Abstract
Sex in fungi is driven by peptide pheromones sensed through seven-transmembrane pheromone receptors. In Cryptococcus neoformans, sexual reproduction occurs through an outcrossing/heterothallic a- sexual cycle or an inbreeding/homothallic – unisexual mating process. Pheromone receptors encoded by the mating-type locus (MAT) mediate reciprocal pheromone sensing during opposite-sex mating and contribute to but are not essential for unisexual mating. A pheromone receptor-like gene, CPR2, was discovered that is not encoded by MAT and whose expression is induced during a- mating. cpr2 mutants are fertile but have a fusion defect and produce abnormal hyphal structures, whereas CPR2 overexpression elicits unisexual reproduction. When heterologously expressed in Saccharomyces cerevisiae, Cpr2 activates pheromone responses in the absence of any ligand. This constitutive activity results from an unconventional residue, Leu222, in place of a conserved proline in transmembrane domain six; a Cpr2L222P mutant is no longer constitutively active. Cpr2 engages the same G-protein activated signalling cascade as the Ste3a/α pheromone receptors, and thereby competes for pathway activation. This study established a new paradigm in which a naturally occurring constitutively active G protein-coupled receptor governs morphogenesis in fungi.
Keywords: constitutively active receptor, cryptococcus, GPCR, mating, pheromone
Introduction
Much of a cell's sense of the environment is dependent on cell surface receptors; among these, the seven-transmembrane (7TM) G protein-coupled receptors (GPCRs) are the largest superfamily and respond to an array of environmental cues as diverse as light, odorants, flavours, nutrients, neurotransmitters and hormones (Pierce et al, 2002). In addition, more than half of all clinically relevant drugs act as agonists or antagonists of GPCRs and, thus, these receptors are also of great pharmacological importance (Drews, 1996). Multicellular eukaryotes have an enormous GPCR repertoire; approximately 800 are encoded in the human genome. Therefore, it is a daunting challenge to link each GPCR to potential ligands and their attendant signalling cascades. Despite considerable effort, approximately 120 of the 367 nonodorant receptors of humans have remained orphans (Civelli, 2005). Surprisingly, several have been found to be constitutively active, and thus signal in a ligand-independent fashion (Rosenkilde et al, 2006).
The hypothesis that some GPCRs can signal in the absence of any external chemical ligand was first supported by studies of the opioid and β2-adrenergic receptors (Koski et al, 1982; Cerione et al, 1984). Many constitutively active mutant (CAM) GPCRs have been generated artificially by mutagenesis; on the other hand, naturally occurring point mutations that increase constitutive activity have also been identified and found to be linked to human diseases (Van Sande et al, 1995). Moreover, approximately 60 wild-type GPCRs from human, mouse or rat have been shown to exhibit considerable constitutive activity (Seifert and Wenzel-Seifert, 2002). Therefore, constitutively active GPCRs may be of great physiological importance in both natural physiology and disease states.
7TM receptors are able to direct cellular developmental decisions. In mammals, these receptors transduce signals through three families of MAPKs leading to cell differentiation, proliferation, apoptosis and development (Chang and Karin, 2001). In Drosophila, frizzled and smoothened are two upstream 7TM receptors functioning, respectively, in the Wnt and Hedgehog signalling pathways that are critical for embryonic development (Chen and Struhl, 1996; Wodarz and Nusse, 1998). And in the single-celled budding yeast Saccharomyces cerevisiae, 7TM receptors sense pheromones from a mating partner and control cell fate decisions involving sexual versus asexual growth (Burkholder and Hartwell, 1985; Hagen et al, 1986).
Fungi harbour a simplified GPCR repertoire. The ascomycete S. cerevisiae expresses only three GPCRs: two (Ste2 and Ste3) sense the mating pheromones and the other (Gpr1) senses sugars (Burkholder and Hartwell, 1985; Hagen et al, 1986; Kraakman et al, 1999). On account of this relative simplicity, the pheromone receptors and Gpr1 have been extensively studied and serve as models for GPCR-mediated signalling. The pheromones and pheromone receptors are expressed in a cell-type specific manner in S. cerevisiae. a-cells express the lipopeptide pheromone a-factor and the α-factor receptor Ste2, whereas α cells express the peptide pheromone α-factor and the a-factor receptor Ste3. Both Ste2 and Ste3 are essential for mating; null mutants are pheromone nonresponsive and fail to initiate mating projections.
A different pheromone/pheromone receptor paradigm is observed in basidiomycete fungi. Only a-factor-like lipopeptide pheromones and Ste3-like pheromone receptors are known, and neither α-factor-like pheromone genes nor Ste2-like orthologs are apparent in any of several sequenced genomes. Moreover, the pheromone and pheromone receptor genes have been incorporated into a second MAT locus in many tetrapolar species including the mushrooms Coprinopsis cinerea and Schizophyllum commune and the corn smut fungus Ustilago maydis (Fraser et al, 2007). The basidiomycetous yeast Cryptococcus neoformans, the causative agent of cryptococcal meningitis, has a defined sexual cycle and a simpler bipolar mating system (a and α) in which a single MAT locus spans an approximately 120 kb recombinationally suppressed region (Hull and Heitman, 2002). More than 20 genes are encoded by the Cryptococcus MAT locus, including the homeodomain transcription factors, pheromones and pheromone receptors, and genes involved in the mating signalling cascade (Lengeler et al, 2002). The pheromone receptors Ste3a and Ste3α are well characterized; both are required for pheromone sensing and mating and may have roles in virulence (Chung et al, 2002; Chang et al, 2003). Thus, it was surprising when another pheromone receptor-like gene, CPR2 (Cryptococcus pheromone receptor 2), was discovered to be encoded by a different chromosome unlinked to MAT.
Here, we have addressed the basic functions of Cpr2 and whether it has a function in pheromone sensing. All lines of evidence indicate that wild-type Cpr2 is a constitutively active GPCR that, when expressed, continuously engages the pheromone response pathway independent of any pheromone ligand. In addition, Cpr2 couples and signals through the same G proteins, MAPK cascade, and downstream transcription factor as the Ste3a/α pheromone receptors. Our findings reveal that cells activate the pheromone response pathway in the absence of a mating partner when CPR2 is expressed, and activation of the pathway switches growth from yeast-like cells to polarized filaments. During filamentous growth, Cpr2 further serves to ensure fidelity of nuclear transmission in the dikaryotic state. Thus, the mating pathway represents a cascade in which ligand-dependent GPCRs evoke expression of a ligand-independent GPCR. This study provides the first example of a wild-type constitutively active GPCR that regulates cellular development in the fungal kingdom. Similar regulatory mechanisms are likely to operate in many other fungi and possibly also in multicellular eukaryotes in which GPCR-mediated signalling is versatile and more complex.
Results
Identification of CPR2, a gene encoding a putative pheromone receptor unlinked to MAT
The MAT locus of C. neoformans has been extensively studied and among the 20 or so genes encoded by this locus, the homeodomain proteins and the pheromone/pheromone receptor genes establish the sexual identity of a cell (Chung et al, 2002; Hull et al, 2005). Therefore, it was intriguing when another presumptive pheromone receptor-like gene (CPR2) was found to be located elsewhere in the genome (Chang and Kwon-Chung, ASM General meeting 2001, abstr. F-55).
A search for Ste3-like pheromone receptors in other basidiomyceteous fungi revealed that many species also possess additional pheromone receptor paralogs that are not part of MAT, including C. cinerea, Coprinellus disseminatus, Phanerochaete chrysosporium and Pholiota nameko (Aimi et al, 2005; James et al, 2006). Phylogenetic analysis of these basidiomycetous Ste3-like pheromone receptors revealed that the receptors encoded by MAT are interspersed in an evolutionary tree with receptors encoded by genes outside of MAT (Supplementary Figure S1). Moreover, receptors of closely related species are in general more closely related, suggesting that either these receptors duplicated and diverged post-speciation, or that more recent gene conversion has altered their evolutionary trajectory (Supplementary Figure S1). One interesting feature distinguishing C. neoformans Ste3a/α from Cpr2 is that the two pheromone receptors Ste3a and Ste3α are more closely related to each other than either is to Cpr2. Ste3α shares 33% protein sequence identity with Ste3a in comparison to only 25% with Cpr2 (Supplementary Figure S2). This indicates that the divergence between Cpr2 and the common ancestor of Ste3a and Ste3α occurred either before the two mating types evolved or that Ste3a and Ste3α have undergone more recent gene conversion with each other compared with Cpr2.
CPR2 expression is induced after cell–cell fusion during mating
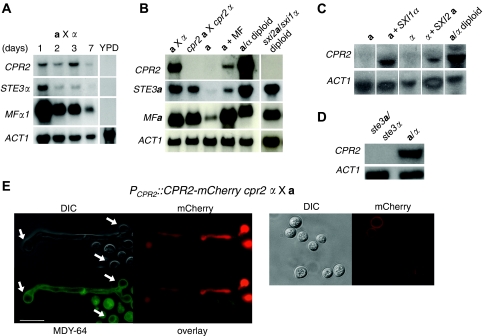
The shared sequence identity between the Ste3-like pheromone receptors and Cpr2 suggested that Cpr2 could have a function in pheromone sensing and signalling. On the basis of earlier studies, several genes encoding pheromone response pathway elements have been shown to be induced during mating (Shen et al, 2002; Chang et al, 2003; Davidson et al, 2003). A C. neoformans mating time course was performed and RNA was extracted to monitor CPR2 expression during the mating process. Northern blot analysis showed that CPR2 expression is induced during mating, with the highest expression observed after 72 h, whereas no CPR2 message was detectable in either a or α cells cultured alone on V8 mating medium (Figure 1A and B and data not shown). In comparison, the STE3α pheromone receptor gene was strongly expressed at 24 h and returned to a basal level of expression after 48 h (Figure 1A). Thus, STE3α and CPR2 expression is temporally regulated.
Figure 1.
CPR2 expression is induced after cell–cell fusion during mating. (A) CPR2 expression during a mating time course. Wild-type serotype D cells were grown in YPD overnight, harvested and crossed on V8 mating medium for 1, 2, 3 and 7 days. The transcript abundance of CPR2, STE3α and MFα1 was measured by northern. ACT1 (actin) served as a loading control. Signals in the lane YPD were derived from α cells alone, which is an independent experiment and therefore separated by a line. (B) CPR2 is expressed during mating and the Sxi1α/Sxi2a complex is required to induce CPR2 expression. CPR2 transcripts were moderately elevated by pheromone and dramatically induced during mating and in diploid cells. Serotype D cells of the indicated genotypes were cultured on V8 mating medium for 72 h and RNA was extracted and subjected to northern analyses with probes to the genes indicated. Signals in the lane sxi2a/sxi1α diploid were derived from an independent RNA gel and are therefore separated by a line. (C) The Sxi1α/Sxi2a complex is sufficient to induce CPR2 expression. Northern analyses were conducted with CPR2 and ACT1 as probes. RNA was extracted from cells cultured on V8 mating medium for 72 h. All lanes were from the same gel. (D) Pheromone signalling is still required to induce CPR2 expression in diploid cells. Conditions for RNA extraction are the same as described in (B) and (C). (E) Cpr2 is highly expressed in hyphae and localizes to the plasma membrane in yeast cells. mCherry was C-terminally fused to Cpr2 under the endogenous CPR2 promoter and expressed in an α cpr2 mutant (YPH342). The strain was crossed to a-cells (JEC20) on V8 mating medium for 48 h. Cells were observed at 1000 × magnification with or without the vacuolar membrane marker dye MDY-64 staining. Arrows indicate the vacuoles based on the DIC and fluorescent images. Left panels are of dikaryotic mating hyphae produced in the mating; right panels are co-cultured yeast cells that have responded to pheromone but not yet undergone cell–cell fusion. Scale bar indicates 10 μm.
To determine whether CPR2 expression was induced before or after cell–cell fusion, mating cells (a × α), a-cells, a-cells treated with synthetic MFα pheromone, and a/α diploid cells were grown on V8 solid medium for 3 days, and RNA was extracted and probed for CPR2 expression. Abundant CPR2 transcripts were detected in a × α mating cells and a/α diploid cells (Figure 1B). In contrast, addition of MFα pheromone to a-cells only modestly increased CPR2 expression. This indicates that, unlike the pheromone genes, whose expression is highly induced by activation of the pheromone response pathway alone (Figure 1B), most CPR2 induction might occur after cell–cell fusion. The sex-determining homeodomain transcription factors Sxi1α and Sxi2a are known to function as a heterodimer after cell–cell fusion and therefore are likely to promote the expression of CPR2. To test this hypothesis, CPR2 expression was analysed in a sxi1α/sxi2a diploid strain and no detectable CPR2 message was observed, showing that Sxi1α and Sxi2a are required for CPR2 expression (Figure 1B). Furthermore, CPR2 is also moderately expressed in haploid a or α cells that ectopically express SXI1α or SXI2a, showing that the Sxi complex is both necessary and in part sufficient to induce CPR2 expression (Figure 1C).
On the other hand, to examine whether autocrine pheromone signalling in diploid cells is a prerequisite for CPR2 expression, CPR2 expression was also monitored in a diploid ste3a/ste3α mutant. Comparing with wild-type diploid cells, the ste3a/ste3α mutant exhibited a dramatic defect in hyphal growth on V8 medium, suggesting that pheromone signalling has a role in supporting filamentous growth (Supplementary Figure S3), which is in accord with earlier studies showing that compatible pheromone signalling is required for filamentous growth in diploid cells of U. maydis (Banuett and Herskowitz, 1989; Spellig et al, 1994). Moreover, no detectable CPR2 expression was observed, showing that pheromone signalling is also important to induce or support CPR2 expression in diploid cells (Figure 1D). We further showed that the lack of CPR2 expression in the ste3a/ste3α mutant is a consequence of decreased or absent levels of the Sxi1a/Sxi2a complex, because the expression of the SXI2a gene was below the limit of detection; overexpression of the CPR2 gene in the ste3a/ste3α mutant partially suppressed the filamentation defect (Supplementary Figure S3).
To visualize Cpr2 expression and protein localization during mating, a Cpr2–mCherry fusion protein expressed from the native CPR2 promoter was introduced into a MATα cpr2 mutant strain. The fusion protein is functional, because it induces filamentation in the ste3α mutant background (see below). As shown in Figure 1E, during mating yeast cells exhibited minimal levels of Cpr2–mCherry, whereas CPR2 expression was dramatically increased in hyphal cells, which exhibit a robust fluorescent signal. This result further supports the conclusion that CPR2 expression is induced after cell–cell fusion. Interestingly, localization of Cpr2–mCherry to the plasma membrane was only observed in yeast cells in which the expression of CPR2 was modestly induced by opposite mating pheromones before cell fusion (Figure 1E; Supplementary Figure S3). In cells that have undergone cell–cell fusion and in the mating hyphae, large vacuoles (based on DIC images and staining with the marker dye MDY-64) were observed and Cpr2 was highly expressed (Figure 1E). These large vacuoles might be a consequence of nutrient deprivation in the mating-inducing medium and the Cpr2–mCherry appears to undergo internalization and is localized inside these structures, which may also suggest that Cpr2 may not need to be plasma membrane-localized to trigger signalling (Figure 1E).
cpr2 mutant produces fewer cell–cell fusion products and generates unusual mating filaments
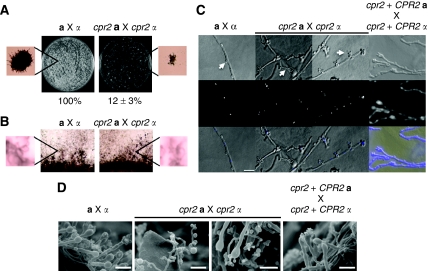
The induced expression of CPR2 during mating suggested that Cpr2 might impact the pheromone response pathway and thus subserve a role in sexual development. cpr2 null mutants in both serotypes (A and D) and mating types (a and α) were generated with dominant drug resistant markers and analysed for their mating abilities. In C. neoformans, mating involves fusion between a and α cells; thus, measuring the number of fusion products generated in a cross serves as a quantitative measurement of fertility. To quantify the fusion efficiency of cpr2 mutants, equal numbers of serotype A cpr2∷NAT α and cpr2∷NEO a-cells were mixed on V8 solid medium for 24 h and the double drug resistant fusion products were selected on yeast extract-peptone-dextrose (YPD) medium containing both nourseothricin and G418. cpr2 mutants exhibited a fusion efficiency of approximately 12% compared with wild type (100%) (Figure 2A). The colonies of the cpr2 × cpr2 fusion products were heterogeneous in size, with many smaller colonies, in contrast to the homogeneous large colonies produced by wild-type cells (Figure 2A). This result suggests that Cpr2 either enhances the efficiency of fusion during the early stages of mating or promotes survival and growth of the dikaryotic (or diploid) cell–cell fusion products.
Figure 2.
cpr2 mutants are fertile, but yield fewer viable dikaryotic fusion products and produce haustorial hyphae during mating. (A) Fusion products from serotype A wild-type or cpr2 × cpr2 mutant crosses were selected on YPD medium containing nourseothricin and G418. Fusion efficiency was determined by counting the number of visible large colonies. Plates were observed at 40 × magnification to distinguish the size differences between colonies derived from wild-type or cpr2 mutant crosses (insets). (B) Serotype A wild-type or cpr2 mutants were crossed on MS medium at 25°C in the dark for 2 weeks and photographed at 40 × magnification. The insets present images of the basidiospore chains. (C) DAPI staining of mating hyphae produced by serotype A wild-type, cpr2, and the cpr2 +CPR2 complemented strains. The arrows indicate the wild-type (left) or abnormal clamp structures and haustorial hyphae (middle panels). These hyphae were photographed at 1000 × magnification and the scale bar indicates 10 μm. See Supplementary Figure S7 for larger images. (D) Scanning electron microscopy images of the wild-type dikaryons and the haustorial hyphae in the cpr2 × cpr2 mutant cross. Bars indicate 10 μm.
Phenotypic analysis of cpr2 a × cpr2 α bilateral crosses showed that, similar to wild-type crosses, abundant dikaryotic mating filaments, basidia and long chains of basidiospores were produced (Figure 2B). This is in contrast to ste3a and ste3α mutants, which exhibit severe cell fusion and mating defects. Thus, CPR2 is not required to initiate the sexual cycle. However, staining of the mating hyphae of wild-type and cpr2 crosses with DAPI revealed that an unusual filamentous structure and abnormal clamp cells were produced during cpr2 × cpr2 crosses. In wild-type crosses, most hyphae observed were dikaryotic with fused clamp connections and largely uniform morphology (Figure 2C). In cpr2 × cpr2 mutant crosses, unusual filamentous structures were observed by both light microscopy and scanning electron microscopy, in addition to the standard dikaryotic mycelia. These unusual filaments were thinner than the dikaryotic filaments, originate from the site where a clamp should have formed, and exhibited a meandering pattern of growth, in contrast to dikaryotic filaments that are more uniform, larger, and exhibit more directed linear growth (Figure 2C and D). These unusual hyphae in the cpr2 mutant crosses share hallmark features with the ‘haustorial hyphae' described in the classic literature (Kwon-Chung, 1998), although their functions are unclear. A large proportion of the clamp cells in the cpr2 mutant cross are abnormal; yeast cells with multiple buds are often observed at the clamps (Figure 2C). This phenotype is attributable to the loss of CPR2 because unusual hyphal structures were only observed in cpr2 × cpr2 bilateral mutant crosses and not in either a WT × α cpr2 or a cpr2 × α WT unilateral mutant crosses and re-introducing the wild-type CPR2 gene into the cpr2 mutants complemented the phenotype (Figure 2C and D).
Cpr2 enhances filamentation and competes with the Ste3α pheromone receptor for signalling
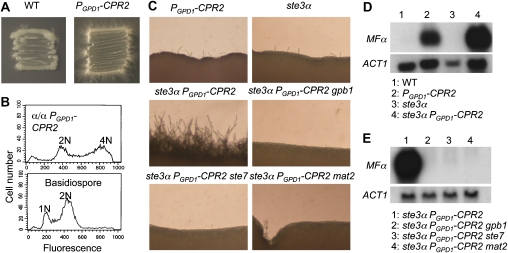
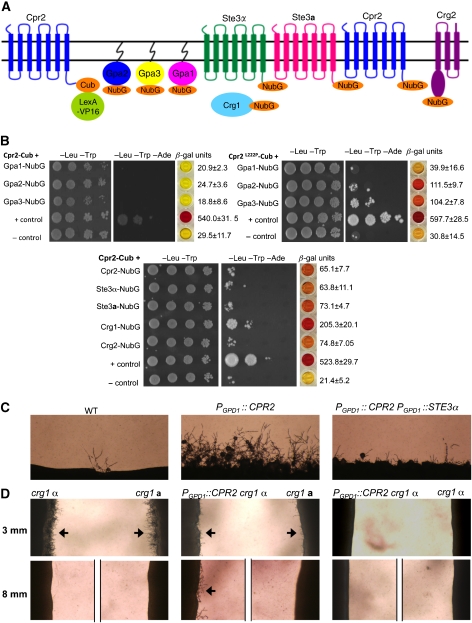
Monokaryotic fruiting is a unisexual mating process that occurs sporadically around the periphery of a colony on V8 or filamentation agar media. Many of the genes involved in pheromone responses and a-α cell mating also affect fruiting (Wickes et al, 1996; Shen et al, 2002; Davidson et al, 2003; Lin et al, 2005). To determine whether Cpr2 has a function in the fruiting process, CPR2 was deleted or overexpressed under the control of the constitutive GPD1 promoter in the wild-type serotype D α strain JEC21 or a related hyperfilamentous strain XL280 (Lin et al, 2005, 2006). Although deletion of CPR2 did not diminish fruiting ability (Supplementary Figure S4), overexpressing CPR2 dramatically enhanced fruiting efficiency in strain JEC21. After 2 weeks of incubation on low-nitrogen medium, filaments were observed along the entire periphery of colonies overexpressing CPR2, in marked contrast to the sporadic filamentation at isolated points around the edges of wild-type colonies (Figure 3A). These filaments were monokaryotic (based on DAPI staining), unfused clamp cells were present and basidia were decorated with short chains of basidiospores, all hallmarks of fruiting. Overexpression of CPR2 also induced fruiting in a-cells (data not shown). To further analyse whether CPR2 overexpression enhances a sexual or asexual sporulation cascade, CPR2 was overexpressed in a wild-type α/α diploid strain. Basidiospores generated by this diploid CPR2 overexpression strain were then isolated and subjected to FACS analysis. Twelve spores that germinated were haploid and had undergone a complete genome reduction (Figure 3B), consistent with a role for CPR2 in activating an α–α unisexual cycle leading to meiotic spore production.
Figure 3.
Overexpression of CPR2 enhances filamentation and activates the pheromone response pathway. (A) CPR2 was overexpressed from the constitutive promoter PGPD1 in strain JEC21. Compared with wild type, fruiting was much more robust in the PGPD1-CPR2 strain (YPH21). Images show colonies after 2 weeks of incubation at 25°C on nitrogen limiting FA medium. (B) FACS analyses showed that the basidiospores generated from a diploid CPR2 overexpression strain (YPH293) underwent a ploidy reduction from diploid to haploid. (C) Overexpression of CPR2 in the ste3α mutant renders cells hyperfilamentous, and this is dependent upon components of the pheromone signalling pathway. Cells were incubated on V8 medium for 24 h and photographed at 100 × magnification. (D, E) By northern analysis the pheromone signalling pathway is activated in the absence of a mating partner in cells overexpressing CPR2. Mutation of components of the canonical pheromone signalling pathway blocks the constitutively active pheromone response in ste3α PGPD1-CPR2 cells. RNA was extracted from cells of the indicated genotypes after 24 h growth on V8 medium at 25°C and probed with the MFα pheromone and ACT1 genes.
As CPR2 overexpression enhances filamentation and sporulation, both hallmarks of sexual reproduction in C. neoformans, we hypothesized that Cpr2 might function as a pheromone receptor. To investigate this hypothesis, CPR2 was overexpressed in a ste3α mutant (ste3α PGPD1∷CPR2) and multiple independent transformants were analysed for CPR2 expression and mating competency. Unexpectedly, the ste3α PGPD1∷CPR2 strain was severely impaired for fusion with wild-type a-cells (approximately 5% fusion efficiency compared with wild type), but it was dramatically self-hyperfilamentous; robust filamentation was observed in as little as 24 h of incubation on V8 mating medium (Figure 3C). This phenotype is reminiscent of a gpa3 mutant, which lacks a Gα subunit of the pheromone response pathway and is self-hyperfilamentous due to constitutive pheromone signalling (Hsueh et al, 2007). The expression level of the pheromone genes (MFα1, 2, 3) was examined in the wild type, PGPD1∷CPR2, ste3α, and ste3α PGPD1∷CPR2 strains by northern blot analysis. Pheromone gene expression was induced in the wild-type CPR2 overexpression strain and massively enhanced in the ste3α PGPD1∷CPR2 background whereas, under the same conditions, these genes were expressed at a very low level in the wild-type or ste3α mutant (Figure 3D). Thus, overexpression of CPR2 triggers pheromone responses in the absence of a mating partner.
To further dissect whether Cpr2 triggers pheromone signalling through the same pathway as the Ste3a/α receptors, genes encoding components of this pathway known to be essential for pheromone response were disrupted in the ste3α PGPD1∷CPR2 strain and multiple independent mutants were analysed. These components include the Gβ subunit Gpb1, the MAPKK gene Ste7, and a transcription factor that functions downstream of the MAPK cascade, Mat2 (Lengeler et al, 2000; Wang et al, 2000; Davidson et al, 2003; X Lin et al, unpublished data). Northern analysis revealed that when genes encoding these components were mutated, pheromone signalling was no longer constitutively activated by Cpr2, and the pheromone genes were not expressed (Figure 3E). In accord, the self-hyperfilamentous phenotype, which is a consequence of a constitutively active pheromone response, was abolished in these strains (Figure 3C).
On the basis of this epistasis analysis, we conclude that Cpr2 elicits the pheromone response through the same Gβγ subunit, MAPK module, and downstream transcription factor as the canonical pheromone response pathway when it is activated through the pheromone receptor Ste3α responding to MFa pheromone. We hypothesize that Cpr2 competes with Ste3α for signalling as Cpr2 overexpression evoked a more pronounced self-filamentous phenotype and higher pheromone gene expression (3.7-fold) when expressed in the ste3α mutant compared with wild-type cells.
Cpr2 is constitutively active due to a substitution of a conserved proline residue in the sixth transmembrane domain
We next addressed whether Cpr2 senses a ligand to trigger the pheromone response pathway. Sequence identity shared between Cpr2 and the Ste3-related pheromone receptors led us to hypothesize that pheromones might serve as agonists for Cpr2. To test this hypothesis, we used S. cerevisiae as a heterologous expression system that has been established earlier to identify ligands of orphan GPCRs (Mentesana et al, 2002). The full-length cDNA clone of CPR2 was expressed in a MATα S. cerevisiae reporter strain that lacks the yeast a-factor receptor Ste3, does not express the α-factor receptor Ste2, and carries a pheromone inducible FUS1-lacZ reporter gene. Deletion of the S. cerevisiae STE3 gene facilitated Cpr2 coupling to the heterotrimeric G protein Gpa1/Ste4/Ste18 which, when activated, engages the pheromone response pathway resulting in induction of β-galactosidase activity.
When Cpr2 was expressed in S. cerevisiae, surprisingly a constitutively active pheromone response was observed even without treating cells with the C. neoformans MFa or MFα synthetic pheromones (Figure 4A). The S. cerevisiae reporter strain is α mating type, and therefore, only produces α factor that should not act on GPCRs related to Ste3 (which respond to lipid-modified peptides) and would be unlikely to act on a highly divergent receptor. This suggested that Cpr2 was either activated by a ubiquitous ligand present in the cultures or Cpr2 is a constitutively active GPCR that functions in a ligand-independent manner.
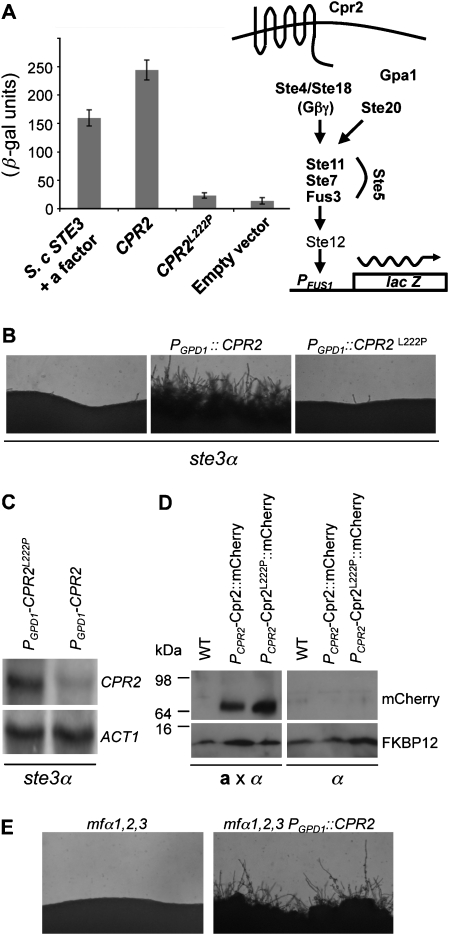
Figure 4.
Cpr2 is a constitutively active receptor that requires the Leu222 residue of TM6 for activity. (A) Cpr2 was heterologously expressed in a S. cerevisiae ste3 mutant carrying the pheromone inducible FUS1-lacZ reporter. Cells expressing S. cerevisiae STE3 treated with yeast a-cell supernatant served as a positive control and cells expressing the empty vector served as a negative control. (B) Leu222 is critical for Cpr2 activity in C. neoformans. Serotype D ste3α cells overexpressing wild-type CPR2 or CPR2L222P were grown on V8 medium for 24 h and photographed at 100 × magnification. (C) RNA was extracted from CPR2 and CPR2L222P overexpression strains (YPH40 and YPH655) and probed with CPR2 and ACT1. (D) Western analysis showed that Cpr2L222P∷mCherry and wild-type Cpr2∷mCherry are equally stable. The fusion proteins were probed with an mCherry antibody and FKBP12 detected with antiserum to S. cerevisiae FKBP12 served as the loading control. (E) CPR2 overexpression induces hyperfilamentation of mfα1,2,3 pheromone-less mutant. Cells of the indicated genotypes (WSC18 and YPH712) were incubated on V8 medium for 4 days and photographed at 40 × magnification.
To address these models, we first carefully analysed the protein sequence of Cpr2, searching for potential residues that could contribute to constitutive activity. It is known that alteration of amino-acid sequences in certain conserved positions in some GPCRs results in CAMs. One well-studied example is the substitution of a conserved proline in the sixth transmembrane domain (TM-VI), which is present in >90% of GPCRs (Baldwin, 1993). Substitution of this proline with leucine substantially increases the ligand-independent activity of many GPCRs, including the pheromone receptors of S. cerevisiae (Ste2, Ste3) and Schizosaccharomyces pombe (Mam2) (Konopka et al, 1996; Stefan et al, 1998; Ladds et al, 2005). Surprisingly, this conserved proline is absent in the TM-VI of wild-type Cpr2, and instead, a leucine is substituted at this position (L222). This observation prompted us to hypothesize that this leucine in TM-VI might be responsible for the constitutive activity of Cpr2. To test this model, a Cpr2L222P allele was generated and heterologously expressed in S. cerevisiae. As shown in Figure 4A, ligand-independent induction of β-galactosidase activity was greatly reduced to a basal level in cells expressing Cpr2L222P, compared with the empty vector control. This result provides evidence that Cpr2 activates the pheromone response pathway through an intrinsic constitutive activity instead of responding to a ubiquitous ligand present in the culture. Moreover, the constitutive activity of Cpr2 requires Leu222 of TM-VI, and a proline residue is typically conserved at this position in most GPCRs.
To investigate whether the requirement of Leu222 for the constitutive activity of Cpr2 also applies to Cpr2 expressed in C. neoformans, the CPR2L222P allele was overexpressed in the ste3α mutant and compared with the self-filamentous phenotype of ste3α cells overexpressing wild-type CPR2. As shown in Figure 4B, CPR2L222P overexpression did not render cells self-filamentous, indicating that Leu222 is critical for CPR2 constitutive activity in C. neoformans. Similar findings were observed in multiple independent transformants. Northern blot analysis confirmed that the CPR2L222P transgene was expressed at or even above the level of the active wild-type CPR2 transgene, showing the difference was not attributable to a lower expression level (Figure 4C). Moreover, when Cpr2–mCherry and Cpr2L222P–mCherry expression was compared by western blot analysis using antibody against mCherry, this analysis showed that Cpr2L222P–mCherry is as stable as Cpr2–mCherry (Figure 4D). Thus, the loss of function of the Cpr2L222P mutant does not appear to be a result of reduced protein stability.
In accord with these lines of evidence supporting the hypothesis that Cpr2 is a wild-type constitutively active receptor, we found that overexpressing CPR2 in the C. neoformans mfα1, 2, 3 pheromoneless mutant still conferred a hyperfilamentous phenotype, excluding the possibility that CPR2 activates the pheromone response pathway in α cells through an autocrine signalling loop involving a pheromone ligand (Figure 4E). Finally, the localization pattern of Cpr2L222P–mCherry is very similar to that of Cpr2–mCherry, which suggests that the internalization of the receptors in the growing hyphae is not caused solely by constitutive Cpr2 activity (Supplementary Figure S5).
Cpr2 and Ste3α interact with the same G proteins and RGS proteins
Epistasis and gene expression analyses support a model in which Cpr2 activates the pheromone response pathway through a conserved Gβ subunit, MAPK module and downstream transcription factor. To further investigate which Gα protein is coupled to Cpr2 and activates signalling, the split-ubiquitin assay was used to dissect these molecular interactions. This assay was developed to detect physical protein–protein interactions between membrane proteins in vivo in S. cerevisiae (Stagljar et al, 1998). In this system, one protein is fused to the C-terminal half of ubiquitin (Cub) coupled to an artificial transcriptional activator (LexA-VP16), and the second protein is fused to a mutated N-terminal half of ubiquitin (NubG). Physical interactions enable re-association of the two halves of split ubiquitin, resulting in cleavage and release of the LexA-VP16 transcription factor and induction of lacZ, HIS3 and ADE2 reporter genes.
Wild-type Cpr2 was fused to Cub (Cpr2–Cub), and the reporter strain was co-transformed with plasmids expressing Gpa1–NubG, Gpa2–NubG or Gpa3–NubG fusion proteins (Figure 5A). Transformants were assayed for growth on synthetic medium lacking histidine or adenine and for β-galactosidase activity. No physical interaction could be detected between Cpr2 and any of the three Gα subunits (Figure 5B). We hypothesized that any interaction between Cpr2 and the coupled Gα subunit might be transient due to the constitutive activity of Cpr2, which may therefore continually dissociate from the interacting Gα subunits. To test this hypothesis, the potential interactions between the three Gα subunits and Cpr2L222P, which lacks ligand-independent activity, were assayed. Remarkably, when Leu222 was substituted with Pro, we observed that Cpr2 L222P now interacted with both Gpa2 and Gpa3 but not with Gpa1 (Figure 5B). Gpa2 and Gpa3 are the G proteins that were recently shown to interact with the pheromone receptor Ste3α to mediate pheromone responses (Hsueh et al, 2007; Li et al, 2007). This result provides an explanation why CPR2 overexpression results in greater signalling in ste3α mutant cells compared with wild-type cells: because Cpr2 and Ste3α interact with the same Gα subunits, the ligand-dependent and ligand-independent receptors compete for access to the signalling cascade. In addition, the results also provide evidence that the Cpr2L222P mutant is stably expressed in S. cerevisiae, and that loss of pheromone signalling activity (Figure 4A) is not attributable to protein destabilization by the proline substitution. To further examine the competition between Cpr2 and Ste3α in C. neoformans, we overexpressed Ste3α in the PGPD1∷CPR2 background and showed that filamentation was dramatically diminished (Figure 5C). This result supports the conclusion that Cpr2 and Ste3α compete for signalling as both receptors interact with the same G proteins.
Figure 5.
Cpr2 physically interacts with Gα subunits, RGS proteins and, GPCRs. (A) A schematic representation of the split-ubiquitin assay with the Cpr2-Cub, Gα-NubG, RGS-NubG, and Ste3a/α-Cub fusion proteins. (B) Cpr2 L222P –Cub interacts with the Gα subunits Gpa2 and Gpa3 and Cpr2–Cub interacts with the RGS protein Crg1, Crg2, dimerizes with itself, Ste3a, and Ste3α. Plasmid pAI-Alg5 expresses a fusion of the endogenous ER protein Alg5 to wild-type Nub, which has strong affinity to Cub and served as a positive control to confirm the correct expression and topology of Cpr2-Cub. Plasmid pDL2-Alg5 expresses Alg5 fused with NubG and served as a negative control. Interactions were measured by both growth and β-galactosidase activity assays. (C) Overexpression of Ste3α in C. neoformans reduces Cpr2-promoted filamentation. Strains of the indicated genotypes were incubated on FA medium for 7 days at 25°C. (D) Confrontation assays were conducted with serotype A strains of the indicated genotypes. Cells of opposite mating type were grown in parallel at distances of 3 or 8 mm from each other on V8 medium and photographed at 40 × magnification after 3 days incubation at 25°C. Arrows indicate conjugation tubes.
The interactions between Cpr2 and the regulator of G protein signalling (RGS) proteins Crg1 and Crg2 were also examined. We reported earlier that the pheromone receptor Ste3α interacts directly with two RGS proteins: Crg1, a DEP domain containing RGS protein and Crg2, which has three C-terminal transmembrane helices (Hsueh et al, 2007). Wild-type Cpr2 was fused with Cub, Crg1 and Crg2 were fused with NubG, and cells expressing both fusion proteins were analysed to test possible interactions. On the basis of growth assays and β-galactosidase expression, wild-type Cpr2 interacts with both Crg1 and Crg2 (Figure 5B). Interestingly, expression of both CRG1 and CPR2 is induced during mating whereas CRG2 is not (Hsueh et al, 2007), suggesting that the Cpr2–Crg1 interaction may be physiologically regulated. To test for genetic interactions between Cpr2 and Crg1, CPR2 was overexpressed in the crg1 mutant. crg1 mutants are supersensitive to pheromones and produce abundant conjugation tubes in a confrontation assay when cells of the opposite mating type are grown in close proximity (approximately 3 mm) on solid medium. However, when the distance between the cells was increased to 8 mm, no conjugation tubes were produced by cells of either mating type. In contrast, the α PGPD1∷CPR2 crg1 mutant still produced conjugation tubes at this increased distance when confronted with an a crg1 mutant (Figure 5D). These results reveal that CPR2 overexpression enhances the ability of the crg1 mutant to respond to pheromone, indicating that expression of a ligand-independent receptor (Cpr2) can enhance the responsiveness of cells to a ligand (pheromone) sensed by a ligand-dependent GPCR.
Cpr2 forms homodimers or heterodimers with itself and Ste3a/α
Many GPCRs can exist as dimers or oligomers and these physical interactions have been shown to regulate trafficking, signalling, ligand-binding properties and cell–cell fusion (Prinster et al, 2005; Levoye et al, 2006). The split-ubiquitin system was used to address the possibility that Cpr2 and Ste3a/α dimerize. Cpr2 was observed to interact with itself, Ste3a, and Ste3α, although the magnitude of these interactions was moderate (Figure 5B). This implies that both homo- and hetero-dimerization of the pheromone receptors and Cpr2 could serve as a mechanism by which their activity may be regulated.
Discussion
The pheromone response pathway is a paradigmatic model for GPCR-mediated signalling pathways that has been studied for decades (Dohlman and Slessareva, 2006). In this study, we investigated the functions of a novel nonmating-type specific pheromone receptor-like gene, CPR2, identified in the human fungal pathogen C. neoformans and conclude that Cpr2 is a wild-type constitutively active GPCR with ligand-independent activity that governs developmental events during the sexual cycle.
Cpr2 and sexual development of C. neoformans
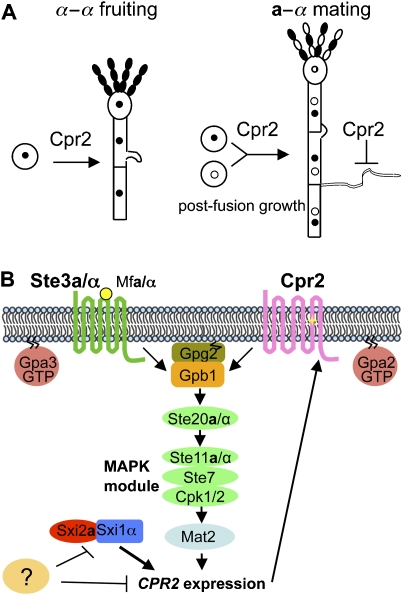
Cpr2 has a function in a-α sexual development (mating) and also promotes unisexual fruiting in C. neoformans (Figure 6A). Although disruption of CPR2 does not impair fruiting under standard conditions, overexpression of CPR2 dramatically enhances and accelerates fruiting. Spores isolated from an α/α diploid CPR2 overexpression strain all underwent a whole genome ploidy reduction (2n to 1n), showing that Cpr2 activates a sexual cycle leading to meiotic spore production. It seems paradoxical that Cpr2 is sufficient, but not necessary for fruiting; however, it may be that Cpr2 has a more prominent function during fruiting in the environment. The conditions that promote fruiting in nature are not known, and in the laboratory, even under optimized media and laboratory conditions, fruiting is an inherently inefficient process. Therefore, searching for conditions that lead to Cpr2 expression is likely to be a fruitful approach to understand this novel developmental cascade involving unisexual mating, and lead to further insights into the roles of Cpr2 in this process.
Figure 6.
Cpr2 activates the pheromone signalling pathway to regulate morphogenesis in C. neoformans. (A) Cpr2 governs yeast to hyphae development in the fruiting process, affects cell–cell fusion efficiency and suppresses abnormal haustorial hyphae formation during mating. (B) A model depicting signalling events leading to Cpr2 expression and subsequently evoked by Cpr2. Upon mating, the pheromone-bound Ste3a/α receptors activate the pheromone signalling pathway and induce a modest level of CPR2 expression. The expression of CPR2 is further dramatically induced after cell–cell fusion by the Sxi1α/Sxi2a heterodimer. Cpr2 is constitutively active and thus sustains pheromone signalling and may support a positive feedback loop, which might be disrupted at a later time point when CPR2 expression is reduced.
In the a-α sexual cycle, unlike the pheromone receptors Ste3a and Ste3α that are critical for mating, cpr2 mutants are still fertile. However, CPR2 contributes to regulate two different morphogenetic processes during mating. First, cpr2 mutants have a defect in cell–cell fusion efficiency, and therefore, CPR2 influences either the efficiency of cell–cell fusion or the growth and survival of the cell–cell fusion products. We think the later is more likely to be the case based on the small, heterogeneous colonies generated after fusion of cpr2 mutants. The second role in mating is that loss of CPR2 causes abnormal filamentous structures to be formed during mating. One major difference between the sexual development of C. neoformans and S. cerevisiae is that in S. cerevisiae, the two nuclei immediately fuse with each other after cell–cell fusion, whereas in C. neoformans, the two nuclei congress but do not fuse after a-α cell fusion and instead form a stable dikaryotic hyphae. In these hyphae, two nuclei, one from each parent, co-exist in a hyphal compartment and the growth of the dikaryons requires coordinate nuclear division and migration. Clamp cells are critical to coordinate correct nuclear migration, ensuring that two nuclei are faithfully segregated (Supplementary Figure S6).
In mushroom fungi, hyphal cells fuse readily without a requirement for pheromone/pheromone receptor recognition; however, pheromone signalling is thought to regulate the fusion step between clamp cells and the hyphae in dikaryons (Casselton and Olesnicky, 1998). In C. neoformans cpr2 mutant crosses, a significant proportion of the clamp cells are abnormal. After forming hyphal branches as the precursor to clamp cells, instead of fusing with the adjacent hyphal cell, the branches continue to elongate or produce multiple buds, resulting in thinner, irregular hyphae that may eventually fuse with other hyphae or neighbouring yeast cells. One plausible role for Cpr2 in dikaryotic hyphae development is to shield the clamp cells from pheromone generated by other cells and hyphae to promote clamp cell fusion and ensure mating fidelity. In the absence of Cpr2, aberrant hyphal morphology may compromise the fidelity of mating.
Leu222 renders Cpr2 constitutively active
Cpr2 is constitutively active when overexpressed in both S. cerevisiae and C. neoformans and activates the same pheromone signalling pathway as the canonical ligand-dependent pheromone receptors Ste3a/α. An unconventional leucine residue in the transmembrane 6 (TM6) domain is required for the ligand-independent signalling activity of Cpr2. For even wild-type ligand-dependent GPCRs, a small fraction of the total receptor pool may exist in the active R* form in the absence of agonist; thus, simply overexpressing some GPCRs is sufficient to elicit a constitutively active response (Milano et al, 1994). However, we do not think that overexpression alone is sufficient in the case of Cpr2-induced pheromone signalling because overexpressing either the canonical pheromone receptor Ste3α or the Cpr2L222P allele did not activate the FUS1-lacZ reporter gene in S. cerevisiae, and overexpression of the Cpr2L222P allele in the C. neoformans ste3α mutant did not cause a hyperfilamentation phenotype (Figure 4).
Both naturally occurring and laboratory generated substitutions of a conserved proline residue in TM6 can render GPCRs constitutively active, and our studies revealed Cpr2 is a naturally occurring example, analogous to laboratory isolated fungal pheromone receptors mutants reported earlier (Konopka et al, 1996; Stefan et al, 1998; Ladds et al, 2005). TM6 of the mammalian thyroid-stimulating hormone receptor is a hotspot for naturally occurring CAMs: several residues in this transmembrane domain, including the conserved proline, have been identified to cause constitutive activity that results in nonautoimmune hyperthyroidism and thyroid adenoma. It is hypothesized that proline causes a kink in the transmembrane helix and is important for receptor conformational changes between the ligand free R and ligand bound R* states (von Heijne, 1991). However, not all GPCRs become constitutively active when this proline residue is missing or mutated (Wess et al, 1993). The Ste3a receptor in C. neoformans is one such example, as it lacks a conserved proline in TM6 but does not display any constitutive activity (data not shown).
Positive feedback loop and cell fate regulation
Positive feedback loops are often incorporated into gene regulatory networks to enable self-perpetuating signalling networks (Ferrell, 2002; Ingolia and Murray, 2007). Examples can be found in organisms ranging from bacteria to multicellular eukaryotes. Mating ability is regulated by a feedback loop in another major fungal pathogen Candida albicans. C. albicans stochastically switches between two cell types—white and opaque, and only opaque cells are mating competent (Miller and Johnson, 2002). A transcription factor, Wor1, has been identified as the master regulator for the white-opaque switching. It is expressed in an all or none fashion; it binds its own promoter region, promotes its own transcription and creates a transcriptional feedback loop (Huang et al, 2006; Zordan et al, 2006). We propose that CPR2 has the potential to introduce a positive feedback loop into the pheromone signalling pathway as Cpr2 can activate pheromone signalling, which in turn generates more CPR2 transcripts (Figure 6B). This may temporarily sustain pheromone signalling after pheromone expression decreases after cell–cell fusion.
Inverse agonist of Cpr2
Most GPCRs are inactive until bound by a ligand that serves as an agonist. Thus, we considered the hypothesis that although being constitutively active, Cpr2 might still be ligand regulated, but in this case, an inverse agonist squelches signalling. Different types of chemicals have been identified as inverse agonists for wild-type constitutively active GPCRs in humans; nonetheless, potential inverse agonists for many constitutively active GPCRs remain unknown (Seifert and Wenzel-Seifert, 2002). Candidates for inverse agonists of Cpr2 could include the MFa and MFα mating pheromones themselves. We explored this hypothesis in the S. cerevisiae heterologous expression system and found that Cpr2 activity was not diminished in the presence of MFa, MFα or both pheromones (data not shown). Furthermore, because the pheromones are farnesylated and other lipid derivatives have been shown to serve as signalling molecules, the potential activity of a series of farnesol derivatives was examined. However, none of these compounds affected the signalling activity of Cpr2 (data not shown, see list of tested compounds in Supplementary data). Therefore, inverse agonists for Cpr2, if these exist physiologically, remain to be identified and are an intriguing hypothesis for future investigation.
Cpr2 cannot replace the function of the pheromone receptor
Duplicated genes often undergo accelerated evolution with one retaining an ancestral function and the other adopting a novel function. On the basis of the phylogenetic analysis, Cpr2, Ste3a and Ste3α diverged from a common ancestor and our results support models in which Cpr2 does not function as a pheromone receptor. Overexpression of Cpr2 in the Ste3α mutant constitutively activates the pheromone signalling pathway; however, these cells largely failed to fuse with a-cells. Similar phenomena have been observed in S. cerevisiae. The P258L substitution in Ste2 dramatically increases FUS1-lacZ reporter expression but does not efficiently trigger projection formation (Konopka et al, 1996). In cells expressing dominant active Gpa1, the expression of the FUS1-lacZ reporter was elevated in the absence of pheromone, whereas the growth of cells was not affected (no cell-cycle arrest) (Guo et al, 2003). The fact that Cpr2 cannot suppress a ste3α mutation to restore mating suggests that Ste3α plays additional roles during mating, possibly involving courtship and morphogenesis oriented in response to a pheromone producing mating partner. Recent studies also showed that in S. cerevisiae an intact receptor-heterotrimeric G protein module, as well as interaction between the pheromone receptors, are critical for successful cell–cell fusion (Shi et al, 2007; Strickfaden and Pryciak, 2008).
Constitutively active pheromone receptors and the evolution of the MAT locus
Approximately 65% of the mushroom fungi have a tetrapolar mating system and one of the two unlinked MAT loci encodes pheromones and pheromone receptors. Nonetheless, approximately 25% of the mushroom fungi are bipolar, with a single MAT locus (Raper, 1966). Studies from two bipolar mushrooms species (P. nameko and C. disseminatus) indicate that mating type co-segregates with the homeodomain MAT locus and the pheromone and pheromone receptors genes are no longer linked to mating type (Aimi et al, 2005; James et al, 2006). Why have these species lost the pheromone/pheromone receptor locus as one of the MAT during evolution? One possible mechanism enabling the switch from tetrapolar to bipolar, as originally proposed by Raper, is that these species became self-compatible at one of the two MAT loci (Raper, 1966). To be self-compatible at the pheromone/pheromone receptor locus, the receptors are either activated by an autocrine signalling loop or constitutively active, locking the pheromone signalling pathway in the ‘on' state. In the laboratory, examples of mutations that alter pheromone receptors to recognize pheromone produced by the same locus have been reported, as well as mutations that lead to constitutive receptor activity (Fowler et al, 2001). For example, in C. cinerea, an R96H mutation in TM3 enables receptor activation in response to pheromone encoded by the same locus and a Q229P mutation in TM6 of another receptor caused constitutive activity (Olesnicky et al, 1999, 2000). Our studies on Cpr2 reveal a paradigmatic example in which constitutively active pheromone receptors are encoded in the genome and could have a function in transitions to self-compatibility.
In summary, Cpr2 is a wild-type constitutively active GPCR that governs morphogenesis of fungal cells. Genes unlinked to MAT encoding pheromone receptor homologs are common in basidiomycetes, and our findings on the function of Cpr2 may be generally applicable to these species as well. In contrast to model budding and fission yeasts, C. neoformans and other filamentous fungi evolved to incorporate more than one Gα subunit for their more complex cell fate decisions. Similarly, in contrast to the budding and fission yeasts that have only canonical pheromone receptors, a constitutively active receptor (Cpr2) has been deployed with the pheromone receptors to effect developmental fates in a fungus that undergoes more complex yeast-hyphae dimorphic transitions during mating. The discovery and elucidation of biological roles for Cpr2 provides a novel mechanism of signalling regulation in fungi, which may have implications for GPCR-mediated signalling cascades throughout the fungal kingdom and even in multicellular organisms.
Materials and methods
Strains, plasmids and media
Strains and plasmids used in this study are listed in Table I. Yeast cells were grown and maintained on YPD and synthetic dextrose media. C. neoformans mating and fruiting assays were conducted on 5% V8 juice agar medium (pH 5 for serotype A and pH 7 for serotype D strains), filament agar (Wickes et al, 1996), or Murashige and Skoog (MS) medium minus sucrose (Sigma-Aldrich).
Table 1.
Strains and plasmids used in this study
| Strain | Genotype | Sources/Reference |
|---|---|---|
| C. neoformans var. neoformans (serotype D, congenic with JEC20 and JEC21) | ||
| JEC20 | MATa | Kwon-Chung et al, 1992 |
| JEC21 | MATα | Kwon-Chung et al, 1992 |
| JEC31 | MATα lys1 | J. Edman |
| YPH50 | MATa ura5 | This study |
| YPH19 | MATα cpr2::NAT | This study |
| YPH20 | MATa cpr2::NAT | This study |
| YPH101 | MATα cpr2::NAT lys1 | This study |
| YPH535 | MATa cpr2::NAT ura5 | This study |
| YPH21 | MATα PGPD1::CPR2 NEO | This study |
| YPH123 | MATα PGPD1::CPR2 NEO ura5 | This study |
| YPH125 | MATa PGPD1::CPR2 NEO | This study |
| WSC43 | MATα ste3α::ADE2 | This study |
| YPH40 | MATα ste3α::ADE2 PGPD1::CPR2 NEO | This study |
| YPH125 | MATα ste3α::ADE2 PGPD1::CPR2 NEO gpb1::NAT | This study |
| YPH523 | MATα ste3α::ADE2 PGPD1::CPR2 NEO ste7::NAT | This study |
| YPH525 | MATα ste3α::ADE2 PGPD1::CPR2 NEO mat2::NAT | This study |
| YPH127 | MATa ste3a::NAT PGPD1::CPR2 NEO | This study |
| YPH286 | MATα/MATα diploid | This study |
| YPH293 | MATα/MATα diploid PGPD1::CPR2 NEO | This study |
| YPH655 | MATα ste3α::ADE2 PGPD1::CPR2L222P NEO | This study |
| YPH552 | MATα ste3α::ADE2 PGPD1::CPR2-mCherry NEO | This study |
| WSC18 | MATα mfα1::ADE2 mfα2,3::URA5 ade2 ura5 | Shen et al, 2002 |
| YPH712 | MATα mfα1::ADE2 mfα2,3::URA5 PGPD1::CPR2 NEO | This study |
| YPH748 | MATα PGPD1::CPR2 NEO PGPD1::STE3α URA5 | This study |
| YPH749 | MATa/α diploid ste3α::ADE2 | This study |
| YPH750 | MATa/α diploid ste3α::ADE2 ste3a::NAT | This study |
| YPH752 | MATa/α diploid ste3α::ADE2 ste3a::NAT PGPD1::CPR2 NEO | This study |
| AI49 | MATa SXI1α URA5 | Idnurm and Heitman, 2005 |
| CHY1014 | MATα PGPD1::SXI2a URA5 | Hull et al, 2005 |
| C. neoformans var. neoformans (serotype D) | ||
| XL280 | MATα | Lin et al, 2006 |
| YPH74 | MATα cpr2::NAT in XL280 background | This study |
| C. neoformans var. grubii (serotype A, congenic to H99 and KN99a) | ||
| H99 | MATα WT | Perfect et al, 1993 |
| KN99a | MATa WT | Nielsen et al, 2003 |
| YSB119 | MATα aca1::NAT ura5 ACA1-URA5 | Bahn et al, 2004 |
| YSB121 | MATa aca1::NEO ura5 ACA1-URA5 | Bahn et al, 2004 |
| CDX12 | MATa cpr2::NAT | This study |
| YPH16 | MATα cpr2::NAT | This study |
| YPH545 | MATa cpr2::NEO | This study |
| YPH657 | MATa cpr2::NAT CPR2::NEO | This study |
| YPH659 | MATα cpr2::NAT CPR2::NEO | This study |
| YPH342 | MATα cpr2::NAT PCPR2::CPR2-mCherry NEO | This study |
| YPH714 | MATα cpr2::NAT PCPR2::CPR2L222P-mCherry NEO | This study |
| YPH276 | MATα crg1::NAT | This study |
| YPH570 | MATa crg1::NAT | This study |
| YPH450 | MATα crg1::NAT PGPD1:: CPR2 NEO | This study |
| S. cerevisiae | ||
| NMY32 | MATa his3Δ200 trp1-901 leu2-3,112 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ (lexAop)8-ADE2 GAL4 | DUALmembrane Kit 2 (Dualsystem Biotech, Zürich, Switzerland) |
| W303-1A | MATa ade2-1 his3-11,15 leu2-3,112 trp-1 ura3-1 can1-100 | Fowler et al, 1999 |
| W303-1B | MATα ade2-1 his3-11,15 leu2-3,112 trp-1 ura3-1 can1-100 | Fowler et al, 1999 |
| SDK45 | MATα ade2-1 his3-11,15 leu2-3,112 trp-1 ura3-1 can1-100 ste3::ADE2 | Fowler et al, 1999 |
| Plasmids used in C. neoformans | ||
| pYH1 | PGPD1:: CPR2 NEO AmpR | This study |
| pYH22 | PGPD1::CPR2-Cherry NEO AmpR | This study |
| pYH33 | PCPR2::CPR2-Cherry NEO AmpR | This study |
| pDX124 | PGPD1:: CPR2L222P NEO AmpR | This study |
| Plasmids used in S. cerevisiae ste3 mutant strain SDK45 for heterologous expression analysis | ||
| pTCFL1 | FUS1::lacZ TRP1 AmpR | This study |
| pYH58 | pPGK::CPR2 cDNA URA3 AmpR | This study |
| pYH59 | pPGK::CPR2 L222PcDNA URA3 AmpR | This study |
| Plasmids used in the split-ubiquitin system in the reporter strain NMY32 | ||
| pYH51 | CPR2-Cub LEU2 KanR | This study |
| pDX74 | CPR2L222P-Cub LEU2 KanR | This study |
| pDX40 | GPA1-NubG TRP1 AmpR | Xue et al, 2006 |
| pDX41 | GPA2-NubG TRP1 AmpR | Xue et al, 2006 |
| pDX42 | GPA3-NubG TRP1 AmpR | Xue et al, 2006 |
| pYH61 | CPR2-NubG TRP1 AmpR | This study |
| pYH62 | STE3α-NubG TRP1 AmpR | This study |
| pYH63 | STE3a-NubG TRP1 AmpR | This study |
| pDX91 | CRG1-NubG TRP1 AmpR | Hsueh et al, 2007 |
| pDX94 | CRG2-NubG TRP1 AmpR | Hsueh et al, 2007 |
Spore dissection and FACS analysis
Spores generated by a diploid CPR2 overexpression strain (YPH293) were isolated by micromanipulation as described (Hsueh et al, 2006). DNA content was determined by FACS analyses (Sia et al, 2000).
Gene disruption and overexpression
An overlap PCR based approach was used to generate all of the gene disruption cassettes with a dominant selectable marker as described earlier (Davidson et al, 2002; Fraser et al, 2003). The overlap PCR products were purified with a QIAGEN gel extraction kit and directly transformed into C. neoformans by biolistic transformation (Davidson et al, 2000a). Primers used for gene disruption are listed in Supplementary Table I. To generate the CPR2 overexpression plasmid pYH1, the wild-type CPR2 gene was amplified from strain JEC21 and cloned into plasmid pKK1 containing the constitutively expressed PGPD1 promoter. The CPR2L222P overexpression plasmid pDX124 was constructed through an overlap PCR approach. The CPR2L222P allele was first amplified with primer pairs JOHE19122/JOHE15828 and JOHE15827/JOHE19123. Primers 15827 and 15828 contain a T to C substitution that changes the target codon from CTT to CCT. The CPR2L222P full-length gene was obtained by second overlap PCR performed with primers JOHE19122/JOHE19123. The PCR products were purified and cloned into plasmid pXL1 containing the PGPD1 promoter (Xue et al, 2006). Both pYH1 and pDX124 were sequenced to confirm no extraneous mutations were introduced during PCR. The wild type or ste3α mutant strains (WSC43) were biolistically transformed with circular plasmid DNA. Transformants were selected on YPD medium containing drug G418. Independent transformants were randomly selected for RNA extraction and northern analysis to examine the level of CPR2 expression.
To generate the diploid ste3a/ste3α mutant, a ste3α ura5 mutant was first fused with strain JEC30 (MATa lys1) on V8 mating medium and fusion products were selected on medium lacking uracil and lysine. The selected colonies were subjected to FACS analysis to confirm ploidy (2n). The wild-type copy of the STE3a gene was then disrupted with the NAT marker in the a/ste3α mutant background following the overlap PCR procedure mentioned above. Transformants were screened by PCR and Southern analysis to confirm the deletion of the gene, resulting in diploid ste3a/ste3α mutant strain YPH750. YPH750 was then transformed with the CPR2 overexpression plasmid pYH1, and the transformants were analysed by northern to examine the overexpression of CPR2, resulting in the ste3a/ste3α PGPD1∷CPR2 strain YPH752. To overexpress the pheromone receptor STE3α, the ORF of STE3α was amplified by primers JOHE20198/JOHE20199 and cloned into the PGPD1 promoter containing plasmid pXL1. This PGPD1∷STE3α overexpression construct was introduced into the CPR2 overexpression strain YPH21. Transformants were screened by northern analysis probing with STE3α, and the confirmed STE3α overexpression strain YPH748 was further subjected to Southern and northern analyses probing with CPR2 to ensure that the integration of the PGPD1∷STE3α construct did not disrupt the PGPD1∷CPR2 locus in the genome.
Expression and localization of Cpr2–mCherry
The Cpr2–mCherry C-terminal fusion construct was generated by an overlap PCR method. The 5′ upstream promoter region and the CPR2 ORF were amplified with primer pair JOHE17102/JOHE16626. The ORF of mCherry was amplified with primer pair JOHE16627/JOHE16673. A second PCR was conducted using both fragments as DNA template with primer pair JOHE17102/JOHE16673. The overlap PCR products were gel-purified and digested with NotI and PacI. Plasmid pXL1 was also digested with NotI and PacI to release the GPD promoter while maintaining the selectable marker NEO. The two DNA fragments were ligated to result in plasmid pYH33 and the insert was sequenced. The serotype A MATα cpr2 mutant strain was biolistically transformed with circular pYH33 to generate strain YPH342. Cpr2–mCherry expression and localization were observed with a ZEISS Axioskop 2 plus fluorescent microscope or a Leica SP5 confocal microscope. The vacuole membrane marker dye MDY-64 (Molecular Probes, Eugene, OR) was used to stain the vacuoles of the cells.
RNA extraction and northern blot analysis
5 ml YPD overnight cultures of the desired strains were harvested, washed with sterile water, and adjusted to a final density of 1 × 107 cells/ml. For mating conditions, equal amounts of a and α cells were first mixed in a test tube. For pheromone induction, synthetic α pheromone (Davidson et al, 2000b) was added to the cultures to reach a final concentration of 10 μM. Cells were spotted onto V8 mating medium and incubated at 25°C (from 24 h to a week). After incubation, cells were removed from V8 solid medium and lyophilized. RNA was extracted with TRIZOL Reagent (Invitrogen) following the manufacturer's instructions. Total RNA (10 μg) was separated by denaturing agarose gel electrophoresis and blotted to Hybond™-N+ nylon membrane (Amersham). Membranes were probed with [32P]-dCTP-radiolabelled DNA fragments and signals were quantified with a Typhoon 9200 imager and Image Quantifier 5.2 software (Molecular Dynamics).
Scanning electron microscopy
Wild-type and cpr2 mutants were crossed on MS medium at 25°C in the dark for 3 weeks. These mating plates was fixed with 0.1 M Na cacodylate buffer (pH 6.8) containing 3% glutaraldehyde by flooding the entire dish for 24 h at 4°C. The plate was washed with 0.1 M Na cacodylate buffer (pH 6.8), and areas of interest were excised into 1-mm blocks from the cultures, and incubated in the fixation buffer for several weeks at 4°C. Samples were then rinsed in three 30 min changes of cold 0.1 M Na cacodylate buffer, and post-fixed in 2% osmium tetroxide in 0.1 M Na cacodylate buffer for 2.5 h at 4°C before being viewed by scanning electron microscopy.
Immunoblotting
Cells from mating and nonmating conditions were harvested from V8 solid medium (48-h incubation) and 5 ml YPD overnight cultures, respectively, and cell lysates were prepared with lysis buffer as described (Bahn et al, 2005). A measure of 25 μg protein of each sample was loaded into a 4–20% Tris–glycine gel (Invitrogen) and transferred to PVDF membrane (Bio-Rad). Cpr2–mCherry and Cpr2L222P–mCherry were detected with a polyclonal antibody against mCherry (Clontech). Rabbit polyclonal antiserum against yeast FKBP12 was used as a loading control.
Cell fusion assays
To perform the cell fusion assays, wild-type strains YSB119 (MATα NAT) and YSB121 (MATa NEO), and cpr2 mutant strains YPH16 (MATα cpr2∷NAT) and YPH545 (MATa cpr2∷NEO) were grown in YPD liquid medium overnight. Cells were washed and adjusted to 2 × 107 cells/ml, mixed, and grown on V8 mating medium in the dark for 24 h. The colonies were removed with cell scrapers, resuspended in sterile water, and plated onto YPD medium containing nourseothricin and G418 to select for cell–cell fusion products that have both drug-resistance markers. Plates were incubated at 30°C for 5 days until colonies were observed. The experiment was conducted in three replicates and the average number of colonies was calculated. The fusion efficiency was determined by comparing the average number of the visible colonies from the cpr2 mutant cross to that from the wild-type cross.
Split-Ubiquitin protein–protein interaction assays
The physical interactions between Cpr2 and Gα subunits, RGS proteins and Ste3a/α were assayed with the split-ubiquitin system (Stagljar et al, 1998). Vectors and yeast strains were provided with the DUALmembrane Kit 2 (Dualsystem Biotech, Switzerland). The CPR2 full length cDNA was amplified with primers JOHE14491/JOHE15357 and cloned into vectors pCCW and pDL2XN, which contain the C-terminal half of ubiquitin (Cub) fused with the artificial transcription factor LexA-VP16 and the mutated N-terminal half of ubiquitin (NubG), respectively. The CPR2L222P allele was generated through an overlap PCR approach as described above in gene disruption and overexpression. Primer pairs JOHE14491/JOHE15828 and JOHE15827/JOHE15357 were used in the first PCR and a second overlap PCR was performed with primers JOHE14491/JOHE15357 to obtain the CPR2L222P full-length cDNA. The PCR products were purified and cloned into vector pCCW. Full-length cDNA for the genes encoding the Gα subunits Gpa1, Gpa2, Gpa3, the RGS protein Crg1, and the pheromone receptors Ste3a/α were all cloned into vector pDL2XN in which NubG is C-terminally fused to the target proteins. NubG was N-terminally fused to Crg2 in vector pDSL-NX. Primers used for these constructs are listed in Supplementary Table I. All plasmids generated were confirmed by DNA sequencing to ensure no extraneous mutations were introduced by PCR. The yeast host strain NMY32 was transformed with plasmids expressing the Cpr2–Cub fusion and the desired NubG fusion proteins. Transformants were analysed for growth on medium lacking histidine or adenine and β-galactosidase activity was determined as a quantitative measurement of the physical interactions.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Tim James, Danny Lew, Xiaorong Lin, Hiro Matsunami, Jen Reedy and Robin Wharton for critical reading and comments on the manuscript, Kasey Carroll and Anna Floyd for technical assistance, Tom Fowler and James Konopka for S. cerevisiae strains, Pat Casey for discussions and lipid derivatives, and Xiaorong Lin for sharing unpublished information on MAT2. We acknowledge use of the C. neoformans serotype A sequencing project (Duke University/Broad Institute) and Valerie Knowlton for SEM assistance. This work was supported by NIH RO1 grant (AI39115) and R21 grant (AI070230).
References
- Aimi T, Yoshida R, Ishikawa M, Bao D, Kitamoto Y (2005) Identification and linkage mapping of the genes for the putative homeodomain protein (hox1) and the putative pheromone receptor protein homologue (rcb1) in a bipolar basidiomycete, Pholiota nameko. Curr Genet 48: 184–194 [DOI] [PubMed] [Google Scholar]
- Bahn YS, Hicks JK, Giles SS, Cox GM, Heitman J (2004) Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot Cell 3: 1476–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, Kojima K, Cox GM, Heitman J (2005) Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell 16: 2285–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin JM (1993) The probable arrangement of the helices in G protein-coupled receptors. EMBO J 12: 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F, Herskowitz I (1989) Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc Natl Acad Sci USA 86: 5878–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder AC, Hartwell LH (1985) The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res 13: 8463–8475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casselton LA, Olesnicky NS (1998) Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev 62: 55–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerione RA, Codina J, Benovic JL, Lefkowitz RJ, Birnbaumer L, Caron MG (1984) The mammalian beta 2-adrenergic receptor: reconstitution of functional interactions between pure receptor and pure stimulatory nucleotide binding protein of the adenylate cyclase system. Biochemistry 23: 4519–4525 [DOI] [PubMed] [Google Scholar]
- Chang L, Karin M (2001) Mammalian MAP kinase signalling cascades. Nature 410: 37–40 [DOI] [PubMed] [Google Scholar]
- Chang YC, Miller GF, Kwon-Chung KJ (2003) Importance of a developmentally regulated pheromone receptor of Cryptococcus neoformans for virulence. Infect Immun 71: 4953–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Struhl G (1996) Dual roles for patched in sequestering and transducing Hedgehog. Cell 87: 553–563 [DOI] [PubMed] [Google Scholar]
- Chung S, Karos M, Chang YC, Lukszo J, Wickes BL, Kwon-Chung KJ (2002) Molecular analysis of CPRalpha, a MATalpha-specific pheromone receptor gene of Cryptococcus neoformans. Eukaryot Cell 1: 432–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelli O (2005) GPCR deorphanizations: the novel, the known and the unexpected transmitters. Trends Pharmacol Sci 26: 15–19 [DOI] [PubMed] [Google Scholar]
- Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, D′Souza C, Wang P, Heitman J (2002) A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148: 2607–2615 [DOI] [PubMed] [Google Scholar]
- Davidson RC, Cruz MC, Sia RA, Allen B, Alspaugh JA, Heitman J (2000a) Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet Biol 29: 38–48 [DOI] [PubMed] [Google Scholar]
- Davidson RC, Moore TD, Odom AR, Heitman J (2000b) Characterization of the MFalpha pheromone of the human fungal pathogen Cryptococcus neoformans. Mol Microbiol 38: 1017–1026 [DOI] [PubMed] [Google Scholar]
- Davidson RC, Nichols CB, Cox GM, Perfect JR, Heitman J (2003) A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol Microbiol 49: 469–485 [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Slessareva JE (2006) Pheromone signaling pathways in yeast. Sci STKE 2006: cm6. [DOI] [PubMed] [Google Scholar]
- Drews J (1996) Genomic sciences and the medicine of tomorrow. Nat Biotechnol 14: 1516–1518 [DOI] [PubMed] [Google Scholar]
- Ferrell JE Jr (2002) Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol 14: 140–148 [DOI] [PubMed] [Google Scholar]
- Fowler TJ, DeSimone SM, Mitton MF, Kurjan J, Raper CA (1999) Multiple sex pheromones and receptors of a mushroom-producing fungus elicit mating in yeast. Mol Biol Cell 10: 2559–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler TJ, Mitton MF, Vaillancourt LJ, Raper CA (2001) Changes in mate recognition through alterations of pheromones and receptors in the multisexual mushroom fungus Schizophyllum commune. Genetics 158: 1491–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Hsueh YP, Findley KM, Heitman J (2007) Evolution of the mating-type locus: the basidiomycetes. In Sex in Fungi, Heitman J, Kronstad JW, Taylor JW, Casselton LA (eds), pp 19–34. Washington, DC: ASM Press [Google Scholar]
- Fraser JA, Subaran RL, Nichols CB, Heitman J (2003) Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell 2: 1036–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Aston C, Burchett SA, Dyke C, Fields S, Rajarao SJ, Uetz P, Wang Y, Young K, Dohlman HG (2003) The yeast G protein alpha subunit Gpa1 transmits a signal through an RNA binding effector protein Scp160. Mol Cell 12: 517–524 [DOI] [PubMed] [Google Scholar]
- Hagen DC, McCaffrey G, Sprague GF Jr (1986) Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a factor: gene sequence and implications for the structure of the presumed receptor. Proc Natl Acad Sci USA 83: 1418–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Idnurm A, Heitman J (2006) Recombination hotspots flank the Cryptococcus mating-type locus: implications for the evolution of a fungal sex chromosome. PLoS Genet 2: e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Xue C, Heitman J (2007) G protein signaling governing cell fate decisions involves opposing Galpha subunits in Cryptococcus neoformans. Mol Biol Cell 18: 3237–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Wang H, Chou S, Nie X, Chen J, Liu H (2006) Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci USA 103: 12813–12818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Boily MJ, Heitman J (2005) Sex-specific homeodomain proteins Sxi1alpha and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot Cell 4: 526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Heitman J (2002) Genetics of Cryptococcus neoformans. Annu Rev Genet 36: 557–615 [DOI] [PubMed] [Google Scholar]
- Idnurm A, Heitman J (2005) Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol 3: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Murray AW (2007) Positive-feedback loops as a flexible biological module. Curr Biol 17: 668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TY, Srivilai P, Kues U, Vilgalys R (2006) Evolution of the bipolar mating system of the mushroom Coprinellus disseminatus from its tetrapolar ancestors involves loss of mating-type-specific pheromone receptor function. Genetics 172: 1877–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka JB, Margarit SM, Dube P (1996) Mutation of Pro-258 in transmembrane domain 6 constitutively activates the G protein-coupled alpha-factor receptor. Proc Natl Acad Sci USA 93: 6764–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski G, Streaty RA, Klee WA (1982) Modulation of sodium-sensitive GTPase by partial opiate agonists. An explanation for the dual requirement for Na+ and GTP in inhibitory regulation of adenylate cyclase. J Biol Chem 257: 14035–14040 [PubMed] [Google Scholar]
- Kraakman L, Lemaire K, Ma P, Teunissen AW, Donaton MC, Van Dijck P, Winderickx J, de Winde JH, Thevelein JM (1999) A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol 32: 1002–1012 [DOI] [PubMed] [Google Scholar]
- Kwon-Chung JK (1998) Filobasidiella Kwon-Chung. In The Yeasts, Kurtzman CP, Fell JW (eds), pp 656–662. Amsterdam: Elsevier Science B.V [Google Scholar]
- Kwon-Chung KJ, Edman JC, Wickes BL (1992) Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun 60: 602–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladds G, Davis K, Das A, Davey J (2005) A constitutively active GPCR retains its G protein specificity and the ability to form dimers. Mol Microbiol 55: 482–497 [DOI] [PubMed] [Google Scholar]
- Lengeler KB, Davidson RC, D'Souza C, Harashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J (2000) Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev 64: 746–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Fox DS, Fraser JA, Allen A, Forrester K, Dietrich FS, Heitman J (2002) Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot Cell 1: 704–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levoye A, Dam J, Ayoub MA, Guillaume JL, Jockers R (2006) Do orphan G-protein-coupled receptors have ligand-independent functions? New insights from receptor heterodimers. EMBO Rep 7: 1094–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shen G, Zhang ZG, Wang YL, Thompson JK, Wang P (2007) Canonical heterotrimeric G proteins regulating mating and virulence of Cryptococcus neoformans. Mol Biol Cell 18: 4201–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Huang JC, Mitchell TG, Heitman J (2006) Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATalpha allele enhances filamentation. PLoS Genet 2: e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Hull CM, Heitman J (2005) Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434: 1017–1021 [DOI] [PubMed] [Google Scholar]
- Mentesana PE, Dosil M, Konopka JB (2002) Functional assays for mammalian G-protein-coupled receptors in yeast. Methods Enzymol 344: 92–111 [DOI] [PubMed] [Google Scholar]
- Milano CA, Allen LF, Rockman HA, Dolber PC, McMinn TR, Chien KR, Johnson TD, Bond RA, Lefkowitz RJ (1994) Enhanced myocardial function in transgenic mice overexpressing the beta 2-adrenergic receptor. Science 264: 582–586 [DOI] [PubMed] [Google Scholar]
- Miller MG, Johnson AD (2002) White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110: 293–302 [DOI] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J (2003) Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect Immun 71: 4831–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesnicky NS, Brown AJ, Dowell SJ, Casselton LA (1999) A constitutively active G-protein-coupled receptor causes mating self-compatibility in the mushroom Coprinus. EMBO J 18: 2756–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesnicky NS, Brown AJ, Honda Y, Dyos SL, Dowell SJ, Casselton LA (2000) Self-compatible B mutants in Coprinus with altered pheromone-receptor specificities. Genetics 156: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR, Ketabchi N, Cox GM, Ingram CW, Beiser CL (1993) Karyotyping of Cryptococcus neoformans as an epidemiological tool. J Clin Microbiol 31: 3305–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ (2002) Seven-transmembrane receptors. Nat Rev Mol Cell Biol 3: 639–650 [DOI] [PubMed] [Google Scholar]
- Prinster SC, Hague C, Hall RA (2005) Heterodimerization of G protein-coupled receptors: specificity and functional significance. Pharmacol Rev 57: 289–298 [DOI] [PubMed] [Google Scholar]
- Raper J (1966) Genetics of Sexuality in Higher Fungi. New York: The Ronald Press [Google Scholar]
- Rosenkilde MM, Benned-Jensen T, Andersen H, Holst PJ, Kledal TN, Luttichau HR, Larsen JK, Christensen JP, Schwartz TW (2006) Molecular pharmacological phenotyping of EBI2. An orphan seven-transmembrane receptor with constitutive activity. J Biol Chem 281: 13199–13208 [DOI] [PubMed] [Google Scholar]
- Seifert R, Wenzel-Seifert K (2002) Constitutive activity of G-protein-coupled receptors: cause of disease and common property of wild-type receptors. Naunyn Schmiedebergs Arch Pharmacol 366: 381–416 [DOI] [PubMed] [Google Scholar]
- Shen WC, Davidson RC, Cox GM, Heitman J (2002) Pheromones stimulate mating and differentiation via paracrine and autocrine signaling in Cryptococcus neoformans. Eukaryot Cell 1: 366–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Kaminskyj S, Caldwell S, Loewen MC (2007) A role for a complex between activated G protein-coupled receptors in yeast cellular mating. Proc Natl Acad Sci USA 104: 5395–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia RA, Lengeler KB, Heitman J (2000) Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet Biol 29: 153–163 [DOI] [PubMed] [Google Scholar]
- Spellig T, Bolker M, Lottspeich F, Frank RW, Kahmann R (1994) Pheromones trigger filamentous growth in Ustilago maydis. EMBO J 13: 1620–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagljar I, Korostensky C, Johnsson N, te Heesen S (1998) A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci USA 95: 5187–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Overton MC, Blumer KJ (1998) Mechanisms governing the activation and trafficking of yeast G protein-coupled receptors. Mol Biol Cell 9: 885–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden SC, Pryciak PM (2008) Distinct roles for two Galpha–Gbeta interfaces in cell polarity control by a yeast heterotrimeric G protein. Mol Biol Cell 19: 181–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sande J, Parma J, Tonacchera M, Swillens S, Dumont J, Vassart G (1995) Somatic and germline mutations of the TSH receptor gene in thyroid diseases. J Clin Endocrinol Metab 80: 2577–2585 [DOI] [PubMed] [Google Scholar]
- von Heijne G (1991) Proline kinks in transmembrane alpha-helices. J Mol Biol 218: 499–503 [DOI] [PubMed] [Google Scholar]
- Wang P, Perfect JR, Heitman J (2000) The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol Cell Biol 20: 352–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wess J, Nanavati S, Vogel Z, Maggio R (1993) Functional role of proline and tryptophan residues highly conserved among G protein-coupled receptors studied by mutational analysis of the m3 muscarinic receptor. EMBO J 12: 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes BL, Mayorga ME, Edman U, Edman JC (1996) Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the alpha-mating type. Proc Natl Acad Sci USA 93: 7327–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Nusse R (1998) Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol 14: 59–88 [DOI] [PubMed] [Google Scholar]
- Xue C, Bahn YS, Cox GM, Heitman J (2006) G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol Biol Cell 17: 667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan RE, Galgoczy DJ, Johnson AD (2006) Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci USA 103: 12807–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information