Abstract
Covalent modification by small ubiquitin-related modifiers (SUMO) regulates p53 transcription activity through an undefined mechanism. Using reconstituted sumoylation components, we purified SUMO-1-conjugated p53 (Su-p53) to near homogeneity. Su-p53 exists in solution as a tetramer and interacts with p300 histone acetyltransferase as efficiently as the unmodified protein. Nevertheless, it fails to activate p53-dependent chromatin transcription because of its inability to bind DNA. With sequential modification assays, we found that sumoylation of p53 at K386 blocks subsequent acetylation by p300, whereas p300-acetylated p53 remains permissive for ensuing sumoylation at K386 and alleviates sumoylation-inhibited DNA binding. While preventing the free form of p53 from accessing its cognate sites, sumoylation fails to disengage prebound p53 from DNA. The sumoylation-deficient K386R protein, when expressed in p53-null cells, exhibits higher transcription activity and binds better to the endogenous p21 gene compared with the wild-type protein. These studies unravel a molecular mechanism underlying sumoylation-regulated p53 function and further uncover a new role of acetylation in antagonizing the inhibitory effect of sumoylation on p53 binding to DNA.
Keywords: acetylation, chromatin transcription, p53, SUMO-1, sumoylation
Introduction
The transcription activity of p53 tumour suppressor protein is regulated by posttranslational modification and its interaction with different proteins. Although modulation of p53 protein stability by oncoprotein association and dissociation appears to be a major way to control p53 activity, a nonproteolytic strategy that is uncoupled from the ubiquitin degradation pathway is also used to regulate p53 function. Covalent modification by small ubiquitin-related modifiers (SUMO) represents one such alteration that fine-tunes p53 transcription activity. In humans, three mature (i.e. protease-processed) SUMO conjugates, SUMO-1 (97 aa), SUMO-2 (92 aa) and SUMO-3 (93 aa), are expressed in most cell types. Conjugation by SUMO-1 generally results in monosumoylated proteins, whereas polysumoylated chains can be observed with SUMO-2 and SUMO-3 linkages. A consensus sumoylation site, ψKxE (ψ, a large hydrophobic residue; x, any amino acid), has been defined, but a number of proteins are sumoylated through noncanonical lysine residues (Johnson, 2004; Meulmeester et al, 2008; Vethantham et al, 2008). Although SUMO-1 and SUMO-2/3 (95% identical to each other and ∼47% identical to SUMO-1) seem interchangeable for conjugation at many lysine residues (Zhang et al, 2008), paralog-specific sumoylation has also been observed (Saitoh and Hinchey, 2000; Meulmeester et al, 2008). Like ubiquitination, sumoylation occurs through an enzymatic cascade involving the heterodimeric SUMO-activating enzyme (SAE1/SAE2 in humans, Aos1/Uba2 in yeast), the SUMO-conjugating enzyme Ubc9, and different SUMO E3 ligases that include five protein inhibitors of activated STATs (PIAS1, PIAS3, PIASxα, PIASxβ and PIASy), RanBP2, and the polycomb protein Pc2. This dynamic linkage is reversed by various SUMO-specific proteases, such as SENP1 predominantly present in the nucleus (Hay, 2007). Sumoylation modulates protein function through nucleocytoplasmic translocation, subnuclear localization, and protein–protein and protein–DNA interaction (Matunis et al, 1996; Hoege et al, 2002; Yang and Sharrocks, 2004; Gill, 2005; Pascual et al, 2005; Baba et al, 2006; Potts and Yu, 2007). Most of these events can potentially sequester transcription factors before they reach their destined chromatin targets. Thus, it is important to distinguish between the indirect and the direct effects of sumoylation on transcriptional control. At present, mechanistic studies of direct sumoylation-linked transcriptional control remain mostly unelucidated.
Sumoylation of p53 occurs at lysine 386 in the C-terminal domain (CTD), which is the regulatory region critical for p53 function. The CTD, spanning amino acids 363–393, is thought to be unstructured and accessible for posttranslational modification in several molecular models of full-length p53 tetramers deduced from X-ray structures of individually resolved DNA-binding, oligomerization and N-terminal activation domains, as well as from NMR, small-angle X-ray scattering and three-dimensional cryoelectron micrograph images of wild-type and mutant p53 tetramers (Kitayner et al, 2006; Okorokov et al, 2006; Tidow et al, 2007). This flexible nature of the CTD, which contains multiple lysine residues (K370, K372, K373, K381, K382, K386), allows p53 to contact DNA nonspecifically, and thus facilitates sequence-specific recognition by its core DNA-binding domain, perhaps through DNA sliding (McKinney et al, 2004). All of these six C-terminal lysine residues are subject to different types of posttranslational modification, including Mdm2-mediated ubiquitination (at all six residues; Rodriguez et al, 2000) and neddylation (at K370, K372, K373; Xirodimas et al, 2004), p300/CBP-mediated acetylation (primarily at K373 and K382; Gu and Roeder, 1997), Set7/9-mediated methylation (K372; Chuikov et al, 2004), Smyd2-mediated methylation (K370; Huang et al, 2006) and Set8-mediated methylation (K382; Shi et al, 2007). Acetylation at K373/K382 and methylation at K372 are linked to p53 activation, whereas other covalent bond formations appear to repress p53 transcription activity. Mice with arginine substitutions of all six lysine residues are viable and appear normal in development, but they exhibit some defects in DNA damage response (Feng et al, 2005; Krummel et al, 2005). Thus, the roles of C-terminal lysine modification remain unclear. In the case of p53 sumoylation, cell-based transfection experiments also give controversial results with either an enhancing (Gostissa et al, 1999; Rodriguez et al, 1999) or no effect (Kwek et al, 2001) of sumoylation on p53 transcription activity. Considering that <5% of p53 is sumoylated in the cell (Melchior and Hengst, 2002) and that p53 is constantly diverted from reaching its chromatin targets by different paths of intracellular trafficking, it is extremely difficult to unambiguously define the transcriptional effects of sumoylation in vivo.
Using an in vitro sumoylation system reconstituted with recombinant human SUMO-1, SAE1/SAE2, Ubc9, PIASxβ and p53, we purified SUMO-1-conjugated p53 (Su-p53) to near homogeneity. Su-p53 exists in solution as a tetramer and interacts with p300 histone acetyltransferase (HAT) as efficiently as the unmodified protein. Nevertheless, it fails to activate p53-dependent transcription in an in vitro chromatin-linked transcription system that we have developed (Thomas and Chiang, 2005; Wu et al, 2006). When the DNA/chromatin-binding activity of Su-p53 was examined, we found that sumoylation prevents the free, but not DNA-bound, form of p53 from associating with its target sequence. Interestingly, acetylation of p53 by p300 restores the DNA-binding activity of Su-p53, indicating that acetylation antagonizes sumoylation-mediated inhibition of p53 function and thereby accounts for a stimulating role of p300-mediated acetylation in p53-dependent chromatin transcription in vivo and in vitro. Our studies not only provide mechanistic insights into sumoylation-regulated p53 transcription activity but also unravel an intricate crosstalk between acetylation and sumoylation in fine tuning the DNA-binding activity of a sequence-specific DNA-binding protein.
Results
Acetylation of p53 C-terminal lysines enhances but is not essential for p53-dependent transcription
In p53-dependent transcription, the acetylation status of p53 at its C-terminal lysine residues and the chromatin both correlate with gene activation and repression in vivo and in vitro (Thomas and Chiang, 2005). To define which acetylation (i.e. p53 versus chromatin) is directly linked to p53-dependent transcription, we first purified two p53 mutant proteins 8KR and Δ30 (Figure 1A). 8KR has arginine substitutions at the six C-terminal lysine residues and also at K305 (acetylated by p300) and K320 (acetylated by PCAF), whereas Δ30 has the C-terminal 30 amino acids deleted. When these mutants and the wild-type protein were tested in a p53/p300-dependent in vitro chromatin transcription system (Thomas and Chiang, 2005) reconstituted with HeLa core histones, human NAP-1, and Drosophila ACF (Figure 1B), using a p53-binding site-containing pWAFMLT chromatin template (Figure 1C) assembled as outlined (Figure 1D), we found that both wild-type and 8KR proteins, but not Δ30, were capable of activating p53-dependent transcription from pWAFMLT chromatin in a dose-dependent manner (Figure 1E). Transcription from the internal control pΔMLP DNA template lacking a p53-binding site remained constant (Figure 1E, lanes 1–10). As acetylation-deficient 8KR, in contrast to Δ30, still induced p300-mediated acetylation on pWAFMLT chromatin (Figure 1F), the results suggest that p300-mediated acetylation of chromatin, rather than p53, is more important for p53-dependent transcription. Although acetylation of p53 does not appear to be directly involved in chromatin transcription, it indeed contributes to the transcriptional activity of p53. To define whether two recently identified acetylation sites at K120 (acetylated by Tip60; Tang et al, 2006) and K164 (also acetylated by p300/CBP; Tang et al, 2008) might contribute to 8KR-mediated activation and whether arginine substitutions of the six C-terminal lysines would give rise to comparable transcription activity as 8KR, we purified two additional mutants 10KR and 6KR (see Figure 1A). We found essentially no differences in transcription activity among 8KR, 10KR and 6KR, all exhibiting approximately two-fold lower activity compared with that of the wild-type protein (Figure 1G). Our data, performed with chromatin assembled with highly purified factors, indicate that C-terminal acetylation by p300, although not essential for p53-dependent transcription, indeed contributes to p53 transcription activity. Moreover, the C-terminal 30 amino acids are crucial for p300-mediated acetylation of p53-targeted chromatin and thus deletion of this regulatory domain impairs p53 transcription activity. Our results are consistent with earlier studies performed with Drosophila S190 extract-assembled chromatin (Espinosa and Emerson, 2001) and with cell-based reporter and chromatin immunoprecipitation (ChIP) assays (McKinney et al, 2004) showing diminished chromatin binding and transcription activity of the Δ30 mutant.
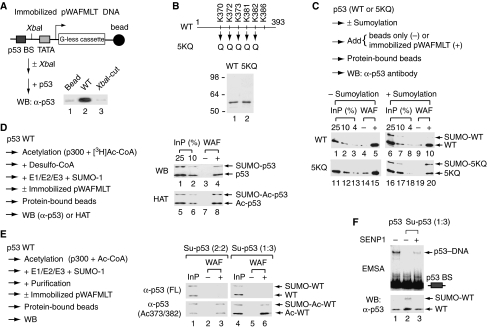
Figure 1.
Acetylation of chromatin but not p53 is directly linked to p53-dependent transcription. (A) Coomassie blue staining of purified p53, 8KR, 10KR, 6KR and Δ30 proteins and their schematic drawing. Numbers indicate the positions of specific amino acid residues and the boundaries of each protein. (B) Coomassie blue staining of purified HeLa core histones (CH), hNAP-1 and ACF proteins. (C) G-less cassette templates used for in vitro transcription assays. (D) Scheme of chromatin assembly for histone acetyltransferase (HAT) assay and in vitro transcription. (E) Wild-type (WT) p53 and 8KR, but not Δ30, efficiently activate transcription from pWAFMLT chromatin. In vitro transcription was performed with pWAFMLT chromatin and the internal control pΔMLP DNA template as described in Materials and methods. (F) Wild-type p53 and 8KR, but not Δ30, support p300-mediated acetylation of pWAFMLT chromatin. In vitro HAT assay was performed with p300 and pWAFMLT chromatin, in the absence (−) or presence of different p53 proteins as indicated. (G) Dose-dependent activation of pWAFMLT chromatin by wild-type p53 and acetylation-defective mutants. Error bars indicate the standard deviation from an average of three independent experiments.
Sumoylated p53 exists in solution as a tetramer with asymmetric C-terminal modification
To explore the functional role of p53 acetylation and its potential interplay with other C-terminal modifications, we established an in vitro sumoylation system reconstituted with recombinant hexahistidine-tagged human E1 (SAE1/SAE2 heterodimer), E2 (Ubc9), E3 (PIASxβ), wild-type p53 and sumoylation-defective K386R, and wild-type SUMO-1 and its conjugation-deficient GA mutant that changes the last glycine in the mature form to alanine (Figure 2A). As expected, sumoylation of p53 under conditions of limiting Ubc9 requires each of the sumoylation components and occurs specifically at K386 (Supplementary Figure 1A and B). The GA mutant of SUMO-1 could not be conjugated efficiently to p53 (also see Supplementary Figure 1A and B) and thus provides a specificity control for p53 sumoylation. Importantly, when a large-scale sumoylation reaction was performed with FLAG-tagged SUMO-1 (f:SUMO-1) and hexahistidine-tagged p53 and E1–E2–E3 enzymes, followed by sequential Ni2+-NTA and anti-FLAG M2 affinity purification (Figure 2B, left scheme), only Su-p53 was purified (Figure 2B, right panel, lane 2). Surprisingly, an approximately equal amount of sumoylated p53 and unmodified p53 was detected in the purified complex. This suggests that Su-p53 exists in solution as a tetramer and not all subunits are equally accessible to the sumoylation enzymes. This biochemical evidence is consistent with molecular modelling of p53 tetramers projecting conformationally distinct C-termini within a p53 tetramer (Kitayner et al, 2006; Okorokov et al, 2006; Tidow et al, 2007). Indeed, the existence of tetrameric Su-p53 was further substantiated by size-exclusion column chromatography (Figure 2C) and glutaraldehyde crosslinking analysis (Figure 2D), both in agreement with the structures of molecularly simulated p53 tetramers, even in the sumoylated form as reported here.
Figure 2.
Purified SUMO-1-conjugated p53 exists in solution as a tetramer with selective C-termini accessible for sumoylation. (A) Coomassie blue staining of purified proteins used for in vitro sumoylation reactions. All recombinant human proteins contain an N-terminal hexahistidine tag. Purification of these hexahistidine-tagged proteins was described in Materials and methods. Molecular size markers (in kDa) are indicated on the left. (B) Sumoylated p53 (Su-p53) contains both SUMO-1-conjugated and unmodified subunits. Purification of Su-p53 was conducted as outlined (left), following a large-scale sumoylation reaction performed with (+) or without (−) the SUMO mix containing either FLAG (f:)- or hexahistidine (6His)-tagged sumoylation components. Purified Su-p53, along with the samples recovered from the mock purification, was visualized by Coomassie blue staining. (C) Su-p53 and unmodified p53 each elutes as a tetramer as determined by Superose 6 size-exclusion column chromatography. The void volume and predetermined protein size markers (in kDa) are indicated above the fraction numbers. (D) Tetrameric p53 and Su-p53 detected by glutaraldehyde crosslinking. An increasing concentration (%) of glutaraldehyde was used for crosslinking homomeric p53 subunits. Products were analysed by Western blotting with anti-p53 full-length antibodies. (E) Purification of Su-p53 with different extent of SUMO-modified subunits. Constant amounts of p53, E1 and E2 enzymes in a standard 1 × reaction were incubated with variable quantities of SUMO-1 and PIASxβ as indicated. The extent of p53 sumoylation, before and after purification (with schematic drawing on the right), was visualized after Coomassie blue staining. (F) Further sumoylation can occur on purified Su-p53. Constant amounts (1 × ) of Su-p53, SUMO-1 and PIASxβ were incubated with an increasing concentration (1–10 × ) of E1 and E2 as indicated. Western blotting was performed with anti-p53 antibodies.
To examine whether only two subunits in a p53 tetramer could be subject to sumoylation, we repeated sumoylation reactions by varying the concentrations of E1 and E2 enzymes and also the incubation time. Intriguingly, a nearly 1:1 ratio of sumoylated and unmodified subunits was constantly observed in various purified complexes (Supplementary Figure 2), suggesting that conformationally distinct C-termini indeed exist in p53 tetramers. Nevertheless, when the amount of SUMO-1 was reduced 50 times or the concentration of PIASxβ increased 10 times, sumoylation at only one or three of the four subunits could be detected in the purified complex (Figure 2E). We also tested whether sumoylation could further occur on purified Su-p53 with 1:1 modified subunits. Indeed, nearly completion of sumoylation reactions on all four subunits could take place in the purified complex (Figure 2F). Our results indicate that, although two subunits in a p53 tetramer are generally accessible for protein modification, sumoylation of one or up to three or four subunits can also occur under different experimental conditions.
Sumoylation inhibits p53 transcription and DNA/chromatin binding, as well as p300-mediated p53 acetylation without impairing its association with p300
As tetramers represent the nuclear form of p53 with a masked nuclear export signal (Kawaguchi et al, 2006; Carter et al, 2007), we expected that sumoylation would play a role in modulating the nuclear activity of p53. Indeed, the chromatin transcription activity of Su-p53, after normalization with an equivalent amount of p53 (Figure 3A), was reduced significantly (Figure 3B, lanes 4 versus 6, and lanes 5 versus 7) and acetylation of Su-p53 and chromatin was barely detected, even though p300 autoacetylation remained robust (Figure 3C, lanes 6 and 7 versus lanes 4 and 5). The loss of Su-p53 transcription activity was not caused by potential inactivation of p53 due to the sequential purification scheme used here (see Figure 2B), as unmodified p53 subjected to the same incubation condition remained fully active in transcription (data not shown). As p300 is critical for p53 transactivation, we examined whether Su-p53 could still interact with this cofactor. As shown in Figure 3D, p300 could interact efficiently with both p53 and Su-p53, as demonstrated by solution interaction assay showing that anti-p300 antibodies pulled down an equivalent amount of p300 as well as p53 and Su-p53 (lanes 1 versus 2, and lanes 4 versus 5). Thus, our failure to detect acetylation on Su-p53 (see Figure 3C, lane 6) suggests that sumoylation at K386 prevents p300-mediated acetylation of p53, likely due to steric hindrance of the bulky SUMO conjugate blocking acetylation at the adjacent lysine residues (K382 and K373).
Figure 3.
Sumoylation inhibits p53-dependent transcription by preventing p53 from binding to DNA/chromatin, correlating with reduced p300-mediated acetylation on p53 and chromatin. (A) Quantification of protein amounts between p53 and Su-p53 by Western blotting. (B) Su-p53 fails to activate p53-dependent transcription from pWAFMLT chromatin. In vitro transcription was performed as described in Materials and methods using different amounts of purified p53 or Su-p53 as indicated. Relative transcription (%) is defined as the signal intensity from pWAFMLT chromatin relative to that of lane 5 (set at 100), after normalization of the transcription signal with that of the pΔMLP internal control. (C) Su-p53 could not be acetylated by p300 and could not efficiently support p300-mediated acetylation on pWAFMLT chromatin. In vitro HAT assay was performed with p300 and pWAFMLT chromatin, in the absence (−) or presence of different amounts of p53 or Su-p53 as indicated and described in Materials and methods. (D) Sumoylation does not affect p53 association with p300. Solution interaction was performed as outlined by incubating p53 or Su-p53 with p300 or buffer alone (−). Asterisk indicates the immunoglobulin heavy chain from anti-p300 antibodies that serve as a good internal control for normalization among different immunoprecipitated (IP) samples. (E) Su-p53 is incapable of binding to pWAFMLT chromatin. In vitro chromatin immunoprecipitation (ChIP) was conducted by incubating p53 or Su-p53 with p300 and pWAFMLT chromatin, followed by formaldehyde crosslinking and micrococcal nuclease digestion before further processing for ChIP analysis as described in Materials and methods. (F) Su-p53 is unable to bind DNA containing a p53-binding site. Electrophoretic mobility shift assay (EMSA) was carried out with a 32P-labelled DNA fragment containing the p53-binding site derived from pWAFMLT (i.e. the distal p53-binding site originally from the human p21 gene). (G) Su-p53 cannot bind to a p53-binding site-containing Hdm2 DNA fragment as determined by EMSA. (H) SENP1 unmasks the DNA-binding activity of Su-p53 after desumoylation. Purified Su-p53 was incubated with wild-type (WT) or a catalytic mutant (R630L/K631M; mt) of SENP1 and then analysed by Western blotting and EMSA using the same pWAFMLT-derived fragment as in (F). (I) Coomassie blue staining of purified WT and mt FLAG-tagged SENP1 used in (H).
To explore the molecular mechanism by which sumoylation blocks p53-dependent chromatin transcription (see Figure 3B), we conducted an in vitro ChIP assay by incubating purified p53 or Su-p53 with pWAFMLT chromatin. Although p53-bound DNA fragments were clearly pulled down by anti-p53 antibody, as detected by PCR amplification with a primer pair spanning the p53-binding site (Figure 3E, lanes 2 and 3 versus lane 1), no DNA fragments were amplified when Su-p53 was incubated with pWAFMLT chromatin (Figure 3E, lanes 4 and 5 versus lanes 2 and 3). The inability of Su-p53 to bind pWAFMLT chromatin in ChIP assays was further confirmed by the use of anti-SUMO-1 antibodies, which again failed to pull down any p53-associated DNA fragments (data not shown). We thus concluded that Su-p53 is incapable of accessing its target site in chromatin. To investigate whether the loss of Su-p53 chromatin-binding activity is due to its failure to bind DNA, we performed an electrophoretic mobility shift assay (EMSA) by incubating an increasing amount of p53 or Su-p53 with a 32P-labelled DNA fragment containing the p53-binding site derived from pWAFMLT. As shown in Figure 3F, Su-p53 was unable to form protein–DNA complexes typically observed with unmodified p53 (lanes 6 and 7 versus lanes 2–5). The loss of Su-p53 DNA-binding activity was also observed with another p53-binding site-containing DNA fragment derived from the Hdm2 gene (Figure 3G), suggesting that SUMO-inactivated p53 binding to DNA is not unique to the pWAFMLT template.
To examine the reversibility of SUMO inhibition, we incubated Su-p53 first with purified wild-type or catalytically inactive SENP1 SUMO isopeptidase (Figure 3I) to remove the SUMO moiety and then repeated EMSA. Indeed, the DNA-binding activity of Su-p53 could be unmasked by cleaving SUMO-1 from p53 tetramers after treatment with enzymatically active SENP1 (Figure 3H). The inability of SENP1 to fully restore the DNA-binding activity of p53 even after a nearly complete removal of the SUMO conjugate (see Figure 3H, bottom panel, lanes 2 versus 3) prompted us to examine whether SENP1 might negatively regulate p53 binding to DNA through direct protein–protein association. Indeed, we found that SENP1 could interact directly with unmodified p53 and block its DNA-binding activity (Supplementary Figure 3). Perhaps, SENP1 has differential effect on the DNA-binding activity of p53 in part regulated by its association with SUMO. This possibility remains to be investigated in the future. Taken together, our results illustrate that sumoylation of p53 negatively regulates its transcription activity by preventing p53 binding to DNA and chromatin.
Acetylation counteracts sumoylation-prohibited sequence recognition by p53
The finding that sumoylation prevents p300-mediated acetylation of p53 (see Figure 3C, lane 6) prompted us to examine whether acetylation by p300 would likewise block sumoylation on p53. To this end, we applied an immobilized DNA-binding assay that allows us to monitor the status of posttranslational modification on p53 before and after DNA binding. As outlined in Figure 4A, removal of the p53-binding site by XbaI digestion eliminated p53 binding to the immobilized template (lanes 2 versus 3), confirming site-specific DNA binding of p53 in this assay. Purified p53 and 5KQ (Figure 4B), an acetylation mimic of p53 containing glutamine substitutions at K370, K372, K373, K381 and K382 (Luo et al, 2004), were then subjected to sumoylation reactions in which the resulting products were assessed for their DNA-binding activity (see outline, Figure 4C). Without sumoylation, wild-type p53 and 5KQ bound equally well to the immobilized template (Figure 4C, lanes 5 and 15). After the sumoylation reaction, only the unmodified and not sumoylated form of wild-type p53 was detected on the immobilized template (Figure 4C, lanes 6 versus 10), consistent with EMSA results showing that Su-p53 failed to bind DNA (see Figure 3F and G). Intriguingly, acetylation-mimic 5KQ was sumoylated (Figure 4C, lanes 11 versus 16), and sumoylated 5KQ protein bound to DNA as efficiently as unmodified 5KQ (Figure 4C, lanes 16 versus 20). This result indicates that acetylation does not block sumoylation and, furthermore, acetylation appears to counteract the inhibitory effect of sumoylation on p53 binding to DNA. To substantiate this, we repeated the experiment with p300-acetylated wild-type p53, in which the acetyl group was 3H-labelled by the HAT reaction that was subsequently terminated by addition of the desulfo-CoA HAT inhibitor before the sumoylation reaction (see outline, Figure 4D). Indeed, sumoylation was observed on acetylated p53 (detected by tritium fluorography, see Figure 4D, lower panel, lane 5) and the doubly modified SUMO-acetylated p53 was able to bind DNA (Figure 4D, lane 8).
Figure 4.
Acetylation overcomes sumoylation-mediated inhibition of sequence recognition by p53. (A) Sequence-specific binding of p53 on immobilized DNA template. Binding of p53 to immobilized pWAFMLT DNA, with or without XbaI digestion, was monitored by Western blotting following the outlined protocol. (B) Coomassie blue staining of purified FLAG-tagged wild-type (WT) p53 and the 5KQ mutant. (C) Sumoylation blocks wild-type p53 but not 5KQ binding to immobilized pWAFMLT DNA. DNA-binding assays were performed as outlined by incubating wild-type p53 or the acetylation-mimic 5KQ mutant with immobilized pWAFMLT DNA (+) or beads alone (−), with (+) or without (−) prior sumoylation as indicated. (D) Acetylation does not prevent subsequent sumoylation of p53 and allows doubly modified p53 binding to DNA. Sequential acetylation and sumoylation reactions were carried out as outlined with the addition of desulfo-CoA in between to terminate the acetylation reaction before the sumoylation reaction. Acetylated p53 was monitored by incorporated 3H on the acetyl group. (E) DNA binding by acetylated Su-p53 containing different SUMO-modified subunits. Purified Su-p53 in 2:2 or 1:3 stoichiometry, as prepared in Figure 2E, with a minor population also acetylated by p300, was incubated in the presence (+) or absence (−) of pWAFMLT DNA. Western blotting performed with anti-p53 full-length (FL) or anti-acetylated K373/382 antibodies was used to monitor the DNA-binding activity of Su-p53 without acetylation (upper panel) or with p300-mediated acetylation at K373/382 (lower panel). (F) SENP1 treatment restores the DNA-binding activity of Su-p53 with 1:3 stoichiometry. Purified Su-p53 (1:3) was incubated in the presence (+) or absence (−) of SENP1 and then analysed by Western blotting and EMSA using the pWAFMLT-derived fragment.
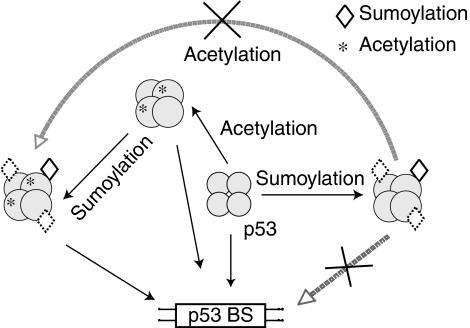
To determine whether sumoylation at one subunit was sufficient to block p53 tetramers binding to DNA and whether acetylation could likewise counteract the inhibitory effect of sumoylation with only one-modified subunit, we performed a similar DNA-binding assay with purified Su-p53 containing 1:3 or 2:2 modified subunits. As shown in Figure 4E, no DNA binding was observed in the majority of nonacetylated Su-p53 with either 2:2 or 1:3 stoichiometry (upper panels), and prior acetylation of Su-p53 at K373 and K382 could alleviate SUMO inhibition of p53 binding to DNA in both configurations (lower panels). As expected, SENP1 treatment of purified Su-p53 with one modified subunit could also restore its DNA-binding activity (Figure 4F). Our findings, summarized in the model presented in Figure 5, illustrate a unidirectional crosstalk between sumoylation and acetylation, likely due to steric hindrance of the bulky SUMO moiety in blocking p300 access to its target sites, whereas the small acetyl group remains permissive for the sumoylation machinery to modify the adjacent K386 residue.
Figure 5.
Model for the crosstalk between sumoylation and acetylation regulating p53 tetramer binding to its target sequence. The subunits modified by acetylation (asterisk) and/or sumoylation (diamond) are arbitrarily assigned to simply reflect the modified state of the C-termini with distinct conformations within p53 tetramers. The p53-binding site is abbreviated as p53 BS.
Sumoylation of p53 in the DNA-bound state fails to dissociate it from its target sequence
Although sumoylation prevents the free form of p53 from binding to DNA, it was unclear whether sumoylation could actively disengage DNA-bound p53 from its target site. To investigate this, we performed sumoylation reactions on the p53-bound DNA template by first incubating p53 or 5KQ with immobilized pWAFMLT, followed by sumoylation reactions performed with either wild-type or GA mutant of SUMO-1 after removal of the unbound proteins (see outline, Figure 6A). Unlike the free form, engaged p53 and 5KQ both stayed bound to DNA after sumoylation (Figure 6A, lanes 1 versus 2) and, as anticipated, the SUMO-1 GA mutant could not be conjugated to either bound protein (Figure 6A, lanes 3 versus 1). The inability of sumoylation to actively dissociate prebound p53 from DNA raised an interesting possibility that a different sumoylation site, distinct from K386 used by the free form of p53, might be activated for SUMO-1 conjugation upon DNA-induced conformational changes. To explore this, we performed sumoylation reactions on DNA-bound p53 containing the wild-type lysine or an arginine substitution at K386 or a putative sumoylation site at K292 (Kwek et al, 2001). As shown in Figure 6B, sumoylation was detected on both wild-type p53 and K292R, but not on the K386R mutant. This experiment demonstrates that sumoylation of DNA-bound p53 still takes place at the same K386 lysine residue as seen with the free form of p53, thus excluding the possibility that a different sumoylation site was used for SUMO conjugation when p53 bound to DNA and thereby accounting for different binding properties between free and DNA-bound p53.
Figure 6.
Sumoylation of p53 in the DNA-bound state occurs at K386 and fails to dissociate p53 from its target sequence. (A) Sumoylation takes place on DNA-bound p53 without leading to its dissociation from the immobilized pWAFMLT template. Sumoylation reactions were performed with wild-type SUMO-1 or the GA mutant on p53- or 5KQ-bound pWAFMLT DNA as outlined. Protein-bound beads (B) and the unbound supernatant (UB) were analysed by Western blotting with anti-p53 full-length antibodies. (B) Sumoylation of DNA-bound p53 occurs at the same K386 residue. Sumoylation of wild-type p53 or the K292R or K386R mutant prebound to immobilized pWAFMLT DNA was performed and analysed as outlined. (C) Enhanced sumoylation on DNA-bound p53 fails to disengage p53 from DNA. DNA-bound p53 was subjected to different extent of sumoylation reactions with 2 times (2 × ) or 5 times (5 × ) more of sumoylation components. Binding assay was conducted as described in (A). (D) Recruitment of mSin3A but not p300 is enhanced by sumoylated p53 bound to DNA. HeLa nuclear extract was incubated with DNA-bound p53, with (+) or without (−) prior sumoylation reactions performed with wild-type (WT) or the GA mutant of SUMO-1. Proteins associated with DNA-bound p53 or sumoylated p53 were then analysed by Western blotting with anti-p53, p300 or mSin3A antibodies.
As sumoylation could occur on more than one subunit of p53 tetramers, it is important to know whether disengagement of DNA-bound p53 requires sumoylation on multiple subunits. To address this issue, we performed sumoylation on DNA-bound p53 with higher concentrations of SUMO components and found that DNA-bound p53 could not be dissociated from the template even with 2:2 or 3:1 modified subunits (Figure 6C, lanes 1 versus 2). Not surprisingly, DNA-bound K386R remained unmodified under high concentrations of sumoylation reactions and associated with the template throughout the entire process (Figure 6C, lanes 3 versus 4). As sumoylation on DNA-bound transcription factors has the potential to recruit transcriptional corepressors (Gill, 2005; Stielow et al, 2008), we further explored whether DNA-bound SUMO-p53 could recruit selective cellular factors. Accordingly, we performed Western blotting to examine which corepressors could be retained on DNA-bound SUMO-p53, after its incubation with HeLa nuclear extract. Although we could not observe SUMO-dependent association of histone deacetylases HDAC1, HDAC2, HDAC6 and SIRT1 (data not shown), we indeed detected an enhanced recruitment of the mSin3A corepressor, but not p300, to DNA-bound SUMO-p53 (Figure 6D). This finding suggests that sumoylation on DNA-bound p53 could potentially regulate p53 transcription activity through recruitment of selective cofactors.
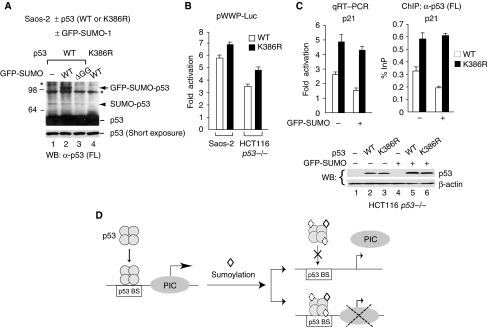
Sumoylation-deficient K386R mutant exhibits higher transactivating activity and binds better to the endogenous p21 gene compared with the wild-type protein
To demonstrate the functional effect of sumoylation in vivo, we first verified K386 as the sumoylation residue by performing transient transfection assays in p53-null Saos-2 cells. As shown in Figure 7A, GFP-tagged SUMO-1 could only be detected on coexpressed p53 with wild type but not diglycine-deleted SUMO-1 or K386R mutant (see the band indicated by an arrow). A small amount of sumoylated p53 formed between endogenous SUMO and exogenous p53, but not K386R, was also observed (see the band depicted by an arrowhead), confirming K386 as the sumoylated residue in vivo. Consistent with our in vitro chromatin transcription experiment showing sumoylation-inhibited p53-dependent transcription (see Figure 3B), the K386R mutant reproducibly exhibited higher transcription activity compared with that of wild-type p53 when p53-targeted human p21 promoter-driven reporter activity was examined in p53-null Saos-2 or HCT116 p53−/− cells (Figure 7B). To examine whether the endogenous p53-targeted p21 gene was likewise regulated, we compared the levels of endogenous p21 transcripts in HCT116 p53−/− cells transfected with a wild-type p53 or K386R expression plasmid, with or without exogenous GFP-SUMO-1 expression, by quantitative RT–PCR and also measured their recruitment to the distal p53-binding site in the p21 gene by ChIP. Consistent with the data shown in Figure 7A, only wild-type p53 but not K386R was sumoylated in HCT116 p53−/− cells (data not shown) and, as expected, the sumoylation-deficient K386R mutant indeed has higher transactivating activity (Figure 7C, left panel) and associates better with the endogenous p21 gene (right panel) when a comparable amount of p53 and K386R proteins were expressed (bottom panels). Collectively, these data support the mechanistic studies performed in vitro unraveling an important role of sumoylation in regulating p53 target gene transcription (Figure 7D).
Figure 7.
Sumoylation-deficient p53 exhibits higher transcription activity and binds better to the endogenous p21 gene than the wild-type protein. (A) Detection of sumoylated p53 in p53-null Saos-2 cells by exogenous expression of p53 or the K386R mutant, with or without coexpressed GFP-tagged wild-type (WT) SUMO-1 (GFP-SUMO) or the diglycine-deleted ΔGG mutant. (B) Sumoylation-defective K386R exhibits higher transactivating activity compared with that of wild-type p53 in enhancing human p21 promoter-driven reporter (pWWP-Luc) activity in p53-null Saos-2 and HCT116 cells. (C) K386R activates endogenous p53-targeted p21 gene expression more efficiently than wild-type p53, correlating with enhanced K386 association with the p21 gene. RNA, chromatin, and cell lysates were harvested from HCT116 p53−/− cells exogenously expressing wild-type p53 or K386R, with (+) or without (−) coexpressed GFP-SUMO-1, and analysed by quantitative RT–PCR (left panel), ChIP (right panel) and Western blotting (bottom panel), respectively. (D) Model for sumoylation-regulated p53 target gene transcription. The transcription preinitiation complex (PIC) assembled on a p53-regulated promoter (left) could be dissociated from (upper right) or remained bound to (lower right) the promoter depending on sumoylation on either free or DNA-bound p53. A poised PIC, after sumoylation on DNA-bound p53 tetramers, is likely in an inactive state.
Discussion
Covalent modification on amino-acid residues offers a simple and often nondisruptive way to fine-tune protein activity through conformational changes that regulate protein–protein and protein–nucleic acid interactions. One of the best examples is illustrated by stress-induced modification occurring on the p53 tumour suppressor protein, which is a short-lived, sequence-specific transcription factor modulating cell-cycle progression, DNA repair, apoptosis and senescence (Riley et al, 2008). However, conclusions based on studies performed in vivo and in cultured cells, particularly the biological effects of phosphorylation (Appella and Anderson, 2001), acetylation (Luo et al, 2004) and sumoylation (Melchior and Hengst, 2002) on p53, are sometimes contradictory, in part due to the complexity of the varying cellular environment and the fact that modification typically takes place in a small population of the total protein within a narrow time window.
To overcome this difficulty, we developed in vitro-reconstituted modification assays that allow us to purify sumoylated p53 to near homogeneity and thus define unambiguously the biochemical property of this modification in regulating p53 function. A number of important findings have been uncovered from this study (see the models presented in Figures 5 and 7D). First, sumoylation occurs in tetrameric p53 with flexible C-terminal conformations inherent in different subunits, providing biochemical evidence supporting molecular modelling of the unstructured C-terminal region inferred from individually resolved subdomains of p53 and the cryoEM structure of p53 tetramers. Second, sumoylation plays a direct role in p53-dependent transcription by inhibiting p53 binding to DNA and chromatin, thereby suppressing p53 target gene transcription. Third, we have discovered a novel role of C-terminal acetylation, which acts primarily to overcome sumoylation-inhibited DNA binding, thereby restoring p53's ability to bind DNA and to support transcription. Although acetylation has been implicated in the regulation of p53 binding to DNA, recruitment of coactivators, enhanced protein stability and dissociation of Mdm2/Mdmx (reviewed in Tang et al, 2008), most of these cell-based assays cannot distinguish direct or indirect effects of acetylation on p53 binding to targeted chromatin and on transcription complex assembly. In fact, recent experiments performed in living cells also suggest that acetylation of p53 at its C-terminal lysine residues, K120, or K164, is primarily implicated in corepressor dissociation without truly altering p53 binding to its target sequences (Tang et al, 2008), consistent with our in vitro studies reported here. Fourth, although acetylation does not prevent subsequent sumoylation on the p53 tetramer, sumoylation appears to block acetylation on the adjacent C-terminal lysine residues, likely due to steric hindrance and conformational changes induced by the bulky SUMO moiety. Fifth, unlike its effect on the free form of p53, sumoylation does not actively dissociate DNA-bound p53 from its target site, even though the same lysine residue (K386) is used for SUMO conjugation in both cases. Sixth, the sumoylation-defective K386R mutant indeed exhibits higher transcription activity compared with the wild-type protein, correlating with its enhanced binding to the endogenous p21 gene. Seventh, sumoylation on DNA-bound p53 allows recruitment of the mSin3A transcriptional corepressor (this study) that in turn recruits other inhibitory activity and prolongs the half-life of p53 on targeted chromatin (Zilfou et al, 2001), thereby providing a molecular insight into mSin3A-potentiated p53 repression of pluripotency-associated gene expression (Lin et al, 2005; Wilkinson et al, 2008). Our failure to detect SUMO-dependent recruitment of histone deacetyltransferases by DNA-bound SUMO-p53 using HeLa nuclear extract indicates that formation of mSin3A corepressor complex is a multi-step process and additional control mechanisms or stress-induced signalling events typically seen in living cells are necessary to form a stable p53–mSin3A–HDAC complex.
The tetramer is the active form of p53 engaged in transcriptional regulation and appears to be the entity modified by phosphorylation (Shieh et al, 1999), acetylation (Čes̆ková et al, 2006) and sumoylation (this study), maybe through formation of unique conformations accessible to the protein modification machinery. It would be interesting to examine whether other types of posttranslational modification, such as methylation, ubiquitination and neddylation also occur in a tetramer-specific manner, which may provide a molecular basis for the crosstalk among different types of modification seen with p53. In our HAT assay, very little if any acetylation was detected on Su-p53 (Figure 3C), suggesting that acetylation and sumoylation take place on the same subunits in a tetramer and the steric hindrance imposed by the bulky SUMO moiety at K386 prevents p300-mediated acetylation at adjacent K382 and K373. Although two of the four subunits in a p53 tetramer seem to be preferentially sumoylated, SUMO modification can also occur on only one or more than two subunits with a similar DNA-binding effect. Although subunit exchange may theoretically generate a small amount of wild-type p53 tetramers in purified Su-p53 and account for a low level of transcription from pWAFMLT chromatin (see Figure 3B, lanes 6 and 7), it does not seem to be kinetically favoured. The barely detectable signal in Su-p53 acetylation reactions indicates that sumoylation may stabilize the unstructured C-terminal region and the entire protein, thereby increasing the stability of an intrinsically unstable p53 tetramer (see the p53 structure review by Joerger and Fersht, 2008). Future structural analysis of Su-p53 will provide molecular insight into this intriguing issue.
The crosstalk among different types of p53 modification that has been reported includes phosphorylation-regulated p53 acetylation (Ou et al, 2005; Yang and Seto, 2008), methylation-triggered p53 acetylation (Ivanov et al, 2007; Kurash et al, 2008), ubiquitination-excluded p53 acetylation (Ito et al, 2002; Li et al, 2002; Le Cam et al, 2006), ubiquitination-enhanced p53 sumoylation (Carter et al, 2007) and sumoylation-inhibited p53 acetylation (this study). Clearly, the interplay among different covalent modifications creates an intricate circuit and a ‘protein code' for regulating p53 function and the activity of many other transcriptional regulators and components of the general transcription machinery and general cofactors (Thomas and Chiang, 2006). The uncovering of SUMO-inhibited p53 binding to DNA is consistent with the findings in other SUMO-regulated DNA-binding proteins, such as thymine DNA glycosylase (Baba et al, 2006), Sox2 (Tsuruzoe et al, 2006) and heat shock transcription factor 2 (Anckar et al, 2006). Conceivably, sumoylation has a more active role in modulating the DNA-binding activity of selective transcription factors. It is important to note that, although sumoylation blocks the free form of p53 from accessing its target sequence, it fails to actively disengage prebound p53 from DNA. An intriguing role of sumoylation on DNA-bound p53 is likely to recruit coregulator complexes recognizing the SUMO mark on p53, as seen in our mSin3A recruitment (Figure 6D) and with other transcription systems (Gill, 2005; Stielow et al, 2008). Perhaps, DNA-induced conformational changes provide a better configuration for sumoylation to occur on poised factors as reported for proliferating cell nuclear antigen (Parker et al, 2008). These possibilities remain to be investigated in the future.
Materials and methods
Plasmid construction
Plasmid pF:p53(8KR)-11d, used for producing human p53(8KR) protein, was created by site-directed mutagenesis changing nucleotides coding for K305 and K320 to arginine (R) in pF:p53(6KR)-11d, which was generated by swapping the p53 cDNA insert, amplified by PCR from pcDNA3-p53(6KR) (Rodriguez et al, 2000) with an NdeI site-containing sense primer and a BamHI site-containing antisense primer, with the TBP insert in pF:TBP-11d (Chiang et al, 1993). Plasmid pF:p53(10KR)-11d was similarly constructed in pF:p53(8KR)-11d by site-directed mutagenesis altering nucleotides coding for K120 and K164 to arginine. pF:p53(Δ30)-11d and pF:p53(K386R)-11d were generated by swapping the cDNA insert in pF:TBP-11d between NdeI and BamHI sites with the coding sequence for amino acids 1–363 and 1–393 of p53 amplified from pF:p53-11d (Thomas and Chiang, 2005) and pcDNA3-p53(K386R) (Rodriguez et al, 2000), respectively. Bacterial expression plasmids p6His:SAE1-11d, p6His:SAE2-11d, p6His:Ubc9-11d, p6His:SUMO1-11d and pF:SUMO1-11d were cloned as described above by amplifying individual cDNA from human cDNA libraries and swapping with the TBP insert in p6His:TBP-11d (Chiang et al, 1993) or pF:TBP-11d. p6His:SUMO1(GA)-11d, changing the coding nucleotide for the last amino acid of human SUMO-1 from glycine to alanine, and pF:p53(K292R)-11d were created by site-directed mutagenesis in p6His:SUMO1-11d and pF:p53-11d, respectively. p6His:p53-11d, p6His:p53(5KQ)-11d, p6His:PIASxβ-11d, pF:SENP1(WT)-11d and pF:SENP1(R630L/K631M)-11d were constructed by cloning each cDNA insert from pF:p53-11d, pcDNA3-p53(5KQ) (Luo et al, 2004), a PIASxβ plasmid (Liu et al, 1998), FLAG-SENP1 and FLAG-SENP1(R630L, K631M) (Cheng et al, 2004), respectively, into the hexahistidine- or FLAG-tagged pET-11d vector as described above. The BglII-EcoRI fragment containing either wild-type or mutant FLAG-SENP1 cDNA was then isolated from pF:SENP1(WT)-11d and pF:SENP1(R630L/K631M)-11d, and cloned into pVL1392 (Invitrogen) at the same enzyme-cutting sites to generate pVL-F:SENP1(WT) and pVL-F:SENP1(R630L/K631M), respectively.
Mammalian expression plasmids pcDNA3-F:p53 and pcDNA3-F:p53(K386R) were constructed by isolating the p53 cDNA insert from pF:p53-11d and pF:p53(K386R)-11d, respectively, between BglII and EcoRI sites and then cloning into pcDNA3 (Invitrogen) between BamHI and EcoRI sites. The other plasmids, pcDNA3-p53, pcDNA3-p53(K386R) (Kwek et al, 2001), pCS2-GFP:SUMO-1, pCS2-GFP:SUMO-1(ΔGG), pCS2-GFP (Potts and Yu, 2007) and pWWP-Luc (Somasundaram et al, 1997) have been described.
Proteins and antibodies
Recombinant FLAG-tagged p53, 8KR, 10KR, 6KR, Δ30, K292R and K386R proteins were purified from bacterial BL21(DE3)pLysS strain harbouring pF:p53-11d, pF:p53(8KR)-11d, pF:p53(10KR)-11d, pF:p53(6KR)-11d, pF:p53(Δ30)-11d, pF:p53(K292R)-11d and pF:p53(K386R)-11d, respectively, as described (Chiang and Roeder, 1993). Wild-type and mutant human SENP1 proteins were purified from insect Sf9 cells infected with baculovirus generated from pVL-F:SENP1(WT) and pVL-F:SENP1(R630L/K631M), respectively, following the published protocol (Wu et al, 1999). Purification of HeLa core histones, FLAG-tagged hNAP-1, ACF (reconstituted from Acf1 and FLAG-tagged ISWI), and FLAG-tagged p300 was conducted according to the published procedures (Thomas and Chiang, 2005). For purification of hexahistidine-tagged p53, 5KQ, SAE1, SAE2, Ubc9, SUMO-1 and SUMO-1(GA), 30 ml of sonication supernatant prepared from bacterial strain harbouring p6His:p53-11d, p6His:p53(5KQ)-11d, p6His:SAE1-11d, p6His:SAE2-11d, p6His:Ubc9-11d, p6His:SUMO1-11d and p6His:SUMO1(GA)-11d, respectively, based on the described protocol (Chiang and Roeder, 1993), was incubated with 0.2 ml of Ni2+-NTA agarose beads (Qiagen) in the presence of 5 mM imidazole at 4°C overnight. After washing the beads three times with BLB (Chiang and Roeder, 1993) plus 5 mM imidazole and once with BC100 containing 20 mM imidazole, bound proteins were eluted from the beads after incubation with 100 mM imidazole-containing buffer at 4°C for 30 min. Elutions were repeated for a total of three times with sequentially increased (i.e. 100, 200 and 400 mM) imidazole concentrations. Hexahistidine-tagged human PIASxβ was purified from bacterial inclusion bodies as described (Wang et al, 2008).
For antibodies, most of them were purchased from commercial vendors: α-p53(FL-393), Santa Cruz (sc-6243); α-p53(DO-1), Santa Cruz (sc-126); α-acetyl-p53(Ac373/382), Upstate (06-758); α-p300(C-20), Santa Cruz (sc-585); α-p300(N-15), Santa Cruz (sc-584); α-SUMO-1(FL-101), Santa Cruz (sc-9060); α-FLAG M2, Sigma (F9291); α-mSin3A(AK-11), Santa Cruz (sc-767); α-β-actin, Sigma (A5441).
In vitro sumoylation
Unless otherwise specified, a 20-μl reaction (equivalent to the ‘1 × ' reaction in Figures 2F and 6C) containing 200 ng of 6His:p53, 100 ng each of 6His:SAE1 and 6His:SAE2, 25 ng of 6His:Ubc9, 40 ng of 6His:PIASxβ, and 330 ng of 6His:SUMO-1 (WT or GA) in 50 mM Tris–HCl, pH 7.5 including 5 mM MgCl2 and 3 mM ATP, was incubated at 30°C for 1 h. The reaction was terminated by adding an equal volume of 2 × protein sample buffer containing 10 mM β-mercaptoethanol and heated at 95°C for 5 min. Products were usually analysed by Western blotting with anti-p53(FL-393) antibodies. Sumoylation with purified Su-p53 (Figure 2F) was conducted as described above, except that 40 ng of Su-p53 and increasing amounts (2–10 × ) of E1 and E2 were used in the reactions.
Purification of Su-p53
For purification of Su-p53, a large-scale sumoylation reaction (equivalent to the ‘1 × ' reaction in Figure 2E and in Supplementary Figure 2) was performed in a 400-μl reaction containing 20 μg of 6His:p53, 40 μg of f:SUMO-1, 2 μg each of 6His:SAE1 and 6His:SAE2, 5 μg of 6His:Ubc9 and 0.8 μg of 6His:PIASxβ in 50 mM Tris–HCl, pH 7.5 including 5 mM MgCl2 and 3 mM ATP at 30°C for 60 min. A measure of 900 μl of BC100 plus 0.03% of NP-40, 0.5 mM PMSF and 1 mM DTT (called BC100-NPD) was then added to the reaction tube, along with 50 μl of Ni2+-NTA beads (50% slurry). Binding was conducted at room temperature for 1 h with rotation. Bound proteins were washed twice with BC100-NPD plus 5 mM imidazole (0.5 ml for each wash), and eluted in 0.3 ml of 400 mM imidazole-containing BC100-NPD after incubation at room temperature for 30 min. The eluate was diluted with 1.2 ml of BC100-NPD, and incubated with 50 μl of M2-agarose beads (Sigma, 50% slurry in BC100) with rotation at room temperature for 1 h. After washing the beads twice with BC100-NPD (0.5 ml each time), Su-p53 was eluted by adding 120 μl of FLAG peptide (0.5 mg/ml)-containing BC100-NPD after incubation with rotation at room temperature for 30 min. Purified Su-p53 contains ∼10 ng/μl of total p53.
For purification of acetylated Su-p53, 20 μg of 6His:p53 was first incubated in a 200-μl HAT reaction (see below) with 1 μg of p300 and 5 μM of acetyl-CoA at 30°C for 60 min. The supernatant, collected after microcentrifugation at 4°C, 14 000 r.p.m. for 5 min, was concentrated to 40 μl using an Amicon Ultra-4 filter unit (Millipore, catalog # UFC801024) and used for the sumoylation reaction, followed by protein purification, as described above.
In vitro transcription and HAT assays
pWAFMLT chromatin was assembled in vitro with purified HeLa core histones, hNAP-1 and ACF, and then used for in vitro transcription by stepwise additions of p53, p300, acetyl-CoA and HeLa nuclear extract, along with pΔMLP DNA, followed by [α-32P]CTP and other ribonucleoside triphosphates as described (Thomas and Chiang, 2005). In vitro HAT assay was also performed according to the published procedure (Thomas and Chiang, 2005).
Size-exclusion column chromatography
Purified p53 or Su-p53 (250 ng each) was loaded on a Superose 6 gel filtration column (GE Healthcare), equilibrated in BC100 buffer (Chiang et al, 1993) at a flow rate 0.4 ml/min. Samples (0.4 ml/tube) were collected, TCA-precipitated, and then analysed by Western blotting.
Glutaraldehyde crosslinking
A 10-μl reaction containing 20 ng of p53 or Su-p53 was incubated with a different concentration (0, 0.001, 0.003, 0.01 or 0.03%) of glutaraldehyde in 25 mM HEPES-Na (pH 8.4) containing 5 mM MgCl2 and 50 mM KCl at 30°C for 20 min. The reaction was stopped by adding an equal volume of 2 × protein sample buffer and then analysed by Western blotting.
Solution interaction assay
Purified p53 or Su-p53 (30 ng) was incubated with 120 ng of p300 in a 150-μl reaction containing BC100 plus 0.03% NP-40 at 4°C for 1 h. Anti-p300 N-15 antibodies (0.5 μg) were then added for 30-min incubation at room temperature, followed by the inclusion of 10 μl (bed volume) of Protein A-Sepharose beads for another 30 min. Bound proteins were washed twice with BC100 plus 0.03% NP-40 (0.3 ml for each wash) and analysed by Western blotting with α-p53(FL-393) or α-p300(C20) antibodies after eluting with 30 μl of 2 × protein sample buffer.
For p53 and SENP1 interaction, 500 ng of 6His:p53 was incubated with 150 ng of f:SENP1 in a 100-μl reaction containing BC100 plus 0.03% NP-40 at room temperature with rotation for 1 h. A measure of 10 μl of Ni2+-NTA beads was then added into the reaction and further incubated at room temperature for 30 min. The bead-bound proteins were analysed by Western blotting with α-FLAG antibodies.
Electrophoretic mobility shift assay
EMSA was performed as described (Hou et al, 2002) in a 10-μl reaction containing 30 ng of poly(dAT), different amounts (0, 5, 10, 25 or 50 ng) of p53 or Su-p53, and 1 fmole of a 32P-labelled p53-binding site-containing DNA fragment spanning −137 to +75 (relative to the transcription start site) of pWAFMLT or derived from Hdm2 intron 1 by PCR amplification of 293 genomic DNA using the primer pair, 5′-GGTTGACTCAGCTTTTCCTCTTG-3′ and 5′-GGAAAATGCATGGTTTAAATAGCC-3′ (Jackson and Pereira-Smith, 2006).
SENP1 treatment
Purified Su-p53 or p53 (10–20 ng) was incubated with 35 ng of wild-type or R630 L/K631 M SENP1 protein in a 10-μl reaction containing 20 mM Tris–HCl (pH 7.5) and 150 mM NaCl at 37°C for 60 min. A quarter of the reaction mix (2.5 μl) was then used for EMSA or Western blotting as shown in Figures 3H, 4F and Supplementary Figure 3B.
In vitro ChIP assay
ChIP performed in vitro with purified proteins was conducted as described (Thomas and Chiang, 2005) with some minor modifications. Briefly, purified p53 or Su-p53 (80 or 160 ng), together with 80 ng of p300 and acetyl-CoA (final 80 μM), was incubated with 150 ng of pWAFMLT chromatin in a 100-μl reaction at 30°C for 30 min. After adding CaCl2 to final 3 mM, the protein-bound chromatin was digested with six units of micrococcal nuclease (Roche) at room temperature for 2 min, followed by 1-min formaldehyde (1%) crosslinking that was subsequently terminated by adding glycine to final 0.125 M. The fixed chromatin solution was pre-cleared with Protein A-Sepharose beads and equally divided into three parts for incubation, respectively, with 1 μg of α-p53(FL-393), α-SUMO-1, or mock, at 4°C overnight. The protein–DNA complex was pulled down with Protein A-Sepharose beads and analysed as described (Thomas and Chiang, 2005).
Immobilized template binding assay
A DNA fragment (∼560 bp), containing the p53-binding site, major late promoter and the entire G-less cassette derived from pWAFMLT, was prepared by PCR amplification with an upstream primer (5′-AACTCGACTGCAGCATATGTATCATACACATACG-3′) and a biotinylated downstream primer (5′ biotin-CGATTCATTAATGCAGCTGG-3′) annealing to the flanking vector sequences. Immobilization of the biotinylated fragment was performed as described (Wu et al, 2003), except that Streptavidin MagneSphere® Paramagnetic Particles (Promega, # Z5481) were used as the magnetic beads. Unless otherwise specified, a 10-μl reaction containing 5 ng of immobilized pWAFMLT DNA, with or without XbaI digestion, and 25 ng of wild-type p53 or 5KQ, with or without prior sumoylation, was incubated in EMSA buffer containing 500 ng of poly(dAT) at 30°C for 1 h. The beads, separated by a magnetic stand, were washed twice with BC100 plus 0.1 mg/ml of BSA and 0.03% NP-40. A measure of 12 μl of 1 × protein sample buffer was then added to the beads and heated at 95°C for 5 min. The amounts of eluted proteins were analysed by Western blotting.
For binding assays involving sequential acetylation and sumoylation reactions (Figure 4D), a 40-μl HAT reaction containing 8 μg of p53, 200 ng of p300 and 2 μCi of [3H]acetyl-CoA was incubated in 40 mM Tris–HCl, pH 8.0 and 85 mM KCl at 30°C for 1 h. Desulfo-CoA was then added to final 2.5 mM to stop the HAT reaction, in which 1 μl (i.e. 200 ng of p53) was used for each sumoylation reaction, followed by immobilized template binding as described above. For binding assay after sequential modifications and protein purification (Figure 4E), 20 μl of purified acetylated Su-p53 was incubated with 5 ng of immobilized pWAFMLT template in a 30-μl EMSA reaction containing 500 ng of poly(dAT) at 30°C for 1 h. Five percentage of protein-bound beads were used for Western blotting.
For binding assays requiring sumoylation on DNA-bound proteins (Figure 6A–C), a 10-μl reaction containing 500 ng of wild-type p53 or mutant p53 (5KQ, K292R or K386R) was incubated with 5 ng of immobilized pWAFMLT template in EMSA buffer containing 500 ng of poly(dAT) at 30°C for 1 h. An equivalent amount of DNA-bound proteins, after quantification by Western blotting, was used for sumoylation reactions as aforementioned. Bound and unbound proteins were then collected and analysed by Western blotting with α-p53(FL-393) antibodies. For cofactor association involving DNA-bound sumoylated p53 (Figure 6D), 400 μg of HeLa nuclear extract was incubated in 100 μl of BC100 plus 0.03% NP40 with DNA-bound sumoylated p53, prepared by five-fold scale-up reactions, at room temperature with rotation for 1 h. After washing three times with BC100 plus 0.1 mg/ml of BSA and 0.03% NP-40, the bead-bound proteins were analysed by Western blotting.
Transfection and reporter gene assay
Human p53-null Saos-2 osteosarcoma and HCT116 p53−/− colon cancer cells were maintained in DMEM plus 10% fetal bovine serum. For exogenous p53 expression, Saos-2 cells (50% confluent in a 6-well plate) were transfected with 2 μg of DNA mix containing 0.6 μg of pcDNA3-F:p53 or pcDNA3-F:p53(K386R), 0.8 μg of pCS2-GFP:SUMO-1, pCS2-GFP:SUMO-1(ΔGG) or pCS2-GFP, and 0.6 μg of pcDNA3, using 100 μl of Fugene 6/DMEM (Roche) following manufacturer's protocols. Cells were treated with MG132 (final 5 μM), 40 h posttransfection, for 4 h and then harvested for Western blot analysis. For reporter gene assays, Saos-2 or HCT116 p53−/− cells (50% confluent in a 24-well plate) were transfected with 5 ng of pcDNA3-p53 or pcDNA3-p53(K386R), together with 0.2 μg each of pWWP-Luc and pcDNA3, and 20 μl of Fugene 6/DMEM. Cells were harvested 48 h later for Western blotting and luciferase assay as described (Lee and Chiang, 2009).
Quantitative RT–PCR and in vivo ChIP
For RNA analysis, HCT116 p53−/− cells (50% confluent in a 6-well plate) were transfected with 2 μg of DNA mix containing 0.6 μg of pcDNA3-F:p53, pcDNA3-F:p53(K386R) or pcDNA3, in the absence or presence of 0.8 μg of pCS2-GFP:SUMO-1 or pCS2-GFP, using 100 μl of Fugene 6/DMEM. Cells were harvested 40–44 h later for RNA isolation and quantification by qRT–PCR as described (Wu et al, 2006) using the following primer pairs annealing to p21, 5′-CAAGCTCTACCTTCCCACGG-3′ and 5′-GCCAGGGTATGTACATGAGG-3′ (Thomas and Chiang, 2005), or β-actin, 5′-GGTCTCAAACATGATCTGGGTC-3′ and 5′-AAATCTGGCACCACACCTTC-3′ (Wu et al, 2006). Unless otherwise specified, fold activation is defined as the ratio of signals in cells transfected with and without the p53 plasmid, each normalized first with the β-actin signal.
For ChIP assays, HCT116 p53−/− cells grown in two 150-mm plates were transfected with eight times more DNA as described above. Cells were then fixed in 1% formaldehyde at 37°C for 10 min. Reactions were terminated by the addition of glycine to 125 mM. The protein-bound chromatin was then isolated and processed for ChIP assays as described (Wu et al, 2006) using primer pairs amplifying the distal p53-binding region in the p21 gene, 5′-GTGGCTCTGATTGGCTTTCTG-3′ and 5′-CTGAAAACAGGCAGCCCAAG-3′, or a 470-nt region downstream of the transcription start site of the control AchR gene (Jackson and Pereira-Smith, 2006), 5′-CCTTCATTGGGATCACCACG-3′ and 5′-GGAGATGAGTACCAGCAGGTTG-3′. All the ChIP signals have been normalized with the AchR control.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure Legends
Acknowledgments
We thank W Gu for pcDNA3-p53(5KQ), AV Gudkov for pcDNA3-p53 and pcDNA3-p53(K386R), RT Hay for pcDNA3-p53(6KR) and pcDNA3-p53(K386R), PR Potts and H Yu for pCS2-GFP, pCS2-GFP:SUMO-1 and pCS2-GFP:SUMO-1(ΔGG), K Shuai for a PIASxβ plasmid, B Vogelstein for HCT116 p53−/− cells, and ETH Yeh for FLAG-SENP1 and FLAG-SENP1(R630L, K631M). We are particularly grateful to Elizabeth E Johnson for help in generating p53 expression plasmids and to Alison S Chiang for constructing SUMO, SAE1, SAE2, Ubc9, PIASxβ and SENP1 expression plasmids. We also thank DA Boothman, T Kodadek, C Mendelson and H Yu for comments on the manuscript. This work was supported in part by grants CA103867 and CA124760 from the National Institutes of Health and is Report CSCN # 026 from University of Texas Southwestern Medical Center Simmons Comprehensive Cancer Center.
References
- Anckar J, Hietakangas V, Denessiouk K, Thiele DJ, Johnson MS, Sistonen L (2006) Inhibition of DNA binding by differential sumoylation of heat shock factors. Mol Cell Biol 26: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appella E, Anderson CW (2001) Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 268: 2764–2772 [DOI] [PubMed] [Google Scholar]
- Baba D, Maita N, Jee J-G, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M (2006) Crystal structure of SUMO-3-modified thymine-DNA glycosylase. J Mol Biol 359: 137–147 [DOI] [PubMed] [Google Scholar]
- Carter S, Bischof O, Dejean A, Vousden KH (2007) C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nature Cell Biol 9: 428–435 [DOI] [PubMed] [Google Scholar]
- Čes̆ková P, Chichger H, Wallace M, Vojtesek B, Hupp TR (2006) On the mechanism of sequence-specific DNA-dependent acetylation of p53: the acetylation motif is exposed upon DNA binding. J Mol Biol 357: 442–456 [DOI] [PubMed] [Google Scholar]
- Cheng J, Wang D, Wang Z, Yeh ET (2004) SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol Cell Biol 24: 6021–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C-M, Ge H, Wang Z, Hoffmann A, Roeder RG (1993) Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J 12: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C-M, Roeder RG (1993) Expression and purification of general transcription factors by FLAG epitope-tagging and peptide elution. Peptide Res 6: 62–64 [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D (2004) Regulation of p53 activity through lysine methylation. Nature 432: 353–360 [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Emerson BM (2001) Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol Cell 8: 57–69 [DOI] [PubMed] [Google Scholar]
- Feng L, Lin T, Uranishi H, Gu W, Xu Y (2005) Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol 25: 5389–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G (2005) Something about SUMO inhibits transcription. Curr Opin Genet Dev 15: 536–541 [DOI] [PubMed] [Google Scholar]
- Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G (1999) Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J 18: 6462–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Roeder RG (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90: 595–606 [DOI] [PubMed] [Google Scholar]
- Hay RT (2007) SUMO-specific proteases: a twist in the tail. Trends Cell Biol 17: 370–376 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Hou SY, Wu S-Y, Chiang C-M (2002) Transcriptional activity among high and low risk papillomavirus E2 proteins correlates with E2 DNA binding. J Biol Chem 277: 45619–45629 [DOI] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL (2006) Repression of p53 activity by Smyd2-mediated methylation. Nature 444: 629–632 [DOI] [PubMed] [Google Scholar]
- Ito A, Kawaguchi Y, Lai C-H, Kovacs JJ, Higashimoto Y, Appella E, Yao T-P (2002) MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J 21: 6236–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov GS, Ivanova T, Kurash J, Ivanov A, Chuikov S, Gizatullin F, Herrera-Medina EM, Rauscher F III, Reinberg D, Barlev NA (2007) Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol 27: 6756–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JG, Pereira-Smith OM (2006) p53 is preferentially recruited to the promoters of growth arrest genes p21 and GADD45 during replicative senescence of normal human fibroblasts. Cancer Res 66: 8356–8360 [DOI] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR (2008) Structural biology of the tumor suppressor p53. Annu Rev Biochem 77: 557–582 [DOI] [PubMed] [Google Scholar]
- Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Ito A, Appella E, Yao T-P (2006) Charge modification at multiple C-terminal lysine residues regulates p53 oligomerization and its nucleus-cytoplasm trafficking. J Biol Chem 281: 1394–1400 [DOI] [PubMed] [Google Scholar]
- Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z (2006) Structural basis of DNA recognition by p53 tetramers. Mol Cell 22: 741–753 [DOI] [PubMed] [Google Scholar]
- Krummel KA, Lee CJ, Toledo F, Wahl GM (2005) The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci USA 102: 10188–10193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, Wall D, Li E, Gaudet F (2008) Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell 29: 392–400 [DOI] [PubMed] [Google Scholar]
- Kwek SS, Derry J, Tyner AL, Shen Z, Gudkov AV (2001) Functional analysis and intracellular localization of p53 modified by SUMO-1. Oncogene 20: 2587–2599 [DOI] [PubMed] [Google Scholar]
- Le Cam L, Linares LK, Paul C, Julien E, Lacroix M, Hatchi E, Triboulet R, Bossis G, Shmueli A, Rodriguez MS, Coux O, Sardet C (2006) E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell 127: 775–788 [DOI] [PubMed] [Google Scholar]
- Lee A-Y, Chiang C-M (2009) Chromatin adaptor Brd4 modulates E2 transcription activity and protein stability. J Biol Chem 284: 2778–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Luo J, Brooks CL, Gu W (2002) Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem 277: 50607–50611 [DOI] [PubMed] [Google Scholar]
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y (2005) p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol 7: 165–171 [DOI] [PubMed] [Google Scholar]
- Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K (1998) Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci USA 95: 10626–10631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W (2004) Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci USA 101: 2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 135: 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney K, Mattia M, Gottifredi V, Prives C (2004) p53 linear diffusion along DNA requires its C terminus. Mol Cell 16: 413–424 [DOI] [PubMed] [Google Scholar]
- Melchior F, Hengst L (2002) SUMO-1 and p53. Cell Cycle 1: 245–249 [PubMed] [Google Scholar]
- Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F (2008) Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell 30: 610–619 [DOI] [PubMed] [Google Scholar]
- Okorokov AL, Sherman MB, Plisson C, Grinkevich V, Sigmundsson K, Selivanova G, Milner J, Orlova EV (2006) The structure of p53 tumour suppressor protein reveals the basis for its functional plasticity. EMBO J 25: 5191–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y-H, Chung P-H, Sun T-P, Shieh S-Y (2005) p53 C-terminal phosphorylation by CHK1 and CHK2 participates in the regulation of DNA-damage-induced C-terminal acetylation. Mol Biol Cell 16: 1684–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JL, Bucceri A, Davies AA, Heidrich K, Windecker H, Ulrich HD (2008) SUMO modification of PCNA is controlled by DNA. EMBO J 27: 2422–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK (2005) A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature 437: 759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Yu H (2007) The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol 14: 581–590 [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A (2008) Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412 [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT (2000) Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol 20: 8458–8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT (1999) SUMO-1 modification activates the transcriptional response of p53. EMBO J 18: 6455–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J (2000) Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem 275: 6252–6258 [DOI] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O (2007) Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 27: 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh S-Y, Taya Y, Prives C (1999) DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasundaram K, Zhang H, Zeng Y-X, Houvras Y, Peng Y, Zhang H, Wu GS, Licht GD, Weber BL, El-Deiry WS (1997) Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CIP1. Nature 389: 187–190 [DOI] [PubMed] [Google Scholar]
- Stielow B, Sapetschnig A, Krüger I, Kunert N, Brehm A, Boutros M, Suske G (2008) Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol Cell 29: 742–754 [DOI] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W (2006) Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 24: 827–839 [DOI] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W (2008) Acetylation is indispensable for p53 activation. Cell 133: 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang C-M (2005) E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol Cell 17: 251–264 [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang C-M (2006) The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41: 105–178 [DOI] [PubMed] [Google Scholar]
- Tidow H, Melero R, Mylonas E, Freund SMV, Grossmann JG, Carazo JM, Svergun DI, Valle M, Fersht AR (2007) Quaternary structures of tumor suppressor p53 and a specific p53-DNA complex. Proc Natl Acad Sci USA 104: 12324–12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruzoe S, Ishihara K, Uchimura Y, Watanabe S, Sekita Y, Aoto T, Saitoh H, Yuasa Y, Niwa H, Kawasuji M, Baba H, Nakao M (2006) Inhibition of DNA binding of Sox2 by the SUMO conjugation. Biochem Biophys Res Commun 351: 920–926 [DOI] [PubMed] [Google Scholar]
- Vethantham V, Rao N, Manley JL (2008) Sumoylation regulates multiple aspects of mammalian poly(A) polymerase function. Genes Dev 22: 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W-M, Lee A-Y, Chiang C-M (2008) One-step affinity tag purification of full-length recombinant human AP-1 complexes from bacterial inclusion bodies using a polycistronic expression system. Protein Expr Purif 59: 144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DS, Tsai W-W, Schumacher MA, Barton MC (2008) Chromatin-bound p53 anchors activated Smads and the mSin3A corepressor to confer transforming growth factor β-mediated transcription repression. Mol Cell Biol 28: 1988–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-Y, Lee A-Y, Hou SY, Kemper JK, Erdjument-Bromage H, Tempst P, Chiang C-M (2006) Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev 20: 2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-Y, Thomas MC, Hou SY, Likhite V, Chiang C-M (1999) Isolation of mouse TFIID and functional characterization of TBP and TFIID in mediating estrogen receptor and chromatin transcription. J Biol Chem 274: 23480–23490 [DOI] [PubMed] [Google Scholar]
- Wu S-Y, Zhou T, Chiang C-M (2003) Human Mediator enhances activator-facilitated recruitment of RNA polymerase II and promoter recognition by TATA-binding protein (TBP) independently of TBP-associated factors. Mol Cell Biol 23: 6229–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xirodimas DP, Saville MK, Bourdon J-C, Hay RT, Lane DP (2004) Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118: 83–97 [DOI] [PubMed] [Google Scholar]
- Yang S-H, Sharrocks AD (2004) SUMO promotes HDAC-mediated transcriptional repression. Mol Cell 13: 611–617 [DOI] [PubMed] [Google Scholar]
- Yang X-J, Seto E (2008) Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell 31: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F-P, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Jänne OA (2008) Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol 28: 5381–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilfou JT, Hoffman WH, Sank M, George DL, Murphy M (2001) The corepressor mSin3a interacts with the proline-rich domain of p53 and protects p53 from proteasome-mediated degradation. Mol Cell Biol 21: 3974–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure Legends