Abstract
Atg8 is conjugated to phosphatidylethanolamine (PE) by ubiquitin-like conjugation reactions. Atg8 has at least two functions in autophagy: membrane biogenesis and target recognition. Regulation of PE conjugation and deconjugation of Atg8 is crucial for these functions in which Atg4 has a critical function by both processing Atg8 precursors and deconjugating Atg8–PE. Here, we report the crystal structures of catalytically inert human Atg4B (HsAtg4B) in complex with processed and unprocessed forms of LC3, a mammalian orthologue of yeast Atg8. On LC3 binding, the regulatory loop and the N-terminal tail of HsAtg4B undergo large conformational changes. The regulatory loop masking the entrance of the active site of free HsAtg4B is lifted by LC3 Phe119, so that a groove is formed along which the LC3 tail enters the active site. At the same time, the N-terminal tail masking the exit of the active site of HsAtg4B in the free form is detached from the enzyme core and a large flat surface is exposed, which might enable the enzyme to access the membrane-bound LC3–PE.
Keywords: Atg4, autophagy, crystal structure, deconjugation, LC3
Introduction
Autophagy is the process through which the bulk degradation of cytoplasmic components by the lysosomal/vacuolar system occurs (Klionsky and Ohsumi, 1999), and it has a critical function in numerous biological processes (Mizushima, 2007). In autophagy, a double-membrane structure called an autophagosome sequesters a portion of the cytoplasm and fuses with the lysosome/vacuole to deliver its contents into the organelle lumen (Baba et al, 1994). Genetic approaches in Saccharomyces cerevisiae have identified many autophagy-related (ATG) genes involved in this process (Klionsky et al, 2003), and subsequent biochemical analyses have demonstrated that a novel ubiquitin-like conjugation system called the Atg8 system is essential for autophagosome formation (Ichimura et al, 2000). In the Atg8 system, nascent Atg8 is cleaved at its C-terminal arginine residue by Atg4, a cysteine protease (Kirisako et al, 2000), and the exposed C-terminal glycine is conjugated to phosphatidylethanolamine (PE) by Atg7, an E1-like enzyme (Tanida et al, 1999; Ichimura et al, 2000), and Atg3, an E2-like enzyme (Ichimura et al, 2000). The Atg8–PE conjugate itself possesses membrane tethering and hemifusion ability (Nakatogawa et al, 2007), and has an essential function in autophagosome formation, especially at the membrane expansion step (Nakatogawa et al, 2007; Xie et al, 2008). For the normal progression of autophagy, not only the formation but also the deconjugation of Atg8–PE is required, which is again mediated by Atg4 (Kirisako et al, 2000). Thus, Atg4 has a critical function in autophagosome formation through the reversible modification of Atg8. Although the conjugation mechanism adopted by the Atg8 system is similar to those of ubiquitin-like conjugation systems, the Atg8 system is unique in that Atg8 modifies a lipid, not a protein. As Atg8–PE is anchored to the membranes, Atg4 has to access the membranes prior to Atg8–PE deconjugation. Therefore, Atg4 seems to possess some structural features suitable for membrane targeting, which are not present in deconjugating enzymes for ubiquitin and other ubiquitin-like modifiers. Atg8 is also unique in that it does not have the Gly-Gly sequence conserved among ubiquitin-like modifiers, but conserves the aromatic residue–Gly sequence at the N-terminal side of the scissile bond.
In mammals, an Atg8-like conjugation system, called the LC3 system, has been shown to exist (Kabeya et al, 2000). Similar to the Atg8 system in yeast, the C-terminal peptide of LC3 is cleaved by mammalian Atg4 homologues (Hemelaar et al, 2003; Marino et al, 2003; Kabeya et al, 2004; Tanida et al, 2004). The processed form, called LC3-I, has a glycine residue at its C terminus and resides in the cytosol (Kabeya et al, 2000). By the actions of mammalian Atg7 and Atg3 homologues (Tanida et al, 2001, 2002), LC3-I is further modified to another form, called LC3-II (Kabeya et al, 2000), which is most likely the PE-conjugated form as in the Atg8 system (Kabeya et al, 2004; Tanida et al, 2006). To date, four human homologues of yeast Atg4 have been reported: HsAtg4A/autophagin-2, HsAtg4B/autophagin-1, autophagin-3 and autophagin-4 (Hemelaar et al, 2003; Marino et al, 2003). Among them, HsAtg4B is the sole enzyme reported to efficiently cleave LC3 precursors and LC3–PE (Kabeya et al, 2004). Thus, HsAtg4B is considered to have an important function in mammalian autophagy by processing and deconjugating LC3.
We previously reported the crystal structure of HsAtg4B in its free form (Sugawara et al, 2005). The active site of HsAtg4B is masked by the regulatory loop, suggesting its conformational change on complex formation with LC3 (Sugawara et al, 2005). However, the molecular mechanism of LC3 recognition by HsAtg4B, especially recognition of the membrane-anchored LC3–PE, remains elusive. Here, we report the crystal structures of a catalytically inert HsAtg4B mutant in complex with the processed and unprocessed forms of LC3. The structures showed that a large conformational change is induced at the regulatory loop upon complex formation with LC3. Further, they showed that the N-terminal tail of HsAtg4B also undergoes a large conformational change, which seems to be crucial for targeting the enzyme to the membrane-bound LC3–PE.
Results and discussion
Overall structure of the HsAtg4B–LC3 complex
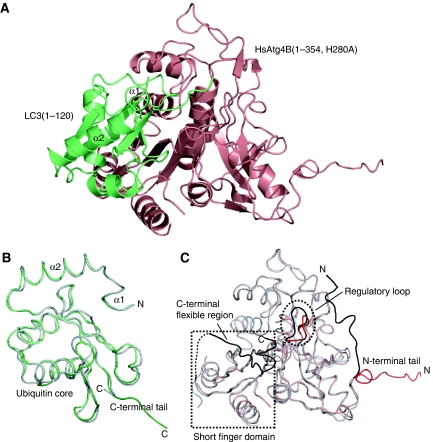
We first tried to crystallize the full-length HsAtg4B complexed with LC3, but could not obtain any crystals. To date, the structure of free HsAtg4B has been reported in two crystal forms (Sugawara et al, 2005; Kumanomidou et al, 2006), both of which showed that the C-terminal 39 residues (residues 355–393) are highly flexible, and only 19 residues (residues 355–373) and 16 residues (residues 362–377) were observed and modelled in each form, though with high B-factors. Therefore, we used a C-terminal deletion mutant of HsAtg4B(1–354) for crystallization with LC3 (Satoo et al, 2007). We obtained crystals of HsAtg4B(1–354) with a substitution of His280 with alanine (a catalytically inert form) in complex with LC3(1–120), a processed form, and determined the structure by the molecular replacement method (Figure 1A; Table I). Apart from certain regions, the folds of the HsAtg4B and LC3 in the complex were almost identical to those of each free form (Figure 1B and C). The r.m.s.d. of Cα atoms between free and LC3-bound HsAtg4B was 1.1 Å for 295 residues, which exclude the N-terminal tail, whereas that between free and HsAtg4B-bound LC3 was 0.9 Å for 112 residues. LC3 is comprised of three regions: the N-terminal domain consisting of two α-helices, the ubiquitin-like core and the C-terminal tail (Sugawara et al, 2004). On complex formation with HsAtg4B, the only large conformational change was observed in the C-terminal tail, which has an extended β-conformation in the complex (Figure 1B). The interaction sites for HsAtg4B are localized in the C-terminal tail and its surrounding regions, and the N-terminal domain has no interaction with HsAtg4B. HsAtg4B consists of the classical papain-like domain and the short finger domain, a domain unique to Atg4 family proteases (Sugawara et al, 2005). The ubiquitin core of LC3 is bound to the interface between the two domains of HsAtg4B, and LC3 extends its C-terminal tail to the active site of HsAtg4B located at the centre of the papain-like domain. When the structure of free HsAtg4B is superimposed on that of the LC3-bound form, the C-terminal region (residues 362–377) partially overlaps with the bound LC3 (Figure 1A and C); therefore, the C-terminal region should be displaced by LC3 upon complex formation. The buried surface between the ubiquitin core of LC3 and HsAtg4B is 1679 Å2, whereas that between the tail of LC3 and HsAtg4B is 944 Å2. It should be noted that upon complex formation with LC3, HsAtg4B undergoes a large conformational change at two regions; the N-terminal tail and the loop masking the catalytic site in free HsAtg4B (Figure 1C). We previously named this loop the ‘inhibitory loop'; however, considering that the role of the loop is not just inhibitory, we have renamed this loop the ‘regulatory loop' in this paper.
Figure 1.
Overall structure of the HsAtg4B–LC3 complex. (A) Ribbon diagram of the HsAtg4B–LC3(1–120) complex. HsAtg4B is coloured salmon red, and LC3 is coloured green. (B) Structural comparison between free and HsAtg4B-bound LC3. Crystal structure of free LC3(1–120) (PDB code 1UGM) is superimposed on the HsAtg4B-bound LC3. HsAtg4B-bound LC3 is coloured green, and free LC3 is coloured grey. (C) Structural comparison between free and LC3-bound HsAtg4B. Crystal structure of free HsAtg4B (PDB code 2CY7) is superimposed on the LC3-bound HsAtg4B. LC3-bound HsAtg4B is coloured salmon red, and free HsAtg4B is coloured grey. The regions with large conformational differences between free and LC3-bound forms are coloured black (free) and red (LC3 bound). All the figures representing molecular structures were generated with PyMOL (DeLano, 2002).
Table 1.
Data collection and refinement statistics
| HsAtg4B construct | 1–354 H280A | 1–354 H280A | 1–354 C74S |
| LC3 construct | 1–120 | 1–124 | 1–124 |
| Beamline | KEK NW12A | SPring8 BL41XU | SPring8 BL41XU |
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 |
| Temperature (K) | 95 | 90 | 90 |
| Resolution range (Å) | 50–1.90 | 50–1.90 | 50–2.05 |
| Outer shell (Å) | 1.97–1.90 | 1.97–1.90 | 2.12–2.05 |
| Observed reflections | 183 688 | 222 721 | 119 677 |
| Unique reflections | 34 587 | 34 039 | 27 207 |
| Completeness (%) | 95.6 (92.5) | 96.4 (69.5) | 90.8 (73.8) |
| Rmerge(I) | 0.056 (0.330) | 0.079 (0.343) | 0.079 (0.268) |
| Resolution range (Å) | 50–1.90 | 50–1.90 | 50–2.05 |
| No. of protein atoms | 3365 | 3401 | 3425 |
| No. of water molecules | 314 | 184 | 153 |
| R/Rfree | 0.192/0.227 | 0.200/0.229 | 0.208/0.252 |
| r.m.s.d. from ideality | |||
| Bond length (Å) | 0.005 | 0.006 | 0.006 |
| Angle (deg) | 1.30 | 1.30 | 1.26 |
The interaction between HsAtg4B and LC3
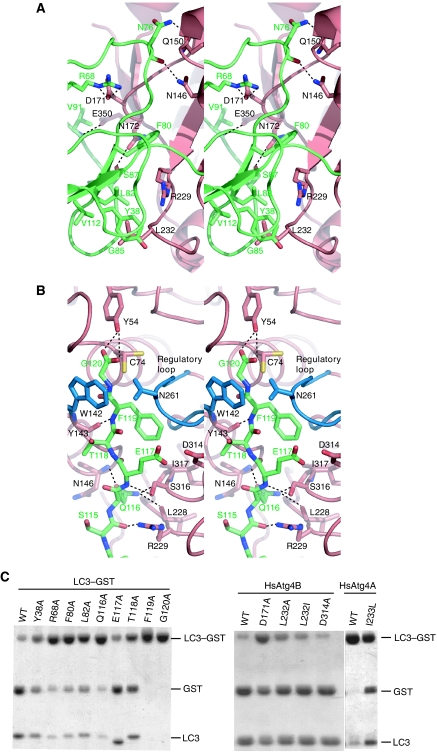
The ubiquitin core of LC3 forms both hydrophilic and hydrophobic interactions with HsAtg4B (Figure 2A). Asn76 of LC3 forms hydrogen bonds with the side chains of Asn146 and Gln150 of HsAtg4B. Ser87 and Val91 of LC3 also form hydrogen bonds with the side chains of Asn172 and Glu350 of HsAtg4B, respectively. Arg68 of LC3 forms a salt bridge with Asp171 of HsAtg4B. In addition to these hydrophilic interactions, hydrophobic interactions are observed at two sites; Phe80 of LC3 is bound to the relatively hydrophobic groove on HsAtg4B, and the side chains of Tyr38, Leu82 and Val112 and the main chain of Gly85 of LC3 form a hydrophobic pocket into which the side chain of Leu232 of HsAtg4B projects. We performed in vitro cleavage assay using LC3–GST as a substrate (Sugawara et al, 2005). As shown in Figure 2C, Tyr38, Arg68, Phe80 and Leu82 of LC3 and Asp171 and Leu232 of HsAtg4B are partially responsible for the processing of LC3–GST. This is consistent with a recent mutational analysis showing that Phe80 and Leu82 of LC3 are required for processing by HsAtg4B (Fass et al, 2007).
Figure 2.
Interaction between HsAtg4B and LC3. (A) Interaction between HsAtg4B and the ubiquitin core of LC3. HsAtg4B is coloured salmon red, and LC3 is coloured green. The side chains of the residues involved in the HsAtg4B–LC3 interaction are shown as stick models. Possible hydrophilic interactions are shown as broken lines. (B) Interaction between HsAtg4B and the C-terminal tail of LC3. Trp142 and the regulatory loop of HsAtg4B are coloured blue, whereas the remainder is coloured as in (A). As the side chain of Cys74 has two conformations, both conformations are shown. Possible hydrophilic interactions are shown as broken lines. (C) In vitro proteolysis assay. Left, LC3 mutants fused to GST were incubated with HsAtg4B. Right, LC3–GST was incubated with HsAtg4B mutants. Procedures are described in detail in Materials and methods.
The C-terminal tail of LC3 also forms hydrophilic interactions with HsAtg4B (Figure 2B). Ser115 and Phe119 of LC3 form hydrogen bonds with the side chain of Arg229 and the carbonyl oxygen of Tyr143 of HsAtg4B, respectively, and the side chain of Gln116 of LC3 forms hydrogen bonds with the main chains of Asn146, Leu228 and Ile317 of HsAtg4B. In vitro cleavage assay showed that Gln116 of LC3 and Arg229 of HsAtg4B are important for the processing of LC3–GST by HsAtg4B (Figure 2C) (Sugawara et al, 2005). Furthermore, Phe119 and Gly120 of LC3 form close contacts with the regulatory loop and the side chain of Trp142 of HsAtg4B, respectively. These two residues are essential for the processing by HsAtg4B (Figure 2C) (Sugawara et al, 2005).
In contrast to the high processing activity of HsAtg4B against LC3, HsAtg4A cannot process LC3 efficiently, although it efficiently processes GATE-16 (Kabeya et al, 2004). All the residues of HsAtg4B that form direct interactions with LC3 are also conserved in HsAtg4A except for Leu232, the position of which is occupied by an isoleucine (Ile233). The substitution of Ile233 with leucine enhanced the processing activity of HsAtg4A against LC3–GST (Figure 2C, right), suggesting that this residue is one of the determinants of human Atg4 homologue specificities. However, the existence of more pivotal site(s) responsible for this specificity is speculated as the substitution of Leu232 of HsAtg4B with an isoleucine only slightly reduced the processing activity of HsAtg4B against LC3–GST (Figure 2C, right).
Structures of the HsAtg4B–LC3 precursor complex
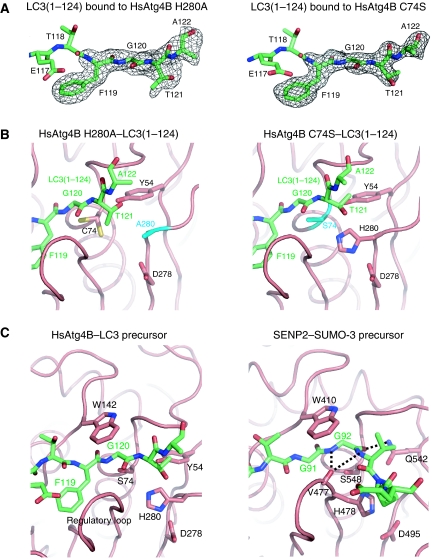
We next determined the structures of LC3(1–124), which has four residues (Thr-Ala-Leu-Ala) at the C terminus of the processing site, bound to HsAtg4B(1–354) with a mutation at either His280 (H280A) or Cys74 (C74S) (Table I). The electron densities of residues 123 and 124 of LC3(1–124) were disordered, so they were excluded from the model (Figure 3A). Figure 3B shows the catalytic site structures of LC3(1–124)-bound HsAtg4B with either a H280A (left) or C74S (right) mutation. Although overall structure is almost identical, some conformational differences are observed at their catalytic sites. When the structure of LC3(1–124)-bound HsAtg4B with a H280A mutation is superimposed on that of free HsAtg4B, the C-terminal region of LC3 (residues 121–122) and the side chain of Cys74 are in steric hindrance with the side chain of His280, suggesting that their conformation in the H280A mutant is affected by the H280A mutation. Therefore, we consider the structure with the C74S mutation to be the more reliable model for the HsAtg4B–LC3 precursor complex. In contrast to the substantial interaction of Phe119 and Gly120 of LC3 with HsAtg4B, Thr121 and Ala122 of LC3 have little interaction with the enzyme. Definite binding pockets for PE are not present around the C-terminal region of LC3. This is consistent with the observation that HsAtg4B has high specificity against residues N-terminal to the scissile bond but not against the C-terminal moiety.
Figure 3.
Structure of HsAtg4B complexed with a LC3 precursor. (A) Annealed Fo−Fc electron density map for the C-terminal region (residues 119–122) of LC3(1–124). Left, LC3(1–124) bound to the HsAtg4B H280A mutant; right, LC3(1–124) bound to the HsAtg4B C74S mutant. Structure of the C-terminal region of LC3 is also shown as a stick model, in which carbon, nitrogen and oxygen atoms are coloured green, blue and red, respectively. (B) Structure of the C-terminal tail of LC3(1–124) and the catalytic site of HsAtg4B. Left, LC3(1–124) bound to HsAtg4B H280A mutant, right, LC3(1–124) bound to the HsAtg4B C74S mutant. HsAtg4B is shown as a ribbon model, whereas LC3 is shown as a stick model. The side chains of the residues comprising the catalytic triad and Tyr54 of HsAtg4B are also shown as a stick model. Carbon, nitrogen, oxygen and sulphur atoms are coloured green, blue, red and yellow, respectively. Mutated catalytic residues are coloured cyan. (C) Structural comparison between the HsAtg4B–LC3 precursor and SENP2–SUMO-3 precursor complexes. Left, structure of the HsAtg4B–LC3 precursor complex; right, structure of the SENP2–SUMO-3 precursor complex. HsAtg4B and SENP2 are shown as a ribbon model, whereas LC3 and SUMO-3 precursors are shown as a stick model. The side chains of the important residues at the catalytic site are also shown as a stick model. Atom colouring is the same as in (A, B).
Recently, the structures of SENP proteases in complex with SUMO precursors have been reported (Reverter and Lima, 2006; Shen et al, 2006). Although SENP protease has little sequence homology with HsAtg4B, the structures around the catalytic sites of both are similar. Figure 3C shows a comparison of the catalytic site structures of the SENP2–SUMO-3 precursor complex in a productive form (right) (Reverter and Lima, 2006) and the HsAtg4B–LC3(1–124) complex (left). In the SENP2–SUMO-3 precursor complex, Trp410 of SENP2 functions as a clamp to position the Gly-Gly motif (Gly91 and Gly92) and the scissile bond of SUMO-3, so that the scissile bond has the cis configuration of the amide nitrogens, which is further stabilized by hydrogen bonds between the amide nitrogens and the carbonyl oxygen of SENP2 Val477. A hydrogen bond between the carbonyl oxygen of SUMO-3 Gly92 and the side chain of SENP2 Gln542 also contributes to the stabilization of the cis configuration. In the case of the HsAtg4B–LC3(1–124) complex, Trp142 of HsAtg4B functions as a clamp and positions Phe119 and Gly120 of LC3 in a similar manner to that observed in the SENP2–SUMO-3 precursor complex. This is in good agreement with the results of mutational studies showing that Trp142 is crucial for the processing activity of HsAtg4B (Sugawara et al, 2005). However, neither the cis configuration nor the hydrogen bonds of the scissile bond have been observed in the HsAtg4B–LC3(1–124) complex. This might be a unique feature of the HsAtg4B–LC3 precursor complex as Gln542 of SENP2, which is conserved as Gln/Asn among most deubiquitinating enzymes (DUBs) and ubiquitin-like specific proteases (ULPs), is not conserved and a Tyr residue (Tyr54) occupies the position in HsAtg4B.
Conformational changes of HsAtg4B upon complex formation with LC3
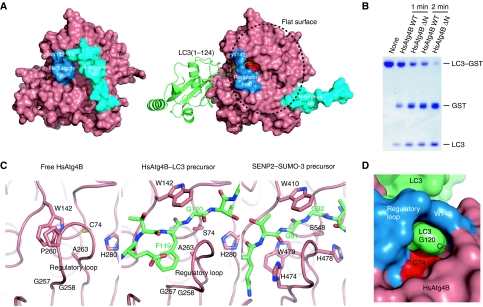
As shown in Figure 1C, two regions of HsAtg4B undergo large conformational changes upon complex formation with LC3. These conformational changes are observed in both LC3(1–120)- and LC3(1–124)-bound HsAtg4B. Figure 4A shows surface models of free (left) and LC3-bound (right) HsAtg4B. LC3(1–124) bound to HsAtg4B is shown as a ribbon model. In free HsAtg4B, both the N-terminal tail and the regulatory loop cover the catalytic site, and Cys74 is buried (Figure 4A, left). The regulatory loop and Trp142 cover the entrance of the catalytic site, whereas the N-terminal tail, especially Tyr8, covers the exit of the catalytic site. Upon LC3 binding, the regulatory loop and the side chain of Trp142 undergo a large conformational change to form a groove, along which the LC3 tail accesses the catalytic cysteine, Cys74 (Figure 4A, right). Furthermore, the N-terminal tail also undergoes a large conformational change that exposes a large flat surface around the exit of the active site of HsAtg4B (Figure 4A, right; circled). HsAtg4B lacking the N-terminal tail showed higher processing activity against LC3–GST than did wild-type HsAtg4B (Figure 4B), indicating that the N-terminal tail actually suppresses the activity of HsAtg4B by blocking the exit of the active site of HsAtg4B.
Figure 4.
Conformational change in HsAtg4B upon complex formation with LC3. (A) Surface model of free (left) and LC3(1–124)-bound (right) HsAtg4B. W142 and the regulatory loop are coloured blue, and the N-terminal tail is coloured cyan. The catalytic Cys74 is coloured red. LC3(1–124) is shown as a ribbon model. (B) In vitro proteolysis assay using N-terminal tail-deleted HsAtg4B. LC3–GST was incubated with either wild-type or N-terminally truncated HsAtg4B and was subjected to SDS–PAGE analysis. (C) Structural comparison between free and LC3(1–124)-bound HsAtg4B and SENP2–SUMO-3 precursor complexes. Left, free HsAtg4B; middle, the HsAtg4B–LC3 precursor complex; right, the SENP2–SUMO-3 precursor complex. The three structures are shown in the same orientation. Model colouring is the same as in Figure 3. (D) Surface model of LC3(1–120) bound to HsAtg4B. Atom colouring is the same as in (A).
Structural basis for the recognition of the LC3 tail by HsAtg4B
The regulatory loop of HsAtg4B is topologically conserved in DUBs and ULPs; however, in these proteases, its conformation is fixed in an open form and it does not cover the active site of the enzymes. Figure 4C shows a comparison of the active site structures of free (left), LC3(1–124)-bound HsAtg4B (middle) and the SENP2–SUMO-3 precursor productive complex (right). In the case of SENP2, the conformation of the loop equivalent to the regulatory loop of HsAtg4B is fixed in an open conformation by His474 and Trp479, which push against the root of the loop. In contrast, only small residues, Gly257, Gly258 and Ala263, are located at the root of the regulatory loop in HsAtg4B. Their side chains are too small to fix the conformation of the regulatory loop in the open state. Instead, the side chain of Trp142 interacts with Pro260 in the regulatory loop and fixes its conformation in the closed form (Figure 4C, left). Thus, the regulatory loop, in cooperation with Trp142, masks the entrance of the catalytic site in the free form. When LC3 is bound to HsAtg4B, the location of the side chain of Trp142 is replaced by Phe119 of LC3 that pushes the root of the regulatory loop to the open conformation in a similar role to that of His474 and Trp479 in SENP2. This results in the formation of a groove at the entrance of the active site. The available space in the groove is too narrow to accommodate any residue other than the glycine at the N-terminal side of the scissile bond (Figure 4D). Thus, Phe119 and Gly120 of LC3 are essential for hydrolysis by HsAtg4B, and such specificity is endowed by the regulatory loop. The aromatic residue–Gly sequence at the N-terminal side of the scissile bond is well conserved among Atg8 family proteins. This is in contrast to ubiquitin and ubiquitin-like modifiers that have a conserved Gly-Gly sequence at the N-terminal side of the scissile bond, which is essential for hydrolysis by DUBs and ULPs. The aromatic residue–Gly sequence is one of the determinants by which Atg4 family proteases distinguish Atg8 family proteins from other ubiquitin-like modifiers. However, this sequence is not enough for hydrolysis by HsAtg4B as a peptide derived from the C-terminal tail region (residues 116–124) of LC3 was not hydrolysed by HsAtg4B (Sugawara et al, 2005), indicating that the tail region by itself cannot open the auto-inhibited structure formed by the regulatory loop and Trp142. Although the binding site for the tail region of LC3 is auto-inhibited, that for the ubiquitin core region of LC3 is accessible. Therefore, we propose the following model; LC3 first binds to HsAtg4B with its ubiquitin core by the interaction shown in Figure 2A. Using this interaction as a scaffold, the tail region of LC3 opens the regulatory loop and binds to the active site of the enzyme for processing. Recently, the structure of UfSP1, a processing and deconjugating enzyme for the ubiquitin-like modifier Ufm1, was reported (Ha et al, 2008). UfSP1 shows higher structural similarity to HsAtg4B than to other deconjugating enzymes, and its loop equivalent to the regulatory loop of HsAtg4B has two conformations in the crystal. The possibility exists that UfSP1 also utilizes the loop for regulation of its protease activity in a manner similar to HsAtg4B.
Interaction of the N-terminal tail with the WXXL-binding site of LC3
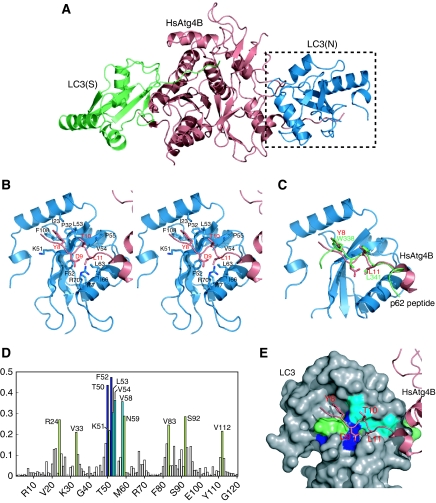
The N-terminal tail of HsAtg4B, which masks the exit of the catalytic site of free HsAtg4B, is detached from the enzyme core in the complex. The N-terminal tail in the open conformation interacts with a crystallographically adjacent, non-substrate LC3 molecule in all three HsAtg4B–LC3 complex structures (Figure 5A, LC3(N)). The N-terminal tail adopts an extended β-conformation and forms an intermolecular β-sheet with β2 of non-substrate LC3. The side chains of Tyr8 and Leu11 of the N-terminal tail interact with the hydrophobic pockets comprised of Ile23, Pro32, Lys51, Phe52, Leu53, Val54, Leu63, Ile67 and Phe108 of LC3 (Figure 5B). In addition, an ionic interaction is formed between Asp9 of the N-terminal tail and Arg70 of LC3 (Figure 5B). Interestingly, this interaction mode is very similar to that observed between Atg8/LC3 and the WXXL motif (Noda et al, 2008). The WXXL motif is a recently identified motif that is specifically recognized by Atg8/LC3, and is conserved in receptor proteins involved in selective autophagy such as p62, the receptor protein for ubiquitinated aberrant proteins (Ichimura et al, 2008; Noda et al, 2008), and Atg19, the receptor protein for aminopeptidase I (Noda et al, 2008). Figure 5C shows a superimposition of LC3 complexed with the WXXL motif-containing region of p62 and the N-terminal region of HsAtg4B. Tyr8 of HsAtg4B and Trp338 of p62 are bound to one hydrophobic pocket of LC3, whereas Leu11 of HsAtg4B and Leu341 of p62 are bound to another hydrophobic pocket in a quite similar manner.
Figure 5.
Interaction of the N-terminal tail with the WXXL-binding site of LC3. (A) Overall structure of HsAtg4B bound to both substrate and non-substrate LC3. Substrate LC3 is designated as LC3(S) and coloured green, whereas non-substrate LC3 is designated as LC3(N) and coloured blue. (B) Stereo-view of the N-terminal tail of HsAtg4B bound to the WXXL-binding site of LC3. The side chains of the residues involved in the interaction between HsAtg4B and LC3 are labelled and shown with a stick model. Model colouring is the same as in (A). (C) Structural comparison of the N-terminal tail–LC3 interaction with the p62–LC3 interaction. The structure of LC3 complexed with a p62 peptide is superimposed on that complexed with the N-terminal tail peptide. As the structure of LC3 is almost identical to each other, only that bound to the N-terminal tail peptide of HsAtg4B is shown. (D) Chemical shift perturbations of the LC3 backbone amide groups upon complex formation with the N-terminal-tail peptide. The combined 1H and 15N chemical shift differences, calculated using the equation Δp.p.m.=[(ΔδHN)2+(ΔδN/5)2]1/2 were plotted against residue numbers. (E) Mapping of chemical shift perturbation results on the crystal structure of LC3 bound to HsAtg4B. The residues with Δp.p.m. >0.4 are shown in blue, 0.4>Δp.p.m.>0.3 in cyan and 0.3>Δp.p.m. >0.2 in green.
To clarify whether the interaction between LC3 and the N-terminal tail of HsAtg4B observed in the crystal is due to a crystal packing artefact or not, NMR analysis was performed using 15N-labelled LC3 and a peptide derived from the N-terminal region (residues 1–18) of HsAtg4B. The peptide was titrated into a solution of 15N-labelled LC3 (Figure 5D). The residues that showed large chemical shift perturbations on the addition of the peptide coincided with those involved in the interaction with the N-terminal tail of HsAtg4B in the crystal (Figure 5E). These results clearly support the notion that the interaction observed in the crystal is also maintained in solution. Thus, the open conformation of the N-terminal tail in the HsAtg4B–LC3 complex seems to be induced by its interaction with the WXXL-binding site of non-substrate LC3.
Conclusions
In this study, we reported the LC3-bound structure of HsAtg4B and identified important residues of both LC3 and HsAtg4B for processing. Further, we showed that the conformation change of the regulatory loop is responsible for the specificity of HsAtg4B against the unique Phe-Gly sequence of LC3 that is distinct from the Gly-Gly sequence conserved among ubiquitin-like modifiers. Besides the regulatory loop, the N-terminal tail showed a conformational change from the open to the closed conformation upon complex formation with LC3. As only the open conformation seems to be favourable for membrane targeting of HsAtg4B that would be required for LC3–PE deconjugation, the conformation of the N-terminal tail could serve for the regulation of the deconjugation activity. The open conformation of the N-terminal tail was stabilized by the interaction with non-substrate LC3 in vitro. Such interaction might also occur in vivo and the open conformation might be induced under LC3-rich conditions. Alternatively, some unidentified factor(s) might be responsible for such stabilization in vivo. Recently, the activity of HsAtg4A and 4B was shown to be regulated by reactive oxygen species in vivo (Scherz-Shouval et al, 2007). To understand the regulation mechanism in LC3–PE deconjugation process, further studies both in vivo and in vitro would be required.
Materials and methods
Expression and purification
Expression and purification of the HsAtg4B(1–354) H280A mutants, LC3(1–120) (LC3-I) and LC3(1–124), for crystallization were described earlier (Satoo et al, 2007). Expression and purification of the HsAtg4B(1–354) C74S mutant for crystallization were performed using the same procedure as that for the H280A mutant. Proteins used for proteolysis assays were prepared as follows. Single-mutant HsAtg4Bs (D171A, L232A, L232I and D314A) and LC3-Is (Y38A, R68A, F80A, L82A, Q116A, E117A, T118A, F119A and G120A), as well as the HsAtg4A I233A mutant, were prepared by PCR-mediated site-directed mutagenesis and sequenced to confirm their identities. Wild-type and mutant LC3-Is with a GST tag fused to the C terminus (LC3–GSTs) were inserted into the pET11a vector (Novagen). They were then expressed in Escherichia coli BL21 DE3 and purified on a glutathione-Sepharose 4B column. Further purification was performed using a HiTrap CM cation-exchange column (GE Healthcare) and a Superdex 75 gel-filtration column (GE Healthcare). Wild-type and single-mutant HsAtg4As and HsAtg4Bs, as well as N-terminally truncated HsAtg4B (residues 25–393), were inserted into the pGEX6p vector. They were then expressed in E. coli BL21 DE3 and purified according to the procedure described for the HsAtg4B(1–354) H280A mutant (Satoo et al, 2007). They were concentrated to 0.01, 0.1 and 1.0 mg/ml for HsAtg4Bs, HsAtg4As and LC3–GSTs, respectively, in 20 mM Tris–HCl (pH 8.0) and 150 mM NaCl. Proteins used for NMR spectroscopy were prepared as follows. The N-terminal tail peptide of HsAtg4B (residues 1–18) was inserted into the pGEX6p-LC3-I vector, and expressed as a fusion to GST–LC3-I in E. coli BL21 DE3. After purification on a glutathione-Sepharose column, GST–LC3-I was cleaved from the peptide with HsAtg4B. Further purification of the peptide was performed using a Superdex 75 gel-filtration column. The purified peptide was concentrated to 1.6 mM in 25 mM sodium phosphate (pH 7.0), 100 mM NaCl and 0.02 mM NaN3. LC3-I uniformly labelled with 15N was prepared by growing E. coli in M9 medium containing 1 g/l 15NH4Cl (Shoko) followed by purification according to the same procedure described for non-labelled LC3-I (Satoo et al, 2007). The purified 15N-labelled protein was concentrated to 0.5 mM in 92% H2O/8% D2O containing 25 mM sodium phosphate (pH 7.0), 100 mM NaCl and 0.02 mM NaN3.
Structure determination and refinement
Crystallization and diffraction data collection for the HsAtg4B(H280A)–LC3(1–120) and HsAtg4B(H280A)–LC3(1–124) complexes have been described earlier (Satoo et al, 2007). Crystallization and diffraction data collection for the HsAtg4B(C74S)–LC3(1–124) complex were performed by the same procedure as that for the HsAtg4B(H280A)–LC3(1–124) complex. The structure of the HsAtg4B(H280A)–LC3(1–120) complex was determined by the molecular replacement method using the MOLREP program (Vagin and Teplyakov, 1997). The site of HsAtg4B was first determined using the free HsAtg4B structure (PDB code 2CY7) as a search model, and that of LC3(1–120) was determined using the LC3-I structure (PDB code 1UGM) as a search model. Crystallographic refinement was performed using the CNS program (Brunger et al, 1998). For each cycle, the model was rebuilt manually using the COOT molecular modelling program (Emsley and Cowtan, 2004) in several steps alternated with cycles of automated refinement using data to 1.9Å resolution. Using the HsAtg4B(H280A)–LC3(1–120) complex structure as an initial model, the structures of the HsAtg4B(H280)–LC3(1–124) and HsAtg4B(C74S)–LC3(1–124) complexes were determined by several cycles of crystallographic refinement using CNS and model building using COOT. The refinement statistics are summarized in Table I.
In vitro proteolysis assay
All proteolysis assays were performed by mixing the enzyme and substrate at a volume ratio 1:1 at room temperature. For LC3 mutant analyses, wild-type and mutant LC3–GSTs (1 mg/ml) were mixed with 0.01 mg/ml HsAtg4B and incubated for 10 min. For HsAtg4B mutant analyses, wild-type and mutant HsAtg4Bs (0.01 mg/ml) were mixed with wild-type LC3–GST (1 mg/ml) and incubated for 10 min. For the analysis of the N-terminal truncated mutant of HsAtg4B, wild-type and N-terminally truncated HsAtg4B (0.01 mg/ml) were mixed with LC3–GST (1 mg/ml) and incubated for 2 min. For HsAtg4A mutant analysis, wild-type and mutant HsAtg4A (0.1 mg/ml) were mixed with LC3–GST (1 mg/ml) and incubated for 10 min. All the reactions were stopped by boiling with the SDS sample buffer and analysed by SDS–PAGE and Coomassie Brilliant Blue staining.
NMR spectroscopy
Two- and three-dimensional NMR experiments were carried out on Varian UNITY inova spectrometers operating at 800, 600 and 500 MHz. Spectra were processed by NMRPipe (Delaglio et al, 1995) and data analysis was assisted by the Sparky program (Kneller et al, 1997). Main chain resonance assignments were carried out using standard spectra. For chemical shift perturbation upon binding N-terminal tail, 1H–15N HSQC spectra of 15N-labelled LC3 were obtained by the addition of 0.2, 0.4, 0.6, 0.8, 1.0, 1.5 and 2.0 M equivalents of the N-terminal tail peptide, respectively. The combined 1H and 15N chemical shift changes (Δp.p.m.) by the addition of 2.0 M equivalents of N-terminal tail peptide were calculated using the equation: Δp.p.m.=[(ΔδHN)2+(ΔδN/5)2]1/2, where ΔδHN and ΔδN are the changes in the chemical shifts along the 1H and 15N axes, respectively.
Accession code
The coordinates and structure factors of the HsAtg4B(H280A)–LC3(1–120), HsAtg4B(H280A)–LC3(1–124) and HsAtg4B(C74S)–LC3(1–124) complexes have been deposited in the Protein Data Bank with accession codes 2Z0D, 2Z0E and 2ZZP, respectively.
Acknowledgments
We are grateful to Dr H Nakatogawa for valuable comments on the paper. The synchrotron radiation experiments were performed at the beamline NW12A at KEK, Japan, and at the beamline BL41XU at SPring8, Japan. This study was supported by Grants-in-Aid for Scientific Research on Priority Areas, the National Project on Protein Structural and Functional Analyses, and Targeted Proteins Research Program from the Ministry of Education, Science and Culture of Japan. This study was carried out under the NIBB Cooperative Research Program (4-148).
References
- Baba M, Takeshige K, Baba N, Ohsumi Y (1994) Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol 124: 903–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54(Part 5): 905–921 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister G, Zhu W, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific LLC [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Part 12, Part 1): 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fass E, Amar N, Elazar Z (2007) Identification of essential residues for the C-terminal cleavage of the mammalian LC3: a lesson from yeast Atg8. Autophagy 3: 48–50 [DOI] [PubMed] [Google Scholar]
- Ha BH, Ahn HC, Kang SH, Tanaka K, Chung CH, Kim EE (2008) Structural basis for Ufm1 processing by UfSP1. J Biol Chem 283: 14893–14900 [DOI] [PubMed] [Google Scholar]
- Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL (2003) A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem 278: 51841–51850 [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y (2000) A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M (2008) Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem 283: 22847–22857 [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T (2004) LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 117(Part 13): 2805–2812 [DOI] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y (2003) A unified nomenclature for yeast autophagy-related genes. Dev Cell 5: 539–545 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Ohsumi Y (1999) Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol 15: 1–32 [DOI] [PubMed] [Google Scholar]
- Kneller DG, Goddard TD (1997) SPARKY 3.105 edit. University of California: San Francisco, CA [Google Scholar]
- Kumanomidou T, Mizushima T, Komatsu M, Suzuki A, Tanida I, Sou YS, Ueno T, Kominami E, Tanaka K, Yamane T (2006) The crystal structure of human Atg4b, a processing and de-conjugating enzyme for autophagosome-forming modifiers. J Mol Biol 355: 612–618 [DOI] [PubMed] [Google Scholar]
- Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C (2003) Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem 278: 3671–3678 [DOI] [PubMed] [Google Scholar]
- Mizushima N (2007) Autophagy: process and function. Genes Dev 21: 2861–2873 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130: 165–178 [DOI] [PubMed] [Google Scholar]
- Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F (2008) Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13: 1211–1218 [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD (2006) Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat Struct Mol Biol 13: 1060–1068 [DOI] [PubMed] [Google Scholar]
- Satoo K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F (2007) Crystallization and preliminary crystallographic analysis of human Atg4B–LC3 complex. Acta Crystallogr Sect F Struct Biol Cryst Commun 63(Part 2): 99–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z (2007) Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26: 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Tatham MH, Dong C, Zagorska A, Naismith JH, Hay RT (2006) SUMO protease SENP1 induces isomerization of the scissile peptide bond. Nat Struct Mol Biol 13: 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F (2004) The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells 9: 611–618 [DOI] [PubMed] [Google Scholar]
- Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F (2005) Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J Biol Chem 280: 40058–40065 [DOI] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E (1999) Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol Biol Cell 10: 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E (2002) Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem 277: 13739–13744 [DOI] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Ueno T, Kominami E (2001) The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem 276: 1701–1706 [DOI] [PubMed] [Google Scholar]
- Tanida I, Ueno T, Kominami E (2004) Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J Biol Chem 279: 47704–47710 [DOI] [PubMed] [Google Scholar]
- Tanida I, Wakabayashi M, Kanematsu T, Minematsu-Ikeguchi N, Sou YS, Hirata M, Ueno T, Kominami E (2006) Lysosomal turnover of GABARAP-phospholipid conjugate is activated during differentiation of C2C12 cells to myotubes without inactivation of the mTor kinase-signaling pathway. Autophagy 2: 264–271 [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Cryst 30: 1022–1025 [Google Scholar]
- Xie Z, Nair U, Klionsky DJ (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 19: 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]