site showed clear electron density for the bound nonapeptide RPPGFSPFR (Figure 4B). The backbone of the bradykinin ligand was in a very similar conformation as the backbone of the modelled endogenous peptide (RMSD of 0.24 Å). The conformation of all side chains could be assigned except for the C-terminal arginine (position 9).
Figure 4B visualizes the hydrogen bonds present between bradykinin and the protein. Most protein–ligand interactions are made with residues 3–8 of bradykinin, and almost exclusively with the backbone of the peptide, explaining the lack of sequence specificity of OppA (Lanfermeijer et al, 2000; Doeven et al, 2004). There is only one well-confined side chain pocket, and that is accommodating side chain 5 of bradykinin. None of the other side chains reside in confined pockets but, instead, they are all in contact with the voluminous binding cavity. This situation is dramatically different compared with OppA from S. typhimurium, in which each side chain of the bound ligand is located in a separate confined pocket.
The well-defined pocket for side chain 5 of bradykinin is very hydrophobic, and it is lined by side chains of Phe450, Trp453, Val454, Trp473, Tyr491 and Phe493 (Figure 3E). The pocket accommodates a phenylalanine side chain in the crystal structure complexed with bradykinin, and an isoleucine side chain in the structure with the mixture of endogenous peptides. The isoleucine side chain was the only well defined side chain of the endogenous peptides. There is some flexibility in the shape of the pocket allowing the different side chains to fit inside. The phenylalanine side chain of bradykinin expels one of two ordered water molecules that are present in the structure with endogenous peptides, and causes displacement of Phe493.
Another striking difference with the homologous Opps (Tame et al, 1994; Nickitenko et al, 1995; Levdikov et al, 2005) is the absence in the OppA* structure of salt bridges between the protein and the N and C termini of the bound peptide. The lack of constraints to the position of the peptide termini, combined with the exceptionally voluminous binding cavity forms the molecular basis for OppA's ability to accept peptides of a broad range of lengths.
OppA* in the open conformation
In total, five structures of OppA* in open conformations were determined. Co-crystallizations of OppA* with each of four different peptides yielded structures of an open-liganded state (see legend to Figure 5 for the peptide sequences). In addition, a structure of OppA* in an open-unliganded state was obtained for selenomethionine-substituted OppA* (selenomethionine incorporation in proteins expressed in L. lactis will be published elsewhere; Berntsson et al, 2009). The five structures are very similar (RMSD 0.29–0.62 Å), and the only difference between the open-unliganded and open-liganded structures is the absence of a ligand in the latter. In the open conformation, the two domains are separated from each other, leaving the ligand-binding cavity accessible to the solvent. The open state is related to the closed state by a rigid body rotation of about 19° around the segments connecting the two domains (the hinge) (Figure 2B). The rotation is due predominantly to changes in the backbone torsion angles of two residues: Ala299 changes the angles (ϕ/ψ) by 5°/19° going from the open to the closed state, and for Met542 the corresponding changes are 12°/4°. Stereo figures of the closed and open conformation of OppA* are shown in Supplementary Figure S4.
Figure 5.
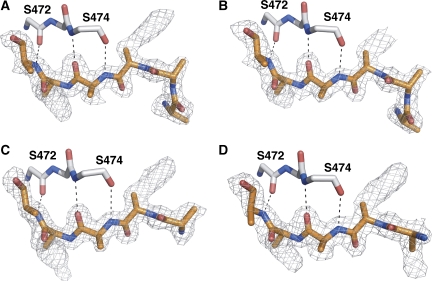
Peptide density in the OppA* structures co-crystallized with (A) Leu-enkephalin (YGGFL, PDB code 3DRH), (B) an octapeptide (RDMPIQAF, PDB code 3DRI), (C) pTH-related peptide (AVSEHQLLHDKGKSIQ, PDB code 3DRJ) and (D) neuropeptide S (SFRNGVGSGVKKTSFRRAKQ, PDB code 3DRK). A peptide (poly-alanine) has been modelled into all densities (orange) but could not be fitted to the known sequences. Ser472 and Ser474, the only two residues that form hydrogen bonds to the peptides, are shown in grey. Notably, the pentapeptide Leu-enkephalin occupies six positions, indicative of the binding in different registers.
Apart from the addition of the different peptides, the crystallization conditions that yielded the structures of OppA* in the open and closed conformation were identical. A possible explanation for the different conformations in the presence of different peptides is their binding affinity. Whereas bradykinin and the endogenous peptides bind to OppA* with high affinity (KD=0.1 μM; Supplementary Figure S3), two of the peptides that yielded structures of the open-liganded conformation are known to bind with much lower affinities (KD of 37.7 μM for the octapeptide (Lanfermeijer et al, 1999) and 50–100 μM for YGGFL (unpublished data)); for the other two peptides, the affinities are not known. It is likely that the crystallization conditions favoured the open conformation when low-affinity ligands were used.
OppA* in the open conformation exposes the residues that are important for ligand binding to the solvent. Unexpectedly, the four structures in the open-liganded state showed a similar electron density in the binding site, irrespective of which peptide was present (Figure 5). The electron density allowed for modelling of the backbone of five or six residues, depending on the peptide present. The additional parts of the longer peptides were probably flexible in the crystals, not producing defined electron density. The interactions with the peptides in the open conformation are provided solely by domain II, with the backbone CO and NH groups of Ser472 and Ser474 forming three hydrogen bonds with the peptide backbones. None of the side chain densities was sufficiently defined to allow for amino-acid assignments. Small differences in side chain density were observed between the different data sets, but in no case the sequence of the peptide could be assigned. The poorly defined electron density of the side chains can be explained by binding of the peptides in different registers, with similar affinities, leading to a heterogeneous population of side chains at any single position. Binding in multiple registers to the open conformation is to be expected, because (i) there are interactions with the backbone of the ligand only (three hydrogen bonds with the backbone CO and NH groups of Ser272 and Ser474 in domain II) and (ii) there are no interactions to fix the positions of the N and C termini of the peptides.
Register shifts in molecular dynamics simulations
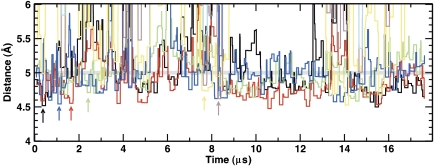
Molecular dynamics simulations of OppA* were performed with a coarse grain representation of the system (Marrink et al, 2007; Monticelli et al, 2008), using the open-liganded structure and the octapeptide (RDMPIQAF) as ligand. During the simulations, the protein maintained its overall shape and structure, and the peptide remained in the binding pocket. However, the peptide showed a high degree of mobility and sampled different binding positions. To determine which peptide residues interacted with specific residues in the binding cavity, the distances between different protein sites and each of the backbone positions of the peptide were calculated. Figure 6 shows the distances between each of the peptide backbone positions and Ser474 from the protein; in the structure with bound bradykinin, Ser474 hydrogen bonds with Gly4 of bradykinin. The interaction of RDMPIQAF with the protein through residue Ser474 was maintained but different peptide residues interacted with the Ser474 during the simulation. Frequent interactions of Ser474 were observed with Ala7 (red), Ile5 (blue), Gln6 (green) and Phe8 (black). Short-lived interactions where also observed with Met3 (brown) and Pro4 (yellow). Consecutive residues were seen to interact with Ser474 site and this shift in the register occurs at a submicrosecond timescale. Snapshots at different time points in the simulation are shown in Supplementary Figure S5A–D.
Figure 6.
Molecular dynamics simulation using the open conformation and the octapeptide RDMPIQAF. The distance of the backbone of Ser474 to each peptide backbone position was calculated (Arg-1: violet; Asp-2: cyan; Met-3: brown; Pro-4: yellow; Ile-5: blue; Gln-6: green; Ala-7: red; Phe-8: black). Due to the coarse grain nature of the simulation, a distance of less than 5 Å (indicated with a grey line) accounts for a hydrogen bond in the atomistic description of the protein. The distances have been binned into windows of 150 ns for clarity.
Discussion
The structures of OppA* described here immediately explain how the protein can bind peptides of a wide range of lengths (4–35 residues) (Detmers et al, 2000). The binding cavity of OppA* that is formed in the closed state is exceptionally large (Figure 3) and the bound nonapeptide bradykinin occupies only ∼30% of the available volume. The structure with endogenous peptides showed some electron density protruding from both the N- and C-terminal ends of the modelled octapeptide, albeit not well defined, suggesting that longer peptides were present in some of the proteins in the crystal. Indeed, analysis of the bound peptides by mass spectrometry revealed that peptides of up to 26 residues long were present. Because OppA* makes specific interactions with a stretch of six peptide residues only, longer peptides could have residual secondary structure in their N- and C-terminal extensions located in the large cavity.
Previous biochemical data on peptide binding to OppA* (Lanfermeijer et al, 1999, 2000; Detmers et al, 2000) had been interpreted on the basis of the crystal structure of the homologous OppA from S. typhimurium. It was concluded that OppA* engulfed only the first six residues of a nonapeptide ligand and that the additional three residues protruded out of the binding cavity and were in contact with the bulk solvent (Detmers et al, 2000). It is now clear that nonapeptides are completely buried, because the binding cavity of OppA* is dramatically larger than that of OppA from S. typhimurium. If the entire cavity of ∼4900 Å3 was packed with peptide it would fit a molecular mass of 3854 Da (Creighton, 1993), corresponding to 35 amino acids (assuming an average residue molecular mass of 110 Da). This would require the packing of the peptide to be as tightly as a typical globular protein, which seems unlikely for hydrated and unstructured peptides with diverse amino-acid composition. Therefore very long peptides may protrude from the cavity, possibly resembling maltose-binding protein (MBP) where oligosaccharides longer than maltotetraose must stick out of the binding cavity (Quiocho et al, 1997). In MBP, these oligosaccharides still induce domain closure that is required for transport. Domain closure is very likely also compulsory for peptide transport by the Opp system, as OppA* does not interact with the transmembrane domain in the absence of ligand, that is, in its open conformation (Doeven et al, 2008).
Similar to the homologous proteins from B. subtilis, S. typhimurium and E. coli (Tame et al, 1994; Nickitenko et al, 1995; Levdikov et al, 2005), the substrate specificity of OppA* is determined almost exclusively by hydrogen bonds of the protein with the peptide backbone (Figure 4). Apart from the side chain at position 5 of bradykinin, all side chains of the peptide are located in the large aqueous binding cavity. There are no side chain-specific interactions with the protein, which explains the protein's indifference towards the exact amino-acid sequence of the peptide (Lanfermeijer et al, 2000; Doeven et al, 2004).
The side chain of residue 5 of bradykinin is the only side chain that is located in a well-defined pocket, which has a pronounced hydrophobic character. It is likely to favour binding of hydrophobic side chains. Indeed, when OppA* was crystallized with an undefined mixture of endogenous peptides bound, most side chains were not defined by electron density because of the chemical heterogeneity of the side chains, but the electron density of the side chain in pocket 5 had well-defined electron density of the sec-butyl group of isoleucine (Figure 4A). This is consistent with the presence of isoleucine residues in the majority of the endogenous peptides bound to OppA* as determined by tandem mass spectrometry. However, the position of the isoleucines in the different endogenous peptides is highly variable (Supplementary Table II). The Venus fly-trap mechanism (Mao et al, 1982) provides an explanation for these observations. The open and closed conformations of the proteins exist in a state of equilibrium, both with and without a ligand bound. Binding of the ligands shifts the equilibrium to the closed conformation, trapping the substrate between the two domains (Tame et al, 1994, 1996; Nickitenko et al, 1995; Bjorkman and Mowbray, 1998; Trakhanov et al, 2005). Peptides can bind in different registers to the open conformation of OppA*, but a peptide will stabilize the closed conformation best if the most favourable interactions with the protein are formed, with pocket 5 apparently preferring an isoleucyl side chain. To be able to bind in different registers, the N and C termini of the peptides must not be fixed in specific positions, which is consistent with the absence of salt bridges between peptides and OppA*. It also requires that there is enough space in the binding cavity to accommodate the N- or C-terminal parts of longer peptides, which is consistent with the exceptionally large volume of the cavity in OppA*. The crystal structures of OppA* in the open conformation, and molecular dynamics simulations provide additional indications that peptides bind to the open conformation of OppA* using different registers.
Why has the binding site in OppA* evolved to have a single well-defined (hydrophobic) side chain pocket, which preferentially accepts an isoleucine side chain, whereas other peptide binding proteins such as OppA from S. typhimurium are promiscuous at every position? The OppABCDF transporter from L. lactis can bind and transport very long peptides, thus giving the organism a selective advantage over its competitors, which transport small peptides of defined length only. To be able to transport long as well as short peptides, salt bridges with the peptides' termini are absent in OppA*. In contrast, homologous OppAs from other organisms use salt bridges (Tame et al, 1994; Levdikov et al, 2005; Tanabe et al, 2007), and these contribute to the binding affinity. Probably, it was necessary to evolve a single, more specific side chain pocket to obtain similar high-affinity binding in OppA*. The choice of a pocket preferring isoleucine makes perfect sense, because L. lactis is auxotrophic for branched chain amino acids. The protein thus preferentially binds peptides that contain at least one amino acid that the organism needs most. Although the isoleucine side chain is clearly preferred in pocket 5, the selectivity is not absolute, because also other hydrophobic side chains, such as phenylalanyl in bradykinin, can be accepted. In this manner, peptides without isoleucine are not completely excluded, and can also be taken up.
The endogenous peptides co-purified with OppA* were very rich in prolines. Apparently, proline-rich peptides bind with high affinity to the protein. The backbone ϕ and ψ angles of the bound peptides (both the endogenous ones and bradykinin) are compatible with the presence of prolines in 4–5 positions (Supplementary Figure S6), Therefore, proline-containing peptides are not expected to have different binding enthalpies than proline-free peptides. However, the entropic energy loss on peptide binding is likely to be smaller for the proline-rich peptides, explaining their high-affinity binding. Physiologically, the ability to bind proline-rich peptides with high affinity is advantageous for L. lactis, because milk caseins are rich in proline residues (9.8% of the residues in α-, β- and κ-casein) and the capacity to synthesize proline is too low to support high growth rates.
In conclusion, the peptide's amino-acid composition has a function in selection by OppA*. Proline-rich peptides containing at least one isoleucine are bound with high affinity. The position of the isoleucine residue in the peptide is not critical, because the ligand can sample different binding registers until the optimal position is found in which the isoleucine ends up in the single hydrophobic pocket. In this manner, the mechanism for peptide selection is based on amino-acid composition rather than the exact sequence.
Materials and methods
Expression and purification of OppA*
Constitutive expression of OppA* in L. lactis AMP2/pAMP21 and protein purification were carried out as described previously (Lanfermeijer et al, 1999), with minor changes. For a detailed protocol, see Supplementary data.
Identification of co-purified endogenous peptides
OppA* was purified together with endogenous peptides as described previously (Lanfermeijer et al, 1999). OppA* (500 nmol) in 20 mM MES pH 6.0, 150 mM NaCl was denatured by adding SDS to a final concentration of 0.4%. The sample was incubated at 37°C for 15 min and subsequently purified on a C18 tip as described (Rappsilber et al, 2003), prior dilution with trifluoroacetic acid (TFA) to a final concentration of 0.1%. The peptides obtained were separated on a Acclaim PepMap100 C18 capillary column (3 μm, 100 Å, 150 mm × 75 μm) at a flow rate of 300 nl/min mounted on an nanoflow liquid chromatography UltiMate™ 3000 Nano-LC System (LC Packings/DIONEX). For elution, a gradient from 3 to 45% acetonitrile in 0.05% TFA was performed in 30 min. Column effluent was mixed 1:4 v/v with a matrix solution of 2 mg/ml α-cyano-4-hydroxycinnamic acid (LaserBio Labs). Fractions of 12 s were spotted on a blank MALDI target with a Probot system (LC Packings/DIONEX). Mass spectrometric data acquisition was performed in positive ion mode. During acquisition, peptides with signal-to-noise level of 40 or more were selected for MS/MS analysis.
Peptide identification was carried out using Mascot (version 2.1; Matrix Science) and ProteinPilot (version 2.0.1; Applied Biosystems) searching against the sequence of the L. lactis genome (Wegmann et al, 2007). All peptides identified with probability higher than 99%, were accepted.
Light scattering
An aliquot of 200 μl of OppA* (∼0.5 mg/ml) was run at a flow rate of 0.5 ml/min on a Superdex 200 10/300GL gel filtration column (GE Healthcare) in 20 mM MES pH 6.0, 150 mM NaCl using an Agilent 1200 series isocratic pump at room temperature. Detectors were used for absorbance at 280 nm (Agilent), static light scattering (miniDawn TREOS Wyatt) and differential refractive index (a Optilab Rex Wyatt). For data analysis, the ASTRA software package version 5.3.2.10 was used (Wyatt), with a value for the refractive index increment (dn/dc)protein of 0.179 ml/mg (Folta-Stogniew and Williams, 1999; Slotboom et al, 2008).
Crystallization and structure determination
Crystals were grown by vapour diffusion in hanging drops. The drops consisted of 1 μl protein (10 mg/ml OppA*, 9 mM Na-MES, pH 6.0, 9 mM NaCl and 1 mM peptide) and 1 μl reservoir solution (0.2 M NaCl, 0.1 M Na-HEPES, pH 7.0 and 20% PEG 6000). For co-crystallization with peptides, the peptide solution (10 mM stock in milliQ water) was mixed 1–10 with protein solution yielding final concentrations of 10 mg/ml OppA, 1 mM peptide, 9 mM Na-MES, pH 6.0 and 9 mM NaCl. Bradykinin (RPPGFSPFR) was also present in the crystallization conditions of the protein with bound endogenous peptides. OppA* stripped of endogenous peptides were used for all crystallization trials except for the one yielding crystals with endogenous ligands. Crystals that yielded the open conformation structures were grown in the presence of four different peptides: Leu-enkephalin (YGGFL), an octapeptide (RDMPIQAF), pTH-related protein 1–16 (AVSEHQLLHDKGKSIQ) and neuropeptide S (SFRNGVGSGVKKTSFRRAKQ). Diffracting crystals of OppA* with endogenously bound peptide, as well as bradykinin, were obtained after 12 weeks of incubation at room temperature. Crystals of OppA* in the open conformation were obtained after 8 h of incubation at room temperature. Crystals were soaked in mother liquor supplemented with 42% PEG 6000 for 30 s and then flash cooled in liquid nitrogen. Heavy atom derivatives were obtained in the same manner, with the addition of 1 M NaBr to the cryo-solution. Data were collected at the beamlines BM16, ID29 and ID14-1 at ESRF, Grenoble (Supplementary Table I). Data processing and reduction were carried out using MOSFLM and program of the CCP4 package (Leslie, 1992; Collaborative Computational Project, 1994).
The structure of OppA* with endogenously bound peptide was solved by MAD data from the bromide derivative combined with sulphur-SAD data. The data were collected at ESRF ID29 (peak wavelength: 0.919 Å; inflection: 0.920 Å; remote: 0.916 Å; S-SAD: 1.775 Å). Initial heavy atom sites were identified using SHELXD (Schneider and Sheldrick, 2002) and phases were calculated using SHARP (Fortelle and Bricogne, 1997). In total, 21 bromide and 11 sulphur sites were refined in SHARP and improved by solvent flattening/flipping, using DM (Cowtan, 1994) and SOLOMON (Abrahams and Leslie, 1996). The phase-combined electron density map was of sufficient quality to let Resolve (Terwilliger, 2000) automatically build 80% of the polypeptide chain. Subsequently, ARP/wARP (Perrakis et al, 1999) used the model together with the native data and automatically traced 520 residues. The resulting electron density maps allowed the loops, which were not built by ARP/wARP, to be manually traced, using Coot (Emsley and Cowtan, 2004) and refined using Refmac5 (Murshudov et al, 1997). Secondary structure elements were determined using DSSP (Kabsch and Sander, 1983).
The structures of OppA* with bound bradykinin and OppA* in the open conformation were solved with molecular replacement using Phaser (McCoy et al, 2005) with the structure of OppA* with bound endogenous peptide as a search model. The output model from Phaser was refined using Coot and Refmac.
Data analysis
Binding pocket volumes were calculated with the program Voidoo from the Uppsala Software Factory (Kleywegt and Jones, 1994), using the probe-occupied volume for calculation. The program was run 10 times with the exact same parameters (probe radius 1.4 Å, grid spacing 0.5 Å, atomic fattening factor 1.1 Å, grid shrink factor 0.9), but with randomly rotated copies of the molecule, to average out random errors.
Structural alignment was produced using the protein structure comparison service SSM at European Bioinformatics Institute (Krissinel and Henrick, 2004). Individual structural alignments to calculate identities were carried out using TM-align (Zhang and Skolnick, 2005).
For detailed information of the molecular dynamics simulations, see Supplementary data.
Accession codes
The coordinates have been deposited in the Proteins Data Bank with accession codes 3DRF, 3DRG, 3DRH, 3DRI, 3DRJ and 3DRK.
Supplementary Material
Supplementary data
Acknowledgments
We thank Dr Hjalmar Permentier for help with mass spectrometry analysis, and the ESRF for providing excellent beamline facilities. This study was supported by grants from Marie Curie Early Stage Training (EST; to RP-AB), The Netherlands Organisation for Scientific Research (NWO; Vidi grant to D-JS), EU (E-MeP & EDICT programmes) and the Netherlands Proteomics Centre (NPC).
References
- Abrahams J, Leslie A (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr 52: 30–42 [DOI] [PubMed] [Google Scholar]
- Berntsson RP-A, Oktaviani NA, Fusetti F, Thunnissen A-MWH, Poolman B, Slotboom D-J (2009) Selenomethionine incorporation in proteins expressed in Lactococcus lactis. Protein Sci (e-pub ahead of print 25 February 2009; doi:10.1002/pro.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemans-Oldehinkel E, Doeven MK, Poolman B (2006) ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett 580: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Bjorkman A, Mowbray S (1998) Multiple open forms of ribose-binding protein trace the path of its conformational change. J Mol Biol 279: 651–664 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cowtan K (1994) DM: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31: 34–38 [Google Scholar]
- Creighton TE (1993) Proteins: Structures and Molecular Properties, p 229. New York: Freeman [Google Scholar]
- Detmers F, Lanfermeijer F, Abele R, Jack R, Tampe R, Konings W, Poolman B (2000) Combinatorial peptide libraries reveal the ligand-binding mechanism of the oligopeptide receptor OppA of Lactococcus lactis. Proc Natl Acad Sci USA 97: 12487–12492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doeven MK, Abele R, Tampe R, Poolman B (2004) The binding specificity of OppA determines the selectivity of the oligopeptide ATP-binding cassette transporter. J Biol Chem 279: 32301–32307 [DOI] [PubMed] [Google Scholar]
- Doeven MK, van den Bogaart G, Krasnikov V, Poolman B (2008) Probing receptor–translocator interactions in the oligopeptide ABC transporter by fluorescence correlation spectroscopy. Biophys J 94: 3956–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Folta-Stogniew E, Williams K (1999) Determination of molecular masses of proteins in solution: implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J Biomol Tech 10: 51–63 [PMC free article] [PubMed] [Google Scholar]
- Fortelle E, Bricogne G (1997) Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol 276: 472–494 [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22: 2577–2637 [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones T (1994) Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr 50: 178–185 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Kunji ERS, Fang G, Jeronimus-Stratingh CM, Bruins AP, Poolman B, Konings W (1998) Reconstruction of the proteolytic pathway for use of beta-casein by Lactococcus lactis. Mol Microbiol 27: 1107–1118 [DOI] [PubMed] [Google Scholar]
- Lanfermeijer F, Detmers F, Konings W, Poolman B (2000) On the binding mechanism of the peptide receptor of the oligopeptide transport system of Lactococcus lactis. EMBO J 19: 3649–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfermeijer F, Picon A, Konings W, Poolman B (1999) Kinetics and consequences of binding of nona- and dodecapeptides to the oligopeptide binding protein (OppA) of Lactococcus lactis. Biochemistry 38: 14440–14450 [DOI] [PubMed] [Google Scholar]
- Leslie AGW (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4+ ESF-EAMCB Newsletter on Protein Crystallography, vol 26 [Google Scholar]
- Levdikov VM, Blagova EV, Brannigan JA, Wright L, Vagin AA, Wilkinson AJ (2005) The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. J Mol Biol 345: 879–892 [DOI] [PubMed] [Google Scholar]
- Mao B, Pear M, McCammon J, Quiocho F (1982) Hinge-bending in L-arabinose-binding protein. The Venus's flytrap model. J Biol Chem 257: 1131–1133 [PubMed] [Google Scholar]
- Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH (2007) The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B 111: 7812–7824 [DOI] [PubMed] [Google Scholar]
- McCoy A, Grosse-Kunstleve R, Storoni L, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61: 458–464 [DOI] [PubMed] [Google Scholar]
- Monnet V (2003) Bacterial oligopeptide-binding proteins. Cell Mol Life Sci 60: 2100–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink SJ (2008) The MARTINI coarse-grained force field: extension to proteins. J Chem Theory Comput 4: 819–834 [DOI] [PubMed] [Google Scholar]
- Murshudov G, Vagin A, Dodson E (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nickitenko A, Trakhanov S, Quiocho F (1995) 2 Å resolution structure of DppA, a periplasmic dipeptide transport/chemosensory receptor. Biochemistry 34: 16585–16595 [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin V (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Quiocho F, Ledvina P (1996) Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol 20: 17–25 [DOI] [PubMed] [Google Scholar]
- Quiocho F, Spurlino JC, Rodseth LE (1997) Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure 5: 997–1015 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Schneider T, Sheldrick G (2002) Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr 58: 1772–1779 [DOI] [PubMed] [Google Scholar]
- Slotboom DJ, Duurkens RH, Olieman K, Erkens GB (2008) Static light scattering to characterize membrane proteins in detergent solution. Methods 46: 73–82 [DOI] [PubMed] [Google Scholar]
- Tame J, Murshudov G, Dodson E, Neil T, Dodson G, Higgins C, Wilkinson AJ (1994) The structural basis of sequence-independent peptide binding by OppA protein. Science 264: 1578–1581 [DOI] [PubMed] [Google Scholar]
- Tame J, Sleigh S, Wilkinson AJ, Ladbury J (1996) The role of water in sequence-independent ligand binding by an oligopeptide transporter protein. Nat Struct Biol 3: 998–1001 [DOI] [PubMed] [Google Scholar]
- Tanabe M, Mirza O, Bertrand T, Atkins HS, Titball RW, Iwata S, Brown KA, Byrne B (2007) Structures of OppA and PstS from Yersinia pestis indicate variability of interactions with transmembrane domains. Acta Crystallogr D Biol Crystallogr 63: 1185–1193 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2000) Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr 56: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakhanov S, Vyas N, Luecke H, Kristensen D, Ma J, Quiocho F (2005) Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity. Biochemistry 44: 6597–6608 [DOI] [PubMed] [Google Scholar]
- Tynkkynen S, Buist G, Kunji E, Kok J, Poolman B, Venema G, Haandrikman A (1993) Genetic and biochemical characterization of the oligopeptide transport system of Lactococcus lactis. J Bacteriol 175: 7523–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann U, O'Connell-Motherway M, Zomer A, Buist G, Shearman C, Canchaya C, Ventura M, Goesmann A, Gasson MJ, Kuipers OP, van Sinderen D, Kok J (2007) Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 189: 3256–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J (2005) TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res 33: 2302–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data